Abstract
Red foliated cotton is a typical dominant mutation trait in upland cotton (Gossypium hirsutum). Although mutants have been described, few responsible genes have been identified and characterized. In this study, we performed map-based cloning of the red foliated mutant gene (Re) derived from the cross between G. hirsutum cv. Emian22 and G. barbadense acc. 3–79. Through expression profiling, metabolic pathway analysis, and sequencing of candidate genes, Re was identified as an MYB113 transcription factor. A repeat sequence variation in the promoter region increased the activity of the promoter, which enhanced the expression of Re. Re expression driven by the 35S promoter produced a red foliated phenotype, as expected. When the gene was driven by a fiber elongation-specific promoter, promoter of α-expansin 2 (PGbEXPA2), Re was specifically expressed in 5- to 10-day post-anthesis fibers rather than in other tissues, resulting in brown mature fibers. Re responded to light through phytochrome-interacting factor 4 and formed a dimer with transparent testa 8, which increased its expression as well as that of anthocyanin synthase and UDP-glucose:flavonoid 3-o-glucosyl transferase, and thus activated the entire anthocyanin metabolism pathway. Our research has identified the red foliated mutant gene in cotton, which paves the way for detailed studies of anthocyanin and proanthocyanidin metabolism and pigment accumulation in cotton and provides an alternative strategy for producing brown fiber.
The ectopic expression of Re, an MYB113 transcription factor resulting in red foliated cotton, produces brown fiber cotton by activating the anthocyanin and proanthocyanidin metabolic pathways.
Introduction
The red coloration of upland cotton (Gossypium hirsutum) plants, resulting from excessive anthocyanin accumulation in light, is a very prominent mutant phenotypic trait. Anthocyanins respond to a range of environmental and developmental signals; they are frequently induced by sunlight and other stresses and are believed to protect plants against UV irradiation, low temperature, pathogen invasion, and herbivore feeding (Tanaka et al., 2008; Nakabayashi et al., 2014; Santos-Buelga et al., 2014; Liang and He, 2018; Xu and Rothstein, 2018). Environmental factors, especially light, play a vital role in the synthesis and accumulation of anthocyanins (Takos et al., 2006; Li et al., 2016a, 2016b, 2016c; Liu et al., 2021).
Chemically, glycosylated anthocyanins are synthesized through the flavonoid pathway and stored in vacuoles (Winkel-Shirley, 2001; Shi and Xie, 2014; Jiang et al., 2015; Luo et al., 2018). The biosynthetic pathway of anthocyanin has been well characterized in plants (Martin et al., 1991; Holton and Cornish, 1995). A series of structural genes (e.g. CHS: chalcone synthase, CHI: chalcone isomerase, F3H: flavanone 3-hydroxylase, DFR: dihydroflavonol 4-reductase, ANS: anthocyanin synthase, ANR: anthocyanidin reductase, UFGT: UDP-glucose:flavonoid 3-o-glucosyl transferase) of the flavonoid pathway have been discovered in various plant species (Winkel-Shirley, 2001; Koes et al., 2005). In addition, some genes involved in the transport and accumulation of anthocyanins, such as glutathione-S transferase (GST), have also been reported (Marrs et al., 1995; Mueller et al., 2000; Kitamura et al., 2004). MYB subgroup 6 (MYB-Sg6) family genes with conserved KPRPR[S/T]F motif regulators are widely considered to be involved in regulating the anabolic metabolism of anthocyanins (Dubos et al., 2010; Wang et al., 2010; Xu et al., 2021) at the transcription level. In Arabidopsis thaliana, the Production of Anthocyanin Pigment 1 (PAP1/MYB75) and its homologous genes (PAP2/MYB90, MYB113, and MYB114) encoding R2R3-MYB proteins have been identified as major regulators of anthocyanin biosynthesis. Overexpression of PAP1 by activation tagging results in the accumulation of anthocyanins in all major organs of Arabidopsis (Borevitz et al., 2000). PAP1 homologs have been demonstrated to control the anthocyanin pathway in various plants, such as tomato (Solanum lycopersicum), carrot (Daucus carota), grapevine (Vitis vinifera), and apple (Malus × domestica) (Borevitz et al., 2000; Schwinn et al., 2006; Espley et al., 2007; Ballester et al., 2010; Albert et al., 2015). In addition, MYB transcription factors (TFs), basic-helix-loop-helix (bHLH) TFs, and WD40 family proteins always combine into a ternary complex (MBW) to regulate the expression of structural genes (Ramsay and Glove, 2005; Gonzalez et al., 2008; Lu et al., 2021). Studies in Arabidopsis have shown that Transparent Testa Glabra1 (TTG1, a WD40 protein), Transparent Testa 8 (TT8), a bHLH TF, and MYB75/MYB113/MYB114 can form an MBW complex to regulate the expression of genes such as DFR and UFGT (Gonzalez et al., 2008; Wei et al., 2019).
Cotton is a major fiber crop, and brown-fiber cotton is an interesting combination of fiber and pigment. The genetic basis of brown fiber (G. hirsutum) was revealed by linkage and association mapping (Wen et al., 2018). Yan et al. (2018) upregulated the expression of GhTT2-3A in fibers at the secondary wall-thickening stage to produce brown mature fibers. Liu et al. (2020) located and identified the inheritance of the Lgf locus that controls the green fluff of upland cotton. By comparing white-fiber, brown-fiber, and green-fiber cotton, Li et al. (2020) found that the metabolism and pigmentation of phenylpropane compounds showed different patterns between brown-fiber and green-fiber cotton.
In cotton, three anthocyanin-related loci, R1, R2, and Rd, have been reported to be responsible for anthocyanin accumulation in red-colored cotton. The R1 locus is related to red stem and foliar organs, the R2 locus is related to red petal spots, and these two loci are homeologous (Killough and Horlacher, 1933; Stephens, 1974). Rd, which produces a red dwarf plant, is incompletely dominant (Percy et al., 2015). The red plant (R1) gene is a traditional genetic marker in cotton (Zhao et al., 2009). The R1 gene was mapped to an interval of 3.4 cM between simple sequence repeat (SSR) markers NAU4956 and NAU6752 on chromosome 16; a carotenoid synthase gene, GhPSY (Gohir. D07G079100, GenBank accession no. KF923933) was identified as the candidate gene for the red foliated phenotype (Cai et al., 2014). In another study, Gao et al. (2013) induced anthocyanin production in the hairy roots of Antirrhinum majus and cotton by ectopic expression of Red Leaf Cotton 1, Gohir. D7G082100) that was homologous to PAP1. By using linkage mapping in a recombinant inbred line (RIL) population constructed by T586 × Yumian1, combined with transcription analysis and genetic transformation, GhPAP1D (Gohir. D7G082100) was found to be the determinant gene of R1 phenotypes (Li et al., 2019a, 2019b).
In this study, F2 populations were constructed by using G. hirsutum cv. Emian22 (E22) and a red foliated mutant line that appeared in the cross between E22 and Sea-island cotton 3–79 (Nie et al., 2015), and the red foliated mutant gene (Re) was mapped. Through metabolic pathway analysis and expression assessment, the candidate gene of Re was identified, and its function was verified by genetic transformation. We also ectopically expressed Re in cotton fibers to change the fiber color, and anthocyanins and proanthocyanidins (PAs) accumulated in the fibers of transgenic plants to produce brown fiber. We thus developed a strategy to generate brown color fiber in cotton.
Results
Phenotype and inheritance of the red foliated mutant
In our previous study, a red foliated cotton mutant, designated temporarily as Re, was produced from the backcross progenies between G. hirsutum cv. E22 and Gossypium barbadense acc. 3–79 (Supplemental Figure S1a). The main organs of the whole mutant plant were a purple-red color (Supplemental Figure S1b). Two F2 segregation populations were constructed using the two parents, E22 and ReS9 (Supplemental Figure S1c). In the segregation population, three phenotypes appeared, including the red foliated plant, the intermediate type, and the green plant (Supplemental Figure S1, c and d), and the segregation ratio fit a Mendelian 1:2:1 inheritance (Supplemental Table S1).
Mapping of the Re gene
To primarily map the Re gene, 494 markers that covered 26 chromosomes uniformly in 10-cM units were selected (Supplemental Table S2), and polymorphisms were screened between the two parents (Figure 1A) and the two bulks. A total of 92 markers were polymorphic between the two parents (Supplemental Table S3), but none of them were polymorphic between the red and green pools. Furthermore, all the markers in the genetic linkage map were used for polymorphism screening, and a total of 7 markers exhibited polymorphisms: MON_C2-0108, MON_DC40065 and NAU3676 on Chr16, HAU2675 and BNL3558 on Chr18, MON_CGR6270 on Chr11, and JESPR175 on Chr13. To clarify the linkage between these seven markers and the Re gene, these markers were used to genotype recessive plants from the F2 population with 952 plants in 2014. Only the marker MON_DC40065 on Chr16 was linked to Re (Figure 1B), and the genetic distance was 1.378 cM. To finely map the gene, more markers were needed. The markers surrounding MON_DC40065 within 5 cM in the map were anchored to Gossypium raimondii, the only genome sequence available at that time, and the genome range of 8,839,721–15,295,694 bp on Chr01 was aligned (Supplemental Figure S2). A total of 470 SSR markers were developed in this region (Supplemental Table S4), and 8 markers were polymorphic between the parents and the two bulks. After linkage analysis, markers Chr01_8M-004, Gr01_9-13M_056, and MON_DC40065 were mapped next to the gene. To narrow down the mapping region, genes were searched in the markers’ corresponding genome sequence, 8,030,000–9,640,000 bp on Chr01 of G. raimondii. A total of 116 genes were found in 8.04–9.65 Mb, and 126 markers were developed based on the 3′-untranslated region (3'-UTR) and 5′-UTR sequence of these genes (Supplemental Table S5), resulting in one polymorphic marker, G089200.1-3′. At this time, the upland cotton genome was released, and the 8.04–9.65-Mb genomic sequence on Chr01 of G. raimondii was aligned to 8.15–9.64 Mb on D07 of upland cotton. An additional 80 SSR markers were developed (Supplemental Table S6), and only the marker Gh_D07_8-9M_77 was polymorphic.
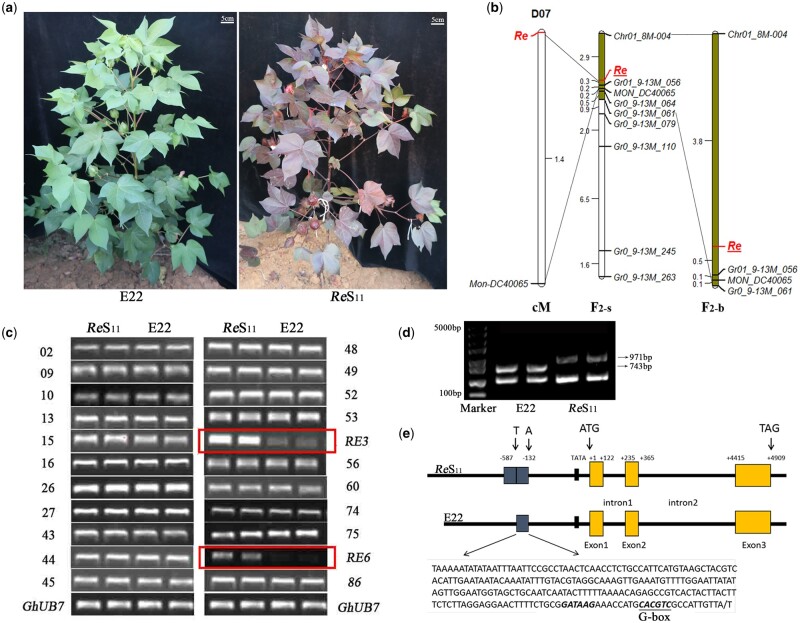
Figure 1.
Map-based cloning of Re. A, Field photos of E22 and ReS11.ReS11 showed a red color in the main tissues, except for the fiber. B, Linkage mapping of Re. The F2-s population contained 318 F2 recessive plants obtained in 2013 and 2014. The F2-b population contained a total of 1,898 F2 recessive plants obtained in 2013, 2014, and 2015. C, Expression analysis of some candidate genes in the interval by RT-PCR. Two genes, RE3 and RE6, showed differences in expression. GhUB7 was used as an internal control gene. D, Large fragment differences were detected in the Re promoter region. The small fragment below represents a homologous amplified sequence. E, Schematic diagram of the structure of Re between E22 and ReS11. A 288-bp duplication was identified upstream of the start codon of Re in ReS11, and a G-box was identified in the repetitive fragment.
To date, a total of 11 markers have shown polymorphism between the parents and the bulks (Supplemental Table S7). First, a total of 318 F2 recessive plants obtained in 2013 and 2014 were used to conduct linkage mapping, which was defined as the F2-s population. Re was located between markers Chr01_8M-004 and Gr01_9-13M_056, with genetic distances of 2.9 and 0.3 cM, respectively, and physical distances of ∼1.5 Mb (Figure 1B). In 2015, a large F2 population was developed, and a total of 1,898 F2 recessive plants combining the former populations (defined as the F2-b population) were genotyped with Chr01_8M-004, Gr01_9-13M_056, Gr01_9-13M_061, and MON_DC40065 (Figure 1B). Finally, Re was located between the markers Chr01_8M-004 and Gr01_9-13M_056, the genetic distances were 3.8 and 0.5 cM, respectively, over a physical distance of ∼1.5 Mb (Figure 1B).
Cloning of the Re gene
There were 88 genes within 8.15–9.64 Mb on the upland cotton D07 chromosome, where the linkage markers Chr01_8M-004, Gr01_9-13M_056, and MON_DC40065 corresponded to (Supplemental Table S8). Seven genes were associated with the process of pigment synthesis and transport through Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, including RE1 to RE7 (Supplemental Table S9). Reverse transcription-polymerase chain reaction (RT-PCR) of all 88 genes between E22 and ReS11 showed that the expression levels of both RE3 and RE6 were substantially different (Figure 1C). RE3 was shown to encode a GST. RE6 was annotated as MYB TF 113 (MYB113), which belongs to the MYB-Sg6. These two genes were considered as candidate genes for Re.
These two genes were cloned and sequenced, but no sequence differences were found in 2,000-bp upstream, 1,000-bp downstream, or in the coding region of RE3 and RE6 between E22 and ReS11. This confusing result made us doubt the RT-PCR result. Genes tend to have multiple copies in allotetraploid cotton, and the expression difference could be the result of nonspecific amplification. The sequencing result of the RT-PCR products of RE3 was specific; however, two results of RE6 were identified because of the four single nucleotide polymorphism sites (Supplemental Figure S3a). One gene sequence was consistent with Gh_D07G0770 (RE3), and another was consistent with Gh_D07G2413 (RE8) that we did not identify at first (Supplemental Figure S3a). RE8 was not in the 8.15–9.64 Mb on chromosome D07 but was located in a scaffold that belonged to chromosome D07. RE8 was also annotated as MYB113, and RE8, RE6, and RE7 (Gh_D07G0771) had a high sequence similarity, which caused RE8 to fail to be assembled into the genome correctly. The latest upland cotton genome confirmed our hypothesis (Wang et al., 2019). Two specific primers based on the differential sequences of RE6 or RE8 were designed. The updated results showed that the expression of RE8 in ReS11 was substantially higher than that of E22 and RE6 showed no expression difference between ReS11 and E22 (Supplemental Figure S3c).
The sequencing of RE8 showed that the cDNA sequences were the same between parents. The most obvious difference was that ReS11 had an additional 288-bp repeat sequence in the 5′-UTR (Figure 1, D and E; Supplemental Figure S3b), which contains a G-box (Figure 1E). The repeat fragment was detected in F2 plants and was not detected in any of the green plants; it cosegregated with the red phenotype. Combined with the metabolic pathway analysis, differences in expression levels, and sequence differences, we speculated that both RE3 and RE8 should be involved in the metabolic pathway of anthocyanins. However, RE8 should be the main reason for the production of red foliated cotton, and was identified as the final candidate gene of Re.
Functional verification of the Re gene
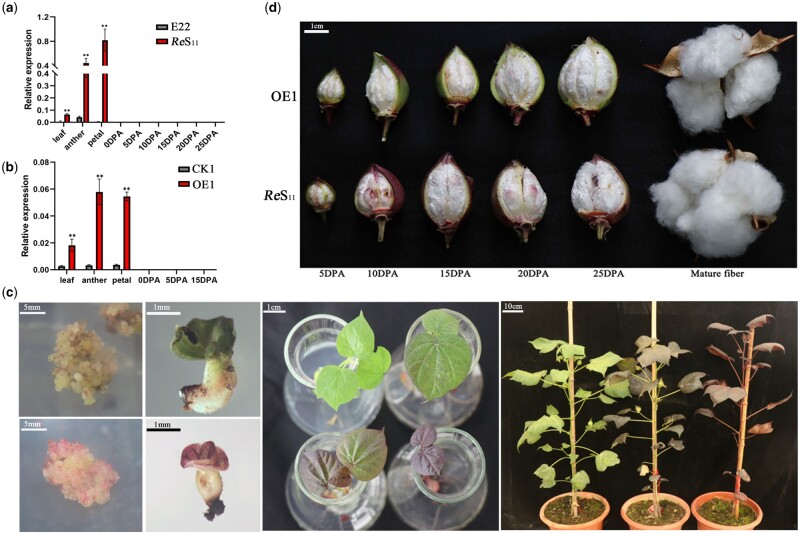
Subcellular localization showed that Re functions in the nucleus (Supplemental Figure S4). The tissue expression pattern of Re between parents showed that Re had a higher expression level in leaves, petals, and anthers of ReS11 than those in E22 (Figure 2A), and almost no expression in the ovules and the fibers at different developmental stages.
Figure 2.
Overexpression of Re shows a red foliated phenotype but not in the fiber. A, Tissue expression pattern of Re between E22 and ReS11.Re was highly expressed in the main tissues of ReS11, except in the fiber. DPA is the abbreviation of days post-anthesis. B, Tissue expression patterns of overexpression lines CK1 and OE1. Re was highly expressed in the main tissues of OE1, except for in the fiber. For (A) and (B), error bars represent the ±sd (three biological replicates), and the statistical significance was calculated using a t test (**P < 0.01, *P < 0.05). C, Anthocyanins accumulated at different stages during genetic transformation. From left to right are the calli, seedling regeneration, rooting culture and transgenic T0 plants from the transformation of 35S:Re. Normal light green and obvious red calli were observed from the callus stage. The regenerated seedlings showed red or green color (transgenic negative control). Finally, transgenic plants with different shades of red were obtained. D, Neither OE1 nor ReS11 produced a red color in the developing or the mature fibers.
Re was driven by the constitutive promoter 35S and transformed into the cotton high-efficiency transformation receptor Jin668 (Li et al., 2019a, 2019b). From the callus tissue stage, obvious anthocyanin accumulation was observed (Figure 2C). Seedlings differentiated from the callus also showed obvious red color, and transgenic T0 plants showing different degrees of red color were obtained, which may be caused by different copy numbers of transgenes or physiological differences (Figure 2C). The overexpression lines showed significant anthocyanin accumulation in leaves, stems, sepals, petals, and anthers, but the fiber was still white (Figure 2, C and D). The tissue expression pattern of Re between the overexpression negative line (CK1) and the positive line (OE1) was tested, and Re had a higher expression in the leaves, anthers, and petals in OE1 than in CK1, but still had a low expression in ovules and fibers at different developmental stages (Figure 2B). Failure to express Re in fibers may be the main reason why the mature fibers of ReS11 and OE1 were still white (Figure 2D).
Specific expression of Re in fiber produces brown fiber
To explore the potential of accumulating pigments in fibers to produce color fibers, the fiber elongation-specific promoter PGbEXPA2 was used to specifically express the Re gene in 5–10 d post-anthesis (DPA) fibers (Li et al., 2015). As expected, in the PGbEXPA2:Re transformed T0 plants, most organs did not show anthocyanin accumulation (Figure 3A); in contrast, a clear purple-red color produced by pigment accumulation was observed in the developing fibers (Figure 3B). Compared with the negative control, a high expression of Re in 5–10 DPA fibers was detected in the positive transgenic plants (Figure 3C). Interestingly, the final mature fiber turned out to be brown (Figure 3B; Supplemental Figure S5).
Figure 3.
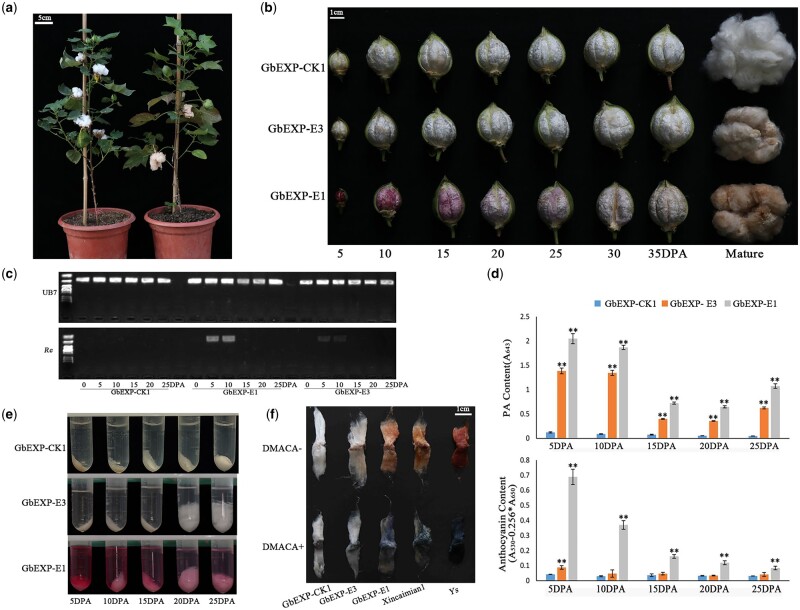
Specific expression of Re in fiber producing a brown fiber cotton. A, A fiber-specific transgenic T0 line of Re (right) and a transgenic negative control (left) are shown. The main tissues of the Re-expression line remained green, and the mature fiber was brown. B, Fiber colors at different developmental stages of three transgenic lines driven by fiber-specific promoters. GbEXP-CK1 was used as the transgenic negative control line. GbEXP-E3 did not show a red color during development, and the mature fiber was brown; GbEXP-E1 appeared red during development, and the mature fiber was brown. C, High and specific expression of Re in 5–10DPA fibers of GbEXP-E1, and lower but detectable expression in fibers of GbEXP-E3. D, Determination of the relative content of PA and anthocyanin in the fibers. Error bars represent ±sd (three biological replicates). PA and anthocyanin content of GbEXP-E1 and GbEXP-E3 were compared with that of GbEXP-CK1, and the statistical significance was calculated using a t test (**P < 0.01, *P < 0.05). E, Extraction of anthocyanins from 5 to 25 DPA fibers. F, Staining of mature fiber by DMACA. XC1 and Ys are light brown and dark brown fiber cotton, respectively, which were used as controls. The darker brown the fiber was, the deeper the blue color was after staining with DMACA.
We analyzed the relative content of PAs and anthocyanins in the fibers at different stages to explore the changes in related substances in the fiber during the process of pigment accumulation. The transgenic line GbEXP-E3 that did not show color during fiber development and finally showed a light brown color, and the transgenic line GbEXP-E1 that showed red fiber during development and finally showed a brown color, were chosen, and the transgenic negative plant GbEXP-CK1 was chosen as the control (Figure 3B). Compared with GbEXP-CK1, the relative contents of PAs and anthocyanins in GbEXP-E1 and GbEXP-E3 were greatly increased, with those higher in GbEXP-E1 than in GbEXP-E3. Anthocyanin and PA contents peaked in 5 and 10 DPA fibers (Figure 3D). Then, anthocyanins gradually decreased with the development of fibers, and the fiber color also faded gradually (Figure 3, B, D, and E). The PA content reached the lowest level at 20 DPA, and the relative content gradually increased at 25 DPA (Figure 3D). This may be because the fiber starts to dehydrate at this time, resulting in an increase in the relative content of PA in fibers.
Studies have shown that the production of brown cotton is due to the accumulation of PAs in the fiber (Feng et al., 2014; Yan et al., 2018). In this experiment, the mature fiber finally appeared brown instead of the expected red. We first stained mature fibers with 4-dimethylaminocinnamaldehyde (DMACA). The light brown cotton material Xincaimian1 (XC1) and the dark brown cotton material Ys were used as controls (Wen et al., 2018). As a result, the mature fibers of GbEXP-E3 and GbEXP-E1 both showed a clear blue color (Figure 3F). The darker the brown fiber was, the darker the blue color that was observed. Combined with the final content changes in anthocyanin and PA, we speculate that the accumulation of PAs produces the brown fiber phenotype.
Re is involved in regulating multiple genes in the pathway of anthocyanin synthesis
The chlorophyll and anthocyanin contents that directly affect the color of the leaves were measured. No significant difference in chlorophyll content was identified in the leaves of the parents and overexpression lines (Supplemental Figure S6a). The anthocyanin content in leaves of ReS11 and OE1 was significantly higher than that of green plants (Supplemental Figure S6b). Considering that Re caused changes in PA content in the fibers, it was worth exploring whether the PA content in the leaves had changed. No difference was detected between the leaves of E22 and ReS11, but PA content in OE1 leaves significantly increased compared with CK1 (Supplemental Figure S7).
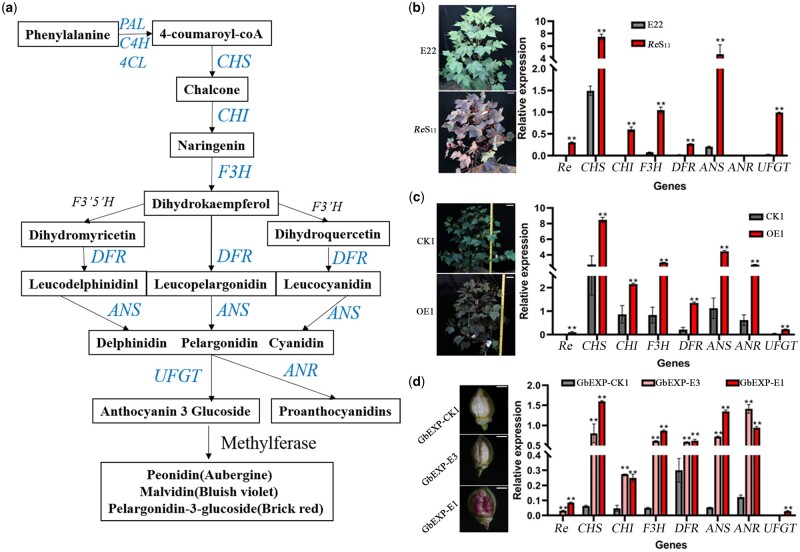
The anthocyanin biosynthesis pathway is relatively conserved (Figure 4A). Through RT-quantitative PCR (RT-qPCR), the expression levels of seven major structural genes (CHS, CHI, F3H, DFR, ANS, ANR, and UFGT) and Re were tested in the parents and overexpression lines. Except for ANR, the other six genes showed a significant expression increase in ReS11 compared with E22 (Figure 4B), which was consistent with the result of large amounts of anthocyanins but not PA accumulating in ReS11. ANR is the key enzyme for the conversion of delphinium, pelargonidin, and cyanidin into PA (Xie et al., 2004; Liu et al., 2013), and UFGT is responsible for converting these three intermediate pigments into anthocyanins of different colors (Boss et al., 1996; Winkel-Shirley, 2001; Figure 4A). Compared with CK1, besides the other five structural genes, the expression of UFGT and ANR in OE1 increased, which caused both the anthocyanin and PA contents to increase (Figure 4C). The PA content only showed a difference between CK1 and OE1, which may be caused by the different background expression levels of these structural genes in E22 and the transgenic receptor Jin668. The expression of several structural genes in E22, except for CHS, was relatively low but was still relatively high in CK1 when compared with the internal reference gene Gh_Ub7 (Figure 4, B and C).
Figure 4.
Expression levels of the main genes in the anthocyanin synthesis pathway. A, Schematic diagram of the main pathways of anthocyanin metabolism in plants drawn based on existing research (Koes et al., 2005). B, The expression of six structural genes, except ANR, was significantly higher in ReS11 than in E22. Field photos of E22 and ReS11 are on the left, and the scale bar represents 10 cm. C, Seven structural genes in OE1 showed significantly higher expression than in CK1. Field photos of CK1 and OE1 are on the left, and the scale bar represents 10 cm. D, The expression of seven structural genes in 5 DPA fibers of three transformed lines driven by fiber-specific promoters. Six structural genes, except UFGT, in GbEXP-E3 and GbEXP-E1 showed significantly higher expression than in GbEXP-CK1, and UFGT only showed significantly increased expression in GbEXP-E1. Five DPA Fiber photos of three transformed lines are on the left, and the scale bar represents 5 mm. The relative expression level was obtained by RT-qPCR. For (B–D), error bars represent ±sd (three biological replicates), and the statistical significance was calculated using a t test (**P < 0.01, *P < 0.05).
When testing the gene expression in 5 DPA fibers driven by fiber-specific promoter, among the first six structural genes, both GbEXP-E3 and GbEXP-E1 showed a significant increase compared with GbEXP-CK1 (Figure 4D). The expression of UFGT only significantly increased in GbEXP-E1 but not in GbEXP-E3, which was consistent with the accumulation of anthocyanins in GbEXP-E1 and GbEXP-E3 (Figures 3, B, E, and 4, D). We also noticed that the expression of ANR in GbEXP-E3 was higher than that of GbEXP-E1 (Figure 4D), which seemed to have a certain antagonistic effect on the expression of ANR and UFGT.
Comprehensive analysis of the expression of structural genes suggests that overexpression of Re can induce an overall increase in the expression of the major structural genes in the anthocyanin metabolism pathway and ultimately produce a large amount of anthocyanin accumulation.
Transcriptome analysis of the cotton anthocyanin metabolism network
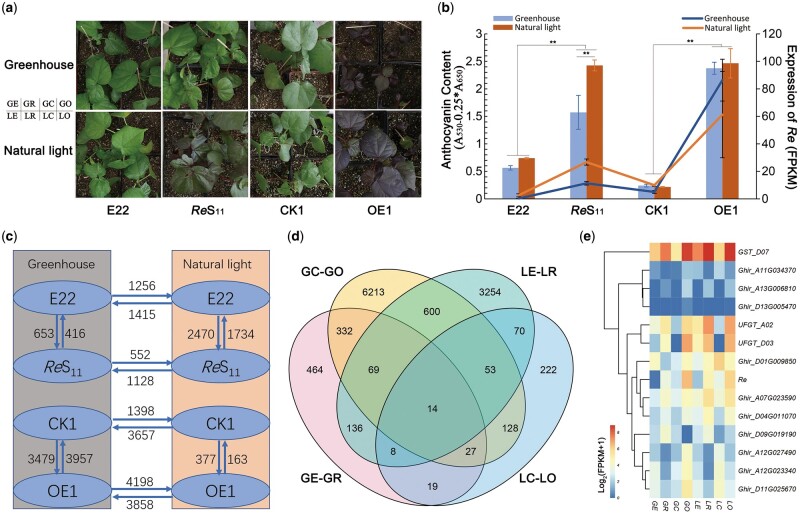
In the field, we observed that the overlapping part of ReS11 leaves appeared greenish, and continuous rainy days also made the red leaves lighter. The accumulation of anthocyanins may be light induced. A bagging experiment was carried out in the field by covering a branch of the ReS11 plant with a kraft brown paper bag for 7 d and using the branch of the same plant at the same position as a control (Supplemental Figure S8). As expected, the bagged branches showed a significant reduction in anthocyanin accumulation compared with the unbagged branches (Supplemental Figure S8). On the bagged branches, the bagged part and the unbagged stems showed obvious red and green dividing lines (Supplemental Figure S8), indicating that light plays a vital role in the accumulation of anthocyanins. To explain the mechanism of this phenomenon, ReS11, E22, CK1, and OE1 were planted and grown under natural light and greenhouse conditions for 3 weeks, and the leaves were taken for anthocyanin content determination and transcriptome analysis. We used “G, L” to represent “greenhouse” and “natural light” conditions, respectively, and “R, E, C, O” to represent ReS11, E22, CK1, and OE1, respectively (Figure 5A); eight combinations are: GR, GE, GC, GO, LR, LE, LC, and LO. LR showed the accumulation of anthocyanin visible to the naked eye, while GR showed the same green color as E22 (Figure 5A). OE1 showed a consistent red color in both environments (Figure 5A). ReS11 and OE1 also showed significantly higher anthocyanin contents than E22 and CK1 in both environments (Figure 5B). Although the anthocyanin content was still relatively high in GR, it was significantly lower than in LR, and the other lines did not show significant differences in the two environments (Figure 5B). Transcriptome data showed that the fragments per kilobase of exon per million mapped reads (FPKM) value of Re in LE, LR, and LC was twice that in the greenhouse, while LO did not show a higher expression than GO (Figure 5B). As a constitutive promoter, the 35S promoter is likely not affected by light.
Figure 5.
Light affects Re expression and anthocyanin accumulation. A, Leaf colors of ReS11, E22, CK1 and OE1 under greenhouse and natural light conditions. ReS11 leaves turned green under greenhouse conditions. B, Re expression and relative anthocyanin content of leaves of four lines in two environments. The bar graph represents the relative content of anthocyanin, and the broken line graph represents the relative expression of Re. Error bars represent ±sd (three biological replicates), and the statistical significance was calculated using a t test (**P < 0.01). C, DEG numbers between pairwise comparisons. The pointing of the arrow represents increased expression, and the number represents the DEGs. D, Venn diagram of overlapping DEGs between GC–GO, LE–LR, GE–GR, and LC–LO. In which, “G, L” represent “greenhouse” and “natural light” conditions, respectively; “R, E, C, O” represent ReS11, E22, CK1, and OE1, respectively. E, Heatmap of the expression levels of the 14 overlapping DEGs.
In both environments, Re maintained a higher expression level in OE1 than in ReS11 (Figure 5B), but the anthocyanin content remained unchanged and was basically the same as ReS11 (Figure 5B), showing the upper limit of the accumulation of anthocyanins in cotton leaves. We also observed the relatively weak growth of GO (Figure 5A), which may result from too much energy depletion from accumulating numerous anthocyanins. However, LO maintained the same growth trend as LC, which indicated that adequate exposure to natural light can compensate for the negative effects caused by the excessive accumulation of anthocyanins. And anthocyanins accumulation can provide protection from excess light, an advantage of outdoor growth of red foliated cotton.
Based on the transcriptome data, we first compared differentially expressed genes (DEGs) between each pair of lines (Figure 5C). The number of DEGs between GE and GR was 1,069, while that between LE and LR was 4,204 (Figure 5C; Supplemental Table S10), reflecting the substantial influence of natural light on ReS11 gene expression. The number of upregulated genes between GR and LR was twice the number of downregulated genes (1,128/552) (Figure 5C; Supplemental Table S10), reflecting that natural light mainly promotes the expression of related genes. LC–LO had the fewest DEGs. The two lines were derived from segregating offspring and had similar genetic backgrounds, and the growth of LC and LO was basically the same under natural light. More DEGs were identified when GC and LO were compared to GO (Figure 5C). The anthocyanin contents of GE–GR, LE–LR, LC–LO, and GC–GO all showed significant differences (Figure 5B), and 14 DEGs were identified among all four comparisons (Figure 5, D and E). In addition to Re, there were three familiar genes with known functions, UFGT (Ghir_A02G015500, Ghir_D03G005110) and GST (Ghir_D07G008160) (Figure 5E).
To identify some light-induced factors, we first compared the DEGs of GR–LR, GO–LO, GE–LE, and GC–LC, and 247 DEGs were identified among all four groups of comparisons (Supplemental Figure S9a; Supplemental Table S10). These 247 DEGs may represent the basic genes of upland cotton under light regulation (Supplemental Figure S9a). KEGG results showed that these DEGs were mainly enriched in metabolic pathways and circadian rhythm pathways (Supplemental Figure S9b), including genes related to the photoreceptor pathway, such as COP1 (Ghir_A10G000590) and TOC1 (Ghir_A05G042880), and some genes participating in flavonoid metabolism, such as FLS (flavonol synthase/flavanone 3-hydroxylase, Ghir_D04G002380) and CHS (Ghir_D09G000030) (Supplemental Table S10). In the 493 GR–LR-specific DEGs (Supplemental Figure S9a), regulatory factors should respond to light and induce Re, and 253 genes showed upregulated expression (Supplemental Figure S9c). Among the 253 upregulated genes, one PIF4 (Ghir_D09G000770) gene was included (Supplemental Table S10). The PIF4 TF has been reported to be involved in the phytochrome B signaling pathway and may regulate gene expression by binding to the G-box motif (Pedmale et al., 2016; Liu et al., 2021).
Weighted gene coexpression network analysis of the core gene set of upland cotton anthocyanin metabolism
Based on transcriptome data, weighted gene coexpression network analysis (WGCNA) was used to conduct a gene coexpression analysis to determine the core gene set in the anthocyanin metabolic pathway in upland cotton. All DEGs between pairwise comparisons were divided into 34 modules (Supplemental Figure S10a), where the MEdarkgrey module showed the highest correlation with anthocyanin content in leaves (Supplemental Figure S10b). KEGG results showed that genes in MEdarkgrey were mainly involved in metabolism, such as the metabolism of glutathione and phenylalanine (Supplemental Figure S10c). This module contained a number of critical structural genes involved in flavonoid metabolism, such as PAL (Ghir_A04G008120, Ghir_D04G012180), UFGT (Ghir_D03G005110), DFR (Ghir_A06G000790), GST (Ghir_D07G008160, Ghir_A07G008080), and GT6 (UDP-glucose flavonoid 3-O-glucosyltransferase 6, Ghir_D07G020010) (Supplemental Figure S10, d and e; Supplemental Table S11). This module can be considered the core gene set involved in anthocyanin synthesis in upland cotton. However, Re was classified into MEsteelblue instead of MEdarkgrey, which also showed a relatively high correlation with anthocyanin content in leaves (Supplemental Figure S10b), and the Re coexpression genes did not include structural genes in the flavonoid metabolic pathway (Supplemental Figure S10, f and g; Supplemental Table S12). This may illustrate that Re can increase expression of some structural genes in the core gene set, which leads to increased anthocyanin synthesis.
The regulatory network of Re
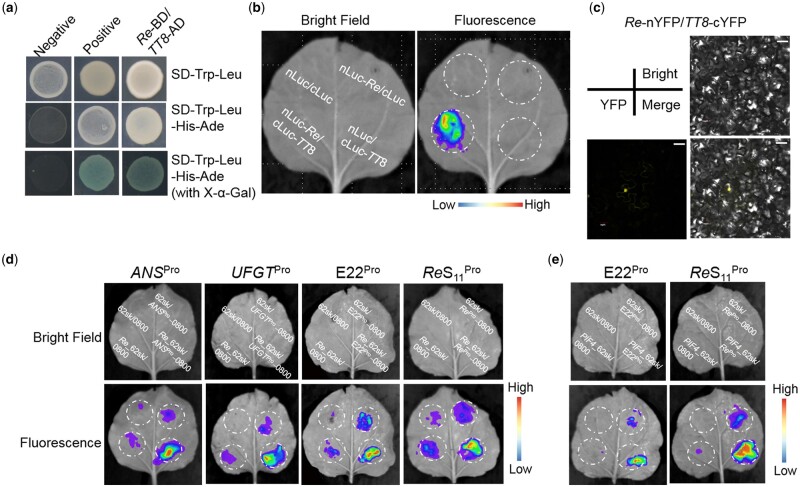
Compared with E22, a 228-bp sequence repeat was found in the promoter region of Re in ReS11 (Figure 1, D and F), which may enhance the driving ability of Re and lead to a higher expression of Re in ReS11. Luciferase (LUC) driven by Re demonstrated a stronger signal than that driven by E22Pro, which verified our hypothesis (Supplemental Figure S11).
Through a yeast two-hybrid experiment, Re was found to interact with TT8 (Figure 6A). To verify these interactions, LUC complementation imaging (LCI) and bimolecular fluorescence complementation (BiFC) assays were performed (Figure 6, B and C). To identify the downstream genes directly regulated by Re, we constructed promoter vectors of the seven structural genes (CHSpro, CHIpro, F3Hpro, DFRpro, ANSpro, ANRpro, and UFGTpro) and GST (GSTpro), which appeared in the core gene set. Through a dual-LUC reporter system, Re was verified to bind to ANSpro and UFGTpro and drive the expression of these genes (Figure 6D; Supplemental Figure S12). Studies have shown that some MYB TFs can bind to their own promoter regions (Espley et al., 2009), and Re was found to be able to bind to both Re and E22pro (Figure 6D). As a light-responsive factor, PIF4 may regulate the expression of Re, which was confirmed by LUC experiments (Figure 6E).
Figure 6.
Re interacts with TT8 to regulate the expression of Re, ANS, and UFGT. A, Yeast two-hybrid assays showing the interaction of Re and TT8. Yeast cells were plated on SD–Trp–Leu, SD–Trp–Leu–His-Ade, and SD–Trp–Leu–His–Ade (with X-α-Gal) media. B, LCI assays of nLuc-Re with cLuc-TT8. C, BiFC assays between Re-nYFP and TT8-cYFP. The scale bar represents 30 μm. D, LUC assays of Re_62sk with ANSPro_0800, UFGTPro_0800, ReS11Pro_0800, and E22Pro_0800. E, LUC assays of PIF4_62sk with ReS11Pro_0800 and E22Pro_0800.
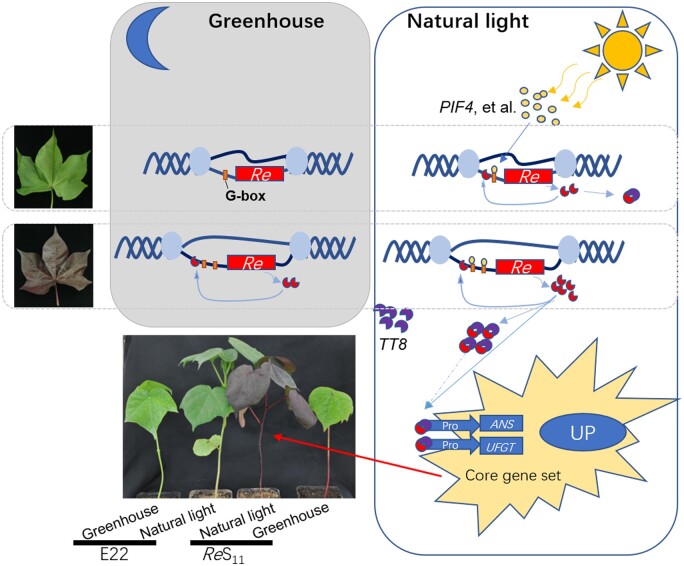
Combining the above results, the possible metabolic network of Re was drawn (Figure 7). Under natural light, the expression of light-responsive factors such as PIF4 increases, and PIF4 binds to the G-box in the promoter region of Re to drive the expression of Re. The repetitive sequence in the promoter region of ReS11 contains two G-boxes, which enhance the expression of Re driven by PIF4. Re can also bind to its own promoter, which further increases Re expression. Re can promote the expression of ANS and UFGT, two downstream genes of the anthocyanin metabolism pathway, leading to the upregulation of the entire core gene set of anthocyanin metabolism (Figure 7).
Figure 7.
Regulation model of Re. The box on the left represents the greenhouse condition, and the box on the right represents the natural light condition. Different shapes and colors represent gene structure or transcription products, as shown in the figure. The solid line represents the identified pathway, and the dashed line represents the inferred pathway.
Re can bind to TT8 to form a dimer. In plants, MYB75 and TT8 can form a complex to regulate anthocyanin metabolism (Gonzalez et al., 2008; Wei et al., 2019). However, TT8 did not show a substantial expression difference and was not identified as DEG between E22 and ReS11 (Supplemental Table S10), and TT8 likely acts as a basic factor in this pathway.
Discussion
In this study, a red foliated cotton mutant was found in the offspring from the cross between sea-island cotton and upland cotton. The color was controlled by a single dominant gene, as confirmed by phenotypic statistics. Re was identified as a MYB113 TF. The location range of Re overlapped with the classic red plant mutant R1; combined with the phenotype of ReS11, Re is candidate gene for R1. Our red foliated cotton is a completely different mutant phenotype derived from the cross between upland cotton and sea-island cotton, while the existing red plant cotton is derived from the cotton multidominant line T586 (Kohel, 1985). In this process, we found that nonspecific amplification can obscure the detection of gene expression, and sequencing the RT-PCR products is necessary especially for polyploids. The incorrect assembly of the reference genome also led to uncertainty in the selection of candidate genes in the positioning interval, although we were lucky to find the final candidate gene. MYB-Sg6 genes are widely involved in anthocyanin synthesis and metabolism by regulating structural genes in the anthocyanin metabolic pathway (Nesi et al., 2001; Jian et al., 2019, 2021; Xu et al., 2021), which helped us to identify candidate genes.
The RT-qPCR results showed that failure to express Re in the fiber is the main reason for the lack of anthocyanin accumulation in the fiber. Through the fiber-specific promoter driving the high expression of Re in 5–10 DPA fibers, we successfully obtained transgenic lines that produced significant anthocyanin accumulation in the developing fiber and a final mature brown fiber rather than a red fiber. This may be because the anthocyanins that accumulate in the fiber at the early stage are transformed during the fiber maturation and dehydration process. The final phenotype of the fiber was very similar to that observed in the study by Yan et al. (2018), in which the upregulated expression of GhTT2-3A in fibers at the secondary wall-thickening stage resulted in brown mature fibers. The final manifestation of the fiber was the excessive accumulation of PA, which is also considered to be the main substance that produces brown cotton (Feng et al., 2014; Yan et al., 2018). Both Re and GhTT2 participated in the regulation of PA in the fiber, and we found that the expression of GhTT2 in GbEXP-E3 also increased significantly (Supplemental Figure S13). Re can directly regulate the expression of ANS, and the products of ANS (the anthocyanidins delphinidin, pelargonidin, and cyanidin) are the precursors of anthocyanins and PAs. This explains why Re can simultaneously cause changes in anthocyanin and PA contents. How Re and GhTT2 coordinate the metabolism of anthocyanins and PA in upland cotton is worth exploring. Through WGCNA and transcriptome data analysis, as well as experimental verification by LUC, LCI, and BiFC assays, we proposed the regulatory pathway of anthocyanins involved in Re. PIF4 may not be the only light response factor that regulates Re, as there were a large number of upregulated DEGs between GR and LR; TT8 likely plays a role as a basic factor in collaborating with Re.
In addition to the known structural genes, we found that MYB4 (Ghir_A03G013820, Ghir_D02G015000) also maintained a complex relationship with the genes in MEdarkgrey (Supplemental Figure S10d). Studies in Arabidopsis have shown that MYB4 plays a dual role in anthocyanin synthesis and participates in resistance to ultraviolet radiation B (Jin et al., 2000; Wang et al., 2020). Uclacyanin 1 (Ghir_A06G016540, Ghir_D06G017430) also occupies an important position in the core gene set, and study has shown that uclacyanin can affect the metabolism of flavonoids in rice (Oryza sativa; Zhang et al., 2020). In addition, some core set genes were closely related to the expression of these structural genes, but their functions are not yet known. Among the coexpressed genes of Re, one gene, MYB308 (Ghir_D09G017850), was functionally annotated (Supplemental Figure S10, f and g; Supplemental Table S12). Study in apple has shown that MYB308 is involved in anthocyanin metabolism and tolerance to cold (An et al., 2020). These results show that anthocyanin metabolism in upland cotton is much more complicated than we understand at the moment.
By constitutive overexpression of Re, transgenic lines with obvious pigmentation in most organs of the plant were obtained, showing that a single gene can play a decisive role in the synthesis and accumulation of secondary metabolites. Excessive accumulation of anthocyanins delays the growth and development of cotton, especially in an environment with insufficient light, such as a greenhouse. When natural light is sufficient, the growth of red cotton plants is basically the same as that of green plants. This shows that cotton can make full use of excess light energy to accumulate secondary metabolites under natural conditions.
Materials and methods
Plant materials
In the backcross inbred lines population derived from the cross between E22 and sea-island cotton (G. barbadense) 3–79, a red foliated mutant was found (Supplemental Figure S1, a and b). This line was continuously self-pollinated for nine generations, and the red plants were selected in each generation for the next generation of selfing. A typical red plant (ReS9) was selected as the parent to cross with E22 to generate the F2 mapping populations. The phenotype was judged accurately at the seedling stage (Supplemental Figure S1c), and only the recessive individuals were transplanted to the field. In the segregation population, three phenotypes appeared, including red foliated cotton, the intermediate type, and the green plant (Supplemental Figure S1d); from the F2 linkage populations, 92, 227, and 1,580 recessive individuals, respectively, were planted in Wuhan, Hubei Province, China, from 2013 to 2015. ReS11 was obtained after two generations of inbred ReS9.
Linkage mapping
Polymorphic markers between parents were screened for linkage markers of the Re according to the bulked segregation method (Michelmore et al., 1991). DNA from 10 individuals with red leaves and green leaves was mixed to build the red bulk and the green bulk. The primers for polymorphism detection between parents were based on our high-density interspecific map (Li et al., 2016a, 2016b, 2016c). SSR loci were scanned by the MicroSAtellite identification tool (http://pgrc.ipk-gatersleben.de/misa/) based on the sequence of the G. raimondii (Wang et al., 2012). UTR primers were designed based on the 3′-UTR and 5′-UTR sequence information of genes in the candidate interval. The polymorphism of primers among the RB and the GB and the two parents was tested by 6% (w/v) denaturing gels and 8% (w/v) nondenaturing gels. The genotype was analyzed by Joinmap 3.0 mapping software (Van Ooijen and Voorrips, 2001). MapMaker version 3.0 software was used for linkage analysis with the target gene Re (Lincoln et al., 1992), and MapChart version 2.2 was used to obtain the genetic map (Voorrips, 2002).
Cloning and sequencing
The primers of the candidate genes were designed based on the TM-1 reference genome (Wang et al., 2019). The genomic DNA of samples used in the test was extracted as described by Paterson et al. (1993). Amplified target gene fragments were subsequently ligated into the pGEM-T Easy cloning vector (Biotech Co. Ltd, Promega, Beijing, China), and the inserts were sequenced using vector M13 primers (5′-CCCAGTCACGACGTTGTAAAACG-3′, 5′-GCGGATAACAATTTCACACAGGA-3′) by Wuhan Qingke Biotechnology Co., Ltd. The final sequences were analyzed with DNAMAN software (https://www.lynnon.com/pc/framepc.html). The cis-acting element was predicted by PlantCARE (Rombauts et al., 1999).
RNA extraction and RT-qPCR
All samples were taken from the experimental field of Huazhong Agricultural University, Wuhan, China. To ensure that the samples were not different due to conditions such as light and temperature, the sampling time was set at 9–10 a.m. on sunny days. Cotton bolls were tagged on the day of flowering as 0 DPA. Bolls of 5, 10, 15, 20, and 25 DPA were taken, frozen with liquid nitrogen immediately after removing the cotton husks, and stored in an ultralow temperature freezer at −80°C. The fiber from which the ovule was removed was ground into powder in liquid nitrogen. Total RNA was extracted from the samples using the RNAprep Pure Plant Kit (TIANGEN Biotech, Beijing, China). For each sample, 3 μg of RNA was reverse transcribed into cDNA using M-MLV reverse transcriptase (Promega). For RT-qPCR analysis, we followed the same steps as Li et al. (2020).
Transcriptome sequencing and WGCNA
The seeds of ReS11, E22, CK1, and OE1 were uniformly germinated. After the cotyledons of cotton seedlings were flattened, half of the plants were transferred to outdoor planting (Wuhan, June), and half were placed in the greenhouse (16-h light/8-h night). The greenhouse used LED lights with a temperature of 26°C (16-h light/8-h night). The brand and model of the LED lights were PAK301608, the color temperature of which is 6,500 k. The spectrum of the white LED lamp ranges from ∼400 to 750 nm. Three weeks later, the leaves were taken and cut into two halves along the main leaf vein; half of the leaves were used for transcriptome sequencing, and the other half was used for anthocyanin content determination, with three biological replicates for each sample. Total RNA was sequenced with the Illumina HiSeq 2000 system. The clean RNA-seq reads were mapped to the TM-1 reference genome (Wang et al., 2019) by HISAT version 2.0 (Kim et al., 2019). FeatureCounts was used to calculate the transcript levels of annotated genes (Liao et al., 2014). DEseq2 in R was used to identify DEGs, with an absolute value of log2[fold change] > 1. The FPKM value was calculated by cufflinks (version 2.2.1) for gene expression levels. The coexpression network was constructed by WGCNA with default settings, the average value of FPKM was used as input data, and the relative content of anthocyanin in leaves was used as phenotypic data. The networks were visualized by Cytoscape_version 3.0.0 (Otasek et al., 2019).
Vector construction and cotton transformation
The cloned coding sequence (CDS) of Re was inserted downstream of the CaMV35S promoter in the pk2GW7 vector. The resultant overexpression vector was transferred into an Agrobacterium tumefaciens strain (GV3101). The fiber-specific expression vector PGbEXPA2 was provided by Li et al. (2016a, 2016b, 2016c), and the CDS region of Re was inserted downstream of the specific expression promoter. The high-efficiency transformation line Jin668 was used as the transformation receptor as described in a previous protocol (Li et al., 2019a, 2019b). In this experiment, except for the T0 generation transgenic lines shown in Figures 2 and 3, A, T2 generation plants were analyzed.
Quantification of anthocyanin and PA contents
Anthocyanins in cotton leaves were extracted and quantified as previously described (Gao et al., 2013). The absorbances at 530 and 657 nm were determined using a multimode plate reader (PerkinElmer, Waltham, MA, USA), and the relative level of anthocyanin was calculated as A530–(0.25*A657) (Rabino and Mancinelli, 1986). The relative PA content determination was performed according to the method of Li et al. (2020). DMACA was used to stain and visualize PA in cotton fibers (Xiao et al., 2007).
Yeast two-hybrid assays
To detect the proteins that interact with Re, a Matchmaker Gold Yeast Two-Hybrid Library Screening System Kit (Clontech, Mountain View, CA, USA; cat. no. 630489) was used. Because of self-activation, the CDS of Re was truncated and cloned into the BD vector pGBKT7 to construct Re-BD as bait, which was introduced into the yeast strain Y2HGold. This bait was used to screen a Y187 library from cotton fibers and leaves. The genes captured by the bait were sequenced and point-by-point verified to confirm the initial interaction. The primers used are listed in Supplemental Table S13, and the information of vectors is listed in Supplemental Table S14.
Subcellular localization and dual-LUC reporter, LCI, and BiFC assays
The CDS of Re was cloned into the N-terminal fusion green fluorescent protein (GFP) vector pMDC43. The vector was then transformed into leaves of Nicotiana benthamiana by agroinfiltration. The green fluorescence of 35S::GFP-Re in the cells was detected after 48 h using an Olympus FV1200 confocal microscope (488-nm excitation wavelength, 44% transmissivity, 100-nm collection bandwidth and gain was 1), and the RFP fluorescence signal of nuclear marker was also detected (559-nm excitation wavelength, 35% transmissivity, 100-nm collection bandwidth and gain was 1).
The promoters of ReS11, E22, CHS, CHI, F3H, DFR, ANS, ANR, UFGT, and GST were cloned into the pGreen II 0800-LUC vector. The CDSs of Re and PIF4 were cloned into the pGreen II 62sk vector. Agrobacterium-infected N. benthamiana was treated in the dark for 24 h, and then the fluorescence intensity of the N. benthamiana leaves was assessed with an in vivo imager. Empty pGreen II 0800-LUC/62sk was used as control. For LCI assays, the CDSs of Re and TT8 were cloned into the JW771 and JW772 vectors, respectively. The LCI assays were conducted according to a previously used method (Chen et al., 2008; Ye et al., 2020). For BiFC assays, the CDS of Re was cloned into the N-terminal fusion yellow fluorescent protein (YFP) vector pCAMBIA1301 via infusion reactions to obtain Re-nYFP. The CDS of TT8 was cloned into the C-terminal fusion YFP vector pCAMBIA1301 to obtain TT8-cYFP. The vectors were transformed into A. tumefaciens strain GV3101 and injected into N. benthamiana leaves by a syringe for transient expression. A confocal microscope (Olympus FV1200) was used to observe the fluorescence in N. benthamiana leaf cells approximately 60 h later (515-nm excitation wavelength, 29% transmissivity, 100-nm collection bandwidth and gain was 1). The primers used are listed in Supplemental Table S13, and the information vectors are listed in Supplemental Table S14.
Statistical analysis
Student’s t test was performed using SPSS version 17.0. Difference was considered significant at P < 0.05 and highly significant at P < 0.01.
Data availability
The raw RNA-Seq data in this study is available in the BioProject under the accession number PRJNA752503.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: RE3 (XM_016889388), Re (MH746529), RE6 (XM_016889354), CHS (XM_016823419), CHI (XM_016810061), ANS (NM_001327309), ANR (XM_016832763), UFGT (XM_016885447), DFR (KF749429), F3H (XM_041083987), PIF4 (XM_016812587), Gh_TT2 (XM_016861402), and GhUB7 (DQ116441).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. A single red foliated cotton mutant appeared in the hybrid population of E22 and 3–79.
Supplemental Figure S2. Alignment relationship of markers linked to Re between G. hirsutum and G. raimondii.
Supplemental Figure S3. Sequencing results of DEGs between E22 and ReS11.
Supplemental Figure S4. Subcellular localization of Re.
Supplemental Figure S5. The specific expression of Re in the fiber producing different shades of brown cotton.
Supplemental Figure S6. Determination of relative content of anthocyanin and chlorophyll in leaves.
Supplemental Figure S7. Determination of relative content of PA in leaves.
Supplemental Figure S8. The effect of natural light on leaf color.
Supplemental Figure S9. DEGs under natural light and greenhouse conditions.
Supplemental Figure S10. WGCNA of DEGs.
Supplemental Figure S11. LUC of Re and E22pro.
Supplemental Figure S12. LUC of Re and six structural genes.
Supplemental Figure S13. Relative expression of GhTT2 in fiber-specific transgenic lines of Re.
Supplemental Table S1. Phenotypic statistics of F2 populations in the field.
Supplemental Table S2. Distribution of the 494 primers used for polymorphic analysis.
Supplemental Table S3. Polymorphic rates of the primers selected from the genetic map.
Supplemental Table S4. The 470 pairs of SSR primers.
Supplemental Table S5. The 126 pairs of primers based on the 3'UTR and 5'UTR sequence of genes.
Supplemental Table S6. The 80 pairs of SSR primers on D07.
Supplemental Table S7. The markers involved in gene mapping.
Supplemental Table S8. RT-PCR primers for candidate genes.
Supplemental Table S9. The genes that are relevant to biosynthesis and transport of pigment metabolic pathways.
Supplemental Table S10. DEGs between pairwise comparisons.
Supplemental Table S11. Co-expression analysis of genes in MEdarkgrey module.
Supplemental Table S12. Genes co-expressed with Re.
Supplemental Table S13. Primers for vector construction and functional verification.
Supplemental Table S14. Vectors used in this study.
Supplementary Material
Acknowledgments
The computations in this article were run on the bioinformatics computing platform of the National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University.
Funding
This work was financially supported by the Genetically Modified Organisms Breeding Major Project of China (No. 2016ZX08009001).
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Nian Wang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Beibei Zhang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Tian Yao, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Chao Shen, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China; College of Biological and Food Engineering, Guangdong University of Petrochemical Technology, Maoming, Guangdong 525000, China.
Tianwang Wen, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China; Key Laboratory of Crop Physiology, Ecology and Genetic Breeding, Ministry of Education, College of Agronomy, Jiangxi Agricultural University, Nanchang, Jiangxi 330045, China.
Ruiting Zhang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Yuanxue Li, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Yu Le, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Zhonghua Li, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Xianlong Zhang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Zhongxu Lin, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
N.W. completed the main experiment and manuscript writing. B.B.Z completed the population construction and genetic mapping. T.Y., Y.X.L., Y.L., and Z.H.L. performed the experiments. C.S. T.W.W., and R.T.Z. provided guidance in molecular marker design and experimental operation. X.L.Z. and Z.X.L. revised the manuscript. Z.X.L. conceived and designed the project.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Zhongxu Lin (linzhongxu@mail.hzau.edu.cn).
References
- Albert NW, Griffiths AG, Cousins GR, Verry IM, Williams WM (2015) Anthocyanin leaf markings are regulated by a family of R2R3-MYB genes in the genus Trifolium. New Phytol 205: 882–893 [DOI] [PubMed] [Google Scholar]
- An J, Wang X, Zhang X, Xu H, Bi S, You C, Hao Y (2020) An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol J 18: 337–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester AR, Molthoff J, Vos Rd, Hekkert BtL, Orzaez D, Fernández-Moreno JP, Tripodi P, Grandillo S, Martin C, Heldens J, et al. (2010) Biochemical and nolecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol 152: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv shiraz grape berries and the implications for pathway regdation. Plant Physiol 111: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, Zhang X, Niu E, Zhao L, Li N, Wang L, Ding L, Guo W (2014) GhPSY, a phytoene synthase gene, is related to the red plant phenotype in upland cotton (Gossypium hirsutum L.). Mol Biol Rep 41: 4941–4952 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec Lc (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, et al. (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21: 168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Li Y, Wang S, Zhang L, Liu Y, Xue F, Sun Y, Wang Y, Sun J (2014) Molecular analysis of proanthocyanidins related to pigmentation in brown cotton fibre (Gossypium hirsutum L.). J Exp Bot 65: 5759–5769 [DOI] [PubMed] [Google Scholar]
- Gao Z, Liu C, Zhang Y, Li Y, Yi K, Zhao X, Cui ML (2013) The promoter structure differentiation of a MYB transcription factor RLC1 causes red leaf coloration in Empire Red Leaf Cotton under light. PLoS One 8: e77891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, John ML, Alan ML (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53: 814–827 [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian W, Cao H, Yuan S, Liu Y, Lu J, Lu W, Li N, Wang J, Zou J, Tang N, et al. (2019) SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic Res 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Sun Q, Zhang T, Liu W, Wang N, Chen X (2021) MdMYB114 regulates anthocyanin biosynthesis and functions downstream of MdbZIP4-like in apple fruit. J Plant Physiol 257: 153353. [DOI] [PubMed] [Google Scholar]
- Jiang W, Yin Q, Wu R, Zheng G, Liu J, Dixon RA, Pang Y (2015) Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. J Exp Bot 66: 7165–7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killough DT, Horlacher WR (1933) The inheritance of virescent yellow and red plant colors in cotton. Genetics 18: 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37: 104–114 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnol 37: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Kohel RJ. (1985) Genetic analysis of fiber color variants in cotton. Crop Sci 25: 793–797 [Google Scholar]
- Li J, Wang M, Li Y, Zhang Q, Lindsey K, Daniell H, Jin S, Zhang X (2019a) Multi-omics analyses reveal epigenomics basis for cotton somatic embryogenesis through successive regeneration acclimation process. Plant Biotechnol J 17: 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang W, Gao J, Yin K, Wang R, Wang C, Petersen M, Mundy J, Qiu JL (2016a) MYB75 Phosphorylation byMPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28: 2866–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Jin X, Wang H, Zhang X, Lin Z (2016b) Structure, evolution, and comparative genomics of tetraploid cotton based on a high-density genetic linkage map. DNA Res 23: 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ouyang X, Zhang Z, He L, Wang Y, Li Y, Zhao J, Chen Z, Wang C, Ding L, et al. (2019b) Over-expression of the red plant gene R1 enhances anthocyanin production and resistance to bollworm and spider mite in cotton. Mol Genet Genomics 294: 469–478 [DOI] [PubMed] [Google Scholar]
- Li Y, Tu L, Pettolino FA, Ji S, Hao J, Yuan D, Deng F, Tan J, Hu H, Wang Q, et al. (2016c) GbEXPATR, a species-specific expansin, enhances cotton fibre elongation through cell wall restructuring. Plant Biotechnol J 14: 951–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tu L, Ye Z, Wang M, Gao W, Zhang X (2015) A cotton fiber-preferential promoter, PGbEXPA2, is regulated by GA and ABA in Arabidopsis. Plant Cell Rep 34: 1539–1549 [DOI] [PubMed] [Google Scholar]
- Li Z, Su Q, Xu M, You J, Khan AQ, Li J, Zhang X, Tu L, You C (2020) Phenylpropanoid metabolism and pigmentation show divergent patterns between brown color and green color cottons as revealed by metabolic and gene expression analyses. J Cotton Res 3: 27 [Google Scholar]
- Liang J, He J (2018) Protective role of anthocyanins in plants under low nitrogen stress. Biochem Biophys Res Commun 498: 946–953 [DOI] [PubMed] [Google Scholar]
- Liao Y, Gordon KS, Wei S (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Lincoln SE, Daly MJ, Lander ES (1992) Constructing genetics maps with MAPMAKER/EXP 3.0: a tutorial and reference manual. A whitehead institute for biomedical research technical report, 3, Whitehead Institute, Cambridge, USA
- Liu D, Liu X, Su Y, Xiao Z, Kai G, Teng Z, Zhang J, Liu D, Zhang Z (2020) Genetic mapping and identification of Lgf loci controlling green fuzz in upland cotton (Gossypium hirsutum L.). Crop J 9: 777–784 [Google Scholar]
- Liu Y, Shi Z, Maximova S, Payne MJ, Guiltinan MJ (2013) Proanthocyanidin synthesis in Theobroma cacao: genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase. BMC Plant Biol 13: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang Y, Fan K, Li Z, Jia Q, Lin W, Zhang Y (2021) PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) negatively regulates anthocyanin accumulation by inhibiting PAP1 transcription in Arabidopsis seedlings. Plant Sci 303: 110788. [DOI] [PubMed] [Google Scholar]
- Lu N, Rao X, Li Y, Ji H, Dixon RA (2021) Dissecting the transcriptional regulation of proanthocyanidin and anthocyanin biosynthesis in soybean (Glycine max). Plant Biotechnol J 19: 1429–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Dai C, Li Y, Feng J, Liu Z, Kang C (2018) Reduced anthocyanins in petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J Exp Bot 69: 2595–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs KA, Alfenlto MR, Lloyd AM (1995) A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375: 397–400 [DOI] [PubMed] [Google Scholar]
- Martin C, Prescott A, Mackay S, Bartlett J, Vrijlandt E (1991) Control of anthocyanin biosynthesis in flowers of Antirrhinurn majus. Plant J 1: 37–49 [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Christopher DG, Rebecca AS, Virginia W (2000) AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol 123: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, et al. (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J 77: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X, Tu J, Wang B, Zhou X, Lin Z (2015) A BIL population derived from G. hirsutum and G. barbadense provides a resource for cotton genetics and breeding. PLoS One 10: e0141064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otasek D, John HM, Jorge B, Alexander RP, Barry D (2019) Cytoscape automation: empowering workflow-based network analysis. Genome Biol 20: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Brubaker CL, Wendel JF (1993) A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep 11: 122–127 [Google Scholar]
- Pedmale UV, Huang SC, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PAB, Sridevi P, Nito K, Nery JR, et al. (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy R, Hendon B, Bechere E, Auld D (2015) Qualitative Genetics and Utilization of Mutants. American Society of Agronomy (ASA), Crop science Society of America (CSSA) and Soil Science Society of America (SSSA), Madison, WI, pp 155–186 [Google Scholar]
- Rabino I, Mancinelli AL (1986) Light, temperature, and anthocyanin production. Plant Physiol 81: 922–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay NA, Glove BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Rombauts S, Déhais P, Montagu MV, Rouzé P (1999) PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res 27: 295–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Buelga C, Mateus N, Freitas FD (2014) Anthocyanins. Plant pigments and beyond. J Agric Food Chem 62: 6879–6884 [DOI] [PubMed] [Google Scholar]
- Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Xie D (2014) Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Patent Biotechnol 8: 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens SG (1974) Geographic and taxonomic distribution of anthocyanin genes in New World cottons. J Genet 61: 128–141 [Google Scholar]
- Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142: 1216–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54: 733–749 [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW, Voorrips RE (2001) JoinMap 3.0, Software for the calculation of genetic linkage maps. Plant Research International, Wageningen, The Netherlands [Google Scholar]
- Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93: 77–78 [DOI] [PubMed] [Google Scholar]
- Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, et al. (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Wang M, Tu L, Yuan D, Zhu D, Shen C, Li J, Liu F, Pei L, Wang P, Zhao G, et al. (2019) Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet 51: 224–229 [DOI] [PubMed] [Google Scholar]
- Wang X, Wu J, Guan M, Zhao C, Geng P, Zhao Q (2020) Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J 101: 637–652 [DOI] [PubMed] [Google Scholar]
- Wei Z, Cheng Y, Zhou C, Li D, Gao X, Zhang S, Chen M (2019) Genome-wide identification of direct targets of the TTG1-bHLH-MYB complex in regulating trichome formation and flavonoid accumulation in Arabidopsis thaliana. Int J Mol Sci 20: 5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T, Wu M, Shen C, Gao B, Zhu D, Zhang X, You C, Lin Z (2018) Linkage and association mapping reveals the genetic basis of brown fibre (Gossypium hirsutum). Plant Biotechnol J 16: 1654–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Zhang Z, Yin M, Luo M, Li X, Hou L, Pei Yan (2007) Cotton flavonoid structural genes related to the pigmentation in brown fibers. Biochem Biophys Res Commun 358: 73–78 [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Dixon RA (2004) Anthocyanidin reductases from Medicago truncatula and Arabidopsis thaliana. Arch Biochem Biophys 422: 91–102 [DOI] [PubMed] [Google Scholar]
- Xu Z, Rothstein SJ (2018) ROS-Induced anthocyanin production provides feedback protection by scavenging ROS and maintaining photosynthetic capacity in Arabidopsis. Plant Signal Behav 13: e1451708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Yang Q, Feng K, Yu X, Xiong A (2021) DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol J 18: 1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Wang Y, Li Q, Zhang Z, Ding H, Zhang Y, Liu H, Luo M, Liu D, Song W, et al. (2018) Up-regulation of GhTT2-3A in cotton fibres during secondary wall thickening results in brown fibres with improved quality. Plant Biotechnol J 16: 1735–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Qiao L, Luo X, Chen X, Zhang X, Tu L (2020) Genome-wide identification of cotton GRAM family proteins reveals that GRAM31 regulates fiber length. J Exp Bot 72: 2477–2490 [DOI] [PubMed] [Google Scholar]
- Zhang Y, He R, Lian J, Zhou Y, Zhang F, Li Q, Yu Y, Fenga Y, Yang Y, Lei M, et al. (2020) OsmiR528 regulates rice-pollen intine formation by targeting an uclacyanin to influence flavonoid metabolism. Proc Natl Acad Sci USA 117: 727–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Cai C, Zhang T, Guo W (2009) Fine mapping of the red plant gene R1 in upland cotton (Gossypium hirsutum). Sci Bull 54: 1529–1533 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-Seq data in this study is available in the BioProject under the accession number PRJNA752503.