Abstract
We analyzed the genetic diversity of 531 Sinorhizobium meliloti strains isolated from nodules of Medicago sativa cultivars in two different Italian soils during 4 years of plant growth. The isolates were analyzed for DNA polymorphism with the random amplified polymorphic DNA method. The populations showed a high level of genetic polymorphism distributed throughout all the isolates, with 440 different haplotypes. Analysis of molecular variance allowed us to relate the genetic structure of the symbiotic population to various factors, including soil type, alfalfa cultivar, individual plants within a cultivar, and time. Some of these factors significantly affected the genetic structure of the population, and their relative influence changed with time. At the beginning of the experiment, the soil of origin and, even more, the cultivar significantly influenced the distribution of genetic variability of S. meliloti. After 3 years, the rhizobium population was altered; it showed a genetic structure based mainly on differences among plants, while the effects of soil and cultivar were not significant.
Alfalfa (Medicago sativa) and its symbiont Sinorhizobium meliloti have a long history of coexistence and coevolution. In every region where alfalfa has been cultivated for centuries, the natural nodulating population of S. meliloti plays a major role in satisfying the nitrogen requirements of the plants. Thus, it is important to investigate the genetic structure of natural populations of S. meliloti and their dynamics in relation to the host plant.
In recent years, the use of molecular techniques has stimulated the development of rapid and simple methods for characterizing natural microbial populations. Studies utilizing restriction fragment length polymorphism-PCR, multilocus enzyme electrophoresis, 16S ribosomal DNA analysis, repetitive extragenic palindromic-PCR, and DNA reassociation (1, 2, 4–7, 12, 15, 17) have revealed extensive genetic variability of microbial communities in natural soil. This variability has been widely investigated in the Rhizobiaceae (2, 5, 10, 16, 19), and there is some evidence of genetic exchange in populations (18). In previous work, we exploited random amplified polymorphic DNA (RAPD) techniques, combined with a powerful statistical analysis (analysis of molecular variance [AMOVA]), to describe the genetic structure of natural S. meliloti populations (8, 9). Greater understanding of the genetic structure and dynamics of indigenous populations of S. meliloti would be of great agricultural interest in view of the influence that an established or varying bacterial population can have on the nodulation efficiency of particular plant cultivars.
Little is known about the evolution of natural bacterial populations through the years in relation to a host plant. Bromfield et al. (1) showed that the M. sativa cultivar had an influence on the frequency of certain phage types in a natural S. meliloti population. Rooney-Varga et al. (12) described a seasonal modification of a natural population of sulfate-reducing bacteria in the rhizosphere of Spartina alterniflora linked to vegetative or reproductive plant growth. Studies considering the time factor and host plant biology will be of great interest for some perennial crops, like alfalfa, and for many other forage legumes.
In the present study, we used RAPD analysis to evaluate the dynamics (in a 4-year period) of the genetic structure of natural populations of S. meliloti nodulating three cultivars of M. sativa with different genetic backgrounds in two Italian soils. The aims of the study were to establish whether and to what extent the nodulating populations changed during the experiment and to evaluate the influence of environmental factors, such as the soil, cultivar, and plant genotype. The growing technique developed in this work (13) allowed us to monitor the bacterial population nodulating the same plant during the 4 years of the culture experiment. Indeed, estimation of the effect of a single plant is very important in relation to breeding for improved symbiosis in alfalfa.
MATERIALS AND METHODS
Sampling procedure. (i) Soils.
Two soils were used in this study. A sandy loam soil from Lodi (northern Po Valley, Lombardy, Italy) contained 6.5% clay, 27.5% silt, 66% sand, 1.6% organic matter, and 0.11% total N; the pH was 6.2. A clay soil from Monterotondo (central Italy) contained 38.1% clay, 32% silt, 29.9% sand, 2.13% organic matter, and 0.14% total N; the pH was 7.3. The soils came from perennial meadows that had never had Medicago species in their plant populations; the level of S. meliloti in these soils was estimated to be 10 cells/g of soil (as determined by the most-probable-number method with the plant infection technique; three replicates were used for each soil).
(ii) Plants.
Three cultivars of M. sativa with different degrees of fall dormancy and with no common genetic background were used: M. sativa cv. Oneida (Stanford Seed Co., Buffalo, N.Y.), a dormant type; M. sativa cv. Lodi (bred at the Istituto Sperimentale Colture Foraggere, Lodi, Italy), an intermediate type; and M. sativa cv. Estival (Pioneer Hi-Bred, Johnston, Iowa), a nondormant type. Seeds were surface sterilized and put in petri dishes to germinate. Plantlets were transplanted individually into soil-filled polyvinylchloride tubes (diameter, 3 cm; height, 80 cm) with 22 2-cm-diameter holes that were used for nondisruptive root sampling. Each tube was put into a larger tube (diameter, 5 cm) filled with the same soil, giving a final density equivalent to 500 plants/m2. Forty plants per soil-cultivar combination were transplanted in spring 1994. The growth trial was conducted in Lodi, Italy, at the Istituto Sperimentale per le Colture Foraggere in a cold greenhouse (−3.5 to 8°C in January; 17 to 34°C in July) with no N fertilization. Phosphorus and potassium (equivalent to 120 kg of P2O5 per ha and 180 kg of K2O per ha) were distributed at the beginning of the trial and in the early spring of each year; irrigation was not limiting, and water was provided according to the vigor of each plant. During the productive seasons from 1995 to 1997, the plants were cut about every 30 days (six or seven cuts per year).
(iii) Sampling.
In November 1994 at the end of the sowing year, in May, August, and November 1995 (first, fifth, and seventh cuts, respectively, of the first productive year), and in September 1997 (end of the trial), each inner tube containing a plant was extracted, and the roots and nodules in the intertube soil layer (i.e., part of the secondary root system) were removed and weighed; in no case was the tap root removed. After each root survey, the plants and the corresponding soil were once again put into the outer tube. Six plants per soil-cultivar combination were chosen, and from these plants three to seven nodules were used for isolation of S. meliloti strains, as previously described (8), so that each strain was derived from a different nodule. Thus, 30 to 40 strains per soil-cultivar combination were obtained. Here we report the data obtained from strains isolated during three periods: November 1994, August 1995, and September 1997.
Because of the high mortality of cultivar Estival (a nondormant type) during the 1996-1997 winter, the last sampling in September 1997 was limited to cultivars Lodi and Oneida.
RAPD analysis.
Amplification reactions were performed directly with cellular lysates as previously reported (8) by using random primers 1247 (5′-AAGAGCCCGT), RF2 (5′-CGGCCCCTGT), OPB7 (5′-GGTGACGCAG), and OPJ10 (5′-AAGCCCGAGG); primers OPB7 and OPJ10 were obtained from Operon Technologies Inc., Alameda, Calif. The amplified bands were separated by 2% (wt/vol) agarose gel electrophoresis, and the patterns were analyzed with a scanner-densitometer (model GDS 2000; Ultra-Violet Products, Cambridge, United Kingdom).
Statistical analysis.
RAPD markers generated by the four primers were used to calculate a Euclidean distance matrix as described by Excoffier et al. (3). The genetic distances for each pair of individuals were analyzed with AMOVA to estimate the variance components and to partition the variation among soils, cultivars, individual plants within a cultivar, nodules within a single plant, and years. All analyses were performed with the ARLEQUIN software (version 1.1) (14). To test for the significance of each variance component, a permutation analysis of the null distribution was carried out by using 100,000 permuted matrices. Pairwise analyses of soils, cultivars, and soil-cultivar combinations were performed by using the AMOVA approach with two variance components: between and within subgroups. The null distribution of the between-subgroup variance was tested by 5,000 permutations.
Unweighted pair group with mathematical average dendrograms were drawn by using the SAHN clustering portion of the NTSYS-pc 2.02 software (11) and the Euclidean distance matrices produced by ARLEQUIN.
RESULTS
We isolated and analyzed 188 S. meliloti strains in November 1994, 189 strains in August 1995, and 154 strains in September 1997; thus, a total of 531 strains were isolated and analyzed. For the analyses, we used four, five, six, and nine polymorphic bands with primers RF2, OPB7, OPJ10, and 1247 respectively; thus, a total of 24 RAPD markers ranging from 800 to 2,400 bp long were used.
The aim of the analysis was to assess the genetic variability in the bacterial population in order to understand whether and to what extent it is influenced by external factors, such as the soil, the alfalfa cultivar, the genotype of the individual plant, or time.
None of the RAPD markers used was specific for strains from a particular soil or cultivar or from a particular year. Therefore, the differences among the sources of variation considered were completely due to the variable frequency of the markers. A high level of interstrain variability was a general feature of the whole bacterial population; in fact, the 24 RAPD markers generated as many as 440 different haplotypes among the 531 strains analyzed.
Soil effect.
To avoid any imprinting from a particular alfalfa cultivar or agronomic practices, the two soils used in this work did not have a previous history of alfalfa cultivation. Probably for this reason, they showed very low densities of S. meliloti after they were removed from the perennial meadows. With the 1994 and 1995 samples, the bacterial populations of the two soils were not genetically distinguishable when the total populations were considered (Table 1, analysis A). However, significant soil effects were found when the between-soil source of variation was considered for each cultivar (Table 1, analyses B through D). In 1994 (the sowing year), a soil effect was particularly evident for cultivar Oneida (about 23% of the total strain variation); it should be noted that the cultivar Oneida samples also exhibited the greatest difference between soils for root dry matter and nodule biomass in the intertube soil layer (Table 2). At the end of the trial in 1997, the difference between soils was not significant both in the general analysis (Table 1, analysis A) and for the two surviving cultivars (Table 1, analyses C and D).
TABLE 1.
Results of AMOVA showing the soil effect on the genetic variability of nodulating S. meliloti populations
| Analysis | Source of variationa | November 1994
|
August 1995
|
September 1997b
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variance component | % of total | P value | Variance component | % of total | P value | Variance component | % of total | P value | ||
| A | Total population, among soils | −0.008 | −0.20 | 0.50036 | −0.199 | −5.03 | 0.70240 | 0.046 | 1.54 | 0.33372 |
| B | cv. Estival, between soils | 0.362 | 9.56 | 0.00185 | 0.170 | 4.61 | 0.03608 | |||
| C | cv. Lodi, between soils | 0.221 | 7.56 | 0.00200 | 0.439 | 11.67 | 0.02157 | 0.070 | 2.16 | 0.16448 |
| D | cv. Oneida, between soils | 0.931 | 22.66 | 0.00212 | 0.187 | 5.94 | 0.00891 | 0.039 | 1.45 | 0.20127 |
The results of only the first level of the hierarchical analysis are reported.
In September 1997, the effects were analyzed only for cultivars Lodi and Oneida.
TABLE 2.
Total aerial dry matter yield, root dry matter, and nodule fresh weight in the intertube soil layer: individual plant means and among-plant coefficients of variation
| Soil | Cultivar | 1994
|
1997
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aerial dry matter yield
|
Root dry matter
|
Nodule fresh wt
|
Aerial dry matter yield
|
Root dry matter
|
Nodule fresh wt
|
||||||||
| Mean (g/plant) | CVa | Mean (g/plant) | CV | Mean (g/plant) | CV | Mean (g/plant) | CV | Mean (g/plant) | CV | Mean (g/plant) | CV | ||
| Sandy loam | Estival | 4.28 | 29.68 | 0.31 | 55.36 | 0.49 | 40.35 | ||||||
| Lodi | 3.27 | 27.96 | 0.27 | 56.50 | 0.40 | 57.07 | 48.18 | 30.54 | 3.23 | 30.20 | 4.45 | 50.59 | |
| Oneida | 3.69 | 27.24 | 0.34 | 56.38 | 0.43 | 58.55 | 41.54 | 27.61 | 1.74 | 55.24 | 1.92 | 75.23 | |
| Mean | 3.75 | 30.55 | 0.30 | 56.82 | 0.44 | 51.84 | 44.35 | 29.89 | 2.37 | 51.10 | 2.99 | 73.86 | |
| Clay | Estival | 4.55 | 23.14 | 0.30 | 55.32 | 0.27 | 55.23 | ||||||
| Lodi | 3.02 | 23.54 | 0.23 | 82.35 | 0.22 | 83.48 | 52.21 | 30.79 | 2.08 | 55.86 | 3.37 | 58.18 | |
| Oneida | 3.24 | 26.96 | 0.16 | 114.73 | 0.17 | 87.22 | 46.77 | 30.95 | 0.95 | 75.86 | 1.67 | 73.73 | |
| Mean | 3.60 | 30.88 | 0.23 | 81.56 | 0.22 | 75.12 | 49.20 | 31.17 | 1.45 | 75.18 | 2.43 | 74.03 | |
CV, coefficient of variation.
Cultivar effect.
The genetic differences among the bacterial populations nodulating the three cultivars during the first and second years ranged from 9 to 15% of the total variance in the whole population (Table 3, analysis A), but the cultivar effect was not significant. However, there were significant differences among cultivars within each soil (Table 3, analyses B and C) in the first 2 years.
TABLE 3.
Results of AMOVA, showing the effects of cultivar, soil, and single plants on the genetic variability of nodulating S. meliloti populations
| Analysisa | Source of variation | November 1994
|
August 1995
|
September 1997b
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variance component | % of total | P value | Variance component | % of total | P value | Variance component | % of total | P value | ||
| A | Total population, among cultivars | 0.375 | 9.46 | 0.13427 | 0.620 | 14.93 | 0.06482 | −0.033 | −1.14 | 1.00000 |
| Soils within cultivar | 0.537 | 13.56 | <0.00001 | 0.353 | 8.50 | <0.00001 | 0.134 | 4.55 | 0.00346 | |
| Plants within cultivar-soil combination | 3.049 | 76.98 | <0.00001 | 3.181 | 76.57 | <0.00001 | 2.846 | 96.58 | <0.00001 | |
| B | Among cultivars | 0.761 | 21.59 | <0.00001 | 1.027 | 25.69 | <0.00001 | 0.054 | 1.96 | 0.19316 |
| Plants within cultivar | 0.182 | 5.15 | <0.00001 | 0.541 | 13.52 | <0.00001 | 0.429 | 15.42 | <0.00001 | |
| Nodules within plant | 2.584 | 73.26 | <0.00001 | 2.431 | 60.79 | <0.00001 | 2.299 | 82.62 | <0.00001 | |
| C | Among cultivars | 0.834 | 19.48 | <0.00001 | 0.734 | 17.08 | <0.00001 | −0.043 | −1.42 | 0.73560 |
| Plants within cultivar | 0.419 | 9.78 | <0.00001 | 0.603 | 14.02 | <0.00001 | 0.592 | 19.35 | <0.00001 | |
| Nodules within plant | 3.029 | 70.73 | <0.00001 | 2.963 | 68.90 | <0.00001 | 2.512 | 82.07 | <0.00001 | |
Analysis B was performed with sandy loam soil, and analysis C was performed with clay soil.
In September 1997, the effects were analyzed only for cultivars Lodi and Oneida.
At the end of the trial, the comparison among cultivars was limited to cultivars Lodi and Oneida because of the high mortality of cultivar Estival (a nondormant type) during the 1996-1997 winter; in the 1997 samples, the variance component effect was not significant for the global analysis and for each soil type (Table 3).
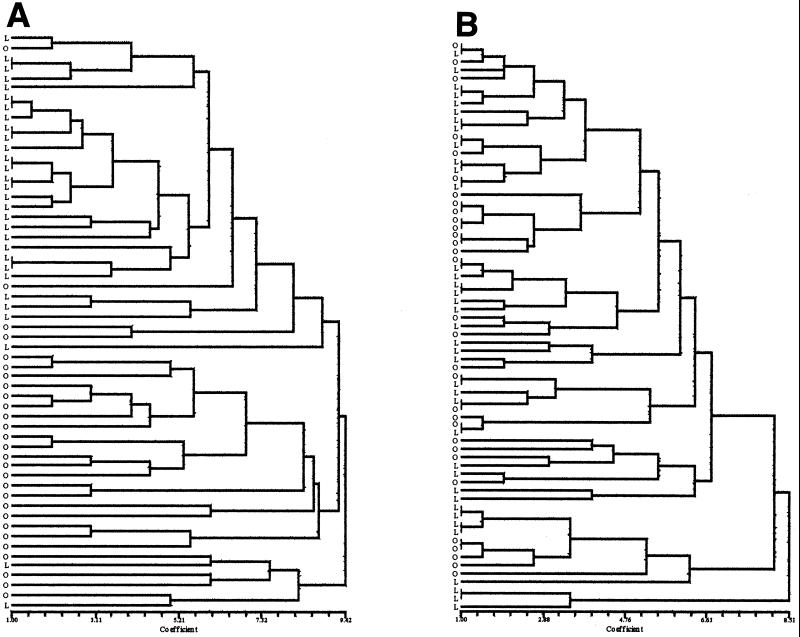
Within cultivars, the genetic variance values due to single plants were always high and significant (Table 3). However, in the 1994 and 1995 samples they were lower than the variation values due to the cultivar effect (Table 3). In contrast, in the final 1997 samples, the variance component due to the plants within cultivars was much greater than that among cultivars. This result was confirmed by comparing the two dendrograms showing the genetic relatedness among S. meliloti strains isolated from nodules of cultivars Lodi and Oneida in 1994 and 1997 (Fig. 1) (because of the similar behaviors of the two soils, only dendrograms for the clay soil are shown in Fig. 1). In 1994, most of the strains isolated from cultivar Lodi or Oneida clustered together, though the genetic diversity was high, while at the end of the trial in 1997, the strains isolated from the two cultivars appeared to be almost completely mixed.
FIG. 1.
Unweighted pair group with mathematical average dendrograms of S. meliloti strains isolated from clay soil in 1994 (A) and 1997 (B). L, cultivar Lodi; O, cultivar Oneida. Each letter indicates a different isolate.
Plant effect.
The plant effect was examined by AMOVA separately for each soil-cultivar combination (Fig. 2). In 1994, the genetic differences among strains isolated from different plants ranged from 4 to 10% of the total variance in the sandy loam soil and from 7 to 16% in the clay soil. Interestingly, we obtained lower root dry matter and nodule fresh weight values with a much higher among-plant coefficient of variation in the clay soil than in the sandy loam soil (Table 2). In the 1995 samples, cultivars Lodi and Estival grown in the sandy loam soil showed the largest and smallest plant effects, respectively, on the variance among nodulating strains. Forage production in this period was highly dependent on symbiotic fixation, as demonstrated by the fact that 51 to 62% of the total nodule biomass was produced in this period, compared to 35 to 40% of the total shoot dry matter and 20 to 23% of the total root dry matter (data not shown). At the end of the trial in 1997, cultivar Lodi grown in clay soil had the greatest plant effect on the genetic variability of S. meliloti; at the same time, it had high root dry matter and nodule biomass values with a large interplant coefficient of variation (Table 2). When the overall plant effect throughout the different sampling periods was considered, there appeared to be a trend toward increased genetic variability, particularly when the sowing year was compared with the final year of the trial when similar sampling procedures were used (Fig. 2; Table 3, analyses B and C).
FIG. 2.
Among-plant variance components in each soil-cultivar combination for the 1994, 1995, and 1997 samples. Percentages of total variance were computed by AMOVA. SL, sandy loam soil; C, clay soil.
The variance component attributable to the bacteria within each plant was always the major source of variation in all of the samples (Table 3, analyses B and C).
Dynamics of the bacterial population.
To test the changes in the symbiotic S. meliloti population after 4 years of cultivation, we compared the isolates obtained from the same plant in 1994 and 1997, when root sampling was carried out once (at the end of the productive season) without disturbing the natural nodulation pattern. The variance component attributable to time (Table 4) was always highly significant, ranging from 36 to 42% in cultivar Lodi and from 17 to 35% in cultivar Oneida.
TABLE 4.
Results of AMOVA showing the dynamics of bacterial populations nodulating the same plant in soil-cultivar combinations: comparison of isolates from the 1994 and 1997 samples
| Soil and cultivar | Source of variationa | Variance component | % of total | P value |
|---|---|---|---|---|
| Sandy loam soil, cv. Lodi | Among years | 2.062 | 41.85 | 0.00222 |
| Sandy loam soil, cv. Oneida | Among years | 1.349 | 35.04 | 0.00230 |
| Clay soil, cv. Lodi | Among years | 1.710 | 35.95 | 0.00188 |
| Clay soil, cv. Oneida | Among years | 0.698 | 17.39 | 0.00233 |
Data for only the first level (among years) of the hierarchical analysis are reported.
DISCUSSION
The main aim of this study was to investigate the genetic structure of nodulating S. meliloti populations by analyzing RAPD markers. In particular, we wished to assess whether the genetic structure was influenced by putative sources of variation, such as the type of soil and the alfalfa cultivar, as well as by the genotypes of the individual plants within cultivars (alfalfa is an allogamous species) and time (alfalfa is a perennial crop usually cultivated as a meadow for three to five years).
The genetic structure of the whole S. meliloti population at each sampling time (synchronic analysis) was apparently not affected by the cultivar or soil in the first 2 years. However, the cultivar and soil effects became highly significant when each soil or cultivar was considered separately. The differences found at the beginning of the trial could have been due more to the fact that the soils were geographically separated than to the type of soil. This result suggests that neither the cultivar nor the soil alone is directly responsible for supporting a genetically different nodulating population. Rather, growth of a particular cultivar in a particular soil could result in differences in the symbiotic populations. A consequence of this hypothesis would be that in order to estimate the genetic diversity of the symbiotic population of a soil, a single alfalfa cultivar is probably not sufficient. Differences in symbiotic populations between locations or between cultivars have been found previously under different experimental conditions (1). The effects of single plants in the sowing year could be related to the different rates at which the root systems colonized the intertube soil layers used for sampling; in fact, the plant effect on the symbiotic population was greater in the clay soil, which also showed the greatest variation among plants for root and nodule biomass (Table 2). This is in agreement with a cultivar effect which we observed in a previous study in which a small subset of strains from the sowing year and first productive year samples was analyzed with two primers (9). At the summer 1995 sampling (fifth cut), root reconstitution had just occurred after the roots and nodules in the intertube soil layer had been removed after the first cut. For this reason, it is impossible to distinguish between the effect attributable to differences in the reconstitution rates of roots and nodules and the effect due to a direct influence on the microbial symbiont. However, strong rhizosphere effects, varying in magnitude with the season, have been reported for subclover (6). In 1997, the natural nodulation pattern was not affected by traumatic events like root removal. The genetic variance of the S. meliloti symbiotic population was completely dependent on the differences among and within plants; in fact, both soil and cultivar effects were not significant in all analyses (Tables 1 and 3). A possible explanation for this result is that the plant partner tended to become more important in the tube plot environment. Considering the changes that occurred in the symbiotic population of each soil-cultivar combination from the beginning to the end of the trial (Table 4), it can be hypothesized that the strains best adapted to the relationship with each plant progressively emerged in all soils for all cultivars. The effect of individual plants on the genetic variability of the symbiotic population appeared to increase from the beginning to the end of the trial. The causes of this could be variations in the plant growth rate, particularly the growth rate of the root system, and genetic isolation inside each tube plot (genetic drift). It should be remembered that the plants investigated and the tube plot soil were always the same throughout the years. Thus, it can be hypothesized that individual alfalfa plants are capable of influencing the symbiotic population.
From the analyses shown in Tables 1 and 3, it is clear that most of the genetic diversity was found within the symbiotic populations of individual plants. This means that an alfalfa plant enters into symbiosis with genetically different strains, suggesting that the alfalfa-S. meliloti relationship is general rather than specific.
In previous analyses of Italian S. meliloti populations, we found significant effects of the soil (8) or cultivar (9) on the genetic diversity. However, in both cases, the bacteria were isolated during a 1-year analysis. A comparison of the previous data with those presented here indicates the importance in population analyses of monitoring rhizobial populations for longer periods of time.
The present findings emphasize the importance of the plant partner, considered both as a cultivar and as an individual plant within a cultivar, in determining the genetic structure of a symbiotic microbial population. On the other hand, the microbial partner maintains very high genetic diversity even within single plants, at least for the time period which we investigated. Both these aspects should be taken into account when methods for improving symbiosis in alfalfa are planned.
ACKNOWLEDGMENT
This research was supported by MIPA Progetto Biotecnologie Avanzate.
REFERENCES
- 1.Bromfield E S P, Shina I B, Wolynetz M S. Influence of location, host cultivar, and inoculation on the composition of naturalized populations of Rhizobium meliloti in Medicago sativa nodules. Appl Environ Microbiol. 1986;51:1077–1084. doi: 10.1128/aem.51.5.1077-1084.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demezas D H, Reardon T B, Watson J M, Gibson A H. Genetic diversity among Rhizobium leguminosarum bv. trifolii strains revealed by allozymes and restriction fragment length polymorphism analyses. Appl Environ Microbiol. 1991;57:3489–3495. doi: 10.1128/aem.57.12.3489-3495.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Excoffier L, Smouse P E, Quattro J M. Analysis of molecular variance inferred from metric distance among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laguerre G, Mavingui P, Allard M R, Charnay M P, Louvrier P, Mazurier S I, Rigottier-Gois L, Amarger N. Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl Environ Microbiol. 1996;62:2029–2036. doi: 10.1128/aem.62.6.2029-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung K, Strain S R, de Bruijn F J, Bottomley P J. Genotypic and phenotypic comparisons of chromosomal types within an indigenous soil population of Rhizobium leguminosarum bv. trifolii. Appl Environ Microbiol. 1994;60:416–426. doi: 10.1128/aem.60.2.416-426.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung K, Yap K, Dashti N, Bottomley P J. Serological and ecological characteristics of a nodule-dominant serotype from an indigenous soil population of Rhizobium leguminosarum bv. trifolii. Appl Environ Microbiol. 1994;60:408–415. doi: 10.1128/aem.60.2.408-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mavingui P, Laguerre G, Berge O, Heulin T. Genetic and phenotypic diversity of Bacillus polymixa in soil and in the wheat rhizosphere. Appl Environ Microbiol. 1992;58:1894–1903. doi: 10.1128/aem.58.6.1894-1903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paffetti D, Scotti C, Gnocchi S, Fancelli S, Bazzicalupo M. Genetic diversity of an Italian Rhizobium meliloti population from different Medicago sativa cultivars. Appl Environ Microbiol. 1996;62:2279–2285. doi: 10.1128/aem.62.7.2279-2285.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paffetti D, Daguin F, Fancelli S, Gnocchi S, Lippi F, Scotti C, Bazzicalupo M. Influence of plant genotype on the selection of nodulating Sinorhizobium meliloti strains by Medicago sativa. Antoine Leeuwenhoek, 1997;7:1–6. doi: 10.1023/a:1000591719287. [DOI] [PubMed] [Google Scholar]
- 10.Piñero D, Martinez E, Selander R K. Genetic diversity and relationships among isolates of Rhizobium leguminosarum biovar phaseoli. Appl Environ Microbiol. 1988;54:2825–2832. doi: 10.1128/aem.54.11.2825-2832.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohlf F J. NTSYS-pc. Numerical taxonomy and multivariate analysis system, version 2.02. New York, N.Y: Exeter Software; 1990. [Google Scholar]
- 12.Rooney-Varga J N, Devereux R, Evans R S, Hines M E. Seasonal changes in the relative abundance of uncultivated sulfate-reducing bacteria in a salt marsh sediment and in the rhizosphere of Spartina alterniflora. Appl Environ Microbiol. 1997;63:3895–3901. doi: 10.1128/aem.63.10.3895-3901.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rotili P, Gnocchi G, Scotti C, Zannone L. posting date. Some aspects of breeding methodology in alfalfa. In: Busbice T, Rotili P, Bingham T, editors. Proceedings of The Alfalfa Genome Conference. 1999. www.naaic.org/TAG/TAGpapers/rotili/rotili.html Madison, Wis. [Online.] www.naaic.org/TAG/TAGpapers/rotili/rotili.html. . [Google Scholar]
- 14.Schneider S, Kueffer J M, Roessli D, Excoffier L. Arlequin ver. 1.1: a software for population genetic data analysis. Geneva, Switzerland: Genetics and Biometrics Laboratory, University of Geneva; 1997. [Google Scholar]
- 15.Segovia L, Piñero D, Palacios R, Martinez-Romero E. Genetic structure of a soil population of nonsymbiotic Rhizobium leguminosarum. Appl Environ Microbiol. 1991;57:426–433. doi: 10.1128/aem.57.2.426-433.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strain S R, Leung K, Whittam T S, De Bruijn F J, Bottomley P J. Genetic structure of Rhizobium leguminosarum biovar trifolii and viciae populations found in two Oregon soils under different plant communities. Appl Environ Microbiol. 1994;60:2772–2778. doi: 10.1128/aem.60.8.2772-2778.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinuesa P, Rademarker J L W, de Bruijn F J, Werner D. Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary Islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S–23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl Environ Microbiol. 1998;64:2096–2104. doi: 10.1128/aem.64.6.2096-2104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whittam S T. Sex in the soil. Curr Biol. 1992;2:676–678. doi: 10.1016/0960-9822(92)90140-6. [DOI] [PubMed] [Google Scholar]
- 19.Young J P W, Wexler M. Sym plasmid and chromosomal genotypes are correlated in field population of Rhizobium leguminosarum. J Gen Microbiol. 1988;134:2731–2739. [Google Scholar]