Abstract
Calcitonin gene-related peptide (CGRP) signaling inhibitors have shown efficacy in both the acute and preventive treatment of migraine. Telcagepant, a first-generation CGRP receptor antagonist, was effective but failed in clinical trials due to hepatotoxicity. Subsequently, although 4 next-generation CGRP receptor antagonists (rimegepant, zavegepant, atogepant, and ubrogepant) were being advanced into late-stage clinical trials, due to telcagepant’s failure, more confidence in the liver safety of these compounds was needed. DILIsym v6A, a quantitative systems toxicology (QST) model of drug-induced liver injury (DILI), was used to model all 5 compounds and thus to compare the 4 next-generation CGRP receptor antagonists to telcagepant. In vitro experiments were performed to measure the potential for each compound to inhibit bile acid transporters, produce oxidative stress, and cause mitochondrial dysfunction. Physiologically based pharmacokinetic models were produced for each compound in order to appropriately estimate liver exposure. DILIsym predicted clinical elevations of liver enzymes and bilirubin for telcagepant, correctly predicting the observed DILI liability of the first-generation compound. By contrast, DILIsym predicted that each of the 4 next-generation compounds would be significantly less likely to cause DILI than telcagepant. Subsequent clinical trials have validated these predictions for each of the 4 compounds, and all 3 of the compounds submitted to FDA to date (rimegepant, ubrogepant, and atogepant) have since been approved by the FDA with no warning for hepatotoxicity. This work demonstrates the potential for QST modeling to prospectively differentiate between hepatotoxic and nonhepatotoxic molecules within the same class.
Keywords: liver injury, quantitative systems toxicology, biological modeling, pharmaceuticals
Inhibition of the calcitonin gene-related peptide (CGRP) pathway has been identified as a potential target for migraine treatment and prevention, which would meet a major unmet medical need (Edvinsson, 2018; Holland and Goadsby, 2018). While several monoclonal antibody treatments targeting CGRP exist (Berman et al., 2020), small molecules present an advantage in that they may be able to engage the CGRP receptors that exist within the brain, as well as can be dosed orally (Mullin et al., 2020). Two of the first-generation small molecules in the CGRP receptor antagonist class, telcagepant and MK-3207, failed during late-stage clinical trials due to liver injury (Hewitt et al., 2011; Ho et al., 2016). Several next-generation drugs have been developed since the failure of telcagepant and MK-3207; 3 compounds, rimegepant (Nurtec ODT), ubrogepant (Ubrelvy), and atogepant (Qulipta), have been approved by the FDA for the treatment of migraine (Al-Hassany et al., 2022).
The identification of liver injury issues with telcagepant and MK-3207 led to the need to investigate potential differences between those 2 drugs and the other CGRP receptor antagonists. Computer modeling, specifically quantitative systems toxicology (QST) modeling, has been investigated as a way of predicting the toxicity potential of a drug (Watkins, 2020). DILIsym, a QST model of drug-induced liver injury (DILI), has demonstrated the ability to differentiate between hepatotoxic and nonhepatotoxic drugs in the same class (Longo et al., 2016; Woodhead et al., 2014) and to identify mechanistic differences among drugs with DILI liabilities within the same class (Woodhead et al., 2019). DILIsym has also been used to prospectively predict the safety of a drug as compared to another drug within that same class that had DILI liabilities (Woodhead et al., 2020). All of these factors make DILIsym a potentially valuable tool for differentiating hepatotoxic from nonhepatotoxic CGRP receptor antagonists.
In this work, DILIsym was used to predict the hepatotoxicity of telcagepant and compare it to 4 next-generation CGRP receptor antagonists: rimegepant, ubrogepant, atogepant, and zavegepant. In vitro hepatotoxicity studies were conducted for all 5 compounds according to standard DILIsym procedure (Longo et al., 2019; Woodhead et al., 2019, 2020). It is worth noting that the simulation work for these compounds was conducted in blinded fashion with assessments of DILI potential made before Phase 3 clinical trial results were completed, making these true prospective predictions.
MATERIALS AND METHODS
DILIsym v6A was used for all hepatotoxicity simulations. Physiologically based pharmacokinetic (PBPK) modeling for telcagepant, rimegepant, and zavegepant were developed in DILIsym v6A; in silico PBPK predictions for ubrogepant and atogepant were generated in GastroPlus 9.5.
PBPK modeling was performed to predict the liver concentration-time profile for each compound. Literature data were used to develop the PBPK model for telcagepant (Han et al., 2010) and to estimate certain PBPK parameters. Clinical trial data were used to develop the PBPK model for rimegepant. The PBPK model for zavegepant was based on animal data and in silico predictions and included adaptations for different dosing routes. Further information on the PBPK modeling for the 5 compounds in this study is provided in the Supplementary Materials. Most notably, a custom PK SimPops was created for rimegepant based on the range of plasma concentrations observed in the clinic; the underlying mechanistic susceptibility variation in v4A_1 was still present in this SimPops. Population variability in PK was not included for the other compounds due to the lack of clinical data for zavegepant, atogepant, and ubrogepant and uncertainty around the variability present in the published data for telcagepant. For atogepant and ubrogepant, no PBPK data were available; as a result, the PBPK for these compounds was developed using the machine learning algorithms in ADMET Predictor (SimulationsPlus, Lancaster, California) as included in GastroPlus 9.5 (SimulationsPlus).
In vitro experiments were conducted on each of the 5 compounds to assess the effect of each drug on the 3 main hepatotoxicity mechanisms represented in DILIsym v6A: reactive oxidative stress (ROS), mitochondrial dysfunction, and bile acid transporter inhibition. For oxidative stress assessments, HepG2 cells were cultured for 6 or 24 h with varying drug concentrations and ROS formation was assayed by dihydroethidium (DHE) fluorescence. Three independent studies in triplicate were conducted on HepG2 cells. Assays were performed by Cyprotex (Macclesfield, UK). A detailed description of the protocol for each compound is provided in the Supplementary Materials.
LC/MS/MS analysis was performed on a parallel culture of HepG2 cells to assess the intracellular concentration of each compound; cell lysate concentration was assayed and subsequently corrected for lysate volume and cell volume.
Parameter values for telcagepant-mediated induction of oxidative stress were identified by simulating the experimental data in DILIsym using a DILIsym dosing scheme meant to represent in vitro conditions (steady-state liver exposure).
Simulation protocols for each compound were chosen based on clinical experience and proposed clinical doses for each compound. The simulations performed were as follows:
Telcagepant: 140 and 280 mg BID
Rimegepant: (1) 75 mg QD, 25 days; (2) 75 mg QD, 25 doses delivered as 5 consecutive days, with one-day rest; (3) 75 mg QD, 14 doses given on alternate days
Ubrogepant: (1) 100 mg QD, 25 days; (2) 200 mg QD, 25 days; (3) 500 mg QD, 25 days; (4) 1000 mg QD, 25 days
Zavegepant: (1) 75 mg QD, oral (PO), 25 days; (2) 750 mg QD, PO, 25 days; (3) 2 mg QD, intranasal (IN), 25 days; (4) 20 mg QD, IN, 25 days; (5) 0.75 mg QD, subcutaneous (SC), 25 days; (6) 7.5 mg QD, SC, 25 days
Atogepant: (1) 60 mg BID, 12 weeks; (2) 120 mg BID, 12 weeks; (3) 300 mg BID, 12 weeks; (4) 600 mg BID, 12 weeks
Simulated alanine transaminase (ALT) elevation frequency and severity, as well as several other clinical and mechanistic outputs, were collected for each simulation. Each of the simulation results were compared with clinical experience where applicable (ie, with telcagepant); the other simulation results were treated as a prospective prediction of liver safety.
Mechanistic simulation results were employed as a way of determining the most important mechanism contributing to any predicted ALT elevations for each compound. The general concept of these simulations has been discussed previously (Longo et al., 2019; Woodhead et al., 2017, 2019). Briefly, simulations are performed on a smaller SimCohorts of responder individuals with each potential mechanism of hepatotoxicity turned off in sequence. A decrease in ALT elevation frequency when a mechanism is off demonstrates that the mechanism in question is contributing to the simulated ALT elevations.
RESULTS
Results of In Vitro Assays
In vitro assays determining the extent of the CGRP signal-blocking compounds’ ability to inhibit bile acid transport, induce mitochondrial toxicity, and generate oxidative stress were interpreted and translated into parameters for input into DILIsym v6A. The presence of a parameter for a given mechanism for a compound should not be taken as proof that the mechanism is a potential liability for the compound; the parameters quantify the potential activity of the compound on each mechanism and as such may not be active at the predicted liver concentration range for the compound. DILIsym simulations integrate these parameters with liver exposure predictions to determine the relevance of these mechanisms for potential DILI.
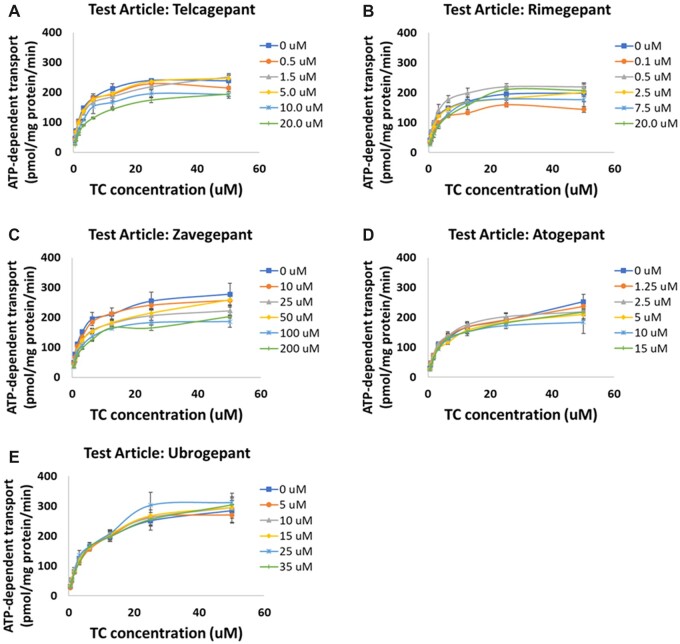
Bile acid transport was inhibited by all compounds to varying degrees. Figure 1 shows the inhibition of the bile salt export pump (BSEP) by each of the CGRP receptor antagonists; Figure 2 shows the inhibition of basolateral bile acid transporters (MRP3 and MRP4). None of the compounds inhibited the sodium taurocholate cotransporting polypeptide (NTCP) (not shown). For each compound, the mode of inhibition for BSEP was assessed; for telcagepant, ubrogepant, and zavegepant, the mode of inhibition was mixed, whereas for rimegepant, the mode of inhibition was competitive. The calculated Ki values along with the calculated α values can be found in Table 1. For MRP3/4 and NTCP, the mode of inhibition was assumed to be mixed inhibition with α = 5 (Woodhead et al., 2020).
Figure 1.
Inhibition of ATP-dependent taurocholate (TC) transport into membrane vesicles mediated by the hepatic bile salt export pump (BSEP) by CGRP receptor antagonist compounds: (A) telcagepant; (B) rimegepant; (C) zavegepant; (D) atogepant; (E) ubrogepant. Ki studies involving assessment of the transporter inhibition at several different concentrations of taurocholate and the test article were performed, and the data were fit using Michaelis-Menten kinetics to determine the Ki and inhibition type for each compound. Note: zavegepant top concentration was ≥10× higher than others and it shows negligible BSEP inhibition (Ki > 340 µM, see Table 1).
Figure 2.
Inhibition of basolateral bile acid transporters by CGRP compounds: (A) inhibition of MRP4 by telcagepant; (B) inhibition of MRP3 by atogepant; (C) inhibition of MRP3 by ubrogepant. Rimegepant and zavegepant did not inhibit basolateral bile acid transporters; telcagepant did not inhibit MRP3; and ubrogepant and atogepant did not inhibit MRP4.
Table 1.
DILIsym Input Parameters for the CGRP Compounds, Including Parameters Related to Mitochondrial Toxicity, ROS Generation, and Bile Acid Transporter Inhibition
| Mechanism | Parameter | Unit | DILIsym Parameter Valuea |
|||||
|---|---|---|---|---|---|---|---|---|
| Telcagepant—High | Telcagepant—Low | Rimegepant | Zavegepant | Atogepant | Ubrogepant | |||
| Mitochondrial dysfunction | Coefficient for ETC inhibition 1 | µM | 3470 | 3470 | 3470 | 1600 | 38 170 | Not used |
| Coefficient for ETC Inhibition 3 | µM | 1.89 | Removed | 1.89 | 2 | 0.1 | 4,217 | |
| Max inhibitory effect for ETC inhibition 3 | Dimensionless | 0.45 | Removed | 0.45 | 1.5 | 0.2 | 0.4 | |
| Uncoupler 1 effect Km | mM | No effect | No effect | No effect | No effect | 15 300 | ||
| Uncoupler 1 effect Vmax | Dimensionless | No effect | No effect | No effect | No effect | 22.5 | ||
| Uncoupler 1 effect Hill | Dimensionless | No effect | No effect | No effect | No effect | 4.3 | ||
| Oxidative stress | RNS/ROS production rate constant 1 | ml/nmol/h | 3.5 × 10−4 | 3.5 × 10−4 | 3.5 × 10−4 | No ROS production | 3.41 × 10−4 | 1.65 ×10−4 |
| Bile acid transporter inhibitionb | BSEP inhibition constant | µM | 19.0 | 19.0 | 27.2 | 341 | 144.2 | No inhibition |
| BSEP inhibition alpha value | Dimensionless | 4.32 | 4.32 | Competitive | 1.368 | 0.64 | No inhibition | |
| NTCP inhibition constant | µM | No inhibition | No inhibition | No inhibition | No inhibition | No inhibition | No inhibition | |
| MRP4 inhibition constant | µM | 42.4 | 42.4 | No inhibition | No inhibition | 42 | 75.3 | |
Values shown in the table for DILIsym input parameters should not be interpreted in isolation with respect to clinical implications, but rather, should be combined with exposure in DILIsym to produce simulations that have predictive and insightful value.
IC50 values were used for transporters other than BSEP, where Ki was measured; alpha value of 5 assumed when IC50 measured.
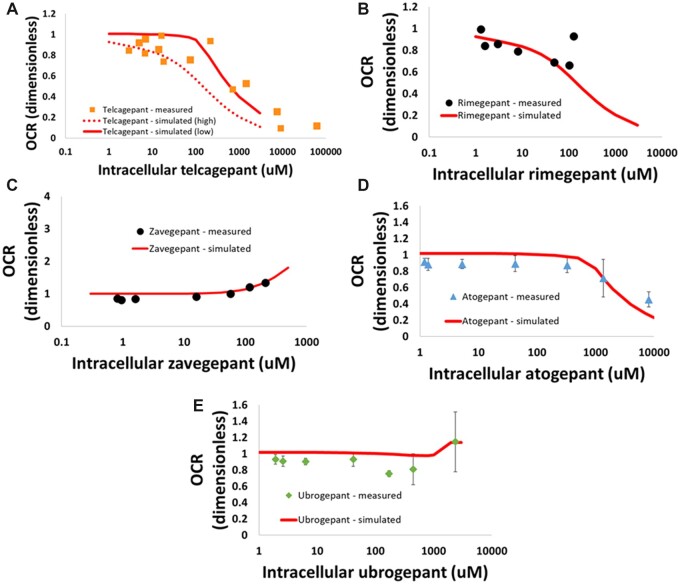
All the CGRP receptor antagonists except zavegepant inhibited the mitochondrial electron transport chain to varying extents as well; zavegepant was determined to be a mild uncoupler of the mitochondrial proton gradient. Figure 3 shows the oxygen consumption rate (OCR) versus intracellular compound concentration graphs for each compound and their attendant fits in MITOsym. Each parameter value calculated in MITOsym was translated into a DILIsym parameter (Woodhead et al., 2019; Yang et al., 2014); these parameters are shown in Table 1. For telcagepant, the in vitro data lent itself to a range of potential representations rather than to a single representation. As a result, 2 alternate parameterizations were used to bound the potential range of ETC inhibition parameter values that could plausibly represent the in vitro data; they are represented as the “high” and “low” parameter sets in Table 1 and Figure 3B. SimPops-based hepatotoxicity simulations were performed with both parameterizations.
Figure 3.
Oxygen consumption rate (OCR) versus measured intracellular compound concentration after a 24-h incubation in HepG2 cells for each of the CGRP receptor antagonist compounds: (A) telcagepant; (B) rimegepant; (C) zavegepant; (D) atogepant; and (E) ubrogepant. Fits to the data in MITOsym used to parameterize the model are shown along with the in vitro data results. For telcagepant, 2 alternate parameterizations were determined to be equally plausible; as a result, both were explored and used in simulations.
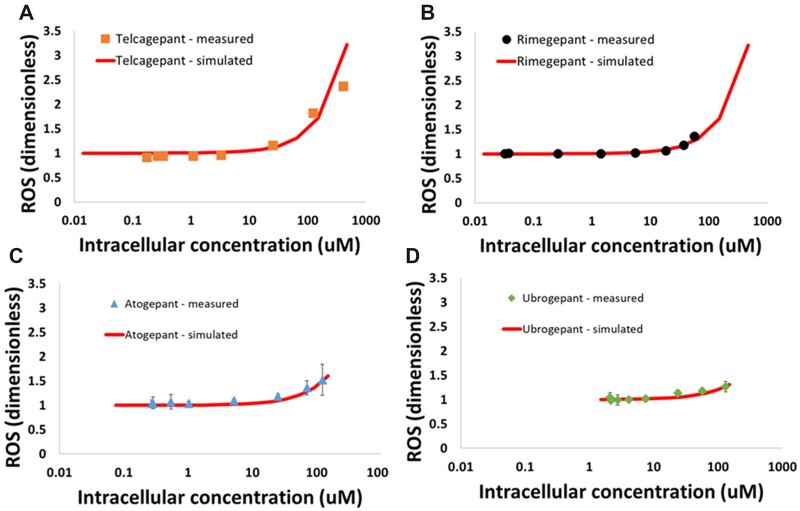
Each of the CGRP antagonists also induced oxidative stress in vitro. Figure 4 shows the relationship between intracellular compound concentration and ROS production for each compound. These data were fit in DILIsym and the resulting parameters are shown in Table 1.
Figure 4.
Reactive oxygen species (ROS) as measured by dihydroethidium (DHE) staining in HepG2 cells after a 24-h incubation with CGRP compounds: (A) telcagepant; (B) rimegepant; (C) atogepant; (D) ubrogepant. Zavegepant did not cause an increase in ROS in the experimental system. Fits to the data in DILIsym used to parameterize the model are also shown.
Simulation Results
Table 2 lists all of the predicted ALT elevation frequencies from the simulations performed in this work. First, DILIsym was used to simulate the telcagepant clinical trials. The 280 mg BID dose predicted ALT elevations for both parameterizations; ALT elevations were predicted using only the “high” parameterization at the 140 mg BID dose. This is similar to clinical experience, where some ALT elevations occurred at both dosing levels. Given the uncertainty in the ETC inhibition data and the resulting representation of telcagepant using outer bounds for hepatotoxicity parameter inputs, an underprediction using one parameterization and an overprediction using the other parameterization would naturally be expected. Notably, the simulations do not include adaptation mechanisms, potentially including mitochondrial biogenesis, that would likely reduce the severity of some of the simulated liver injury; this is why the number of Hy’s Law cases with telcagepant treatment is overestimated by the simulations (Figure 5).
Table 2.
Simulated ALT Elevations in the v4A_1 SimPops for Each of the CGRP Compounds
| Compound | Oral Dosing Protocol | Simulated ALT > 3X ULNa | Observed ALT > 3X ULN in Clinic |
|---|---|---|---|
| Telcagepant—High ETC | 140 mg BID, 12 weeks | 17.5% (50/285) | 1.9% (5/263) |
| 280 mg BID, 12 weeks | 76.1% (217/285) | 3.2% (8/265) | |
| Telcagepant—Low ETC | 140 mg BID, 12 weeks | 0.0% (0/285) | 1.9% (5/263) |
| 280 mg BID, 12 weeks | 7.72% (22/285) | 3.2% (8/265) | |
| Rimegepant | 75 mg QD, alternate day dosing, 14 total doses | 0.35% (1/285) | — |
| 75 mg QD, 5 days on, 1 day off, 25 total doses | 0.7% (2/285) | — | |
| 75 mg QD, daily dosing for 25 days, 25 total doses | 1% (3/285) | — | |
| Zavegepant | 750 mg oral QD, 25 days, 25 total doses | 0.0% (0/285) | |
| 75 mg oral QD, 25 days, 25 total doses | 0.0% (0/285) | ||
| 20 mg IN QD, 25 days, 25 total doses | 0.0% (0/285) | ||
| 2 mg IN QD, 25 days, 25 total doses | 0.0% (0/285) | ||
| 0.75 mg IV QD, 25 days, 25 total doses | 0.0% (0/285) | ||
| 7.5 mg IV QD, 25 days, 25 total doses | 0.0% (0/285) | ||
| Atogepant | 60 mg BID, 12 weeks | 0% (0/285) | |
| 120 mg BID, 12 weeks | 0% (0/285) | ||
| 300 mg BID, 12 weeks | 0.3% (1/285) | ||
| 600 mg BID, 12 weeks | 10.2% (29/285) | ||
| Ubrogepant | 100 mg QD, 15 days | 0% (0/285) | |
| 200 mg QD, 15 days | 0% (0/285) | ||
| 500 mg QD, 15 days | 1.1% (3/285) | ||
| 1000 mg QD, 15 days | 11.6% (33/285) | ||
| 100 mg QD, 25 days | 0% (0/285) | ||
| 200 mg QD, 25 days | 0% (0/285) | ||
| 500 mg QD, 25 days | 1.4% (4/285) | ||
| 1000 mg QD, 25 days | 11.6% (33/285) |
The alternate parameterizations for telcagepant are included in the table; comparisons to clinical data were only available for telcagepant at the time the simulations were conducted and are included in the table as well.
Upper limit of normal (ULN) for ALT in DILIsym is 40 U/L.
Figure 5.
eDISH plots demonstrating the severity of simulated ALT elevations for each of the CGRP compounds for which toxicity was predicted at indicated dosing regimens. eDISH plots show ALT fold change on the x-axis and bilirubin fold change on the y-axis. Individuals in the bottom-right quadrant have ALT elevations but no bilirubin elevations; individuals in the top-right quadrant have both ALT and bilirubin elevations and are considered most at risk for developing severe DILI (Hy’s Law). The simulations do not include several adaptation mechanisms that would likely reduce the severity of the ALT elevations (though not the frequency).
Rimegepant and zavegepant were both predicted to be relatively safe compared with telcagepant. ALT elevations with rimegepant were simulated in some extreme dosing scenarios, especially when the high-PK representation was used; however, as is seen in the eDISH plots of the simulation results (Figure 5), these were generally mild compared with the severe ALT elevations and Hy’s Law cases simulated with telcagepant (Figure 5). For zavegepant, no ALT elevations were predicted at any dose, even those well above the proposed therapeutic doses (Table 2, not shown in Figure 5 because all individuals were in the normal range on the eDISH plot).
Both atogepant and ubrogepant also compared favorably to telcagepant. ALT elevations with both compounds were not simulated at the proposed clinical doses, and only appeared at doses 10-fold higher than the clinical doses (Table 2).
Mechanistic simulation results show that telcagepant hepatotoxicity is driven by ETC inhibition and bile acid transporter inhibition (Table 3). This is distinct from rimegepant, whose predicted minor and mild ALT elevations in the high dosing frequency scenarios were caused by ETC inhibition alone, with only mild contributions from bile acid transporter inhibition and ROS generation (Table 3). Ubrogepant and atogepant simulated ALT elevations at the supratherapeutic doses were largely ROS dependent. For zavegepant, no simulated ALT elevations were observed, so mechanistic simulations were unable to be performed.
Table 3.
Mechanistic Investigation Simulations for Each of the CGRP Compounds Focused on the Select Specific Scenarios That Did Produce ALT Elevations (ie, to Identify When ALT Elevations Occur, What Underlies It)
| Compound and Oral Protocol | Mechanism Off | Mechanisms On | Simulated ALT > 3× ULN |
|---|---|---|---|
| Telcagepant—Original ETC, 140 mg BID, 12 weeks | None | BAi; ETCi; ROS | 50/50 |
| BAi | ETCi; ROS | 21/50 | |
| ETCi | BAi; ROS | 0/50 | |
| ROS | BAi; ETCi | 46/50 | |
| Rimegepant, high PK individual, 75 mg QD, daily dosing for 25 days, 25 total doses | None | BAi; ETCi; ROS | 21/21 |
| BAi | ETCi; ROS | 14/21 | |
| ETCi | BAi; ROS | 0/21 | |
| ROS | BAi; ETCi | 18/21 | |
| Atogepant, 600 mg BID, 12 weeks | None | All | 29/29 |
| ROS | BAi, ETCi | 2/29 | |
| ETCi | BAi, ROS | 24/29 | |
| BAi | ETCi, ROS | 24/29 | |
| Ubrogepant, 1000 mg QD, 25 days | None | All | 33/33 |
| ROS | BAi, ETCi, UC | 0/33 | |
| ETCi | BAi, UC, ROS | 33/33 | |
| UC | BAi, ETCi, ROS | 33/33 | |
| ETCi, UC | BAi, ROS | 33/33 | |
| BAi | ETCi, UC, ROS | 33/33 |
Mechanisms were turned off and on to determine which mechanism contributed the most to the observed ALT elevations. Only those individuals who developed ALT elevations in the initial 285-individual SimPops simulation were used in the mechanistic investigation simulations.
Abbreviations: BAi, bile acid transporter inhibition; ETCi, electron transport chain inhibition; UC, mitochondrial proton gradient uncoupling; ROS, reactive oxygen species generation.
DISCUSSION AND CONCLUSIONS
Telcagepant failed in the clinic due to severe liver injury concerns, and DILIsym simulations conducted herein correctly predicted the hepatotoxicity liability of telcagepant. Taken together, the simulation results in this work predicted that the 4 next-generation CGRP receptor antagonists would be significantly safer than telcagepant with respect to liver injury. Neither rimegepant, ubrogepant, atogepant, nor zavegepant were predicted to cause severe liver injury (ie, Hy’s Law cases) at the tested clinical dose regimens, even when the worst-case scenarios for clinical exposure were considered. At the actual (rimegepant, atogepant, ubrogepant) or anticipated (zavegepant) FDA recommended doses and regimens, the vast majority of individuals fall into the normal range for simulated ALT elevations, with for rimegepant a few individuals passing just beyond the normal ALT threshold but with no bilirubin elevations.
Mechanistic simulation results also underscore the difference between telcagepant and the other CGRP receptor antagonist compounds. Telcagepant hepatotoxicity was driven by a combination of ETC inhibition and bile acid transporter inhibition; this combination of mechanisms is often responsible for predicted serious hepatotoxicity in DILIsym (Longo et al., 2019; Woodhead et al., 2017). Meanwhile, the other compounds were shown to have only a single mechanism—either ETC inhibition or ROS formation—as their biggest potential liability; this suggests that an adaptive response to ROS that is not included in DILIsym or mitochondrial biogenesis could prevent serious injury from occurring from these other, newer CGRP receptor antagonists. The bile acid transporter mechanism is especially interesting; whereas rimegepant, atogepant, and ubrogepant all showed signals in a BA transporter IC50 assay, further exploration in the in vitro systems and with DILIsym revealed several reasons why those compounds do not have bile acid transport inhibition liabilities; for example, rimegepant is a competitive inhibitor of BSEP whereas telcagepant is a mixed inhibitor of BSEP, and for most individuals the concentration of rimegepant in the liver is well below the measured Ki while telcagepant liver concentration is well above the measured Ki. The clinical importance is 2-fold: (1) a competitive mode of inhibition (rimegepant) can be overcome as bile salt acid levels increase and BSEP continues to provide functional export of bile salts, whereas mixed mode (telcagepant), which includes noncompetitive inhibition, cannot be similarly overcome which leads to the build up of bile salts associated with liver injury; and (2) for most individuals, clinical liver concentrations at Cmax are well above the BSEP Ki (telcagepant) and indicates ongoing inhibition and reduction of the majority of bile salt export function, whereas clinical liver concentrations at Cmax for most individuals fall well below the BSEP Ki (rimegepant) which indicates that the majority of bile salt export function remains intact.
The clinical experience with these 4 next-generation CGRP signal-blocking drugs has been largely in line with DILIsym predictions for these compounds. Rimegepant, ubrogepant, and atogepant have all demonstrated clinical efficacy and safety (Al-Hassany et al., 2022) and have since been approved by the FDA for the treatment of migraine (acute for ubrogepant, preventive for atogepant, and dual-therapy acute and preventive for rimegepant). Meanwhile, clinical trial results for zavegepant which continues to advance in clinical trials for the acute (nasal) and preventive (oral) treatment of migraine without a liver safety signal (Al-Hassany et al., 2022; Goadsby et al., 2020; Moreno-Ajona et al., 2020). These results suggest that the QST approach demonstrated by DILIsym can lead to increased confidence in the ability to differentiate between liver safety profiles of drugs in the same class.
In a previous paper, DILIsym was used to compare telcagepant and ubrogepant to MK-3207, another CGRP receptor antagonist that failed in clinical trials due to liver injury. Telcagepant and MK-3207 were predicted to be hepatotoxic by DILIsym in that work, whereas ubrogepant was predicted to be safe (Yamazaki et al., 2013). These simulations were performed independently from the simulations in this work, with a set of in vitro hepatotoxicity assays conducted separately from what was conducted for this report. Although some minor differences between the results from the 2 simulation projects exist, the striking qualitative similarity between the predictions for telcagepant and ubrogepant (ie, that telcagepant could cause severe hepatotoxicity while ubrogepant would be safe) between the 2 independently conducted simulation projects demonstrates the robustness of the DILIsym QST approach.
In summary, DILIsym prospectively predicted improved liver safety of the 4 next-generation CGRP receptor antagonists rimegepant, zavegepant, atogepant, and ubrogepant relative to the hepatoxic first-generation molecule telcagepant, and these predictions have been born out in the clinical trials conducted to date. It is worth noting that the same general methods employed in this manuscript have been used to compare hepatotoxic potential among drugs within other compound classes as well (Woodhead et al., 2019, 2020). Our results support the value of QST modeling in drug development.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
Biohaven, Inc. provided funding for this research.
DECLARATION OF CONFLICTING INTERESTS
Biohaven Pharmaceuticals is the manufacturer of rimegepant and zavegepant, 2 of the compounds analyzed in this paper. C.C. is employed by Biohaven.
Supplementary Material
Contributor Information
Jeffrey L Woodhead, DILIsym Services, Inc., A Simulations Plus Company, Research Triangle Park, North Carolina 27706, USA.
Scott Q Siler, DILIsym Services, Inc., A Simulations Plus Company, Research Triangle Park, North Carolina 27706, USA.
Brett A Howell, DILIsym Services, Inc., A Simulations Plus Company, Research Triangle Park, North Carolina 27706, USA.
Paul B Watkins, Institute for Drug Safety Sciences, UNC-Eshelman School of Pharmacy, University of North Carolina, Chapel Hill, North Carolina 27599, USA.
Charles Conway, Biohaven Pharmaceuticals, Inc., New Haven, Connecticut 06510, USA.
REFERENCES
- Al-Hassany L., Goadsby P. J., Danser A. H. J., MaassenVanDenBrink A. (2022). Calcitonin gene-related peptide-targeting drugs for migraine: How pharmacology might inform treatment decisions. Lancet Neurol. 21, 284–294. [DOI] [PubMed] [Google Scholar]
- Berman G., Croop R., Kudrow D., Halverson P., Lovegren M., Thiry A. C., Conway C. M., Coric V., Lipton R. B. (2020). Safety of rimegepant, an oral CGRP receptor antagonist, plus CGRP monoclonal antibodies for migraine. Headache 60, 1734–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvinsson L. (2018). The CGRP pathway in migraine as a viable target for therapies. Headache 58(Suppl 1), 33–47. [DOI] [PubMed] [Google Scholar]
- Goadsby P. J., Dodick D. W., Ailani J., Trugman J. M., Finnegan M., Lu K., Szegedi A. (2020). Safety, tolerability, and efficacy of orally administered atogepant for the prevention of episodic migraine in adults: A double-blind, randomised phase 2b/3 trial. Lancet Neurol. 19, 727–737. [DOI] [PubMed] [Google Scholar]
- Han T. H., Blanchard R. L., Palcza J., Martucci A., Miller-Stein C. M., Gutierrez M., Panebianco D., Rippley R. K., Lines C., Murphy M. G. (2010). The dose proportionality of telcagepant after administration of single oral and intravenous doses in healthy adult subjects. Arch. Drug Inf. 3, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt D. J., Aurora S. K., Dodick D. W., Goadsby P. J., Ge Y. J., Bachman R., Taraborelli D., Fan X., Assaid C., Lines C., et al. (2011). Randomized controlled trial of the CGRP receptor antagonist MK-3207 in the acute treatment of migraine. Cephalalgia 31, 712–722. [DOI] [PubMed] [Google Scholar]
- Ho T. W., Ho A. P., Ge Y. J., Assaid C., Gottwald R., MacGregor E. A., Mannix L. K., van Oosterhout W. P. J., Koppenhaver J., Lines C., et al. (2016). Randomized controlled trial of the CGRP receptor antagonist telcagepant for prevention of headache in women with perimenstrual migraine. Cephalalgia 36, 148–161. [DOI] [PubMed] [Google Scholar]
- Holland P. R., Goadsby P. J. (2018). Targeted CGRP small molecule antagonists for acute migraine therapy. Neurotherapeutics 15, 304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo D. M., Woodhead J. L., Walker P., Herédi-Szabó K., Mogyorósi K., Wolenski F. S., Dragan Y. P., Mosedale M., Siler S. Q., Watkins P. B., et al. (2019). Quantitative systems toxicology analysis of in vitro mechanistic assays reveals importance of bile acid accumulation and mitochondrial dysfunction in TAK-875-induced liver injury. Toxicol. Sci. 167, 458–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo D. M., Yang Y., Watkins P. B., Howell B. A., Siler S. Q. (2016). Elucidating differences in the hepatotoxic potential of tolcapone and entacapone with DILIsym(®), a mechanistic model of drug-induced liver injury. CPT Pharmacometrics Syst. Pharmacol. 5, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Ajona D., Pérez-Rodríguez A., Goadsby P. J. (2020). Gepants, calcitonin-gene-related peptide receptor antagonists: what could be their role in migraine treatment? Curr. Opin. Neurol. 33, 309–315. [DOI] [PubMed] [Google Scholar]
- Mullin K., Kudrow D., Croop R., Lovegren M., Conway C. M., Coric V., Lipton R. B. (2020). Potential for treatment benefit of small molecule CGRP receptor antagonist plus monoclonal antibody in migraine therapy. Neurology 94, e2121–e2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins P. B. (2020). DILIsym: Quantitative systems toxicology impacting drug development. Curr. Opin. Toxicol. 23-24, 67–73. [Google Scholar]
- Woodhead J. L., Brock W. J., Roth S. E., Shoaf S. E., Brouwer K. L. R., Church R., Grammatopoulos T. N., Stiles L., Siler S. Q., Howell B. A., et al. (2017). Application of a mechanistic model to evaluate putative mechanisms of tolvaptan drug-induced liver injury and identify patient susceptibility factors. Toxicol. Sci. 155, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead J. L., Pellegrini L., Shoda L. K. M., Howell B. A. (2020). Comparison of the hepatotoxic potential of two treatments for autosomal-dominant polycystic kidney disease using quantitative systems toxicology modeling. Pharm. Res. 37, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead J. L., Yang K., Oldach D., MacLauchlin C., Fernandes P., Watkins P. B., Siler S. Q., Howell B. A. (2019). Analyzing the mechanisms behind macrolide antibiotic-induced liver injury using quantitative systems toxicology modeling. Pharm. Res. 36, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead J. L., Yang K., Siler S. Q., Watkins P. B., Brouwer K. L. R., Barton H. A., Howell B. A. (2014). Exploring BSEP inhibition-mediated toxicity with a mechanistic model of drug-induced liver injury. Front. Pharmacol. 5, 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M., Miyake M., Sato H., Masutomi N., Tsutsui N., Adam K.-P., Alexander D. C., Lawton K. A., Milburn M. V., Ryals J. A., et al. (2013). Perturbation of bile acid homeostasis is an early pathogenesis event of drug induced liver injury in rats. Toxicol. Appl. Pharmacol. 268, 79–89. [DOI] [PubMed] [Google Scholar]
- Yang Y., Nadanaciva S., Will Y., Woodhead J. L., Howell B. A., Watkins P. B., Siler S. Q. (2014). MITOsym®: A mechanistic, mathematical model of hepatocellular respiration and bioenergetics. Pharm. Res. 32, 1975–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.