Abstract
Streptococcus thermophilus CNRZ 385 expresses a cell envelope proteinase (PrtS), which is characterized in the present work, both at the biochemical and genetic levels. Since PrtS is resistant to most classical methods of extraction from the cell envelopes, we developed a three-step process based on loosening of the cell wall by cultivation of the cells in the presence of glycine (20 mM), mechanical disruption (with alumina powder), and enzymatic treatment (lysozyme). The pure enzyme is a serine proteinase highly activated by Ca2+ ions. Its activity was optimal at 37°C and pH 7.5 with acetyl-Ala-Ala-Pro-Phe-paranitroanilide as substrate. The study of the hydrolysis of the chromogenic and casein substrates indicated that PrtS presented an intermediate specificity between the most divergent types of cell envelope proteinases from lactococci, known as the PI and PIII types. This result was confirmed by the sequence determination of the regions involved in substrate specificity, which were a mix between those of PI and PIII types, and also had unique residues. Sequence analysis of the PrtS encoding gene revealed that PrtS is a member of the subtilase family. It is a multidomain protein which is maturated and tightly anchored to the cell wall via a mechanism involving an LPXTG motif. PrtS bears similarities to cell envelope proteinases from pyogenic streptococci (C5a peptidase and cell surface proteinase) and lactic acid bacteria (PrtP, PrtH, and PrtB). The highest homologies were found with streptococcal proteinases which lack, as PrtS, one domain (the B domain) present in cell envelope proteinases from all other lactic acid bacteria.
Lactic acid bacteria (LAB) are widely used as starters in fermented milk products due to their properties of milk acidification and flavor development. For these applications, their capacity to grow fast in milk is of major importance. LAB are fastidious microorganisms and require an exogenous source of amino acids or peptides for optimal growth. As milk is poor in these low-molecular-weight compounds, their growth largely depends on their proteolytic system to achieve hydrolysis of caseins (65). The cell envelope proteinase (CEP) is the key enzyme of this process since it is the only enzyme capable of initiating the breakdown of caseins into oligopeptides. The latter are then transported into the bacteria and further degraded by a complex set of intracellular peptidases (12).
The cell envelope proteinases of lactococci, and to a lesser extent those of lactobacilli, have been the subject of intensive biochemical and genetic investigation (for a review, see reference 37). Lactococcal proteinase PrtP is synthesized as an inactive preproenzyme and maturated via an autoproteolytic process involving a chaperone lipoprotein PrtM, and it is anchored to the cell wall. The hydrolysis specificity of CEPs determined on caseins or casein peptides varies among strains, and several classifications have been proposed (5, 18, 21, 22, 24, 40, 66). The differences observed in substrate specificity are only due to the variation, in the CEP sequences, of very few amino acids (8, 61, 68).
The CEPs from LAB and also from pyogenic streptococci are serine proteinases which belong to the subtilisin-like serine proteinase family known as the subtilase family (60). They are multidomain proteins with highly conserved catalytic domains. Among species, more variation exists between these CEPs at their C terminus (presence or absence of the B domain and of the helical domain [59]) and in their way of anchoring to the cell envelopes. Most frequently, LAB possess only one CEP, but the presence of two CEPs has been described in lactobacilli (48, 63).
In spite of the wide utilization of Streptococcus thermophilus in the production of dairy products (yogurt, hard cooked cheese, soft cheese), little is known about its CEP. Most strains of S. thermophilus do not express or express a very low level of CEP (14, 56, 58). A screening of S. thermophilus strains revealed that only 3 among 97 of them possessed a level of proteinase activity close to that of the proteinase-positive lactococcal strain (56). These strains also grew and produced acid in milk faster than the 94 others. For two of them, strains CNRZ 385 and 703, the CEPs were characterized in cell wall fractions and not in pure preparations, as the release treatments tested remained unsuccessful (57). DNA hybridization and immunoblot studies suggested that CEP from S. thermophilus and PrtP from lactococci were not closely related (36, 57).
The present paper describes for the first time the release and purification of a CEP from S. thermophilus CNRZ 385. We determined its biochemical properties and its substrate specificity as well as the sequence of its encoding gene. We propose that this CEP from S. thermophilus be called PrtS.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and transformation.
The strain S. thermophilus CNRZ 385 originated from the Centre National de Recherche Zootechnique culture collection (INRA, Jouy-en-Josas, France). This strain is plasmid free and was previously classified as an H strain, i.e., a strain with a high acidification rate in milk (56). It was grown in M17 medium at 37°C (64) supplemented with 20 g of lactose · liter−1. Escherichia coli TG1 (29) was transformed according to the method of Hanahan et al. (32) except that E. coli cells were grown overnight in Luria-Bertani medium (53) containing 10 mM MgCl2 instead of SOB medium (53) and that KCl was used to replace RbCl in the washing buffers.
Cell wall proteinase release.
S. thermophilus CNRZ 385 was grown in 5 liter of M17 Lac broth (Difco) supplemented with 20 g of lactose · liter−1 and 20 mM glycine. Cells were collected at the end of the exponential growth phase by centrifugation at 8,500 × g at 4°C for 20 min. They were washed twice with 50 mM β-glycerophosphate buffer, pH 7, and then resuspended in 125 ml of 50 mM Bis-Tris buffer, pH 6.5. Alumina powder (2.5-fold the pellet weight) was added, and the mixture was manually crushed for 5 min with a pestle. The alumina powder was removed by centrifugation at 500 × g for 5 min, and the supernatant obtained was centrifuged at 20,000 × g for 15 min at 4°C. The pellet was resuspended in 125 ml of 50 mM Bis-Tris buffer, pH 6.5, supplemented with 2 mM CaCl2 and 1 mg of lysozyme · ml−1 and then incubated for 2 h at 37°C. Cell debris were removed by centrifugation at 4°C for 15 min at 8,500 × g, and the supernatant, referred to as cell wall extract, was used for proteinase purification.
Cell wall proteinase purification. (i) Ultrafiltration.
The cell extract (120 ml) was concentrated by ultrafiltration in 50 mM Bis-Tris buffer, pH 6.5, containing 2 mM CaCl2 through a 100-kDa cut-off cellulose ester membrane (Spectra Por; Spectrum Medical Industries, Los Angeles, Calif.). Thirty milliliters was recovered and checked for proteinase activity with 14C casein as the substrate (see below).
(ii) Ion-exchange chromatography.
The dialyzed extract was loaded on a UnoQ column (Bio-Rad Laboratories, Hercules, Calif.) equilibrated with 50 mM Bis-Tris buffer, pH 6.5. Bound proteins were eluted at a flow rate of 2 ml · min−1 with a four-step linear gradient of NaCl: firstly, 0 to 0.15 M for 5 min; secondly, 0.15 M for 5 min; thirdly, 0.15 to 0.4 M for 40 min; and finally, 0.4 to 1 M for 5 min. Fractions (2 ml) were collected and checked for their [14C]casein hydrolysis activity; those which were active were pooled and concentrated using a 50-kDa Centriplus concentrator (Amicon, Damers, Mass.) for further purification.
(iii) Gel filtration chromatography.
The last purification step was performed on a Superose 6 column (Pharmacia-Amersham, Uppsala, Sweden) with 50 mM Bis-Tris buffer, pH 6.5, containing 0.15 M NaCl. The concentrated sample was applied to the column and eluted at a flow rate of 0.2 ml · min−1, and 0.6-ml fractions were collected. The column was calibrated with thyroglobulin (670 kDa), gamma globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa).
Protein quantification and proteinase activity measurement.
Protein concentrations were determined in the cellular extract and purified fractions by the method of Bradford (6) using the Pierce (Rockford, Ill.) protein assay reagent with bovine serum albumin as the standard.
Throughout the purification, proteinase activity was measured on [14C]casein according to the method of Donnelly et al. (17) as adapted by Monnet et al. (44) with 1 h of incubation at 37°C. It was also assayed by microplate testing with a chromogenic substrate by measuring the release of paranitroaniline from acetyl (Ac)-Ala-Ala-Pro-Phe-paranitroanalide (pNA) as described by Zevaco et al. (70). Standard conditions were 10 min of incubation at 30°C in 0.1 M Tris-HCl, pH 8, containing 5 mM CaCl2 with an appropriate quantity of enzyme.
The substrate specificity of the proteinase was determined with both chromogenic and casein substrates. With chromogenic substrates, the enzyme solution was incubated for 30 min at room temperature with 0.75 mM substrate in 50 mM Bis-Tris, pH 6.5, buffer (for 3-methoxysuccinyl [MS]-Arg-Pro-Tyr-, succinyl [S]-Val-Pro-Phe-, Ac-Ala-Ala-Pro-Phe-, benzoyl-Phe-Val-Arg-, benzyloxycarbonyl-Gly-Pro-Arg-, or benzyloxycarbonyl-Phe-Arg-pNa assays) or at 37°C with 0.5 mM substrate in 0.1 M phosphate buffer, pH 7 (for S-Ala-Glu-Pro-Phe- and 3-MS-Arg-Pro-Tyr-pNa assays), in microplates in the presence of 2 mM CaCl2. With 14C-labeled whole and β-caseins used as described above, the incubation conditions were 15 min at 30°C in the presence of 5 mM CaCl2. With the αS1-casein-(1-23) fragment, synthesized with a Peptide Synthesizer Synergy device (model 432A; Applied Biosystems, Foster City, Calif.), the reaction conditions were 10 μl of pure proteinase solution (100 ng), 40 μM substrate, and 10 mM CaCl2 in buffers with different pH (4, 5.2, 6.2, and 8) and at different temperatures (18, 30, and 37°C) for different incubation times (from 2 to 8 h). The reactions were stopped by the addition of 1% (final concentration) trifluoroacetic acid (TFA). The hydrolysates were then analyzed by reverse-phase high-performance liquid chromatography using a C18 column (Nucleosyl C18; Shandon) in a TFA-acetonitrile (CH3CN) solvent system (solvent A: 0.115% TFA; solvent B: 0.1% TFA–60% CH3CN) and recorded at 214 nm. The column was equilibrated with buffer A at a flow rate of 1 ml · min−1. Elution was carried out using a linear gradient of 0 to 100% solvent B for 30 min. Peaks were collected, dried, and identified by mass spectrometry using a LD-TOF system (model G2025A; Hewlett-Packard, Palo Alto, Calif.).
Biochemical characterization of proteinase. (i) Electrophoresis, blotting, and N-terminal protein sequencing.
The purified fraction was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 8 to 25% gradient acrylamide gels (Pharmacia) according to the method of Laemmli (38). The proteins were silver stained according to the method of Blum et al. (3). The purified enzyme (10 μg) was fixed on a polyvinylidene difluoride ProSorb cartridge (Applied Biosystems) and directly applied onto a sequencer (model 477A; Applied Biosystems).
(ii) Effect of pH and temperature on proteinase activity.
The effect of pH was determined on Ac-Ala-Ala-Pro-Phe-pNa (0.75 mM) as described above at 37°C for 10 min in microplates in the presence of 5 mM CaCl2. pH was tested in the range of 4.5 to 8.5 with the following 0.1 M buffers: sodium acetate, pH 4.5 to 5.5; Bis-Tris, pH 5.5 to 7; and Tris, pH 7 to 8.5.
The temperature effect was determined at temperatures ranging from 15 to 45°C. The enzyme was incubated for 10 min in 0.1 M Tris buffer, pH 8, containing 5 mM CaCl2 at the required temperature with Ac-Ala-Ala-Pro-Phe-pNa (0.75 mM) as the substrate, and the activity was measured as described above.
(iii) Effect of inhibitors and ions on proteinase activity.
The purified enzyme was preincubated for 10 min at room temperature in microplates containing 0.1 M Tris (pH 8) with various inhibitors or ions; Ac-Ala-Ala-Pro-Phe-pNa (0.75 mM) was then added to the reaction mixture, the mixture was incubated for 10 min at 20°C; and proteinase activity was measured as described above. CaCl2 (5 mM) was added to all reactions. Inhibitors tested were phenylmethylsulfonyl fluoride (1 mM), chymostatin (0.1 mM), dithiothreitol (1 mM), l-trans-epoxysuccinyl leucylamide-(4-guanidino)butane (E64) (10 μM), iodoacetic acid (1 mM), EDTA (1 mM), antipain (0.1 mM), and bestatin (0.01 mM); ions tested were CaCl2 (2, 5, and 10 mM) and NaCl (0.6 and 1.2 M).
Genetic characterization of proteinase. (i) Total DNA preparation.
Total DNA of S. thermophilus CNRZ 385 was prepared as described by Pospiech and Neumann (50).
(ii) PCRs.
The S. thermophilus CNRZ 385 proteinase gene was amplified in a two-step PCR process using a Perkin-Elmer DNA thermal cycler (model 480). Oligonucleotides were from Eurogentec (Seraing, Belgium) or Genaxis (Montigny-le-Bretonneux, France).
Firstly, the N-terminal part of the gene was amplified using convergent degenerated oligonucleotides 1 (5′AAYATHGAYAGYAAYAC3′) and 2 (5′YTTRTANCCRCTRTACCA3′), both deduced from the N-terminal sequence of the proteinase. Streptococcal DNA (50 ng) was added to a PCR mixture (2.5 U of Appligene Taq polymerase, 6 μM oligonucleotide 1) and, after 5 min of denaturation at 95°C, oligonucleotide 2 (6 μM) was added. Then, 30 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 37°C, and 30 s of elongation at 72°C were performed. The amplified fragment was purified from 4% Nusieve agarose gel with the Spin-X system (Corning Costar Co., Cambridge, Mass.); it was then cloned in pGEM-T Easy vector (Promega Corp., Madison, Wis.) by transformation of thermocompetent TG1 E. coli cells. This fragment was then sequenced from the recombinant vector as described below. A 57-bp sequence was determined.
Secondly, the whole gene was sequenced by successive inverse PCRs, using two divergent oligonucleotides in the first reaction (5′AAAGTTTGGTACAGYGGYTACA3′ and 5′AATGATCGTRTTACTRTCGATG3′) deduced from the 57-bp sequence obtained in the first step and using the LA PCR in vitro cloning kit (Takara Biomedicals, Shiga, Japan). The latter is a cloning system to specifically amplify long unknown regions of DNA when the DNA sequence of only one end of the region of interest is known. Amplification was accomplished by PCR with restriction enzyme-specific cassettes (double-stranded synthetic oligonucleotides which are ligated to the region of interest) and cassette-specific primers in combination with primers designed from the sequence of interest (34).
(iii) DNA sequencing.
Amplified DNA fragments were extracted from 0.7% agarose gels with the Qiaquick gel extraction kit (Qiagen Inc., Chatsworth, Calif.) and sequenced. The Sanger method of DNA sequencing was carried out on double-strand DNA plasmids and on PCR products with the BigDye Terminator cycle sequencing ready reaction kit (370A DNA sequencer; Applied Biosystems). The reported sequences were determined at least twice for both strands. The DNA and protein sequences were analyzed with the Genetics Computer Group sequence analysis software package from the University of Wisconsin (16) and Mail Fasta (National Center for Biotechnology Information).
Nucleotide sequence accession number.
The GenBank nucleotide accession number for prtS and its flanking regions is AF243528.
RESULTS
PrtS is a serine proteinase highly activated by CaCl2.
The methods used for releasing CEPs from other LAB were not efficient for the S. thermophilus PrtS proteinase; in particular, the easiest method, which consists of incubating the cells in a Ca2+-free buffer, was inefficient (data not shown) (57). Several other classical methods usually used for protein extraction from the cell envelopes, such as cell wall or cell membrane destabilization treatments, or cell breaking processes also remained unsuccessful, as a maximum of 12.5% of initial proteinase activity was recovered (data not shown). We were thus led to develop a specific method to release PrtS from the cell envelopes. It combined the three following treatments: firstly, the loosening of the cell wall during cellular growth by the addition of glycine (20 mM) to the culture medium; secondly, breaking of the cells with alumina powder; and finally, digestion of the cell wall by lysozyme.
PrtS was then purified to homogeneity through a three-step process including ultrafiltration and ion-exchange and gel filtration chromatographies; 60 μg of pure enzyme was recovered from a 5-liter culture, with a purification factor of about 110-fold and an activity recovery of 36%. The molecular mass of PrtS was estimated to be 153 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The 19 N-terminal amino acids (aa) of PrtS were as follows: H2N-Asn-Ile-Asp-Ser-Asn-Thr-Ile-Ile-Thr-Val-Pro-Lys-Val-Trp-Tyr-Ser-Gly-Tyr-Lys.
PrtS was active over a pH range of 5.5 to 8.5 with Ac-Ala-Ala-Pro-Phe-pNa as the substrate, with maximum activity at pH 7.5 and about 70% of the activity remaining at pH 7 and 8. The optimum temperature was 37°C with the same substrate.
PrtS is a serine proteinase, as it was strongly inhibited by serine proteinase inhibitors such as phenylmethylsulfonyl fluoride (100% inhibition) and, to a lesser extent, chymostatin (75% inhibition). Inhibition was also observed with iodoacetic acid (a cystein proteinase inhibitor) (83% inhibition), whereas little or no inhibition of PrtS activity occurred with EDTA (a metal chelating reagent) (30% inhibition), bestatin (an aminopeptidase inhibitor) (no inhibition), or E64 (a strictly cystein proteinase inhibitor) (no inhibition).
We tested PrtS activity with Ac-Ala-Ala-Pro-Phe-pNa as the substrate in the presence of two concentrations of NaCl, 0.6 M (3.5%) or 1.2 M (7%); PrtS was still active and even slightly activated, as 111 and 120% of initial activity were recovered, respectively. CaCl2 highly activated PrtS; initial activity increased by 2- to 10-fold after addition of 2 to 10 mM CaCl2, respectively. Furthermore, after conservation for several weeks at −20°C, PrtS activity was observable only in presence of CaCl2.
PrtS has a substrate specificity close to that of CEPs from other LAB.
Regarding its activity towards both chromogenic and casein substrates, PrtS presents a mixed substrate specificity of lactococcal PI and PIII types.
Of the seven chromogenic substrates tested (Table 1), PrtS was capable of hydrolyzing the four substrates sharing an aromatic amino acid at position P1 and a proline at position P2 (according to the nomenclature of Schechter and Berger [54]), but its activity was severely reduced when a negatively charged amino acid was located at position P3. The other three substrates, which possess a positively charged amino acid at position P1 instead of an aromatic one, were not hydrolyzed. One of the two best hydrolyzed substrates was MS-Arg-Pro-Tyr-pNA (MS-Arg), which is the preferential substrate of PI type lactococcal proteinase (9), whereas S-Ala-Glu-Pro-Phe-pNA (S-Glu), which is more specific of PIII-type lactococcal proteinase (9), was poorly degraded by PrtS.
TABLE 1.
Specificity of PrtS from S. thermophilus CNRZ 385 toward chromogenic substrates
| Substratea | Activity (%) |
|---|---|
| Ac-Ala-Ala-Pro-Phe-pNa | 100 |
| MS-Arg-Pro-Tyr-pNa | 100 |
| S-Val-Pro-Phe-pNa | 89 |
| S-Ala-Glu-Pro-Phe-pNa | 22 |
| Bz-Phe-Val-Arg-pNa | 0 |
| NZbz-Gly-Pro-Arg-pNa | 0 |
| Z-Phe-Arg-pNa | 0 |
Abbreviations: Bz, benzoyl; Z, benzyloxycarbonyl.
PrtS was capable of hydrolyzing whole [14C]casein and also [14C]β-casein at about the same rate (β casein/whole casein hydrolysis ratio of 0.8), which indicates that PrtS, as a PIII-type lactococcal proteinase, does not present a marked preference for β-casein.
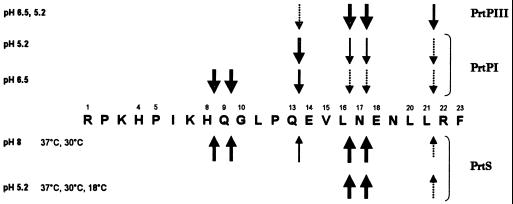
Depending on the reaction conditions (e.g., pH, salinity), PrtS hydrolyzed αs1-casein-(1-23) fragment at different sites (Fig. 1), all corresponding to hydrolysis sites already identified for PI- and PIII-type lactococcal proteinases. Under all conditions tested, hydrolysis occurred preferentially and primarily at bonds 16-17 and 17-18 (as indicated in Fig. 1). Near the optimum pH and temperature, i.e., at pH 8 and 37°C, PrtS cleaved the same bonds as PI-type lactococcal proteinase. At a pH corresponding to that prevailing in cheese (pH 5.2), the proteinase hydrolyzed only three bonds, corresponding to those recognized by the PIII-type lactococcal proteinase (21). At a more acidic pH (pH 4, as observed in yogurts) or in the presence of a high concentration of NaCl (4%, as observed in cheeses), the activity was no longer visible under our measurement conditions. The hydrolysis of 8-9 and 9-10 bonds of the αs1-casein-(1-23) fragment along with the absence of hydrolysis of these two bonds at acidic pH (5.2), which are typical of PI type proteinase, indicate that, qualitatively, PrtS specificity on this substrate is closer to that of PI- than to that of PIII-type lactococcal proteinase (19).
FIG. 1.
Specificity of PrtS from S. thermophilus CNRZ 385 and lactococcal PrtPs toward αs1-casein-(1-23) fragment. The cleavage sites are indicated by arrows. The sizes of the arrows are related to relative cleavage rates. Abbreviations: PrtS, proteinase from S. thermophilus CNRZ 385; PrtPI and PrtPIII, proteinases from L. lactis, HP and AM1, respectively (20, 21).
PrtS is a multidomain protein, maturated and anchored to the cell wall. (i) Sequencing and analysis of prtS gene.
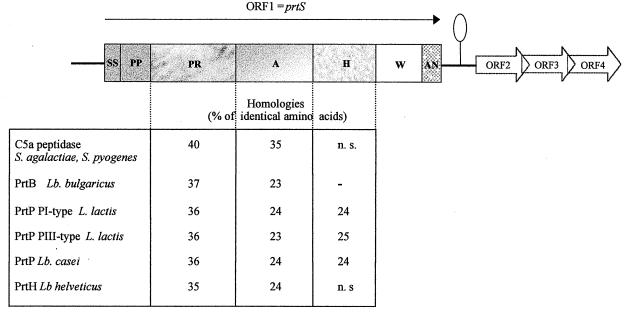
The prtS gene was sequenced first by performing PCR with two degenerated oligonucleotides deduced from the N-terminal amino acid sequence of the protein PrtS. A DNA fragment of 57 bp was amplified; its deduced amino acid sequence fitted perfectly with the N-terminal sequence of PrtS, indicating that the expected gene had been identified. Then, by successive inverse and LA PCRs, a sequence of 6,142 bp containing the entire prtS gene was determined. Sequence analysis of the 6,142-bp fragment revealed one complete open reading frame (ORF1) and three incomplete and overlapping ORFs: ORF2, ORF3, and ORF4 (Fig. 2). ORF1, which corresponded to the prtS gene, was 4,755 bp long and encoded a putative protein of 1,585 amino acids, with a calculated molecular mass of 169 kDa. A potential ribosome binding site complementary to the 3′ end of S. thermophilus 16S rRNA (Genbank sequence accession number X68418) was found 5 bp upstream of the putative ATG start codon (bases 333 to 335) of ORF1. Consensus sequences corresponding to potential promoters were not clearly identified in the overall sequence. About 80 bp downstream of ORF1, an inverted repeat resembling a putative terminator of transcription was present. No ORFs were found in the 330 bp upstream of prtS gene, unlike what was observed for lactococci, where an ORF encoding a maturation lipoprotein (PrtM) was present upstream of prtP (36). No consensus sequence corresponding to the specific binding site of the transcriptional activator Mga of protein M and C5a peptidase from group A streptococci (42), involved in virulence, was found in the 330-bp sequence upstream of prtS.
FIG. 2.
Genetic organization of prtS from S. thermophilus CNRZ 385 and its flanking regions and homologies with other CEPs from LAB and pyogenic streptococci. The oval indicates the Potential terminator of transcription. Abbreviations: SS, signal sequence; PP, propeptide; PR, catalytic domain; A, globular domain; B, PR stabilizing domain; H, helical domain; W, cell wall domain; AN, cell wall anchor; n.s., not significant.
(ii) Homology search.
Comparison of the amino acid sequence of PrtS with the proteins from the databases revealed high homology with CEPs from pyogenic streptococci (C5a peptidase from Streptococcus agalactiae [11] and Streptococcus pyogenes [10], and Csp from S. agalactiae [59]) and LAB (PrtP from lactococci [36, 67] and Lactobacillus casei [33], PrtB from Lactobacillus delbrueckii subsp. bulgaricus [30], and PrtH from Lactobacillus helveticus [48]). PrtS is organized in several structural and/or functional domains (Fig. 2) as recently described by Siezen (59) for the CEP of the above-mentioned species. Compared to the latter, PrtS differs mainly in its organization by the absence of a B domain (located between the A and H domains), as is the case for the pyogenic streptococcal proteinases (Fig. 2).
The first domain, the preprodomain, is removed during exportation and maturation processes. It is composed of a prosequence (109 aa) preceded by a probable classical 35-aa signal peptide common to exported proteins from gram-positive bacteria (45). The cleavage site for the signal peptide was predicted to be located between Ala35 and Asp36 by both the neural networks and the hidden Markov models developed by Nielsen et al. (47). The proof of the prosequence's removal was established firstly because the following amino acids corresponded to those of the N-terminal sequence of the purified protein, i.e., of the mature proteinase. Secondly, the molecular mass of the purified enzyme (153 kDa) was close to that deduced from the whole nucleotide sequence of prtS (169 kDa) without the propeptide (15.6 kDa).
The subsequent domain (PR) (495 aa), corresponding to the N-terminal part of the mature proteinase, is the catalytic domain (Fig. 2). It has homologies with the subtilisin-like proteinases known as subtilases (60) and contains, as that of CEPs from LAB and pyogenic streptococci, a large insert (140 residues) that is absent in subtilisins. The PR domain is the domain best conserved between PrtS and the other CEPs, with amino acid identities ranging from 35% with CEPs from LAB to 40% with C5a peptidase from streptococci (Fig. 2). The PR domain of the cell wall proteinase Csp from S. agalactiae is composed of 495 aa and, among the 356 already reported, 77% are identical to those of PrtS (59).
The central region of PrtS (the A domain [438 aa]) also shows homologies, to a lower extent, with that of the other CEPs, as 23 to 35% of the amino acids were identical (Fig. 2).
The C-terminal part of PrtS is composed of three domains (Fig. 2). The first domain, the H domain (367 aa), is predicted to be rich in α-helix secondary structure (66%) and presents only low homology with PrtP from lactococci and L. casei. The second domain, the W domain (106 aa), has no homologies with CEPs or with other proteins from the databases. It has the usual composition of the cell wall domain of gram-positive bacteria since it contains unusually high levels of Pro-Gly and Ser-Thr residues (16 and 20%, respectively) (26). Finally, the third domain, the AN domain (35 aa), most probably constitutes an anchor, with a typical LPXTG sorting motif (here, LPNTG) (25) followed by a stretch of hydrophobic residues, predicted to constitute an α-helix and ending with a charged tail (55). It has homologies with the corresponding domain of PrtP from lactococci and L. casei (25 to 35% identity) and of the C5a peptidase of S. agalactiae and S. pyogenes (32% identity).
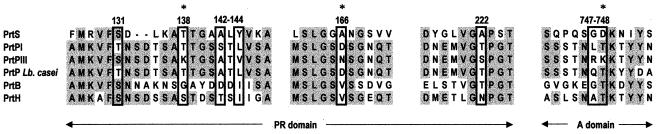
Amino acid alignments of PrtPs with subtilisin allowed the identification of relevant substrate binding regions in CEP sequences (regions 126 to 147, 161 to 171, 216 to 225, 742 to 753) (61). In these regions, the involvement of amino acids at positions 138, 166, and 748 in substrate specificity has been demonstrated by punctual mutagenesis (61). Alignment of the PrtS sequence with those of other LAB CEPs showed a related but different sequence for the proteinase of S. thermophilus, as PrtS possesses original amino acids at positions 126, 128, 132 to 136, 143 to 144, 146, 161, 165, 167, 169, 171, 217 to 219, 222, 224, 743 to 746, 748, and 750 to 751 (Fig. 3). The substrate binding sequences considered are only 40 and 41.5% identical between PrtS and L. lactis PrtPI and PrtPIII, respectively, while they are 93% identical between PrtP from L. casei and L. lactis PrtPI.
FIG. 3.
Multiple sequence alignment of substrate binding regions of PrtS from S. thermophilus CNRZ 385 and other CEPs from LAB and pyogenic streptococci. Boxed letters are amino acids of the substrate binding areas which are different in PrtP from L. lactis SK11 (PIII type) and Wg2 (PI type). Shaded letters are amino acids conserved in the majority of the sequences. Asterisks show amino acids involved in L. lactis PrtP substrate specificity as demonstrated by punctual mutagenesis (61). Residue numbering corresponds to that of mature L. lactis PrtP. Abbreviations: PrtS, proteinase from S. thermophilus CNRZ 385 (this work); PrtPI, proteinase from L. lactis Wg2 (36); PrtPIII, proteinase from L. lactis SK11 (69); PrtP Lb. casei, proteinase from L. casei NCDO 151 (33); PrtB, proteinase from L. delbrueckii subsp. bulgaricus NCDO 1489 (30); PrtH, proteinase from L. helveticus CNRZ 32 (48).
The ORFs identified downstream of prtS gene (ORF2, -3, and -4) potentially encode proteins that display identities with transposases from Streptomyces coelicolor, Bacillus thuringiensis, and Enterococcus faecalis, respectively, which suggests that this region was subjected to rearrangement.
DISCUSSION
PrtS, a member of the subtilase family.
The proteinase PrtS from S. thermophilus is a subtilisin-like serine proteinase (subtilase), and as such, shares with the CEPs from LAB and streptococci a serine base catalysis and highly conserved residues around the catalytic triad (Asp95, Ser196, His424) (60). In the subtilase family, PrtS from S. thermophilus is more homologous to streptococcal proteinase due to its sequence and the absence of a B domain that is present in CEPs from the other LAB. As with the majority of subtilases, PrtS is synthesized as a preproenzyme, which is subsequently maturated. In lactococci and some lactobacilli, a lipoprotein (PrtM) acting as a chaperone is required in this maturation step (69). The gene encoding PrtM is located directly upstream of the prtP gene, and the promoters of the two genes overlap in lactococci and L. casei (33, 36). As in L. delbrueckii subsp. bulgaricus (30) and L. helvetivus (48), no gene potentially encoding a lipoprotein was found in the close vicinity of prtS from S. thermophilus. Thus, we don't know whether a PrtM-like protein is required for PrtS maturation.
Cell envelope anchoring and release of PrtS.
PrtS is a cell wall-associated proteinase, with typical cell wall sorting signals of gram-positive bacteria located at its C terminus, i.e., an LPXTG motif followed by a hydrophobic domain and a charged tail constituting a cell wall anchor (55). This anchor is preceded by a Pro-, Gly-, Thr-, and Ser-rich region, which probably allows PrtS to span the cell wall with strong and tight linkage to the peptidoglycan, as proposed for the fructosyltransferase of Streptococcus salivarius (52). Thanks to its LPXTG motif, PrtS is probably covalently associated to the cell wall; such motifs present in surface proteins of Gram-positive bacteria are specifically recognized by a sortase that amide links the threonine residue to the peptidic cross-bridge of the peptidoglycan (41).
The CEPs from most LAB are released by incubating the cells in a Ca2+-free buffer (43), which induces the loss of weakly bound Ca2+ ions, leading to the exposure of a site susceptible to autoproteolysis (20). This Ca2+-binding site and the autoproteolytic site are most probably located in the B domain of the CEP (8). As PrtS lacks this B domain, it could not be released from the cell envelope in the same way as lactococcal PrtP. The probable cell wall covalent linkage of PrtS could explain the failure of detergent and cell wall-destabilizing treatments to release it. Lytic enzymes were successfully used for the release of lactococcal and lactobacilli PrtP (13, 23), lactobacilli PrtB (63), and C5a peptidase from group B streptococci (4) but allowed only a low level of PrtS release, perhaps because the S. thermophilus cell wall has a different susceptibility or accessibility to lysozyme. Finally, we succeeded in releasing PrtS from the cell wall by cultivating the cells in the presence of glycine followed by mechanical cell disruption and enzymatic treatment of the cell envelopes. Glycine is known to inhibit the peptide cross-linking of the peptidoglycan since it is incorporated in peptidoglycan precursors instead of alanine (31). Cultivation of the cells in the presence of glycine could thus enhance the susceptibility of the cell wall to lysozyme by leading to a more loosely cross-linked peptidoglycan.
Substrate specificity of PrtS.
The substrate specificity of PrtS from S. thermophilus on both chromogenic and casein substrates indicated that PrtS from strain CNRZ 385 is intermediate between the most divergent types, PrtPI and PrtPIII, as is the case for the lactobacillus CEPs (24, 40) and for the majority of lactococcal PrtP (5, 18). The PrtS sequence is consistent with this observation, since the substrate binding residues of PrtS are totally identical to neither those of PrtPI nor those of PrtPIII but are a combination of them, with unique residues as well. Furthermore, the variability in substrate specificity towards the αs1-casein-(1-23) fragment of CEPs is correlated with a variability in the substrate binding residues (18). In this regard, PrtS from strain CNRZ 385 possesses a unique substrate binding site among the CEPs from LAB which have been characterized up to now.
Origin of PrtS.
The presence of a high cell wall proteinase activity is common in L. lactis, whereas it is unusual in S. thermophilus (56). In L. lactis, this property probably results in an adaptation of the bacterium to the milk environment. The lactococcal prtP gene is indeed most frequently located on a plasmid that also carries the genes necessary for lactose utilization (35). In S. thermophilus, this infrequent characteristic could result from a different way that S. thermophilus ensures its growth in milk with regards to nitrogen nutrition. In dairy products, S. thermophilus is in fact often associated, in a symbiotic way, with lactobacilli which are known to be more proteolytic than S. thermophilus (51, 58) and to supply the latter with assimilable nitrogen compounds (2, 49, 58). Furthermore, S. thermophilus is qualitatively less demanding in regard to amino acids than lactococci are (15, 46; C. Letort and V. Juillard, personal communication).
On the basis of the infrequent presence of highly active CEPs in S. thermophilus, we can speculate that PrtS originates from a transfer of a proteinase gene from another bacterium, as suggested by the presence of signals of DNA rearrangement in the close vicinity of the prtS gene.
Role of PrtS.
Strains CNRZ 385 and 703 of S. thermophilus were first characterized by their rapid growth and high acidification rate in milk (1, 56). This rapid growth was then correlated with the presence of high proteinase activity associated with the cell wall of these strains, since a nitrosoguanidine proteinase-negative mutant of strain CNRZ 385 presented a low acidification rate in milk (56). Thus, as observed for the CEPs from the other LAB, the main role of PrtS concerns the amino acid supply to the cell via casein hydrolysis (56).
As already demonstrated for LAB CEPs (7, 27, 28, 62), PrtS, via its substrate specificity, probably has two main technological implications: firstly, in bacterial optimal development and consequently in milk acidification rate; secondly, in cheese ripening and flavor development.
In addition, LAB cell wall proteinases could be involved in the development of dairy product health properties via bioactive peptide production, as has already been demonstrated with antihypertensive peptide production by the L. helveticus proteinase (39).
With a proteinase-negative mutant, under construction, we will specify the functions of PrtS and the consequences of its presence during symbiotic growth and during ripening. In addition, the specificity studies presented in this work need to be widened to include other S. thermophilus strains in order to evaluate the biodiversity of the S. thermophilus cell wall proteinase.
ACKNOWLEDGMENTS
M.D.F.-E. was a recipient of a training grant from the European Union (contract BIO4-CT98-5043).
We thank J. C. Huet for N-terminal protein sequencing, C. Beauvallet for mass spectrometry analysis, C. Ouali for α51-casein-(1-23) fragment synthesis, and A. Lopez for her technical suggestion for gene sequencing. We are grateful to J. C. Gripon for critical reading of the manuscript.
REFERENCES
- 1.Accolas J P, Bloquel R, Didienne R, Regnier J. Propriétés acidifiantes des bactéries lactiques thermophiles en relation avec la fabrication du yoghourt. Lait. 1977;561-562:1–23. [Google Scholar]
- 2.Bautista E S, Dahiya R S, Speck M L. Identification of compounds causing symbiotic growth of Streptococcus thermophilus and Lactobacillus bulgaricus in milk. J Dairy Res. 1966;33:299–307. [Google Scholar]
- 3.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 4.Bohnsack J F, Zhou X, Williams P A, Cleary P P, Parker C J, Hill H R. Purification of the proteinase from group B streptococci that inactivates human C5a. Biochim Biophys Acta. 1991;1079:222–228. doi: 10.1016/0167-4838(91)90129-n. [DOI] [PubMed] [Google Scholar]
- 5.Boutrou R, Sepulchre A, Gripon J C, Monnet V. Simple tests for predicting the lytic behaviour and proteolytic activity of lactococcal strains in cheese. J Dairy Sci. 1998;81:2321–2328. [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Broadbent J R, Strickland M, Weimer B C, Johnson M E, Steele J. Peptide accumulation and bitterness in Cheddar cheese made using single-strain Lactococcus lactis starters with distinct proteinase specificity. J Dairy Sci. 1998;81:327–337. [Google Scholar]
- 8.Bruinenberg P G, De Vos W M, Siezen R J. Prevention of C-terminal processing of Lactococcus lactis SK11 cell-envelope proteinase by engineering of an essential surface loop. Biochem J. 1994;302:957–963. doi: 10.1042/bj3020957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruinenberg P G, Limsowtin G K Y. Diversity of proteolytic enzymes among lactococci. Aust J Dairy Technol. 1995;50:47–50. [Google Scholar]
- 10.Chen C C, Cleary P P. Complete nucleotide sequence of the streptococcal C5a peptidase gene of Streptococcus pyogenes. J Biol Chem. 1990;265:3161–3167. [PubMed] [Google Scholar]
- 11.Chmouriguina I, Suvorov A, Ferrieri P, Cleary P P. Conservation of the C5a peptidase genes in group A and B streptococci. Infect Immun. 1996;64:2387–2390. doi: 10.1128/iai.64.7.2387-2390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen J E, Dudley E G, Pederson J A, Steele J L. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1999;76:217–246. [PubMed] [Google Scholar]
- 13.Coolbear T, Reid J R, Pritchard G G. Stability and specificity of the cell wall-associated proteinase from Lactococcus lactis subsp. cremoris H2 released by treatment with lysozyme in the presence of calcium ions. Appl Environ Microbiol. 1992;58:3263–3270. doi: 10.1128/aem.58.10.3263-3270.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coolbear T, Pillidge C J, Crow V L. The diversity of potential cheese ripening characteristics of lactic acid starter bacteria. 1. Resistance to cell lysis and levels and cellular distribution of proteinase activities. Int Dairy J. 1994;4:697–721. [Google Scholar]
- 15.Desmazeaud M. L'état des connaissances en matière de nutrition des bactéries lactiques. Lait. 1983;63:267–316. [Google Scholar]
- 16.Devereux J, Haerberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donnelly W J, Barry J G, Richardson Y. 14C-Methylated casein as a substrate for plasmin and its application to the study of milk protein transformations. Biochim Biophys Acta. 1980;626:117–126. doi: 10.1016/0005-2795(80)90203-2. [DOI] [PubMed] [Google Scholar]
- 18.Exterkate F A. The lactococcal cell envelope proteinases: differences, calcium-binding effects and role in cheese ripening. Int Dairy J. 1995;5:995–1018. [Google Scholar]
- 19.Exterkate F A, Alting A C. The conversion of the αs1-casein (1-23)-fragment by the free and bound form of the cell-envelope proteinase of Lactococcus lactis subsp. cremoris in conditions prevailing in cheese. Syst Appl Microbiol. 1993;16:1–8. [Google Scholar]
- 20.Exterkate F A, Alting A C. Role of calcium in activity and stability of the Lactococcus lactis cell envelope proteinase. Appl Environ Microbiol. 1999;65:1390–1396. doi: 10.1128/aem.65.4.1390-1396.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Exterkate F A, Alting A C, Slangen C J. Specificity of two genetically related cell-envelope proteinases of Lactococcus lactis subsp. cremoris towards αs1-casein-(1-23)-fragment. Biochem J. 1991;273:135–139. doi: 10.1042/bj2730135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Exterkate F A, Alting A C, Bruinenberg P G. Diversity of cell envelope specificity among strains of Lactococcus lactis and its relationship to charge characteristics on the substrate-binding region. Appl Environ Microbiol. 1993;59:3640–3647. doi: 10.1128/aem.59.11.3640-3647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernández de Palencia P, Peláez C, Requena T, Martín-Hernández M C. Release and partial characterization of cell-envelope proteinases from Lactococcus lactis subsp. lactis IFPL 359 and Lactobacillus casei subsp. casei IFPL 731 isolated from goat's-milk cheese. Z Lebensm Unters Forsch. 1995;201:87–90. [Google Scholar]
- 24.Fernández de Palencia P, Peláez C, Romero C, Martín-Hernández M C. Purification and characterization of the cell wall proteinase of Lactobacillus casei subsp. casei IFPL 731 isolated from raw goat's milk cheese. J Agric Food Chem. 1997;45:3401–3405. [Google Scholar]
- 25.Fischetti V A, Pancholi V, Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins from Gram-positive cocci. Mol Microbiol. 1990;4:1603–1605. doi: 10.1111/j.1365-2958.1990.tb02072.x. [DOI] [PubMed] [Google Scholar]
- 26.Fischetti V A, Pancholi V, Schneewind O. Common characteristics of the surface proteins from gram-positive cocci. Appendix 1. In: Dunny G M, Clear P P, McKay L L, editors. Genetics and molecular biology of streptococci, lactococci, and enterococci. Washington, D.C.: American Society for Microbiology; 1991. pp. 290–294. [Google Scholar]
- 27.Flambard B, Richard J, Juillard V. Interaction between proteolytic strains of Lactococcus lactis influenced by different types of proteinase during growth in milk. Appl Environ Microbiol. 1997;63:2131–2135. doi: 10.1128/aem.63.6.2131-2135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flambard B, Helinck S, Richard J, Juillard V. The contribution of caseins to the amino acid supply for Lactococcus lactis depends on the type of cell envelope proteinase. Appl Environ Microbiol. 1998;64:1991–1996. doi: 10.1128/aem.64.6.1991-1996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibson T J. Studies on the Epstein Barr virus genome. Ph.D. thesis. Cambridge, England: University of Cambridge; 1984. [Google Scholar]
- 30.Gilbert C, Atlan D, Blanc B, Portalier R, Germond J E, Lapierre L, Mollet B. A new cell surface proteinase: sequencing and analysis of the prtB gene from Lactobacillus delbrueckii subsp. bulgaricus. J Bacteriol. 1996;178:3059–3065. doi: 10.1128/jb.178.11.3059-3065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammes W, Schleifer K H, Kandler O. Mode of action of glycine on the biosynthesis of peptidoglycan. J Bacteriol. 1973;116:1029–1053. doi: 10.1128/jb.116.2.1029-1053.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanahan D, Jessee J, Bloom F R. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 1985;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- 33.Holck A, Naes H. Cloning, sequencing and expression of the gene encoding the cell-envelope-associated proteinase from Lactobacillus paracasei subsp. paracasei NCDO 151. J Gen Microbiol. 1992;138:1353–1364. doi: 10.1099/00221287-138-7-1353. [DOI] [PubMed] [Google Scholar]
- 34.Isegawa Y, Sheng J, Sokawa Y, Yamanishi K, Nakagomi O, Shigeharu U. Selective amplification of cDNA sequence from total RNA by cassette-ligation mediated polymerase chain reaction (PCR): application to sequencing 6.5 kb genome segment of hantavirus strain B-1. Mol Cell Probes. 1992;6:467–475. doi: 10.1016/0890-8508(92)90043-w. [DOI] [PubMed] [Google Scholar]
- 35.Kok J. Genetics of proteolytic system of lactic acid bacteria. FEMS Microbiol Rev. 1990;76:2056–2064. doi: 10.1111/j.1574-6968.1990.tb04877.x. [DOI] [PubMed] [Google Scholar]
- 36.Kok J, Leenhouts K J, Haandrikman A J, Ledeboer A M, Venema G. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988;54:231–238. doi: 10.1128/aem.54.1.231-238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunji E R S, Mierau I, Hagting A, Poolman B, Konings W N. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- 38.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Maeno M, Yamamoto N, Takano T. Identification of an hypertensive peptide from casein hydrolysate produced by a proteinase from Lb. helveticus CP790. J Dairy Sci. 1996;79:1316–1321. doi: 10.3168/jds.S0022-0302(96)76487-1. [DOI] [PubMed] [Google Scholar]
- 40.Martín-Hernández M C, Alting A C, Exterkate F A. Purification and characterization of the mature, membrane-associated cell-envelope proteinase of Lactobacillus helveticus L89. Appl Microbiol Biotechnol. 1994;40:828–834. [Google Scholar]
- 41.Mazmanian S, Liu G K, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 42.McIver K S, Heath A S, Greenand B D, Scott J R. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A Streptococcus. J Bacteriol. 1995;177:6619–6624. doi: 10.1128/jb.177.22.6619-6624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mills O E, Thomas T D. Release of cell wall-associated proteinase(s) from lactic streptococci. N Z J Dairy Sci Technol. 1978;13:209–215. [Google Scholar]
- 44.Monnet V, Le Bars D, Gripon J C. Partial characterization and comparison of cell wall proteinases from 5 strains of Streptococcus lactis. Lait. 1987;67:51–61. [Google Scholar]
- 45.Navarre W W, Schneewind O. Surface proteins of Gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neviani E, Giraffa G, Brizzi A, Carminati D. Amino acid requirements and peptidase activities of Streptococcus salivarius subsp. thermophilus. J Appl Bacteriol. 1995;79:302–307. doi: 10.1111/j.1365-2672.1995.tb03141.x. [DOI] [PubMed] [Google Scholar]
- 47.Nielsen H, Engelbrecht J, Brunak S, Von Heijne G. Identification of procaryotic and eucaryotic signal peptides prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 48.Perderson J A, Mileski G J, Weimer B C, Steele J L. Genetic characterization of a cell envelope-associated proteinase from Lactobacillus helveticus CNRZ32. J Bacteriol. 1999;181:4592–4597. doi: 10.1128/jb.181.15.4592-4597.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pette J W, Lolkema H. Yogurt. II. Growth stimulating factor for Streptococcus thermophilus. Neth Milk Dairy J. 1950;4:209–224. [Google Scholar]
- 50.Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 51.Rajagopal S N, Sandine W E. Associative growth and proteolysis of Streptococcus thermophilus and Lactobacillus bulgaricus in skim milk. J Dairy Sci. 1990;73:894–899. [Google Scholar]
- 52.Rathsam C, Jacques N A. Role of C-terminal domains in surface attachment of the fructosyltransferase of Streptococcus salivarius ATCC 25975. J Bacteriol. 1998;180:6400–6403. doi: 10.1128/jb.180.23.6400-6403.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 54.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 55.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 1993;12:4801–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shahbal S, Hemme D, Desmazeaud M. High cell wall-associated proteinase activity of some Streptococcus thermophilus strains (H-strains) correlated with a high acidification rate in milk. Lait. 1991;71:351–357. [Google Scholar]
- 57.Shahbal S, Hemme D, Renault P. Characterization of a cell envelope-associated proteinase activity from Streptococcus thermophilus H-strains. Appl Environ Microbiol. 1993;59:177–172. doi: 10.1128/aem.59.1.177-182.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shankar P A, Davies F L. Proceedings of the XX International Dairy Congress. 1978. Proteinase and peptidase activities of yogurt starter bacteria; p. 473. Paris. [Google Scholar]
- 59.Siezen R J. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie Leeuwenhoek. 1999;76:139–155. [PubMed] [Google Scholar]
- 60.Siezen R J, Leunissen J A M. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 1997;6:501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siezen R J, Bruinenberg P G, Vos P, Van Alen-Boerrigter I J, Nijhuis M, Alting A C, Exterkate F A, De Vos W M. Engineering of the substrate-binding region of the subtilisin-like, cell envelope proteinase of Lactococcus lactis. Protein Eng. 1993;6:927–937. doi: 10.1093/protein/6.8.927. [DOI] [PubMed] [Google Scholar]
- 62.Stadhouders J, Toepoel L, Wouters J T M. Cheese making with prt- and prt+ variants of N-streptococci and their mixtures. Phage sensitivity, proteolysis and flavour development during cheese ripening. Neth Milk Dairy J. 1988;42:183–193. [Google Scholar]
- 63.Stefanisti D, Sakellaris G, Garel J R. The presence of two proteinases associated with the cell wall of Lactobacillus bulgaricus. FEMS Microbiol Lett. 1995;128:53–58. [Google Scholar]
- 64.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas T D, Mills O E. Proteolytic enzymes of starter bacteria. Neth Milk Dairy J. 1981;35:255–273. [Google Scholar]
- 66.Visser S, Exterkate F A, Slangen C J, De Veer J C M. Comparative study of action of cell wall proteinases from various strains of Streptococcus cremoris on bovine αs1-, β-, and κ-casein. Appl Environ Microbiol. 1986;52:1162–1166. doi: 10.1128/aem.52.5.1162-1166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vos P, Simons G, Siezen R J, De Vos W M. Primary structure and organization of the gene for a procaryotic cell envelope-associated serine proteinase. J Biol Chem. 1989;264:13579–13589. [PubMed] [Google Scholar]
- 68.Vos P, Van Alen-Boerrigter I J, Buist G, Haandrikman A J, Nijhuis M, de Reuver M B, Siezen R J, Venema G, de Vos W M, Kok J. Engineering of the Lactococcus lactis serine proteinase by construction of hybrid enzymes. Protein Eng. 1991;4:479–484. doi: 10.1093/protein/4.4.479. [DOI] [PubMed] [Google Scholar]
- 69.Vos P, Van Asseldonk M, Van Jeveren F, Siezen R J, Simons G, de Vos W M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989;171:2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zevaco C, Monnet V, Gripon J C. Intracellular X-prolyl dipeptidyl peptidase from Lactococcus lactis ssp. lactis: purification and properties. J Appl Bacteriol. 1990;68:357–366. [Google Scholar]