Abstract
Fibrillins (FBNs) are the major structural proteins of plastoglobules (PGs) in chloroplasts. PGs are associated with defense against abiotic and biotic stresses, as well as lipid storage. Although FBN2 is abundant in PGs, its independent function under abiotic stress has not yet been identified. In this study, the targeting of FBN2 to PGs was clearly demonstrated using an FBN2-YFP fusion protein. FBN2 showed higher expression in green photosynthetic tissues and was upregulated at the transcriptional level under high-light stress. The photosynthetic capacity of fbn2 knockout mutants generated using CRISPR/Cas9 technology decreased rapidly compared with that of wild-type (WT) plants under high-light stress. In addition to the photoprotective function of FBN2, fbn2 mutants had lower levels of plastoquinone-9 and plastochromanol-8. The fbn2 mutants were highly sensitive to methyl jasmonate (MeJA) and exhibited root growth inhibition and a pale-green phenotype due to reduced chlorophyll content. Consistently, upon MeJA treatment, the fbn2 mutants showed faster leaf senescence and more rapid chlorophyll degradation with decreased photosynthetic ability compared with the WT plants. The results of this study suggest that FBN2 is involved in protection against high-light stress and acts as an inhibitor of jasmonate-induced senescence in Arabidopsis (Arabidopsis thaliana).
A plant lipid-associated protein is required for photoprotection against high-light stress and inhibition of jasmonate-induced senescence.
Introduction
Fibrillins (FBNs) are lipid-associated proteins found in the plastids. They play structural roles in sequestering carotenoids in chromoplasts of fruits and petals, storage of sterol esters in elaioplasts of tapetum cells, and envelopment of various prenyl lipids (e.g. tocopherols [Tocs], plastoquinone [PQ], and phylloquinone) in chloroplasts (Deruere et al., 1994; Ting et al., 1998; Hernandez-Pinzon et al., 1999; Lundquist et al., 2012; Suzuki et al., 2013).
The FBN gene family has been found in photosynthetic organisms, cyanobacteria, and land plants such as Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), tomato (Solanum lycopersicum), cucumber (Cucumis sativus), and Brassica species (Laizet et al., 2004; Singh and McNellis, 2011; Davidi et al., 2015; Lohscheider and Rio Bartulos, 2016). In Arabidopsis, 11 FBN subfamilies consist of 14 genes that have been duplicated and altered during evolution (Laizet et al., 2004; Singh and McNellis, 2011). The subcellular compartmentalization of FBN was determined by analyzing the proteomes of Arabidopsis chloroplasts and isoelectric point (pI)/hydrophobicity analysis of 14 FBNs (Lundquist et al., 2012). These analyses suggest that FBN1a, 1b, 2, 4, 7a, 7b, and 8 are localized in PGs, and FBN3a, 3b, 6, and 10 are present only in the thylakoid. Analyses also suggested that FBN5, which functions to maintain the activity of solanesyl diphosphate synthases in the PQ-9 biosynthesis pathway, is present in the stroma (Kim et al., 2015) whereas others are present between thylakoids and stroma.
More than half of the FBNs are found in the chloroplast plastoglobules (PGs), which are connected to the external lipid leaflet of the thylakoid membrane, showing the potential to exchange lipid metabolites. They include several proteins and lipids, such as prenylquinones, triacylglycerols (TAGs), carotenoids, and phytyl esters (Austin et al., 2006). PGs are dynamically involved in the response to abiotic and biotic stresses, together with thylakoids, and synthesize and transfer PQ, Toc, and TAG (Rottet et al., 2015). Recently, proteomic analysis of lipid-synthesizing enzymes and stress-related functional proteins in PGs has been linked to the developmental process of chloroplasts as well as to the protective functions of various biotic and abiotic stresses (Rottet et al., 2015). However, despite FBNs accounting for 30% of the dry weight of PGs, only a few of the FBNs have been studied (Ytterberg et al., 2006). Soybean (Glycine max) FBN1a, 1b, and 7a have been reported to accumulate under drought and flood stress conditions (Mutava et al., 2015). The FBN2 and FBN4 levels increase after bacterial inoculation (Pseudomonas syringae) (Jones et al., 2006). FBN4 knockdown in apple (Malus × domestica) trees decreases the PQ content in PGs, and these trees are more sensitive to environmental stress than wild-type (WT) plants (Singh et al., 2010, 2012). Because FBNs are located in chloroplasts, they seem to be highly related to light stress. FBN1a and 1b are involved in protection against high-light stress through ABA-mediated signaling (Yang et al., 2006). FBN1a, 1b, and 2 knockdown in Arabidopsis resulted in delayed shoot growth and a deficit in anthocyanin accumulation under high-light conditions (Youssef et al., 2010). These phenotypes were recovered by jasmonate (JA) treatment, which suggests that FBN1-2 might induce JA production mediated by PG accumulation under high-light stress conditions (Youssef et al., 2010). JA is a phytohormone derived from the lipids α-linolenic acid (18:3) and hexadecatrienoic acid (16:3) (Ruan et al., 2019). One of the JA biosynthesis enzymes, allene oxide synthase (AOS), has been identified in PGs by proteomic analysis (Vidi et al., 2006; Ytterberg et al., 2006), suggesting that PG may be related to JA biosynthesis or JA-mediated responses. However, the mechanism of PG involvement in the JA-mediated mechanism remains unknown. Additionally, it has been suggested that FBN1a and 1b may form homodimers or heterodimers during PG formation (Gamez-Arjona et al., 2014a), and FBN6 influences light acclimation and sulfate metabolism (Lee et al., 2020).
Because the amino acid sequence of FBN2 showed moderate identity to those of FBN1a and FBN1b, these proteins are assumed to connect to each other for the same function (Youssef et al., 2010). However, the FBN2 expression was slightly different from those of FBN1a and FBN1b in various plant tissues, and ATTED-II co-expression analysis (Obayashi et al., 2018) showed that FBN2 is highly associated with the genes involved in photoprotection, unlike FBN1a and FBN1b (Supplemental Table S1). This suggests that FBN2 may play an important role in PGs. Nevertheless, few studies have been conducted to delineate the FBN2 functions. In this study, FBN2 was specifically mutagenized using CRISPR/Cas9 genome editing to investigate the effects of light stress and JA treatment. The fbn2 knockout mutants exhibited decreased photosynthetic ability under high-light stress and responded rapidly to JA-mediated senescence. These results suggest that FBN2 is involved in photoprotection and delays senescence under environmental stress.
Results
FBN2 is mainly expressed in photosynthetic green tissues
FBN2 showed 48% and 47% identity with FBN1a and FBN1b, respectively, at the amino acid level (Supplemental Figure S1A). In the phylogenetic tree, we determined whether there was a difference in function between FBN2 belonging to Group 2 and FBN1a and FBN1b belonging to Group 1 (Supplemental Figure S1B; Singh and McNellis, 2011). To investigate the expression of FBN2 in various tissues, we analyzed its transcriptional levels by quantitative reverse transcription PCR (RT-qPCR) using FBN2-specific primers. FBN2 is expressed in various tissues of WT plants, including the leaves, stems, flowers, roots, and siliques. FBN2 showed higher expression in leaves than in other tissues (Figure 1A). To visualize the expression pattern of FBN2 in tissues, we performed β-glucuronidase (GUS) staining analysis of FBN2 promoter:GUS transgenic plants. FBN2 was highly expressed in leaves but not in roots and developing seeds in the siliques, which are nongreen tissues (Figure 1, B–H). The distinctive characteristic was that FBN2 expression was not detected in the pollen, anthers of flowers, and developing seeds (Figure 1, E and G). In contrast, FBN1a and FBN1b were strongly expressed in reproductive tissues, anther locules, and pollen (Supplemental Figure S1C). These results suggest that FBN2 may play an important role in photosynthetic tissues.
Figure 1.
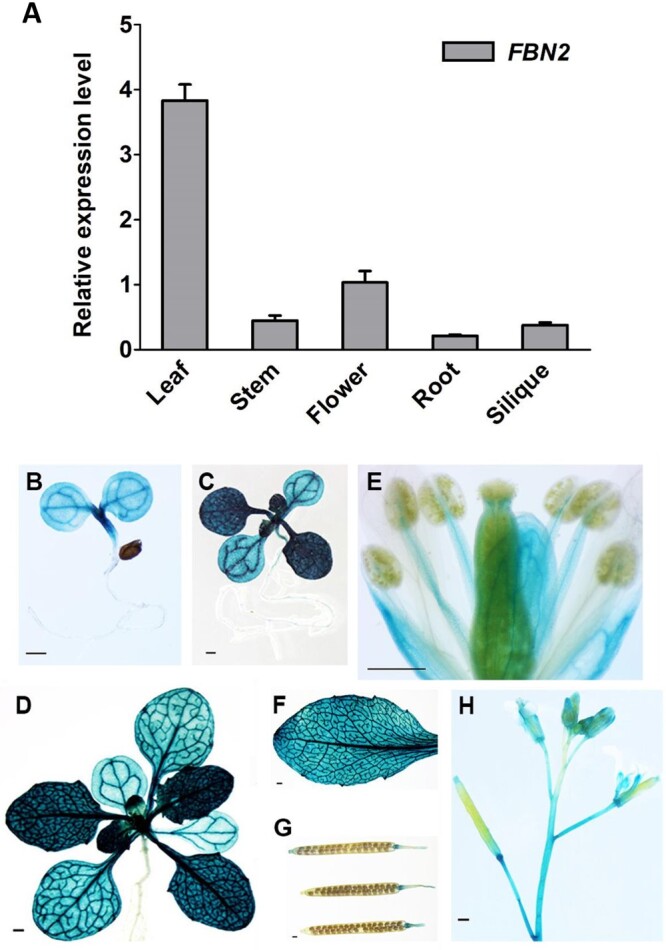
Expression of FBN2 in Arabidopsis. A, RT-qPCR analysis of FBN2 gene expression in various tissues of 5-week-old WT (Col-0) Arabidopsis. Data represent means with SEM from independent samples (n = 3). B–H, GUS staining in transgenic plants of FBN2 promoter:GUS. B, Four-day-old seedling. C, Ten-day-old seedling. D, Twenty four days of plant growth. E, Floral organs. F, Rosette leaf. G, Siliques. H, Upper part of stem in the 5-week-old plant. Scale bar = 1 mm. The GUS staining picture shows the transgenic homozygous T3 generation independent plants. It represents three independent individuals showing the same expression pattern.
FBN2 localizes to the PGs in chloroplasts
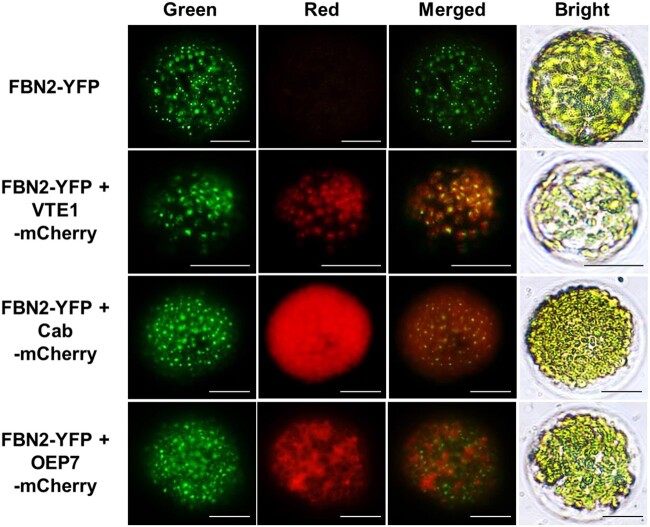
Using proteomic analyses, FBN family members have been identified in chloroplast thylakoids, PGs, and stroma (Ytterberg et al., 2006; Lundquist et al., 2012). FBN2 was reportedly located in PGs along with six other FBNs (FBN 1a, 1b, 4, 7a, 7b, and 8) (Ytterberg et al., 2006). To analyze the subcellular localization of FBN2, we expressed an FBN2-yellow fluorescent protein (YFP) fusion protein under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter in Arabidopsis and Nicotiana benthamiana. In two independent observations, YFP green fluorescence signals were observed as globular spots over a background of red chlorophyll auto-fluorescence in the chloroplasts (Supplemental Figure S2). Similarly, FBN2 fluorescence signals were observed in N. benthamiana protoplasts transiently expressing FBN2-YFP (Figure 2). We co-transformed the Toc cyclase (VTE1)-, chlorophyll a/b-binding protein (Cab)-, and chloroplast outer envelope membrane protein 7 (OEP7)-mCherry constructs with FBN2-YFP to determine whether FBN2 was localized to the PG in the chloroplast. In previous reports, VTE1 was identified as a PG-targeted protein (Vidi et al., 2006; Lee et al., 2020). To observe the distinct PG localization of FBN2 in chloroplasts, transit peptides (TPs) of Cab and OEP7 were used to target the chloroplast stroma and outer membrane, respectively (Kim et al., 2013). The FBN2 (green) fluorescence signals were colocalized with those of VTE1 (red), showing yellow signals in globular spots (Figure 2). The signals partially overlapped with the red Cab signals but not with those of OEP7. These results suggest that FBN2 is localized in PG and maybe another chloroplast sub-compartment, possibly stroma or thylakoid membrane.
Figure 2.
Subcellular localization of FBN2. The 35S:FBN2-YFP construct was transiently expressed in N. benthamiana. VTE1, Cab, and OEP7 fused with mCherry were co-transformed with FBN2 to visualize the PGs, stroma, and outer envelope membrane markers. Protoplasts were isolated from N. benthamiana leaves. YFP and mCherry fluorescence are indicated in green and red, respectively. Scale bar = 20 μm.
Additionally, yeast two-hybrid analysis was performed to investigate whether FBN2 interacts with other PG-containing FBNs and proteins. We confirmed that FBN1a and FBN1b interact with each other, as previously reported (Gamez-Arjona et al., 2014a), but we could not identify a potential FBN2 interactor under our study conditions (Supplemental Figure S3).
Generation of fbn2 knockout mutants by CRISPR/Cas9 genome editing
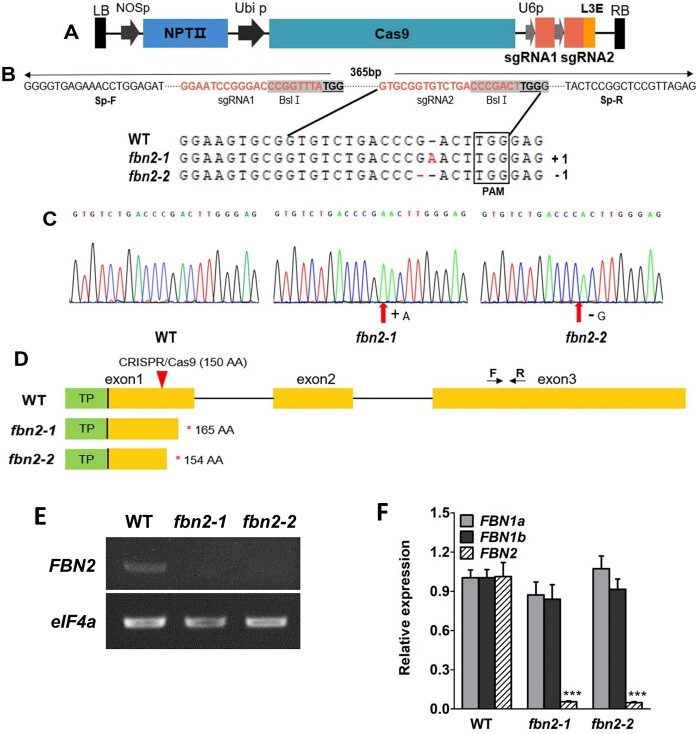
Because an Arabidopsis fbn2 T-DNA mutant could not be obtained from the ABRC stock center, the CRISPR/Cas9 system was used to generate fbn2 knockout mutants to study the function of FBN2 in Arabidopsis. We constructed the FBN2-CRISPR/Cas9 vector with two single guide RNAs (sgRNA)s to target a 78-base pair deletion in the first exon of FBN2 (Figure 3, A and B). Two independent fbn2 mutants were preliminarily screened by DNA-PCR and confirmed by DNA sequencing using specific FBN2 primers in the T3 generation (Figure 3C and Supplemental Table S2). The fbn2 mutants contained one base pair insertion “A” or deletion “G,” designated as fbn2-1 and fbn2-2, respectively (Figure 3, B and C). The fbn2-1 and fbn2-2 mutants contained FBN2 with premature stop codons at 165 and 154 aa, respectively (Figure 3D). To determine whether genome editing affected the expression of FBN2, FBN2 transcripts were confirmed by RT-PCR and RT-qPCR analysis (Figure 3, E and F). The results showed that the transcriptional levels of fbn2 mutants were remarkably lower than that of the WT plants (Figure 3, E and F). The expression levels of FBN1a and FBN1b in the fbn2 mutants were the same as those in the WT (Figure 3F). The FBN2-specific mutation was successfully induced and the mutants did not express the normal FBN2 transcript.
Figure 3.
Generation of fbn2 knockout mutants. A, CRISPR-Cas9 vector containing two guide RNA sequences for FBN2 knockout. B, Two independent mutated nucleotide sequences obtained by genome editing. C, Edited nucleotide sequences are compared with those of the WT using chromatograms from sequencing. D, Predicted fbn2-1 and fbn2-2 proteins. The fbn2-1 and fbn2-2 proteins show premature stop codons. Arrows with F and R indicate the location of the RT-qPCR primers for detecting the FBN2 transcription. E, FBN2 transcript analysis by RT-PCR in WT, fbn2-1, and fbn2-2. F, Comparison of the expression of FBN1a, 1b, and 2 in the WT, fbn2-1, and fbn2-2 mutants. Data represent means with SEM from independent samples (n = 3). Statistically significant differences from the WT are indicated by one-way analysis of variance (ANOVA) with Tukey’s post hoc test (***P < 0.001). NOSp, nopaline synthase promoter; NPTII, kanamycin resistance gene; Ubi p, Ubiquitin promoter; U6p, U6 promoter; L3E, end linker 3; PAM, protospacer adjacent motif; TP, transit peptide.
The fbn2 mutants showed decreased photosynthetic capacity under high-light stress conditions
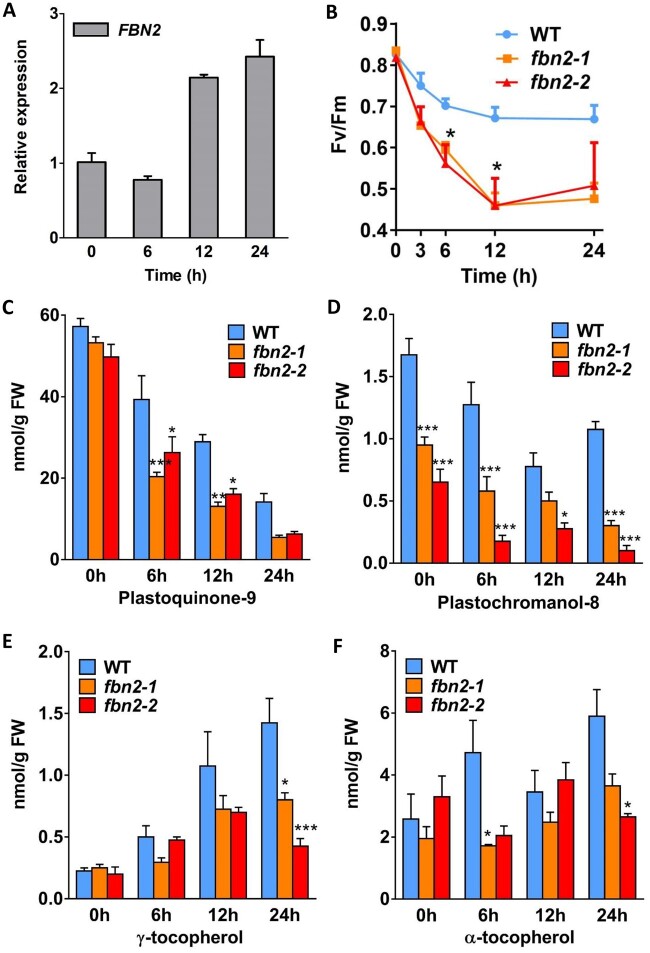
FBNs are involved both in lipoprotein structure formation in plastids as well as photosynthesis during plant development, thus providing tolerance to photooxidative stress and resistance to biotic stress (Singh and McNellis, 2011). The function of FBN2, together with FBN1a and FBN1b, is likely involved in protection against abiotic stress conditions such as high light, drought, and flooding (Youssef et al., 2010; Mutava et al., 2015). However, the independent function of FBN2 in conferring tolerance to high-light stress is unknown. Here, we studied the FBN2 function in photosynthesis under high-light stress conditions. The transcriptional levels of FBN2 increased by over two-fold after 12 and 24 h of high-light stress in the WT (Figure 4A). This result shows that FBN2 was induced under high-light stress. To determine the photosynthetic phenotype of the fbn2 mutants under high-light stress, photosynthetic capacity was measured in terms of the maximum potential efficiency of photosystem II (PSII) photochemistry for 24 h under high-light/cold stress. In the WT plants, photosynthetic capacity was moderately reduced to 0.67 at 12 h, whereas in fbn2-1 and fbn2-2 mutants, it decreased rapidly to 0.5 (Figure 4B). The WT value remained at 0.67 until 24 h, but in fbn2-1 and fbn2-2, it remained near 0.5. Also, the fbn2-1 mutant, complemented by 35S:FBN2 full-length cDNA, recovered ∼88% of photosynthetic capacity compared with the WT at 24 h (Supplemental Figure S4).
Figure 4.
FBN2 expression, photosynthetic capacity, and prenyl lipid content. A, FBN2 expression in WT Arabidopsis plants after 0, 6, 12, and 24 h exposure to high light (850 µmol m−2 s−1 light). B, Maximum quantum yield of PSII (Fv/Fm) in the WT and fbn2 mutants under high-light/cold stress (850 µmol m−2 s−1 light/15°C). C–F, Quantification of PQ-9, PC-8, and Tocs in the WT and fbn2 mutants at 0, 6, 12, and 24 h of exposure to high light (1,000 µmol m−2 s−1 light). Data represent means with SEM from independent samples (n = 4). Statistically significant differences from WT are indicated by one-way ANOVA with Tukey’s post hoc test (*P < 0.05, **P < 0.01, ***P < 0.001). FW, fresh weight.
To analyze the differences among the photosynthetic contributions of FBN2, FBN1a, and FBN1b under high-light stress, fbn1a and fbn1b T-DNA-inserted single mutants were selected (Supplemental Figure S5, A and B). Double mutants were produced by crossing fbn1a and fbn1b (Supplemental Figure S5C). Finally, an fbn1a fbn1b fbn2 triple mutant was produced by fbn2 knockout in the fbn1a fbn1b double mutant using CRISPR-Cas9 (Supplemental Figure S5D). The photosynthetic capacities of these five mutants were compared under high-light/cold stress at 24 h (Supplemental Figure S5E). The Fv/Fm value of fbn1a was 0.65, which did not differ significantly from the 0.68 of WT, but fbn1b was significantly reduced to 0.5. The fbn1a fbn1b double mutant was further reduced to 0.45. Interestingly, fbn2-1 exhibited a lower value, 0.39, than the fbn1a fbn1b double mutant. The photosynthetic capacity of the fbn1a fbn1b fbn2 triple mutant was reduced to 0.31. This suggests that FBN2 plays a major role in photoprotection under high-light stress.
The fbn2 mutants have reduced PQ-9 and PC-8 levels
The prenyl lipids in chloroplast PGs are composed of PQ-9, plastochromanol-8 (PC-8), and Toc, which are known antioxidants that inhibit oxidative damage to thylakoid membranes caused by reactive oxygen species (ROS) that accumulate under high-light stress in plants (Gruszka et al., 2008; Zbierzak et al., 2010). These lipids can also be exchanged between the PGs and thylakoid membranes. To investigate whether the photoinhibition of the fbn2 mutant under high light was related to prenyl lipid accumulation in PGs, we measured the prenyl lipid content in the WT and fbn2 mutants using high-performance liquid chromatography (HPLC) analysis. Under normal light conditions, the PQ-9 content in the fbn2 mutants was not significantly different from that in the WT (Figure 4C), but the content of PC-8, a metabolite of PQ-9, was significantly reduced compared with that in WT plants (Figure 4D). The contents of PQ-9 and PC-8 in the fbn2 mutants were reduced more than those in the WT plants under high-light conditions (Figure 4, C and D). The γ-Toc and α-Toc contents of the WT and fbn2 mutants showed statistically insignificant differences under normal light conditions (Figure 4, E and F). The γ-Toc and α-Toc contents increased with the duration of high-light stress in the WT plants, but lower levels were observed in the fbn2 mutants than in the WT plants under high-light conditions (Figure 4, E and F). These results suggest that FBN2 mutations can affect the accumulations of PQ-9, PC-8, and Toc, which have antioxidant functions that protect the thylakoid membrane from high-light damage.
The fbn2 mutants can accumulate anthocyanin similar to the WT
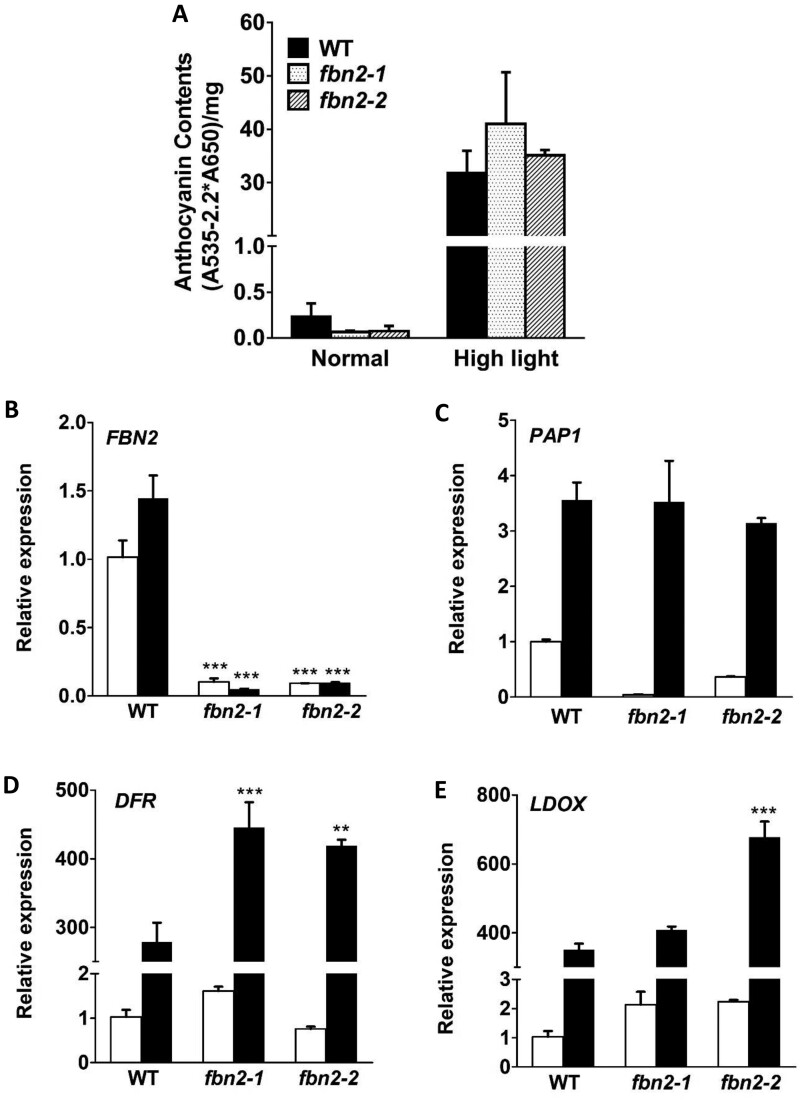
Under high-light stress, one of the mechanisms of photoprotection involves anthocyanin accumulation. The accumulation of anthocyanin, a nonphotochemical pigment, is initiated by stimulation of the photosynthetic machinery. This restores redox balance and reduces oxidative damage (Steyn et al., 2002). The anthocyanin content in the fbn2-1 and fbn2-2 mutants was slightly, but insignificantly, lower than that in the WT plants under normal light intensity and anthocyanin content increased in the WT and two fbn2 mutants under high-light stress (Figure 5A). Under normal conditions, the expression levels of the PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1) anthocyanin transcription factor were lower in fbn2 mutants than in WT plants but increased to the same level under high-light stress (Figure 5, B and C). The expression of the downstream genes, DIHYDROFLAVONOL 4-REDUCTASE (DFR) and LEUCOANTHOCYANIDIN OXIDASE (LDOX), tended to increase in the fbn2 mutants compared with the levels in the WT plants grown at the same time under high-light conditions (Figure 5, D and E).
Figure 5.
Anthocyanin accumulation in fbn2 mutants after 5 days of high-light stress. A, Anthocyanin content under normal and high-light conditions. B–E, Expression of FBN2, PAP1, DFR, and LDOX transcripts under normal and high-light conditions. White and black bars indicate normal and high-light conditions, respectively. The results of biological triplicates were averaged. Data represent means with SEM from independent samples (n = 3). Statistically significant differences from the WT are indicated by one-way ANOVA with Tukey’s post hoc test (**P < 0.01, ***P < 0.001).
FBN2 is required to form osmiophilic PGs in high-light stress
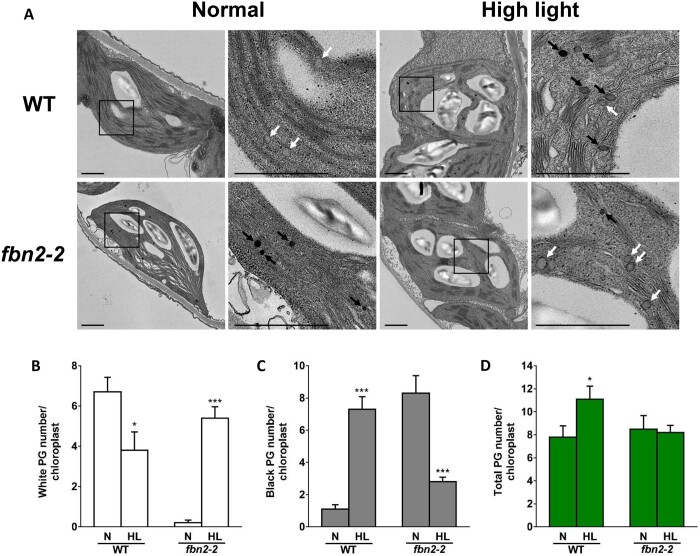
To determine whether FBN2 affects PG formation, chloroplasts in WT and fbn2-2 mutants were analyzed by transmission electron microscopy. Osmiophilic (black) and nonosmiophilic (white) PGs were observed in the chloroplasts (Figure 6A). The WT showed a high number of nonosmiophilic PGs under normal conditions and increased numbers of osmiophilic PGs under high light (Figure 6, B and C). The total PG number of the WT was higher under high light than normal light (Figure 6D). In contrast, the fbn2-2 mutant showed a low number of nonosmiophilic PGs in normal conditions and increased numbers of nonosmiophilic PGs instead of osmiophilic PGs under high-light conditions (Figure 6, B and C). However, the total PG number in the fbn2-2 mutant did not change between the normal and high-light conditions (Figure 5D). PGs accumulate PQ, carotenoids, and TAG, thereby contributing to the osmiophilicity of PG (Dahlin and Ryberg, 1986). Thus, FBN2 may require accumulation of lipids in the PGs for the formation of osmiophilic PG under high light.
Figure 6.
Transmission electron micrograph of chloroplasts in the WT and fbn2 mutants. A, Chloroplast of WT and fbn2-2 mutant leaf stained with osmium tetroxide. White or black arrows indicate nonosmiophilic or osmiophilic PGs. Scale bar = 1 μm. B–D, The numbers of nonosmiophilic (white) or osmiophilic (black) PGs and total PGs per chloroplast. 10 chloroplasts were counted from each leaf of three plants. Data represent means with SEM from independent samples (n = 10). Statistically significant differences from the WT are indicated by t test (*P < 0.05, ***P < 0.001). N, normal; HL, high light.
The fbn2 mutants are more sensitive to methyl JA
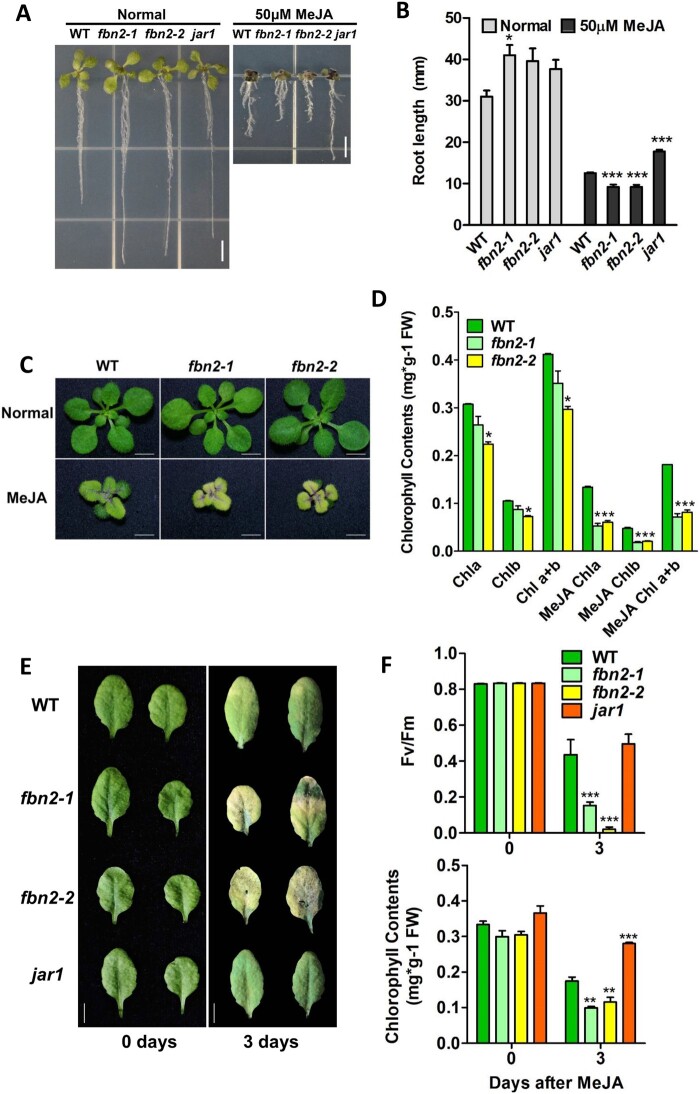
JA-treated plants exhibit a response that inhibits shoot and root growth and promotes anthocyanin accumulation and leaf senescence (Staswick et al., 1998; He et al., 2002; Zhang and Turner, 2008; Qi et al., 2011). Phenotypes, such as senescence and the relationship between PG and JA production, have frequently been mentioned in studies on FBNs and PG-located proteins (Youssef et al., 2010; Lippold et al., 2012; Lundquist et al., 2013; Li et al., 2019). Therefore, we investigated the FBN2-deficient plant response to methyl JA (MeJA), a JA compound. Root length was measured to test JA sensitivity. The roots of the fbn2-1 and fbn2-2 mutants grown on Murashige and Skoog (MS) medium were longer than those of the WT and jasmonate-resistant 1(jar1) mutants. Root growth in all plants was affected by the MS medium containing 50 μM MeJA (Figure 7A) and significantly inhibited in the fbn2-1 and fbn2-2 mutants compared with that in the WT (Figure 7B). In contrast, the roots of the jar1 mutant, which is insensitive to MeJA, grew longer than those of the WT. This shows that fbn2 mutants are very sensitive to MeJA.
Figure 7.
fbn2 mutant sensitivity to MeJA treatment. A, Growth of WT and fbn2 or jar1 mutant seedlings under normal and MeJA (50 μM) treated conditions. B, Root growth under normal and MeJA treated conditions. C, Pale-green phenotype of WT and fbn2 mutants after 14 days of growth on unsupplemented and MeJA (25 μM)-supplemented MS medium. D, Chlorophyll content in the WT and fbn2 mutants under conditions described in (C). E, Leaves of WT and fbn2 or jar1 mutants after 0 and 3 days of MeJA (100 μM) treatment under dark conditions. F, Maximum quantum yield of PSII (Fv/Fm) and chlorophyll content of leaves described in (E). Scale bar = 5 mm (A and E), 2 mm (C). The results of biological triplicates were averaged. Data represent means with SEM from independent samples (n = 10 for B, n ≥ 3 for [D] and [F]). Statistically significant differences from WT are indicated by one-way ANOVA with Tukey’s post hoc test (*P < 0.05, ***P < 0.001). Chl, chlorophyll.
The fbn2 mutants promote JA-induced senescence
The fbn2 mutants were compared with WT plants for growth on MS medium containing 25 μM MeJA. There was no difference in growth between the fbn2 mutants and the WT grown for 14 days on MS medium. However, adding MeJA to the MS medium inhibited the growth of both fbn2 mutants and WT plants, and a pale green phenotype was observed in fbn2 mutants compared with the WT plants (Figure 7C). The chlorophyll content in fbn2 mutants was 65%–70% compared with that in the WT on the normal MS medium, but there was no difference in the color of visible leaves (Figure 7, C and D). Under MeJA treatment, the chlorophyll content decreased in both the WT and fbn2 mutants. A 50% decrease was observed in the fbn2 mutants compared with the content in the WT plants, reflective of the pale green phenotype (Figure 7D).
To investigate whether the MeJA-induced pale-green phenotype of the fbn2 mutant leaves was related to JA-induced leaf senescence, we compared leaf senescence among the WT, fbn2 mutants, and jar1 plants under dark conditions in 100 μM MeJA solution. After 3 days of treatment, the detached leaves of the fbn2 mutants showed greater chlorophyll loss, as reflected by a yellow color, than those of the WT plants (Figure 7E). The photosynthetic ability (Fv/Fm) and chlorophyll content were also significantly lower in the fbn2 mutants than in the WT and jar1 mutants (Figure 7F). These results showed that the fbn2 mutants promoted JA-induced senescence.
Expression analysis of genes involved in JA-induced senescence
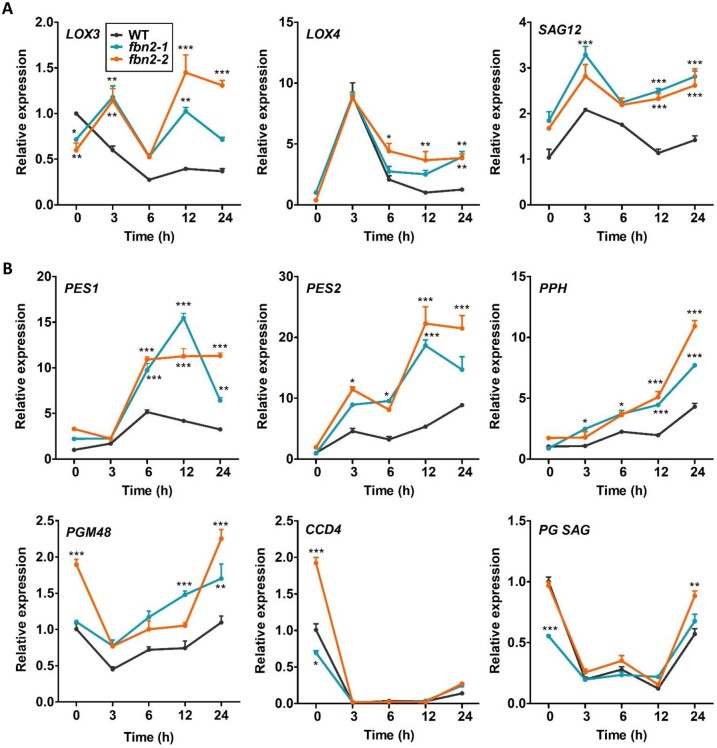
The FBN2 transcription was downregulated under dark conditions, whereas upregulation was observed under high-light conditions (Supplemental Figure S6). After 12 and 24 h of MeJA treatment under dark conditions, FBN2 expression increased compared with that observed under dark conditions only. This suggests that FBN2 may be up-regulated by JA. Changes in the expression of JA biosynthesis and senescence-related genes at different times after MeJA treatment were analyzed (Figure 8). The level of the JA biosynthesis gene, Lipoxygenase 3 (LOX3), decreased at 6 h in the WT plants and increased very slowly thereafter. In contrast, LOX3 levels in the fbn2 mutants increased at 3 h and decreased at 6 h (Figure 8A). Compared with the WT plants, the level of LOX4 increased substantially in the fbn2 mutants at 3 h MeJA treatment and then decreased. However, a high expression level was observed in the fbn2 mutants with respect to the expression in the WT plants after 3 h (Figure 8A). The Senescence-associated gene (SAG12), a JA-induced senescence gene, was more highly expressed after 3 h in the fbn2 mutants than in the WT plants (Figure 8A). Additionally, we analyzed the expressions of Phytyl ester synthase 1 (PES1), PES2, Pheophytin pheophorbide hydrolase (PPH), M48 peptidase (PGM48), and Carotenoid cleavage dioxygenase 4 (CCD4), which are senescence-associated genes present in PGs (Figure 8B). The expression levels of PES1 and PES2 were higher than those in the WT; the levels increased after 3 h, reaching a maximum at 12 h, and then started to decline at 24 h. The expressions of PPH and PGM48 continued to increase over time. The levels of CCD4 and an unknown SAG in PGs rapidly decreased at 3 h and increased at 24 h in both WT and fbn2 mutants. These results show that FBN2 affects the expression levels of JA biosynthesis and senescence-associated genes through the induction of JA.
Figure 8.
Changes in the JA biosynthesis and senescence-related gene expressions in WT and fbn2 mutants after 24 h of MeJA treatment under dark conditions. A, JA biosynthesis genes, LOX3 and LOX4, and senescence-related gene, SAG12. B, PG-localized senescence-associated genes, PES1, PES2, PPH, PGM48, CCD4, and PG SAG. Data represent means with SEM from independent samples (n = 3). Statistically significant differences from WT are indicated by one-way ANOVA with Tukey’s post hoc test (*P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
FBN2 is important in photoprotection independently with FBN1a and FBN1b
FBN2 is known to be a major protein of PGs in chloroplasts along with FBN1s (FBN1a and FBN1b) (Ytterberg et al., 2006; Lundquist et al., 2012). FBN1a, FBN1b, and FBN2 are considered to have functional redundancy, and their roles in abiotic and biotic stresses were investigated by knocking down the three FBNs simultaneously using RNA interference (RNAi) technology (Youssef et al., 2010). However, FBN2 showed a large difference compared with FBN1s in the sequence of the first exon, and only 48% identity was observed at the protein level (Supplemental Figure S1A). The difference in the lipocalin motif sequences at amino acids suggests the possibility of interactions with different peptides or lipid molecules. Proteins similar to Arabidopsis FBN1s are involved in binding with sterol esters in anthers and carotenoids in chromoplasts (Vishnevetsky et al., 1996; Pozueta-Romero et al., 1997; Wang et al., 1997). The FBN1a (BrPAP1), FBN1b (BrPAP2), and FBN2 (BrPAP3) were isolated from Chinese cabbage (Brassica rapa) plants. BrPAP1 is present in anther elaioplasts, BrPAP2 in petal chromoplasts, and BrPAP3 is expressed in photosynthetic green tissues (Kim et al., 2001). Arabidopsis FBN1s showed upregulation of transcripts under cold and drought conditions, while the opposite regulation was observed for FBN2 (Laizet et al., 2004). In our study, FBN2 expression was not observed in anthers and pollen, which is different from the expression of FBN1s in flowers (Supplemental Figure S1C). Since FBN2, FBN1a, and FBN1b were all expressed at similar levels in leaves (Figure 3F and Supplemental Figure S1C), all three genes can be involved in photoprotection. Additionally, FBN2 did not interact with other PG-localized FBNs and proteins in our yeast two-hybrid analysis, although the interaction between FBN1a and FBN1b was consistent with a previous report (Supplemental Figure S3; Gamez-Arjona et al., 2014a). Nevertheless, a recent study reported that FBN2 interacted with FBN1a and 1b in PG by co-immunoprecipitation (Co-IP) and bimolecular fluorescence complementation (BiFC) assays (Torres-Romero et al., 2022), suggesting that FBN2 may interact with FBN1s in vivo. Interestingly, the signals in the self-interaction of FBN2 using BiFC assay were observed in the widespread chloroplast unlike that of FBN1s. Therefore, how FBN2 plays a role in PG along with FBN1s and in other chloroplast sub-compartment remains to be determined in the future. We showed that FBN2 plays an important role in photoprotection against high-light stress because fbn2 single mutants were as sensitive to high light as the fbn1a fbn1b double mutant (Figure 4B and Supplemental Figure S5E). Consistently, the fbn2 T-DNA insertion mutant reduced the maximum quantum yield (Fv/Fm) similar to the fbn1a-1b double and fbn1a-1b-2 triple mutants from another research group (Torres-Romero et al., 2022).
FBN2 is related to prenyl lipids accumulation of PG under high-light stress
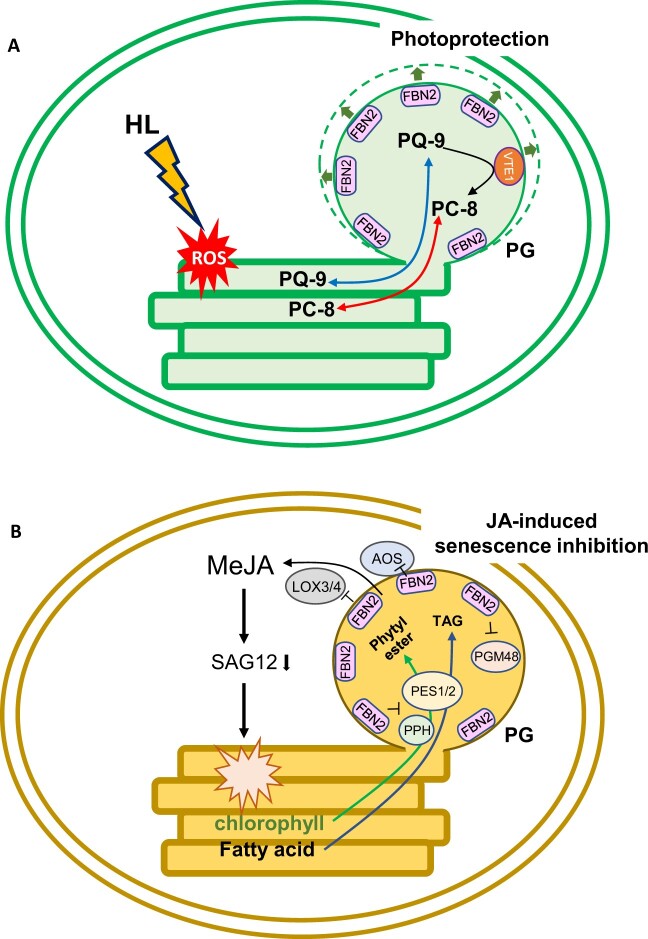
The fbn2 knockout mutant showed a significant decrease in PQ-9, PC-8, and Toc contents compared with the WT plants under high-light stress (Figure 4, C–F). Photoprotection under excess light conditions includes antioxidant regeneration, ROS scavenging, and dissipation of excess excitation energy or electrons. PGs reportedly store antioxidants, such as PQ-9, PC-8, and Toc, and protect thylakoid membranes against ROS (Havaux et al., 2005; Vidi et al., 2006; Eugeni Piller et al., 2014). VTE1 contributes to PC-8 and Toc synthesis in PG with FBNs (Kobayashi and DellaPenna, 2008; Szymanska and Kruk, 2010; Zbierzak et al., 2010). A previous study showed that the FBN2 protein content in the vte1 mutant was lower than that in the WT by western blot analysis (Martinis et al., 2013), suggesting that proteins that are localized to PG can affect each other in their function. Although FBN2 did not directly interact with VTE1 in this study (Supplemental Figure S3), FBN2 may maintain the structure of PG for the storage and synthesis of PQ-9, PC-8, and Toc. Torres-Romero et al. (2022) demonstrated higher ROS production in fbn1-2 mutants than in WT plants under 1-week high-light stress. Therefore, we hypothesized that the photosynthetic capacity in the fbn2 mutants was reduced as cause reduction of the antioxidants, leading to the disenabled removal or scavenging of ROS production by high-light stress. The accumulation of anthocyanins in the fbn2 mutants was similar to that in the WT plants (Figure 5A). In contrast, the fbn1-2 mutants showed decreased accumulation of anthocyanins under other stress conditions (600 µmol m−2 s−1 light intensity for 1 week). Nevertheless, ROS production was still higher in the mutants than in the WT (Torres-Romero et al., 2022). Consequently, we suggest that anthocyanin accumulation does not contribute greatly to the photoprotection of PGs under high-light stress conditions and that other metabolites of PG (e.g. PQ-9, PC-8, and Toc) are more effective. In fact, compared with the WT under high-light stress, the fbn2-2 mutant showed a lack of formation of osmiophilic PGs containing lipid components, such as PQ-9 and PC-8 (Figure 6, A and C). Similar to our results, previous studies have reported that FBN4 knockdown apple tree using RNAi also showed accumulation of nonosmiophilic PGs (Singh et al., 2010). A decrease of PQ, carotenoid, and TAG content in PG may lead to nonosmiophilic PG (Dahlin and Ryberg, 1986). Although we did not directly measure the TAG content, we observed a decrease in prenyl lipids in fbn2 mutants (Figure 4, C–F) leading to the formation of nonosmiophilic PGs. Therefore, FBN2 may contribute to photoprotection by accumulating prenyl lipids, such as PQ-9 and PC8, within PG (Figure 9A).
Figure 9.
Proposed model for FBN2 function in photoprotection and JA-induced senescence inhibition. A, The PC-8 antioxidant synthesized by the enzyme VTE1 from PQ-9 is stored in PG. Then, the PC-8 and PQ-9 may transfer to the thylakoids for protection against ROS production under high-light conditions. It is necessary to expand PG and enzyme function to accumulate metabolites. Therefore, FBN2 may provide structural maintenance of PG for photoprotection. B, PG-localized proteins (PES1/2, PPH, and PGM48) have been associated with senescence, and JA biosynthesis enzymes (AOS and LOXs) have been frequently identified in PG studies. In JA-induced senescence, FBN2 may inhibit these proteins for metabolites accumulation such as phytyl esters and TAGs produced from chlorophyll degradation, as well as for JA initial biosynthesis as a PG structural protein. HL, high light.
FBN2 is associated with inhibition of senescence induced by JA
Plants produce a variety of phytohormones that can adapt to external stimuli. Among phytohormones, JA is associated with various plant developmental processes, including root growth, fruit ripening, senescence, and defense signaling against biotic and abiotic stresses (Staswick et al., 1998; Rao et al., 2000; He et al., 2002; Zhang and Turner, 2008; Qi et al., 2011). The jar1 mutant, which is unable to synthesize JA-lle, is not sensitive to MeJA, and its root growth is not suppressed more than that of WT roots (Staswick et al., 1998). The fbn2 mutants were more sensitive to MeJA than the WT and jar1 plants, showing greater suppression of root growth and a pale green phenotype in leaves (Figure 7, A, C, and E). MeJA treatment promotes chlorophyll degradation and induces leaf senescence, which upregulates SAG expression (He et al., 2002). PG-localized proteins have been investigated for their associations with JA biosynthesis, senescence, and lipid synthesis (van Wijk and Kessler, 2017). In the activity of bc1 complex kinase 1 and 3 (abc1k1/3) double mutants, JA biosynthetic proteins were recruited into the PGs, resulting in a phenotype similar to the senescence-like phenotype under light stress conditions, but the reason and pathway were not clear (Lundquist et al., 2013). In addition, FBN2 protein levels were reduced in abc1k3 mutants compared with the WT (Martinis et al., 2013). Overexpression of the PG-localized metallopeptidase PGM48 promotes natural senescence (Bhuiyan et al., 2016). In contrast, the pes1/2 double mutant retained green leaves during dark-induced senescence because of delayed chlorophyll degradation. PES1/2 synthesizes fatty acid phytyl esters and TAGs in PGs from free phytol and fatty acids produced by chlorophyll breakdown during chlorotic stress or senescence (Lippold et al., 2012). Although FBNs are the most abundant PG proteins, the relationship between FBNs and senescence has not been reported. The fbn2 mutants showed upregulation of JA biosynthesis and senescence-related genes, including LOX3/4, PES1/2, PGM48, SAG12, and PPH, upon MeJA treatment (Figure 8). This may explain why fbn2 mutants promote leaf senescence. Additionally, AOS that has been reported to contribute to initial JA biosynthesis in chloroplasts was found in PG (Ytterberg et al., 2006), and LOX2 was predicted to be positioned on the PG surface with FBN1a (Espinoza-Corral et al., 2021). JA biosynthesis enzymes, such as AOS, LOX2, and allene oxide cyclase (AOC), were identified in Co-IP analysis with FBN2, and three FBNs (FBN1a, 1b, and 2) interacted with AOS in vivo using the BiFC assay (Torres-Romero et al., 2022). In consideration of our results and those of previous studies, we propose the possibility that FBN2 may control the proteins for accumulation of senescence-related metabolites, such as phytyl esters and TAGs, and initial JA biosynthesis as a structural protein of PG under MeJA and dark treatment (Figure 9B).
Materials and methods
Plant materials and growth conditions
All plants, viz. Arabidopsis (A. thaliana) Col-0, T-DNA mutants, and transgenic plants, were grown either in potting soil in growth chambers at 23°C under a 16-h/8-h light/dark cycle, with a light intensity of 100 µmol m−2 s−1, or in 0.5 × MS agar medium with 1% (w/v) sucrose under the same conditions. The 3-week-old WT and fbn2 mutants were exposed to 850 µmol m−2 s−1 light and grown in the same growth chamber condition for up to 24 h at 15°C to induce high-light/cold stress. The duration of the high-light stress treatment varied depending on the experiment. Prenyl lipids were measured at 0, 3, 6, 12, and 24 h intervals, anthocyanin was measured for 5 days, photosynthetic proteins were analyzed at 0, 12, and 24 h intervals, and electron microscopy was performed for 12 h. For JA treatment, the plants were grown on MS agar medium (normal) and the same medium containing 25 or 50 µM MeJA for seedling phenotype observation and root length measurement, which was performed using ImageJ software (http://imagej.nih.gov/ij/).
For the leaf senescence assay, rosette leaves of 4-week-old plants were harvested, treated with 100 μM MeJA or water, and kept under dark conditions for 3–5 days.
Histochemical GUS reporter assay
FBN1a, 1b, and 2 promoters:GUS constructs were created with different promoter regions (all regions about 3 kb in the 3-UTR of the upstream gene and 5′-UTR of FBNs) of FBN1a, FBN1b, and FBN2 using a specific primer (see Supplemental Table S2) cloned into pENTR/D-TOPO to generate pENTR-FBN1a, 1b, and 2 promoters, respectively. Each promoter region was transferred to the pMDC163 destination vector using the Gateway system to make the pMDC163-promoter:GUS constructs. Transgenic Arabidopsis Col-0 plants transformed with these constructs were selected on an MS medium containing hygromycin. In the T1 generation, 15 or more transformants were obtained per construct. Three independent lines for each construct, which were considered to be one copy of a transgene with a 3:1 segregation ratio of hygromycin resistance and sensitivity in progeny, were advanced to the T3 generation. Each transformant, homogenized with the transgene, was subjected to GUS staining. The three independent lines had the same GUS staining pattern for each tissue. The GUS staining was performed using 1 mg mL−1 5-bromo-4chloro-3-indolyl-β-D-glucuronide solution in 100 mM sodium phosphate buffer (pH 7.0), 10 mM EDTA, 0.1% Triton X-100, 100 µg mL−1 chloramphenicol, 2 mM potassium ferricyanide, and 2 mM potassium ferrocyanide. After overnight incubation with the solution at 37°C, the pigments were removed by treatment with 70% ethanol.
Transcript analysis
For transcriptional expression analysis, different Arabidopsis tissues from 5-week-old WT plants and leaves in WT, mutants, and complementation lines under normal or high light or JA treatment were sampled and fixed in liquid nitrogen. Total RNA was extracted from these samples using TRIzol reagent (Favorgen) and treated with RNase-free DNase I (Thermo Fisher, Waltham, MA, USA) to remove genomic DNA contamination. cDNA was synthesized from total RNA using a cDNA synthesis kit (Takara, Shiga, Japan) following the manufacturer’s recommendations. RT-qPCR was performed using SYBR Green premix (Toyobo, Osaka, Japan); the all RT-qPCR primers for housekeeping gene (eIF4a), FBNs (FBN1a, FBN1b, and FBN2), anthocyanin-related genes (PAP1, DFR, and LDOX), and JA- and senescence-related genes (LOX3/4, SAG12, PES1/2, PPH, PGM48, CCD4, and PG SAG) are listed in Supplemental Table S2. Relative expression levels were normalized to the levels of the endogenous control gene eIF4a. The reactions were performed in triplicate. For RT-PCR analysis of fbn1a, fbn1b, fbn2, fbn1a fbn1b double, fbn1a fbn1b fbn2 triple mutants, fbn2-1/FBN2 complementation lines, the specific primers of endogenous control (eIF4a and ACT7), and FBN genes (FBN1a, FBN1b, and FBN2) was designed and listed in Supplemental Table S2.
Subcellular localization
The full-length FBN2 gene amplified from the cDNA of Arabidopsis leaves was cloned into pDONR221 using the Gateway vector and BP Clonase (Invitrogen, Waltham, MA USA). FBN2 was transferred to pEarleygate101 to construct the 35S:FBN2-YFP fusion vector using LR clonase (Invitrogen) (Earley et al., 2006). Full-length VTE1 was amplified from the same cDNA using primers containing Xba1/BamH1 enzyme sites for insertion into the pCAMBIA3300-mCherry vector. The Cab- and OEP7-mCherry vectors were obtained from R&D Bioresources deposited by Hwang Inhwan, Pohang University (Kim et al., 2013). These vectors were introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis Col-0 plants using the floral-dip method (Clough and Bent, 1998), or into N. benthamiana by transient Agrobacterium infiltration (Wydro et al., 2006). Transgenic Arabidopsis plants were selected on MS medium containing DL-phosphinothricin and grown in soil for 2 weeks for imaging. Nicotiana benthamiana plants were maintained for 4–5 days after 1 day of dark conditions and were directly observed from leaves or isolated protoplasts as described (Yoo et al., 2007). The collected samples were analyzed using a Leica TCS SP5 confocal microscope with 40x/1.25 NA Oil objective and excitation/emission at 488/520 nm for YFP and 640/700 nm for chlorophyll autofluorescence or a Nikon Eclipse Ts2R-FL fluorescence microscope with 40× objective.
Generation of fbn2 mutants and complementation of fbn2-1 mutant
Two sgRNAs of FBN2 were designed using the CRISPR-P tool (http://crispr.hzau.edu.cn/CRISPR2/) at 78-bp intervals. These were amplified by the golden gate cloning method (Weber et al., 2011) using the PCR primers listed in Supplemental Table S1. Level 1 vectors were assembled as pICH47751::sgRNA1 and pICH47761::sgRNA2 under the Arabidopsis U6 promoter. The level 2 vector was assembled as a level 1 vector, pICH47732::NOSp::NPTII-OCST, pICH47742::35Sp::Cas9-NOST, and pICH41766, into the pAGM4723 vector. To increase genome editing efficiency in the final vector, the 35S promoter was replaced with a ubiquitin promoter for Cas9 expression. Agrobacterium tumefaciens strain GV3101 containing the final gene-editing construct (pAGM4723-FBN2) was inoculated into Col-0 Arabidopsis plants using the floral dip method. Transgenic plants were selected based on their kanamycin resistance. To identify knockout mutants, the genomic DNA was extracted from the transgenic plants, and PCR was performed using specific primers designed from the sgRNAs region in FBN2.
To complement the fbn2-1 mutant, the 35S:FBN2-YFP vector used for subcellular localization was transformed into the fbn2-1 mutant using the same floral-dip method as described above. FBN2-YFP was cloned under the control of the CaMV35 promoter by removing only the stop codon from the FBN2 full-length cDNA and fusion YFP in an in-frame. Homozygous plants were selected using DL-phosphinothricin for the T4 generation.
Identification of fbn1a and fbn1b single T-DNA mutants and generation of fbn1a fbn1b double and fbn1a fbn1b fbn2 triple mutants
To select fbn1a and fbn1b T-DNA insertion homozygous mutants, the salk_130350 and sail_384_A10 lines were provided from ABRC (https://abrc.osu.edu/). For the T-DNA insertion homozygous lines, LP and RP primers for each line were designed using SIGnAL “T-DNA Express” web (Supplemental Tables S2). T-DNA insertion into the 1st exon of FBN1a in salk_130350 was confirmed by DNA-PCR genotyping to select a homozygous line, which did not express the full-length FBN1a transcript through RT-PCR (Supplemental Figure S5A). In sail_384_A10, a T-DNA-inserted homozygous mutant was selected in the same way, confirming that T-DNA was inserted into the third exon (Supplemental Figure S5B). The lines derived from salk_130350 and sail_384_A10 were named fbn1a and fbn1b, respectively.
The fbn1a fbn1b double mutant was generated by crossing fbn1a and fbn1b. F1 was produced by crossing fbn1a with fbn1b. By selfing F1, an F2 descendant generation was obtained. Thirty-two F2 progenies were cultivated and genotyped by DNA-PCR to select three lines in which two genes were homozygous. For these homozygous lines, RT-PCR confirmed that they were fbn1a fbn1b double knockout lines in which FBN1a and FBN1b genes were not expressed (Supplemental Figure S5C). The fbn1a fbn1b fbn2 triple mutant was prepared in the same manner as the fbn2 mutant. The FBN2 gene was knocked out by CRISPR-Cas9 gene-editing system with two sgRNA ‘GGAATCCGGGACCCGGTTTA’ and ‘GTGCGGTGTCTGACCCGACT’ in the selected fbn1a fbn1b double mutant. Finally, sequencing confirmed a 77 bp deletion and predicted an early stop in the translated amino acid sequence (Supplemental Figure S5D).
Measurement of chlorophyll fluorescence
The maximum fluorescence yield of PSII (Fv/Fm) was measured using a Fluorpen FP 100 instrument (PSI, Czech). The plant leaves were dark-adapted for 15 min before taking measurements using leaf dark clips and 3,000 µmol m−2 s−1 intensity light. The maximum PSII efficiency was calculated as (Fm−Fo)/Fm, where Fm is the maximum chlorophyll fluorescence intensity and Fo is the minimum chlorophyll fluorescence intensity. The measurement was obtained by three repetitions from three individual plants exposed to 850 μmol m−2 s−1 light at 15°C for up to 24 h in a growth chamber.
Measurement of anthocyanin and chlorophyll content
For anthocyanin extraction, the ground plant tissues were treated with an extraction buffer containing 18% isopropanol, 1% hydrochloric acid, and 81% water and boiled for 5 min (Deikman and Hammer, 1995). Chlorophyll extraction was performed by treating the ground tissue with 80% acetone, as described previously (Su et al., 2010). The extract was incubated in the dark at 25°C for at least 3 h, and the chlorophyll content was measured by spectrophotometry (DeNovix DS-11+).
HPLC analysis
For PQ-9 analysis, PC-8, and Tocs, frozen tissues were ground in liquid nitrogen and total lipids were extracted as described previously (Bligh and Dyer, 1959). HPLC (Shimadzu LC-20AD) was used to analyze the samples in ethanol on a C18 reverse-phase column at 30°C, as previously described (Kim et al., 2015). The samples were separated in isocratic mode with methanol at a 1.0 mL/min flow rate. Toc and PC-8 were monitored fluorimetrically at the excitation and emission wavelengths of 290 and 330 nm, respectively. The PQ-9 was detected at a 255 nm wavelength. The compounds were quantified using external calibration standards, and the data were corrected by comparison with the recovery of rac-tocol (Matreya), which was used as the internal standard.
Transmission electron micrograph
The leaf fragments were collected from 3-week-old WT and fbn2 mutant plants treated either with normal or high light (100 or 850 µmol m−2 s−1) and fixed with 2% glutaraldehyde and 2% paraformaldehyde in 0.05 M sodium cacodylate buffer (pH 7.2). Samples were postfixed in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer (pH 7.2) for 2 h, and dehydrated in 0.5% uranyl acetate and graded ethanol. The samples were then embedded in Spurr’s resin. Ultrathin sections (70 nm thick) were obtained using a Leica EM UC7 ultramicrotome and imaged using a Talos L120C transmission electron microscope.
Yeast two-hybrid assay
To analyze the interaction between FBN2 and other PG proteins, full-length genes, except for FBN2, were amplified from cDNA extracted from Arabidopsis leaves without the TP predicted by TargetP version 1.1 (http://www.cbs.dtu.dk/services/TargetP/). These PCR products were cloned into pDONR221 by BP clonase (Invitrogen) using the Gateway system and were transferred to pDEST-pGBKT7 (bait) or pDEST-pGADT7 (prey) vectors using LR clonase (Invitrogen). Yeast transformation was performed according to the Yeast Protocols Handbook (Clontech, Mountain View, CA, USA). The empty and pGBKT7-FBN2 bait vectors were first transformed into a yeast strain (AH109), then the pGADT7 prey vectors were transformed into yeast cells and selected on selective SD/-Leu/-Trp medium. The final colonies were selected and spotted on selective SD/-Ade/-His/-Leu/-Trp media. The images in Supplemental Figure S3 were observed after 3 days at 30°C.
Accession numbers
Sequence data were obtained from GenBank/EMBL libraries under the following accession numbers: FBN1a (At4g04020), FBN1b (At4g22240), FBN2 (At2g35490), PAP1 (At1g56650), DFR (At5g42800), LDOX (At4g22880), LOX3 (At1g17420), LOX4 (At1g72520), PES1 (At1g54570), PES2 (At3g26840), PPH (At5g13800), PGM48 (At3g27110), CCD4 (At4g19170), and PG SAG (At1g73750).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of protein sequences and expression patterns of FBN1a, FBN1b, and FBN2.
Supplemental Figure S2. Subcellular localization of FBN2 in Arabidopsis and N. benthamiana cells.
Supplemental Figure S3. Yeast two-hybrid analysis of the interaction between FBN2 and PG proteins.
Supplemental Figure S4. Complementation of fbn2-1 mutant in Arabidopsis.
Supplemental Figure S5. Comparison of photosynthetic capacity among fbn1a, fbn1b, fbn2-1 single, fbn1a fbn1b double, and fbn1a fbn1b fbn2 triple mutants under high-light stress.
Supplemental Figure S6. Expression of FBN2 under dark and JA treatments.
Supplemental Table S1. Top 20 co-expressed genes with FBN2.
Supplemental Table S2. Oligonucleotides used in this study.
Funding
This work was supported by grants from the Mid-Career Researcher Program of the National Research Foundation of Korea (NRF-2020R1A2C2008175) and New Breeding Technologies Development Program (Project No. PJ016533), the Program of the Rural Development Administration, Korea, and the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry, and Fisheries (IPET) (319107-4), Republic of Korea.
Conflict of interest statement. The authors declare that this work was conducted in the absence of any commercial or financial relationship that could be construed as a conflict of interest.
Supplementary Material
Contributor Information
Inyoung Kim, Department of Molecular Biology, Sejong University, Seoul 05006, South Korea.
Eun-Ha Kim, Department of Agricultural Biotechnology, National Institute of Agricultural Science, Rural Development Administration, Jeonju 54874, South Korea.
Yu-ri Choi, Department of Molecular Biology, Sejong University, Seoul 05006, South Korea.
Hyun Uk Kim, Department of Molecular Biology, Sejong University, Seoul 05006, South Korea; Department of Bioindustry and Bioresource Engineering, Plant Engineering Research Institute, Sejong University, Seoul 05006, South Korea.
I.K., E.H.K., Y.C., and H.U.K. performed the experiments. I.K. and H.U.K. analyzed the data and wrote the paper. All authors read and approved the final manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Hyun Uk Kim (hukim64@sejong.ac.kr).
References
- Austin JR, Frost E, Vidi P-A, Kessler F, Staehelin LA (2006) Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18:1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan NH, Friso G, Rowland E, Majsec K, van Wijk KJ (2016) The plastoglobule-localized metallopeptidase PGM48 is a positive regulator of senescence in Arabidopsis thaliana. Plant Cell 28:3020–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743 [DOI] [PubMed] [Google Scholar]
- Dahlin C, Ryberg H (1986) Accumulation of phytoene in plastoglobuli of SAN-9789(Norflurazon)-treated dark-grown wheat. Physiol Plant 68:39–45 [Google Scholar]
- Davidi L, Levin Y, Ben-Dor S, Pick U (2015) Proteome analysis of cytoplasmatic and plastidic beta-carotene lipid droplets in Dunaliella bardawil. Plant Physiol 167:60–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deikman J, Hammer PE (1995) Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana. Plant Physiol 108:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deruere J, Romer S, d’Harlingue A, Backhaus RA, Kuntz M, Camara B (1994) Fibril assembly and carotenoid overaccumulation in chromoplasts: a model for supramolecular lipoprotein structures. Plant Cell 6:119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45:616–629 [DOI] [PubMed] [Google Scholar]
- Espinoza-Corral R, Herrera-Tequia A, Lundquist PK (2021) Insights into topology and membrane interaction characteristics of plastoglobule-localized AtFBN1a and AtLOX2. Plant Signal Behav 16:1945213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugeni Piller L, Glauser G, Kessler F, Besagni C (2014) Role of plastoglobules in metabolite repair in the tocopherol redox cycle. Front Plant Sci 5:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamez-Arjona FM, de la Concepcion JC, Raynaud S, Merida A (2014a) Arabidopsis thaliana plastoglobule-associated fibrillin 1a interacts with fibrillin 1b in vivo. FEBS Lett 588:2800–2804 [DOI] [PubMed] [Google Scholar]
- Gruszka J, Pawlak A, Kruk J (2008) Tocochromanols, plastoquinol, and other biological prenyllipids as singlet oxygen quenchers-determination of singlet oxygen quenching rate constants and oxidation products. Free Radic Biol Med 45:920–928 [DOI] [PubMed] [Google Scholar]
- Havaux M, Eymery F, Porfirova S, Rey P, Dormann P (2005) Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17:3451–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128:876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Pinzon I, Ross JH, Barnes KA, Damant AP, Murphy DJ (1999) Composition and role of tapetal lipid bodies in the biogenesis of the pollen coat of Brassica napus. Planta 208:588–598 [DOI] [PubMed] [Google Scholar]
- Jones AM, Bennett MH, Mansfield JW, Grant M (2006) Analysis of the defence phosphoproteome of Arabidopsis thaliana using differential mass tagging. Proteomics 6:4155–4165 [DOI] [PubMed] [Google Scholar]
- Kim DH, Xu ZY, Hwang I (2013) Generation of transgenic Arabidopsis plants expressing mcherry-fused organelle marker proteins. J Plant Biol 56:399–406 [Google Scholar]
- Kim EH, Lee Y, Kim HU (2015) Fibrillin 5 is essential for plastoquinone-9 biosynthesis by binding to solanesyl diphosphate synthases in Arabidopsis. Plant Cell 27:2956–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Wu SS, Ratnayake C, Huang AH (2001) Brassica rapa has three genes that encode proteins associated with different neutral lipids in plastids of specific tissues. Plant Physiol 126:330–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, DellaPenna D (2008) Tocopherol metabolism, oxidation and recycling under high light stress in Arabidopsis. Plant J 55:607–618 [DOI] [PubMed] [Google Scholar]
- Laizet Y, Pontier D, Mache R, Kuntz M (2004) Subfamily organization and phylogenetic origin of genes encoding plastid lipid-associated proteins of the fibrillin type. J Genome Sci Technol 3:19–28 [Google Scholar]
- Lee K, Lehmann M, Paul MV, Wang L, Luckner M, Wanner G, Geigenberger P, Leister D, Kleine T (2020) Lack of FIBRILLIN6 in Arabidopsis thaliana affects light acclimation and sulfate metabolism. New Phytol 225:1715–1731 [DOI] [PubMed] [Google Scholar]
- Li J, Yang J, Zhu B, Xie G (2019) Overexpressing OsFBN1 enhances plastoglobule formation, reduces grain-filling percent and jasmonate levels under heat stress in rice. Plant Sci 285:230–238 [DOI] [PubMed] [Google Scholar]
- Lippold F, vom Dorp K, Abraham M, Holzl G, Wewer V, Yilmaz JL, Lager I, Montandon C, Besagni C, Kessler F, et al. (2012) Fatty acid phytyl ester synthesis in chloroplasts of Arabidopsis. Plant Cell 24:2001–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohscheider JN, Rio BC (2016) Plastoglobules in algae: a comprehensive comparative study of the presence of major structural and functional components in complex plastids. Mar Genomics 28:127–136 [DOI] [PubMed] [Google Scholar]
- Lundquist PK, Poliakov A, Bhuiyan NH, Zybailov B, Sun Q, van Wijk KJ (2012) The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol 158:1172–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist PK, Poliakov A, Giacomelli L, Friso G, Appel M, McQuinn RP, Krasnoff SB, Rowland E, Ponnala L, Sun Q, et al. (2013) Loss of plastoglobule kinases ABC1K1 and ABC1K3 causes conditional degreening, modified prenyl-lipids, and recruitment of the jasmonic acid pathway. Plant Cell 25:1818–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinis J, Glauser G, Valimareanu S, Kessler F (2013) A chloroplast ABC1-like kinase regulates Vitamin E metabolism in Arabidopsis thaliana. Plant Physiol 162:652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutava RN, Prince SJK, Syed NH, Song L, Valliyodan B, Chen W, Nguyen HT (2015) Understanding abiotic stress tolerance mechanisms in soybean: a comparative evaluation of soybean response to drought and flooding stress. Plant Physiol Biochem 86:109–120 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Aoki Y, Tadaka S, Kagaya Y, Kinoshita K (2018) ATTED-II in 2018: a plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant Cell Physiol 59:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozueta-Romero J, Rafia F, Houlne G, Cheniclet C, Carde JP, Schantz ML, Schantz R (1997) A ubiquitous plant housekeeping gene, PAP, encodes a major protein component of bell pepper chromoplasts. Plant Physiol 115:1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23:1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, Lee H, Creelman RA, Mullet JE, Davis KR (2000) Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12:1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottet S, Besagni C, Kessler F (2015) The role of plastoglobules in thylakoid lipid remodeling during plant development. Biochim Biophys Acta 1847:889–899 [DOI] [PubMed] [Google Scholar]
- Ruan J, Zhou Y, Zhou M, Yan J, Khurshid M, Weng W, Cheng J, Zhang K (2019) Jasmonic acid signaling pathway in plants. Int J Mol Sci 20:2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Laremore TN, Smith PB, Maximova SN, McNellis TW (2012) Knockdown of FIBRILLIN4 gene expression in apple decreases plastoglobule plastoquinone content. PLoS ONE 7:e47547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Maximova SN, Jensen PJ, Lehman BL, Ngugi HK, McNellis TW (2010) FIBRILLIN4 is required for plastoglobule development and stress resistance in apple and Arabidopsis. Plant Physiol 154:1281–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, McNellis TW (2011) Fibrillin protein function: the tip of the iceberg? Trends Plant Sci 16:432–441 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Yuen GY, Lehman CC (1998) Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J 15:747–754 [DOI] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol 155:349–361 [DOI] [PubMed] [Google Scholar]
- Su SQ, Zhou YM, Qin JG, Yao WZ, Ma ZH (2010) Optimization of the method for chlorophyll extraction in aquatic plants. J Freshwater Ecol 25:531–538 [Google Scholar]
- Suzuki T, Tsunekawa S, Koizuka C, Yamamoto K, Imamura J, Nakamura K, Ishiguro S (2013) Development and disintegration of tapetum-specific lipid-accumulating organelles, elaioplasts and tapetosomes, in Arabidopsis thaliana and Brassica napus. Plant Sci 207:25–36 [DOI] [PubMed] [Google Scholar]
- Szymanska R, Kruk J (2010) Plastoquinol is the main prenyllipid synthesized during acclimation to high light conditions in Arabidopsis and is converted to plastochromanol by tocopherol cyclase. Plant Cell Physiol 51:537–545 [DOI] [PubMed] [Google Scholar]
- Ting JTL, Wu SSH, Ratnayake C, Huang AHC (1998) Constituents of the tapetosomes and elaioplasts in Brassica campestris tapetum and their degradation and retention during microsporogenesis. Plant J 16:541–551 [DOI] [PubMed] [Google Scholar]
- Torres-Romero D, Gomez-Zambrano A, Serrato AJ, Sahrawy M, Merida A (2022) Arabidopsis fibrillin 1-2 subfamily members exert their functions via specific protein-protein interactions. J Exp Bot 73:903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk KJ, Kessler F (2017) Plastoglobuli: plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Annu Rev Plant Biol 68:253–289 [DOI] [PubMed] [Google Scholar]
- Vidi PA, Kanwischer M, Baginsky S, Austin JR, Csucs G, Dormann P, Kessler F, Brehelin C (2006) Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J Biol Chem 281:11225–11234 [DOI] [PubMed] [Google Scholar]
- Vishnevetsky M, Ovadis M, Itzhaki H, Levy M, Libal-Weksler Y, Adam Z, Vainstein A (1996) Molecular cloning of a carotenoid-associated protein from Cucumis sativus corollas: homologous genes involved in carotenoid sequestration in chromoplasts. Plant J 10:1111–1118 [DOI] [PubMed] [Google Scholar]
- Wang TW, Balsamo RA, Ratnayake C, Platt KA, Ting JT, Huang AH (1997) Identification, subcellular localization, and developmental studies of oleosins in the anther of Brassica napus. Plant J 11:475–487 [DOI] [PubMed] [Google Scholar]
- Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S (2011) Assembly of designer TAL effectors by Golden Gate cloning. PLoS ONE 6:e19722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wydro M, Kozubek E, Lehmann P (2006) Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim Polonic 53:289–298 [PubMed] [Google Scholar]
- Yang Y, Sulpice R, Himmelbach A, Meinhard M, Christmann A, Grill E (2006) Fibrillin expression is regulated by abscisic acid response regulators and is involved in abscisic acid-mediated photoprotection. Proc Natl Acad Sci USA 103:6061–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Cho Y, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572 [DOI] [PubMed] [Google Scholar]
- Youssef A, Laizet Y, Block MA, Marechal E, Alcaraz JP, Larson TR, Pontier D, Gaffe J, Kuntz M (2010) Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress. Plant J 61:436–445 [DOI] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier JB, van Wijk KJ (2006) Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol 140:984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbierzak AM, Kanwischer M, Wille C, Vidi PA, Giavalisco P, Lohmann A, Briesen I, Porfirova S, Brehelin C, Kessler F, et al. (2010) Intersection of the tocopherol and plastoquinol metabolic pathways at the plastoglobule. Biochem J 425:389–399 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Turner JG (2008) Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3:e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.