Abstract
Background
Clinical management of women carrying a germline pathogenic variant (PV) in the BRCA1/2 genes demands for accurate age-dependent estimators of breast cancer (BC) risks, which were found to be affected by a variety of intrinsic and extrinsic factors. Here we assess the contribution of polygenic risk scores (PRSs) to the occurrence of extreme phenotypes with respect to age at onset, namely, primary BC diagnosis before the age of 35 years (early diagnosis, ED) and cancer-free survival until the age of 60 years (late/no diagnosis, LD) in female BRCA1/2 PV carriers.
Methods
Overall, estrogen receptor (ER)-positive, and ER-negative BC PRSs as developed by Kuchenbaecker et al. for BC risk discrimination in female BRCA1/2 PV carriers were employed for PRS computation in a curated sample of 295 women of European descent carrying PVs in the BRCA1 (n=183) or the BRCA2 gene (n=112), and did either fulfill the ED criteria (n=162, mean age at diagnosis: 28.3 years, range: 20 to 34 years) or the LD criteria (n=133). Binomial logistic regression was applied to assess the association of standardized PRSs with either ED or LD under adjustment for patient recruitment criteria for germline testing and localization of BRCA1/2 PVs in the corresponding BC or ovarian cancer (OC) cluster regions.
Results
For BRCA1 PV carriers, the standardized overall BC PRS displayed the strongest association with ED (odds ratio (OR) = 1.62; 95% confidence interval (CI): 1.16–2.31, p<0.01). Additionally, statistically significant associations of selection for the patient recruitment criteria for germline testing and localization of pathogenic PVs outside the BRCA1 OC cluster region with ED were observed. For BRCA2 PV carriers, the standardized PRS for ER-negative BC displayed the strongest association (OR = 2.27, 95% CI: 1.45–3.78, p<0.001).
Conclusions
PRSs contribute to the development of extreme phenotypes of female BRCA1/2 PV carriers with respect to age at primary BC diagnosis. Construction of optimized PRS SNP sets for BC risk stratification in BRCA1/2 PV carriers should be the task of future studies with larger, well-defined study samples. Furthermore, our results provide further evidence, that localization of PVs in BC/OC cluster regions might be considered in BC risk calculations for unaffected BRCA1/2 PV carriers.
Supplementary Information
The online version contains supplementary material available at (10.1186/s12885-022-09780-1).
Keywords: Breast cancer, Polygenic risk score, PRS, Risk assessment, BRCA1, BRCA2
Background
Inherited pathogenic variants (PVs) in the BRCA1/2 genes are the most common cause of hereditary breast cancer (BC). Associated lifetime risks for BC development were assessed in a variety of studies, recent estimates range from 60 to 75% for female BRCA1 and from 55 to 76% for female BRCA2 germline PV carriers [1–4]. Clinical management of individuals found to be at high risk for BC focuses on risk reduction and early diagnosis of cancer, and includes early age (25–30 years) BC screening via magnetic resonance imaging (MRI) and mammograms, or risk-reducing mastectomy. Preventive medical treatment, e.g., with tamoxifen or denosumab, in unaffected BRCA1/2 PV carriers is under discussion [5]. Accurate age-dependent estimates of BC penetrance in PV carriers are a crucial prerequisite in genetic counseling for making informed, individualized decisions about whether and when the risks associated with considered preventive measures are in relation to the expected BC risk.
BC incidence of BRCA1 PV carriers rises sharply in the 4th decade of life and then remains at a similar, constant level [2]. The majority of tumors in BRCA1 PV carriers are estrogen receptor-negative (ER-), but their proportion decreases with age [6, 7]. In BRCA2 PV carriers, BC incidence increases rapidly until ages of 40 to 50 years, and then remains almost constant [2]. The majority of tumors are ER-positive (ER+), but the proportion of ER- tumors increases with age [6, 7].
BC risks of female BRCA1/2 PV carriers were found to be affected by a variety of intrinsic and extrinsic factors, such as mammographic density [8, 9], family history of BC and ovarian cancer (OC), variant localization [2], birth cohort [10–12], reproductive history (e.g., age at menarche, first birth, and menopause, number of full-term pregnancies, breastfeeding) [13] and further modifiable factors such as body weight, physical activity, and alcohol consumption [14], among others.
Furthermore, numerous studies demonstrated that the effects of BC susceptibility loci, i.e., common single nucleotide polymorphisms (SNPs), which individually contribute only slightly to individual BC risks, but whose effects can be summed up to polygenic risk scores (PRSs) are able to achieve a clinically relevant degree of BC risk discrimination [8, 15–20]. However, it was also consistently shown that PRS risk stratification is reduced in BRCA1/2 PV carriers in comparison to the general population [21–23]. Moreover, a recent case-only study by Coignard et al. [24] comprising more than 60,000 unselected BC cases and 13,000 cases with BRCA1/2 mutations, provided evidence that several SNPs associated with BC risk in the general population, and are therefore considered in PRS calculations, are in fact associated with the BRCA1/2 mutation status, and hence do not have effects on BC risk in BRCA1/2 PV carriers.
In order to construct a highly informative study sample with respect to modifying factors of BC risk in female BRCA1 PV carriers, Sepahi et al. introduced the approach of investigating patients with “extreme phenotypes” due to age at primary BC diagnosis [25]. The authors analyzed a sample of 133 BRCA1-positive patients who were either diagnosed with primary BC at an age younger than 35 years (early diagnosis, ED) or remained cancer-free until the age of 60 years (late/no diagnosis, LD). The study concluded with the assumption that co-occurring truncating variants in further DNA repair genes associate with ED, although no statistical significance was reached to prove this hypothesis. All patients examined in the study of Sepahi et al. met the inclusion criteria of the German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC) for germline testing (Additional file 1: Table S1), i.e. were highly selected for BC/OC family history and/or early BC/OC onset.
Here, we revisit the approach of Sepahi et al. to assess the utility of PRSs for the discrimination of extreme phenotypes with respect to age at BC onset in female BRCA1/2 PV carriers. We employed SNP sets for PRS computation as developed and evaluated by Kuchenbaecker et al. [22], which to our knowledge are the only published PRS SNP sets to date specifically adapted for BC risk stratification in women with a BRCA1/2 PV, namely an overall BC PRS comprising 88 loci, a PRS specific for ER+ BC comprising 87 SNPs and a PRS specific for ER- BC comprising 53 SNPs. In contrast to Sepahi et al., our sample is not only composed of patients selected by the GC-HBOC inclusion criteria for germline testing, but also by BRCA1/2 PV carriers recruited at the Suzanne Levy-Gertner Oncogenetics Unit at the Sheba Medical Center (Tel Aviv University, Israel). Due to a less restrictive access to an initial screening for Israeli BRCA1/2 founder mutations [26], these individuals are expected to be unselected for family history and/or early BC/OC onset.
Methods
Study sample
As a PV, we defined class 4/5 protein-truncating variants with respect to 5-tier variant classification system suggested by the International Agency for Research on Cancer (IARC). Variant classification was performed according to the guidelines published by the Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) [27] and the American College of Medical Genetics and Genomics (ACMG) [28]. Only female BRCA1/2 PV carriers with a primary BC diagnosis before the age of 35 years (ED group) or cancer-free survival until the age of 60 years (LD group) were considered. No individual in the ED group was diagnosed with OC prior to BC, and no individual in the LD group underwent prophylactic mastectomy before the age of 60 years.
All patients in the study sample were identified through diagnostic germline testing and recruited between 1996 and 2017 by 13 centers of the GC-HBOC and at Suzanne Levy-Gertner Oncogenetics Unit at the Sheba Medical Center (Tel Aviv University). Individuals recruited by the GC-HBOC were selected by the corresponding inclusion criteria for germline testing (Additional file 1: Table S1), whereas the BRCA1/2 PV carriers recruited by the Sheba Medical Center met the broader Israeli inclusion criteria for BRCA1/2 screening [26], and are therefore considered unselected. All patients were tested for PVs in BRCA1/2 and the CHEK2:c.1100delC variant. Individuals with PVs both in BRCA1 and BRCA2 (n=1) and individuals with co-occurring variant CHEK2:c.1100delC (n=1) were excluded.
Regarding the classification of variant localizations within BRCA1/2, we applied – in concordance with Sepahi et al. [25] – the definitions of Rebbeck et al. [29], i.e., BRCA1 variants intersecting with regions c.179–505, c.4328–4945 or c.5261–5563 (with respect to transcript NM_007294.4) were considered to be localized within a breast cancer cluster region (BCCR) and variants intersecting with regions c.1380–4062 were considered to be localized within an ovarian cancer cluster region (OCCR). For BRCA2, PVs intersecting with c.1–596, c.772–1806 or c.7394–8904 (with respect to transcript NM_000059.4) were evaluated as being localized within a BCCR, and PVs intersecting with c.3249–5681 or c.6645-7471 as being localized within an OCCR.
SNP genotyping
For SNP genotyping, we used a customized 48.48 amplicon-based target enrichment panel (Access Array®, Fluidigm, San Francisco, CA, USA). Variants which could not be covered due to technical limitations, were replaced by adjacent SNPs in linkage disequilibrium (Additional file 2: Table S2). Subsequent parallel next-generation sequencing (NGS) of the barcoded amplicons of the samples was performed by employing an Illumina NextSeq500 sequencing device (Illumina, San Diego, CA, USA) as described previously [17]. All DNA samples were centrally analyzed at the Center for Familial Breast and Ovarian Cancer, University Hospital Cologne, Germany. Raw BCL files were demultiplexed using bcl2fastq2 Conversion Software v2.19 available at https://support.illumina.com Sequence reads were mapped to the human reference genome assembly GRCh37, including decoy sequences (hs37d5), using BWA-MEM of Burrows-Wheeler Aligner (BWA) v0.7.15 [30], and target-specific primer sequences were removed using BAMClipper v1.1 [31]. SNP calling was performed on a merged BAM file including all samples using FreeBayes v1.0.0 [32] under specification of common SNPs from dbSNP build 151 [33] as variant input, and under specification of -q 20, i.e., a minimum phred-scaled base quality of 20. Variant calls were filtered for a minimum read depth of 10 and converted to TPED format using PLINK v1.9 [34].
Quality control & PRS computation
Quality controls were performed as previously described [17]. SNP calls and samples were filtered for a minimum call rate of 0.95. One sample was identified as an outlier due to heterogeneity using the Bonferroni-corrected upper and lower 5 percentiles of the empricially estimated normal distribution and excluded. Samples with putative African or putative Asian ancestry were identified by multi-dimensional scaling based on a combined set of 342 overlapping SNPs in the sample and 1000 Genomes data. Four individuals were excluded because African or Asian ancestry could not be ruled out (Additional file 3: Figure S1).
For each person i, an indvidual PRS was derived via
where β is the per-allele log odds ratio and gij is the number of effect alleles in person i for locus j. Missing genotypes were imputed to the average observed genotype in the sample. Alleles with complementary alleles, i.e., either C/G or A/T, were excluded from PRS calculation due to ambiguity. This criterion also applied to SNP rs11571833, which is located within BRCA2. Quality filters resulted in effective SNP set sizes of N=77 for the overall and ER+ BC PRS, and of N=50 for the ER- BC PRS (Additional file 4: Table S3). Of the total 86 SNPs employed for PRS calculations, four showed statistically significant differences in observed allele frequencies when comparing samples recruited by GC-HBOC and by Sheba Medical Center (Bonferroni-corrected Fisher’s exact test p<0.05, Additional file 4: Table S3). PRSs for each SNP set were standardized to have mean 0 and variance 1.
Statistical analysis
Analyses were conducted using the GenABEL v1.8 utilities [35], R v3.6 and PLINK v1.9 [34]. All statistical tests were two-sided with p values below 0.05 considered statistically significant. The association of standardized PRSs with ED, respectively LD, of primary BC was assessed by employing a binomial logistic regression model (outcome 1: ED, outcome 0: LD) under adjustment for selection by the GC-HBOC inclusion criteria for germline testing and localization of BRCA1/2 PVs within the corresponding BCCRs and OCCRs.
A robust sandwich variance estimation approach [36], clustering observations based on family ID, was applied to account for related individuals in the sample.
Results
Quality controls resulted in a curated sample set comprising 295 BRCA1/2 PV carriers out of 293 families as input for the PRS computation (183 BRCA1; 112 BRCA2, Additional file 5: Table S4)). Of the total 295 BRCA1/2 PV carriers, 162 individuals developed BC before the age of 35 years and therefore belonged to the ED group. From the 133 individuals in the LD group (cancer-free survival until the age of 60 years), 41 were known to be diagnosed with BC after the age of 60 years. Key characteristics of the study sample are given in Table 1.
Table 1.
Key characteristics of the study sample. BC: breast cancer; ER: estrogen receptor; OC: ovarian cancer; PV: pathogenic variant; (not) sICGT: (not) selected by the inclusion criteria for germline testing of the German Consortium for Hereditary Breast and Ovarian Cancer. *Diagnosis after age 60
| total | ED | LD | |
|---|---|---|---|
| study sample | 295 | 162 | 133 |
| with BC | 203 | 162 | 41* |
| sICGT | 188 | 114 | 74 |
| ≥1 relative with BC/OC | 170 | 99 | 71 |
| not sICGT | 107 | 48 | 59 |
| BRCA1 PV carriers | 183 | 116 | 67 |
| PV within BCCR | 33 | 19 | |
| PV within OCCR | 21 | 19 | |
| BRCA2 PV carriers | 112 | 46 | 66 |
| PV within BCCR | 12 | 7 | |
| PV within OCCR | 16 | 14 | |
| ER status known, first BC | 136 | 105 | 31 |
| ER+ | 62 | 44 | 18 |
| ER- | 74 | 61 | 13 |
| ER status unknown | 67 | 57 | 10 |
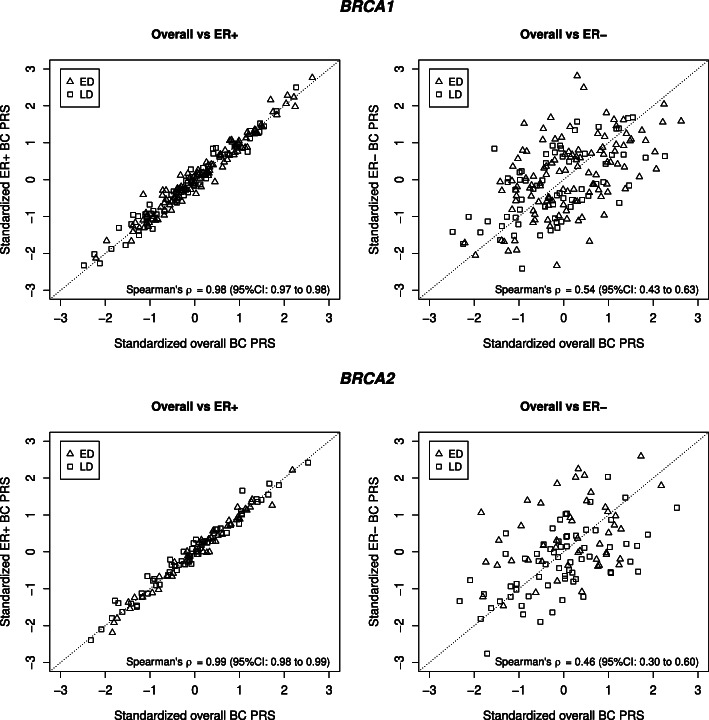
Standardized overall BC and ER+ BC PRSs showed a strong correlation at the individual level (BRCA1 PV carriers: Spearman’s correlation coefficient ρ=0.98 (95% confidence interval (CI): 0.97–0.98); BRCA2 PV carriers: ρ=0.99 (95% CI: 0.98–0.99), Fig. 1). Correlations between the individual standardized overall BC and ER- BC PRSs were considerably lower in comparison, but also statistically significant (BRCA1 PV carriers: ρ=0.54 (95% CI: 0.43–0.63); BRCA2 PV carriers: ρ=0.46 (95% CI: 0.30–0.60), Fig. 1).
Fig. 1.
Standardized polygenic risk score (PRS) values per individual and Spearman’s rank correlation coefficients ρ. (Upper left) Overall breast cancer (BC) PRS versus estrogen receptor-positive (ER+) BC PRS in 183 BRCA1 pathogenic variant (PV) carriers who were either diagnosed with BC before the age of 35 years (early diagnosis, ED) or were unaffected at least until the age of 60 years (late/no diagnosis, LD). (Upper right) Overall BC PRS versus estrogen receptor-negative (ER-) BC PRS in the same sample. (Lower left) Overall BC PRS versus ER+ BC PRS in 112 BRCA2 PV carriers who belonged to either the LD or ED group. (Lower right) Overall BC PRS versus ER- BC PRS in the same sample
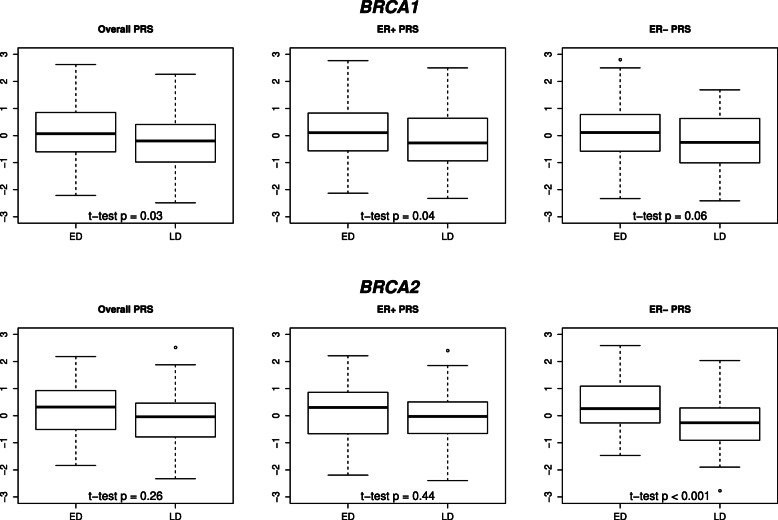
For both BRCA1 and BRCA2 PV carriers, and consistently across all PRSs under consideration, the average PRS was increased in the ED group in comparison to the average PRS in the LD group (Fig. 2). However, these differences reached statistical significance exclusively considering the overall or ER+ BC PRS of the BRCA1 PV carriers (two-sided Welch’s t-test p≤0.04), and the ER- BC PRS of the BRCA2 PV carriers in our study sample (two-sided Welch’s t-test p<0.001).
Fig. 2.
Boxplots and two-sided Welch’s t-test p values of standardized polygenic risk scores (overall breast cancer (BC), estrogen receptor (ER)-positive BC (ER+), ER-negative BC (ER-) PRS) of 183 BRCA1 and 112 BRCA2 pathogenic variant carriers, who were either diagnosed with BC before the age of 35 years (early diagnosis, ED) or were unaffected at least until the age of 60 years (late/no diagnosis, LD)
Binomial logistic regression analyses were employed to assess the association of the standardized PRSs with ED of BC taking selection by the GC-HBOC inclusion criteria and localization of BRCA1/2 PVs, i.e., localization within the BCCRs or OCCRs, into account.
For BRCA1 PV carriers, among the three different PRS SNP sets employed, the standardized overall BC PRS displayed the strongest association with ED (odds ratio (OR) = 1.62; 95% CI: 1.16–2.31, p=0.007, Table 2) in our analysis. Additionally, a statistically significant association between selection by the GC-HBOC inclusion criteria for germline testing and ED with ORs ranging from 2.84 to 3.52 (p≤0.005) was observed. Localization of PVs within the BRCA1 OCCR was statistically significantly associated with LD with ORs ranging from 0.32 to 0.35 (p≤0.02), whereas no statistically significant effect of localization of PVs within the BRCA1 BCCRs was observed.
Table 2.
Binomial logistic regression analysis results. Odds ratios (ORs), 95% confidence intervals (CIs) and p values based on two-tailed z-test for binomial logisitic regression analyses with extremely young age (<35 years) at breast cancer (BC) diagnosis (early diagnosis, ED) as the output considering standardized polygenic risk scores (PRSs), selection by the inclusion criteria for germline testing of the German Consortium for Hereditary Breast and Ovarian Cancer (sICGT) and localization of pathogenic variants (PVs) within the corresponding BC cluster regions (BCCRs) or OC cluster regions (OCCRs). Three PRS SNP sets were employed for PRS computation: overall BC, estrogen-receptor-positive (ER+) BC and ER-negative (ER-) BC PRS [22]. 183 BRCA1 and 112 BRCA2 PV carriers were considered who were either diagnosed with BC before the age of 35 years (ED) or unaffected until the age of 60 years
| OR | 95%CI | p | |
|---|---|---|---|
| BRCA1 PV carriers | |||
| overall BC PRS | 1.62 | 1.16–2.31 | 0.007 |
| inside BCCR | 0.65 | 0.30–1.39 | 0.26 |
| inside OCCR | 0.32 | 0.13–0.75 | 0.01 |
| sICGT | 3.45 | 1.67–7.43 | 0.002 |
| ER+ BC PRS | 1.60 | 1.14–2.30 | 0.01 |
| inside BCCR | 0.65 | 0.30–1.39 | 0.26 |
| inside OCCR | 0.32 | 0.13–0.75 | 0.01 |
| sICGT | 3.52 | 1.70–7.63 | 0.002 |
| ER- BC PRS | 1.38 | 1.00–1.92 | 0.05 |
| inside BCCR | 0.62 | 0.29–1.33 | 0.16 |
| inside OCCR | 0.35 | 0.15–0.81 | 0.02 |
| sICGT | 2.84 | 1.42–5.89 | 0.005 |
| BRCA2 PV carriers | |||
| overall BC PRS | 1.30 | 0.87–1.96 | 0.21 |
| inside BCCR | 2.94 | 0.96–9.55 | 0.06 |
| inside OCCR | 2.02 | 0.75–5.64 | 0.16 |
| sICGT | 2.21 | 0.77–6.80 | 0.15 |
| ER+ BC PRS | 1.21 | 0.81–1.82 | 0.37 |
| inside BCCR | 2.95 | 0.97–9.55 | 0.06 |
| inside OCCR | 1.98 | 0.74–5.49 | 0.17 |
| sICGT | 2.20 | 0.77–6.75 | 0.15 |
| ER- BC PRS | 2.27 | 1.45–3.78 | <0.001 |
| inside BCCR | 2.77 | 0.87–9.38 | 0.07 |
| inside OCCR | 1.88 | 0.66–5.56 | 0.25 |
| sICGT | 2.32 | 0.77–7.48 | 0.13 |
Considering BRCA2 PV carriers, the strongest association with ED was observed for the ER- BC PRS (OR = 2.27; 95% CI: 1.45–3.78, p<0.001), whereas the overall and ER+ BC PRS did not show statistically significant association with ED (Table 2). Regardless of the PRS SNP set employed, no statistically signifcant effect of selection by the GC-HBOC inclusion criteria for germline testing and localization of PVs within the BRCA2 OCCRs or BCCRs was observed.
Discussion
In our study considering female BRCA1/2 PV carriers who either developed BC before the age of 35 years (ED) or remained cancer-free until the age of 60 years (LD), we assessed whether extreme age of BC onset due to the definition of Sepahi et al. [25] can be explained by PRSs, among other known factors. Three subtype-specific PRS SNP sets as proposed by Kuchenbaecker et al. [22] for BC risk discrimination in BRCA1/2 PV carriers were employed, namely an overall BC, an ER+ BC and an ER- BC PRS. All PRS SNP sets investigated consist of a maximum of 88 loci and therefore have the potential to be straightforwardly implemented in routine diagnostic multi-gene panel analyses.
Our findings indicate the highest ability of ED/LD discrimination in BRCA1 PV carriers if applying the overall BC PRS, and a slightly reduced but comparable performance of the ER+ PRS. For BRCA2 PV carriers, the ER- BC PRS showed the highest ability of ED/LD discrimination. The observed associations were statistically significant, allowing us to clearly demonstrate the contribution of PRSs to the development of extreme phenotypes of BRCA1/2 PV carriers with respect to age at primary BC diagnosis.
However, previous studies applying Cox regressions with years of life until first BC diagnosis as the outcome, reported the highest potential for BC risk stratification of the ER- BC PRS for BRCA1 PV carriers and of the overall BC PRS for BRCA2 PV carriers [22, 37]. Kuchenbaecker et al. analyzed data for 15,252 BRCA1 and 8,211 BRCA2 PV carriers from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) to develop and evaluate the PRS SNP sets employed here [22]. It is remarkable, that the authors focused primarily on distinguishing between carriers and noncarriers of a BRCA1/2 PV (which is essentially equivalent to distinguishing between carriers and noncarriers of a pathogenic alteration in a BC risk gene with high penetrance), but not between BRCA1 and BRCA2 PV carriers per se. Separate evaluation, however, concluded that the ER- BC PRS performed best for BRCA1 PV carriers, whereas for BRCA2 PV carriers, the overall BC PRS showed the strongest association. This conclusion was confirmed by Barnes et al. [37], who retrospectively evaluated the overall, ER+ and ER- version of a 313 PRS SNP set adapted to BC risk stratification in women unselected for germline mutation status [16]. This study was also performed on the basis of CIMBA data, and involved almost 19,000 female BRCA1 and more than 12,000 female BRCA2 PV carriers. Besides the different outcomes considered in regression analyses, the deviating conclusions regarding the best-performing PRSs could be due to artificial enrichment with LD cases in our study design. Only 6.1% (928/15,252) of the BRCA1 and 10.1% (833/8,211) of the BRCA2 PV carriers examined by Kuchenbaecker et al. did not receive a primary BC diagnosis until after an age of 60 years or were known to have no disease by that age, and thus, meeting the LD criteria; in the study by Barnes et al. this was true for 6.0% (1143/18,935) and 10.1% (1251/12,339), respectively.
Further, in our sample the proportion of the ER+ subtype among all BC cases of BRCA1 PV carriers in the ED group was 29.4% (20/68) and 55.6% (5/9) in the LD group, with missing data of receptor status for 52 tumors (ED: 48, LD: 4), whereas Mavaddat et al. reported in agreement with previous findings by Foulkes et al. an ER+ proportion of <20% among BC diagnoses before age 40 years, and of approximately 45% and >50% among BC diagnoses between ages 60 and 69 years, and >70 years, respectively [6, 7]. Regarding BRCA2 PV carriers in our sample, the ER+ proportion was 64.9% (24/37) in the ED group, with no data available for 9 tumors, and 59.1% (13/22) in the LD group, with missing data for 6 tumors. In contrast, Mavaddat found a proportion of >80% of ER+ tumors among BC diagnoses between the ages of 30 and 39 years in BRCA2 PV carriers, and of >70%, respectively >60%, for BC diagnoses between 60 and 69 years, respectively >70 years, of age [7]. Thus, the proportion of ER- tumors was lower than expected for BRCA1 PV carriers and higher as expected for BRCA2 PV carriers in our sample, which may also have led to divergent results regarding the comparison of performances between PRS sets compared with previous studies based on CIMBA data.
Our results, based on a sample independent of previous genome-wide association studies (GWASs) for PRS construction, also highlight the urgent need for a careful review of BC PRSs specific to BRCA1/2 PV carriers and their performance. The question is not only whether an ER-specific or overall BC PRS should be used, but also whether the PRS is equally appropriate for risk stratification in young and older women and whether it is even justified to use a common PRS for BRCA1 and BRCA2 PV carriers. Coignard et al. [24] suggested that several SNPs included in PRS computation are associated with BRCA1/2 mutation status rather than with BC risk in general. The authors presented 7 SNPs associated with BRCA1 carrier status and 3 SNPs associated with BRCA2 carrier status, of which 7 (BRCA1-associated: rs13281615, rs704010, rs4784227, rs616488 and rs17530068; BRCA2: rs10759243 and rs6001930) were identical or in linkage disequilibrium to SNPs included in the PRS calculations presented. In addition, Coignard et al. identified two, respectively three, loci that modify BC risks in BRCA1, respectively BRCA2, mutation carriers exclusively, which point towards the need for establishment of divergent PRSs for BRCA1 and BRCA2 PV carriers. However, our sample was too small to contribute to these efforts by investigating the associations of individual SNPs with BC with sufficient statistical power.
The study of Sepahi et al. [25] considering 133 BRCA1 PV carriers fulfilling either the ED or LD criteria, pointed towards an association of germline BRCA1 PVs within the OCCR with LD, although no statistical significance was reached. Our study now shows a statistically significant association based on a sample of 183 BRCA1 PV carriers. Consistent with Sepahi et al., there was no statistically significant effect of localization of PVs within the BRCA1 BCCRs. With regard to the localization of PVs in the BRCA2 gene, no statistically significant associations with ED were found, but this is probably due to the small sample size of only 112 PV carriers.
Besides limited sample size, this study has further limitations, as co-occuring truncating variants in DNA-repair genes were not considered as suggested for discrimination between extreme phenotypes with respect to age at BC onset by Sepahi et al. [25] and possible biases due to patient recruitment in two different countries can not be excluded. Further, adjustment of analyses for year of birth was not feasible, as considered individuals from the ED group were born in 1953 at the earliest, and individuals from the LD group no later than 1957.
Conclusions
Although the optimal PRS SNP set for this purpose remains to be found in future studies with larger sample size, our results show that the PRS is an essential prerequisite for the clinical management of unaffected BRCA1/2 PV carriers, as it helps to identify those women who should be offered prophylactic measures at a comparatively very young age. Furthermore, our results provide further evidence that the localization of pathogenic BRCA1/2 mutations should be considered in BC risk calculations.
Supplementary Information
Additional file 1 Inclusion criteria of the German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC) for germline testing.
Additional file 2 SNPs replaced by SNPs in linkage disequlibrium.
Additional file 3 First three dimensions of multi-dimensional scaling of a combined set of the curated study sample and 2,157 individuals of known ancestry, namely European (EUR), African (AFR), East Asian (EAS), or South Asian (SAS), from the 1000 Genomes data comprising 342 SNPs. Samples that were identified as of putative African or Asian origin from the study sample are shown in red (ExPh OUT).
Additional file 4 SNPs included in polygenic risk score (PRS) computation, corresponding per-allele log odds ratios, and comparison of allele frequencies between patients recruited by Sheba Medical Center, Tel Aviv University (AF Sheba) and patients recruited by the German Consortium for Hereditary Breast and Ovarian Cancer (AF GC-HBOC).
Additional file 5 Prevalence of IARC class 4/5 pathogenic germline variants in the BRCA1 gene (considering transcript NM_007294.4) and the BRCA2 gene (considering transcript NM_000059.4) in the curated final study sample consisting of 295 individuals.
Acknowledgements
We thank all individuals who participated in this study. Computations were partially run on the German Research Foundation (DFG)-funded high-performance computing cluster CHEOPS, which is provided and maintained by the computing center of the University of Cologne (RRZK).
Abbreviations
- BC
Breast cancer
- BCCR
Breast cancer cluster region
- BRCA1
Breast cancer 1 gene
- BRCA2
Breast cancer 2 gene
- CI
Confidence interval
- ED
Early diagnosis
- ER+
Estrogen receptor-positive
- ER-
Estrogen receptor-negative
- GC-HBOC
German Consortium for Hereditary Breast and Ovarian Cancer
- LD
Late diagnosis
- OC
Ovarian cancer
- OCCR
Ovarian cancer cluster region
- OR
Odds ratio
- PRS
Polygenic risk score
- PV
Pathogenic variant
- SNP
Single nucleotide polymorphism
Authors’ contributions
CoE, JB, RS and EH drafted the manuscript. CoE and JB analyzed the data. CoE, JB, CS, KK, RS, EF and EH designed the study. JB, YL and BB contributed in variant interpretation. YL, EF, BW, DN, AR, NA, SW, JudH, AG, GS, VD, JR, JulH and AM provided the DNA samples. JB, BB, KW, ChE and CoE collected the clinical and genetic data. RS, EF and EH supervised and administered the project. All the authors contributed in critical revision of the manuscript. All authors read and approved the final manuscript.
Funding
Genetic analyses were supported by the Köln Fortune Program, Faculty of Medicine, University of Cologne, Germany. The German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC) is funded by the German Cancer Aid (#110837, #70111850) and the Federal Ministry of Education and Research (BMBF), Germany (grant no 01GY1901). The funding source had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; nor the decision to submit the manuscript for publication. Open Access funding enabled and organized by Projekt DEAL, supported by the DFG (German Research Foundation, 491454339).
Availability of data and materials
The dataset produced or analyzed in this study is not publicly available due to privacy reasons but it will be available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
This study was performed in accordance with the Declaration of Helsinki. Ethical approval was granted by the ethics committee of the University of Cologne (07-048). All participants signed a written informed consent before study enrollment.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rita K. Schmutzler, Eitan Friedman, Eric Hahnen and Corinna Ernst contributed equally to this work
Contributor Information
Julika Borde, Email: julika.borde@uk-koeln.de.
Yael Laitman, Email: yael.laitman@gmail.com.
Britta Blümcke, Email: britta.bllumcke@uk-koeln.de.
Dieter Niederacher, Email: niederac@med.uni-duesseldorf.de.
Konstantin Weber-Lassalle, Email: konstantin.weber-lassalle@uk-koeln.de.
Christian Sutter, Email: christian.sutter@med.uni-heidelberg.de.
Andreas Rump, Email: andreas.rump@uniklinikum-dresden.de.
Norbert Arnold, Email: norbert.arnold@uksh.de.
Shan Wang-Gohrke, Email: shan.wang-gohrke@uniklinik-ulm.de.
Judit Horváth, Email: judit.horvath@ukmuenster.de.
Andrea Gehrig, Email: gehrig@biozentrum.uni-wuerzburg.de.
Gunnar Schmidt, Email: schmidt.gunnar@mh-hannover.de.
Véronique Dutrannoy, Email: veronique.dutrannoy-toensing@laborberlin.com.
Juliane Ramser, Email: juliane.ramser@mri.tum.de.
Julia Hentschel, Email: julia.hentschel@medizin.uni-leipzig.de.
Alfons Meindl, Email: alfons.meindl@gmx.de.
Christopher Schroeder, Email: christopher.schroeder@med.uni-tuebingen.de.
Barbara Wappenschmidt, Email: barbara.wappenschmidt@uk-koeln.de.
Christoph Engel, Email: christoph.engel@imise.uni-leipzig.de.
Karoline Kuchenbaecker, Email: k.kuchenbaecker@ucl.ac.uk.
Rita K. Schmutzler, Email: rita.schmutzler@uk-koeln.de
Eitan Friedman, Email: feitan@tauex.tau.ac.il.
Eric Hahnen, Email: eric.hahnen@uk-koeln.de.
Corinna Ernst, Email: corinna.ernst@uk-koeln.de.
References
- 1.Easton DF, Pharoah PD, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, Devilee P, Meindl A, Couch FJ, Southey M, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372(23):2243–57. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips K-A, Mooij TM, Roos-Blom M-J, Jervis S, Van Leeuwen FE, Milne RL, Andrieu N, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. Jama. 2017;317(23):2402–16. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 3.Mavaddat N, Peock S, Frost D, Ellis S, Platte R, Fineberg E, Evans DG, Izatt L, Eeles RA, Adlard J, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. JNCI J Natl Cancer Inst. 2013;105(11):812–22. doi: 10.1093/jnci/djt095. [DOI] [PubMed] [Google Scholar]
- 4.Hartmann LC, Lindor NM. The role of risk-reducing surgery in hereditary breast and ovarian cancer. N Engl J Med. 2016;374(5):454–68. doi: 10.1056/NEJMra1503523. [DOI] [PubMed] [Google Scholar]
- 5.Singer CF. Nonsurgical prevention strategies in BRCA1 and BRCA2 mutation carriers. Breast Care. 2021;16(2):135–9. doi: 10.1159/000507503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foulkes WD, Metcalfe K, Sun P, Hanna WM, Lynch HT, Ghadirian P, Tung N, Olopade OI, Weber BL, McLennan J, et al. Estrogen receptor status in BRCA1-and BRCA2-related breast cancer: the influence of age, grade, and histological type. Clin Cancer Res. 2004;10(6):2029–34. doi: 10.1158/1078-0432.CCR-03-1061. [DOI] [PubMed] [Google Scholar]
- 7.Mavaddat N, Barrowdale D, Andrulis IL, Domchek SM, Eccles D, Nevanlinna H, Ramus SJ, Spurdle A, Robson M, Sherman M, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) Cancer Epidemiol Prev Biomark. 2012;21(1):134–47. doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee A, Mavaddat N, Wilcox AN, Cunningham AP, Carver T, Hartley S, de Villiers CB, Izquierdo A, Simard J, Schmidt MK, et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet Med. 2019;21(8):1708–18. doi: 10.1038/s41436-018-0406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson HD, Zakher B, Cantor A, Fu R, Griffin J, O’Meara ES, Buist DS, Kerlikowske K, van Ravesteyn NT, Trentham-Dietz A., et al. Risk factors for breast cancer for women aged 40 to 49 years: a systematic review and meta-analysis. Ann Intern Med. 2012;156(9):635–48. doi: 10.7326/0003-4819-156-9-201205010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg Å, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–30. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guindalini RSC, Song A, Fackenthal JD, Olopade OI, Huo D. Genetic anticipation in BRCA1/BRCA2 families after controlling for ascertainment bias and cohort effect. Cancer. 2016;122(12):1913–20. doi: 10.1002/cncr.29972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King M-C, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–6. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 13.Toss A, Grandi G, Cagnacci A, Marcheselli L, Pavesi S, De Matteis E, Razzaboni E, Tomasello C, Cascinu S, Cortesi L. The impact of reproductive life on breast cancer risk in women with family history or BRCA mutation. Oncotarget. 2017;8(6):9144. doi: 10.18632/oncotarget.13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lammert J, Grill S, Kiechle M. Modifiable lifestyle factors: opportunities for (hereditary) breast cancer prevention - a narrative review. Breast Care. 2018;13(2):108–13. doi: 10.1159/000488995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lakeman IM, Hilbers FS, Rodríguez-Girondo M, Lee A, Vreeswijk MP, Hollestelle A, Seynaeve C, Meijers-Heijboer H, Oosterwijk JC, Hoogerbrugge N, et al.Addition of a 161-SNP polygenic risk score to family history-based risk prediction: impact on clinical management in non-BRCA1/2 breast cancer families. Journal of Medical Genetics. 2019; 56(9):581–9. [DOI] [PubMed]
- 16.Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, Tyrer JP, Chen T-H, Wang Q, Bolla MK, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. 2019;104(1):21–34. doi: 10.1016/j.ajhg.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borde J, Ernst C, Wappenschmidt B, Niederacher D, Weber-Lassalle K, Schmidt G, Hauke J, Quante AS, Weber-Lassalle N, Horváth J, et al. Performance of breast cancer polygenic risk scores in 760 female CHEK2 germline mutation carriers. JNCI J Natl Cancer Inst. 2021;113(7):893–9. doi: 10.1093/jnci/djaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muranen TA, Mavaddat N, Khan S, Fagerholm R, Pelttari L, Lee A, Aittomäki K, Blomqvist C, Easton DF, Nevanlinna H. Polygenic risk score is associated with increased disease risk in 52 Finnish breast cancer families. Breast Cancer Res Treat. 2016;158(3):463–9. doi: 10.1007/s10549-016-3897-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muranen TA, Greco D, Blomqvist C, Aittomäki K, Khan S, Hogervorst F, Verhoef S, Pharoah PD, Dunning AM, Shah M, et al. Genetic modifiers of CHEK2* 1100delC-associated breast cancer risk. Genet Med. 2017;19(5):599–603. doi: 10.1038/gim.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shieh Y, Hu D, Ma L, Huntsman S, Gard CC, Leung JW, Tice JA, Vachon CM, Cummings SR, Kerlikowske K, et al. Breast cancer risk prediction using a clinical risk model and polygenic risk score. Breast Cancer Res Treat. 2016;159(3):513–25. doi: 10.1007/s10549-016-3953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallagher S, Hughes E, Wagner S, Tshiaba P, Rosenthal E, Roa BB, Kurian AW, Domchek SM, Garber J, Lancaster J, et al. Association of a polygenic risk score with breast cancer among women carriers of high-and moderate-risk breast cancer genes. JAMA Netw Open. 2020;3(7):e208501. doi: 10.1001/jamanetworkopen.2020.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuchenbaecker KB, McGuffog L, Barrowdale D, Lee A, Soucy P, Healey S, Dennis J, Lush M, Robson M, Spurdle AB, et al.Vol. 109. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. 2017; 109(7):djw302. [DOI] [PMC free article] [PubMed]
- 23.Sawyer S, Mitchell G, McKinley J, Chenevix-Trench G, Beesley J, Chen XQ, Bowtell D, Trainer AH, Harris M, Lindeman GJ, et al. A role for common genomic variants in the assessment of familial breast cancer. J Clin Oncol. 2012;30(35):4330–6. doi: 10.1200/JCO.2012.41.7469. [DOI] [PubMed] [Google Scholar]
- 24.Coignard J, Lush M, Beesley J, O’mara TA, Dennis J, Tyrer JP, Barnes DR, McGuffog L, Leslie G, Bolla MK, et al. A case-only study to identify genetic modifiers of breast cancer risk for BRCA1/BRCA2 mutation carriers. Nat Commun. 2021;12(1):1–22. doi: 10.1038/s41467-020-20314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sepahi I, Faust U, Sturm M, Bosse K, Kehrer M, Heinrich T, Grundman-Hauser K, Bauer P, Ossowski S, Susak H, et al. Investigating the effects of additional truncating variants in DNA-repair genes on breast cancer risk in BRCA1-positive women. BMC Cancer. 2019;19(1):1–12. doi: 10.1186/s12885-019-5946-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernstein-Molho R, Laitman Y, Schayek H, Reish O, Lotan S, Haim S, Zidan J, Friedman E. The yield of targeted genotyping for the recurring mutations in BRCA1/2 in Israel. Breast Cancer Res Treat. 2018;167(3):697–702. doi: 10.1007/s10549-017-4551-7. [DOI] [PubMed] [Google Scholar]
- 27.Spurdle AB, Healey S, Devereau A, Hogervorst FB, Monteiro AN, Nathanson KL, Radice P, Stoppa-Lyonnet D, Tavtigian S, Wappenschmidt B, et al. ENIGMA—evidence-based network for the interpretation of germline mutant alleles: An international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Human Mutat. 2012;33(1):2–7. doi: 10.1002/humu.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–23. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, Mazoyer S, Chenevix-Trench G, Easton DF, Antoniou AC, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA. 2015;313(13):1347–61. doi: 10.1001/jama.2014.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Au CH, Ho DN, Kwong A, Chan TL, Ma ES. BAMClipper: removing primers from alignments to minimize false-negative mutations in amplicon next-generation sequencing. Sci Rep. 2017;7(1):1–7. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907. 2012.
- 33.Sherry ST, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):s13742—015. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aulchenko YS, Ripke S, Isaacs A, Van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294–6. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 36.Robitzsch A, Grund S. miceadds: Some Additional Multiple Imputation Functions, Especially for ’mice’. R package version 3.13-12.2022. https://CRAN.R-project.org/package=miceadds.
- 37.Barnes DR, Rookus MA, McGuffog L, Leslie G, Mooij TM, Dennis J, Mavaddat N, Adlard J, Ahmed M, Aittomäki K, et al. Polygenic risk scores and breast and epithelial ovarian cancer risks for carriers of BRCA1 and BRCA2 pathogenic variants. Genet Med. 2020;22(10):1653–66. doi: 10.1038/s41436-020-0862-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Inclusion criteria of the German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC) for germline testing.
Additional file 2 SNPs replaced by SNPs in linkage disequlibrium.
Additional file 3 First three dimensions of multi-dimensional scaling of a combined set of the curated study sample and 2,157 individuals of known ancestry, namely European (EUR), African (AFR), East Asian (EAS), or South Asian (SAS), from the 1000 Genomes data comprising 342 SNPs. Samples that were identified as of putative African or Asian origin from the study sample are shown in red (ExPh OUT).
Additional file 4 SNPs included in polygenic risk score (PRS) computation, corresponding per-allele log odds ratios, and comparison of allele frequencies between patients recruited by Sheba Medical Center, Tel Aviv University (AF Sheba) and patients recruited by the German Consortium for Hereditary Breast and Ovarian Cancer (AF GC-HBOC).
Additional file 5 Prevalence of IARC class 4/5 pathogenic germline variants in the BRCA1 gene (considering transcript NM_007294.4) and the BRCA2 gene (considering transcript NM_000059.4) in the curated final study sample consisting of 295 individuals.
Data Availability Statement
The dataset produced or analyzed in this study is not publicly available due to privacy reasons but it will be available from the corresponding author upon reasonable request.