Abstract
Pediocin PA-1 is a food grade antimicrobial peptide that has been used as a food preservative. Upon storage at 4°C or room temperature, pediocin PA-1 looses activity, and there is a concomitant 16-Da increase in the molecular mass. It is shown that the loss of activity follows first-order kinetics and that the instability can be prevented by replacing the single methionine residue (Met31) in pediocin PA-1. Replacing Met by Ala, Ile, or Leu protected the peptide from oxidation and had only minor effects on bacteriocin activity (for most indicator strains 100% activity was maintained). Replacement of Met by Asp was highly deleterious for bacteriocin activity.
Bacteria produce ribosomally synthesized antimicrobial polypeptides termed bacteriocins. Bacteriocins produced by gram-positive bacteria are usually membrane-permeabilizing cationic peptides with less than 50 amino acid residues (19, 20, 23, 25). These bacteriocins may be divided into two classes; class I contains bacteriocins (often referred to as lantibiotics) with modified residues, and class II contains bacteriocins without modified residues. Within class II, the so-called pediocin-like bacteriocins produced by a variety of lactic acid bacteria constitute a dominant group. At least 14 different bacteriocins belonging to this group are presently known, and pediocin PA-1 (4, 15, 17, 22), leucocin A UAL-187 (13), mesentericin Y105 (14), sakacin P (28), and curvacin A (identical to sakacin A) (16, 28) were the first of these to be identified.
The pediocin-like bacteriocins are characterized by a YGNGV motif and a disulfide bridge in a highly conserved N-terminal region, by high antilisterial activity, and by their membrane-permeabilizing mode of action (6, 7, 20). Some of the pediocin-like bacteriocins (such as pediocin PA-1) also contain a disulfide bridge in the C-terminal region, whereas others (such as sakacin P) do not. The highly conserved N-terminal region is hydrophilic and cationic, and it has been proposed that this region mediates the initial binding of these bacteriocins to target cells through electrostatic interactions (5). The somewhat less conserved C-terminal half is hydrophobic and/or amphiphilic and is thought to penetrate into the hydrophobic part of the target cell membrane, thereby mediating membrane leakage (10, 18). Structural analysis indicates that a 15- to 20-residue stretch from the middle toward the C-terminal end forms an amphiphilic α-helix upon interaction with membranelike structures and that the remaining C-terminal residues are relatively unstructured (12, 30).
Much of the interest in pediocin-like bacteriocins is due to their antilisterial activity and thus to their potential for use as antimicrobial additives in food. Their use as additives requires that they be sufficiently stable and consequently devoid of residues that are prone to potentially damaging chemical modifications. However, several of the pediocin-like bacteriocins contain methionine residues whose sulfur atom may be oxidized, which results in destabilization of the bacteriocin. In this study, we focused on the methionine residue present in the C-terminal half of pediocin PA-1, which presently is perhaps the pediocin-like bacteriocin that is most promising for use as an antimicrobial additive. Pediocin PA-1 variants were constructed in which Met31 was replaced by Ala, Leu, Ile, and Asp, and the effects of these mutations on bacteriocin stability, activity, and target cell specificity were determined.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Wild-type pediocin PA-1 was produced by and purified from Pediococcus acidilactici LMG2351, which was isolated from commercial starter cultures obtained from Christian Hansen Laboratories, Copenhagen, Denmark (22). Mutant pediocin PA-1 and wild-type sakacin P were produced by and purified from a two-plasmid bacteriocin expression system developed recently (3, 9). This system is based on the use of pSAK20 and either pSPP2 (for production of wild-type sakacin P) or pPED2 (for production of mutant pediocin PA-1) introduced into the bacteriocin-deficient strain Lactobacillus sake Lb790. pSAK20 and pSPP2/pPED2 confer resistance to chloramphenicol and erythromycin, respectively, pSPP2 and pPED2 are pLPV111 (3)- based Escherichia coli-Lactobacillus shuttle vectors in which a bacteriocin gene and its cognate immunity gene have been placed under control of a bacteriocin-specific promoter derived from the sakacin A producer L. sake Lb706 (3). pSAK20 is a pVS2 (29)-based plasmid that contains the orf4sapKRTE operon from L. sake Lb706 (2, 3). The orf4sapKRTE operon contains genes encoding proteins necessary for activation of the bacteriocin-specific promoters and for processing and secretion of the prebacteriocins (2, 3, 21).
Epicurian Coli XL1-Blue supercompetent cells (Stratagene) were used for cloning all of the mutated pPED2 plasmids, and plasmids with desired mutations were transformed into L. sake Lb790/pSAK20.
E. coli was grown at 37°C in Luria-Bertani medium (Difco) with vigorous agitation, whereas the lactic acid bacteria were grown at 30°C without agitation. The indicator strains used in the bacteriocin assays were L. sake NCDO 2714 (type strain), Lactobacillus coryneformis subsp. torquens NCDO 2740, Enterococcus faecalis NCDO 581, P. acidilactici NCDO 1859, Pediococcus pentosaceus FBB63B, Leuconostoc mesenteroides subsp. dextranicum NCDO 529, and Carnobacterium piscicola UI49 (27). C. piscicola UI49 was grown in M17 medium (Oxoid) supplemented with glucose and Tween 80 at final concentrations of 0.4% (wt/vol) and 0.1% (vol/vol), respectively. The other lactic acid bacteria were grown in MRS broth (Oxoid). For agar plates, the media were solidified by adding 1.5% (wt/vol) agar. The selective antibiotic concentrations used were 150 μg of erythromycin per ml for E. coli, 10 μg of erythromycin per ml and 10 μg of chloramphenicol per ml for normal growth of plasmid-containing L. sake Lb790, and 2 μg of erythromycin per ml and 5 μg of chloramphenicol per ml for initial selection of L. sake Lb790/pSAK20 transformed with pPED2 variants.
Purification of sakacin P, pediocin PA-1, and mutant pediocin PA-1.
Wild-type and mutant bacteriocins were purified to homogeneity from 400-ml cultures by ammonium sulfate precipitation followed by cation-exchange chromatography, hydrophobic interaction chromatography, and reverse-phase chromatography as described previously (22). The primary structures of the compounds were confirmed by determining the molecular masses with a Voyager-DE RP matrix-assisted laser desorption ionization–time of flight mass spectrometer (Perseptive Biosystems); α-cyano-4-hydroxycinnamic acid was used as the matrix. Typically, the errors in the masses which were determined were ≤1 Da. The purities of the bacteriocins were verified to be greater than 90% by analytical reverse-phase chromatography by using a μRPC SC 2.1/10 C2/C18 column (Pharmacia Biotech) in the SMART chromatography system (Pharmacia Biotech).
The concentrations of purified bacteriocins were determined by measuring UV absorption at 280 nm, and the values were converted to protein concentrations by using molecular extinction coefficients calculated from the contributions of individual amino acid residues.
Bacteriocin assay.
Bacteriocin activity was measured by using a microtiter plate assay system, essentially as described previously (24). The wells of a microtiter plate contained 200 μl of culture medium with bacteriocin fractions at twofold dilutions and an indicator strain at an optical density at 610 nm of about 0.01. The microtiter plate cultures were incubated overnight (12 to 16 h) at 30°C, after which growth of the indicator strain was measured spectrophotometrically at 610 nm with a microtiter plate reader. The MIC was defined as the concentration of bacteriocin that inhibited growth of the indicator strain by 50%.
Plasmid isolation and transformation.
Plasmids were isolated from E. coli and L. sake Lb790 by using the Wizard Plus SV Minipreps DNA purification system (Promega). To ensure lysis of L. sake, lysozyme and mutanolysin were added to the cell resuspension solution included in the Wizard Plus SV Minipreps kit to final concentrations of 5 mg/ml and 15 U/ml, respectively.
Chemocompetent Epicurian Coli XL1-Blue supercompetent cells were transformed by using the protocol provided with a Quick Change site-directed mutagenesis kit (Stratagene). L. sake Lb790/pSAK20 was transformed by electroporation by using a Gene Pulser and Pulse Controller unit (Bio-Rad Laboratories) as described previously (1). L. sake Lb790/pSAK20 cells were made competent by growth in MRS broth supplemented with 1.5% (wt/vol) glycine. After this, the cells were washed with 1 mM MgCl2 and then with 30% (wt/vol) polyethylene glycol 1500 (molecular weight range, 1,300 to 1,600) prior to electroporation.
Site-directed mutagenesis and DNA sequencing.
Mutations in the pediocin PA-1 gene cloned in pPED2 were obtained by using a Quick Change site-directed mutagenesis kit (Stratagene). The PCR were performed with a GeneAmp 2400 PCR system (Perkin-Elmer) by using Pfu DNA polymerase (Stratagene). The 50-μl reaction mixtures each contained about 40 ng of plasmid template, 125 ng of each oligonucleotide primer (Eurogentec), each deoxynucleoside triphosphate (Stratagene) at a final concentration of 0.05 mM, and 2.5 U of Pfu DNA polymerase. After a 1-min hot start at 95°C, 16 cycles of the following program were run: denaturation for 30 s at 95°C, primer annealing for 1 min at 50°C, and polymerization for 12 min at 68°C. The PCR product was digested for 1 h at 37°C with restriction enzyme DpnI (Stratagene) to eliminate the original template and thereby increase mutation efficiency. The DNA sequences of the mutant plasmids were verified by automated DNA sequence determination by using an ABI PRISM 377 DNA sequencer and an ABI Prism Ready Reaction dye terminator cycle sequencing kit (Perkin-Elmer).
Four of the eight oligonucleotide primers used for site-directed mutagenesis had the following general sequence: 5′-CAATAATGGAGCTxyzGCATGGGCTACTGGTGG-3′, where xyz indicates the methionine codon (ATG) which was the target for mutagenesis. This methionine codon was changed to ATT, GCG, CTA, and GAT for the isoleucine, alanine, leucine, and aspartate mutants, respectively. The four other primers used were complementary to the above-mentioned oligonucleotide primers.
RESULTS
Production and purification of sakacin P, pediocin PA-1, and mutant pediocin PA-1 molecules.
The methionine residue in pediocin PA-1 (position 31) (Fig. 1) was in all mutant molecules replaced by another residue, either alanine, isoleucine, leucine, or aspartate (designated ped[M31A], ped[M31I], ped[M31L], and ped[M31D], respectively). The mutants were all produced by using the L. sake Lb790/pSAK20/pPED2 two-plasmid expression system, whereas wild-type pediocin PA-1 was produced by using the natural pediocin PA-1 producer (P. acidilactici LMG2351), as it yielded about five times more bacteriocin than the two-plasmid expression system. Wild-type sakacin P was produced by using the L. sake Lb790/pSAK20/pSPP2 expression system, since it yielded about five times more than the natural sakacin P producer (L. sake LTH673). Moreover, sakacin P produced by the expression system was stable for months, whereas sakacin P produced by the natural producer lost activity during purification and storage as a result of degradation by contaminating extracellular proteases produced by L. sake LTH673 (10).
FIG. 1.
Sequences of pediocin PA-1 and sakacin P. Black areas with white lettering in the sakacin P sequence indicate regions where pediocin PA-1 and sakacin P are identical.
Between 10- and 100-μg portions of bacteriocins and mutant bacteriocins were purified from 400-ml cultures. In the last reverse-phase chromatography step, pediocin PA-1 and sakacin P gave one major symmetrical absorbance peak that contained a peptide with bacteriocin activity and the expected molecular weight (Table 1). The mutant bacteriocins gave more complex absorbance profiles, which contained several, often asymmetric absorbance peaks, presumably because of incorrect formation of disulfide bridges (see Discussion). For each mutant, the fraction with the most bacteriocin activity was collected. Mass spectrometry confirmed that each of these fractions contained the expected mutant bacteriocin (Table 1), and analysis of the fractions by analytical reverse-phase chromatography revealed single major absorbance peaks, verifying the purity of the mutant bacteriocins.
TABLE 1.
Theoretical and experimental molecular masses of pediocin PA-1 and mutant bacteriocin molecules
| Bacteriocin | Theoretical mass (Da)a | Observed mass (Da)b |
|---|---|---|
| Pediocin PA-1 | 4,624.3 | 4,624.1 ± 0.1 |
| Ped[M31A] | 4,564.2 | 4,563.9 ± 0.1 |
| ped[M31D] | 4,608.2 | 4,608.0 ± 0.1 |
| ped[M31I] | 4,606.3 | 4,606.3 ± 0.1 |
| ped[M31L] | 4,606.3 | 4,606.4 ± 0.2 |
The theoretical molecular masses were calculated from the amino acid sequences, assuming that the cysteine residues form disulfide bridges and that methionines (only present in wild-type pediocin PA-1) are not oxidized.
The observed masses are means based on three parallel measurements obtained by mass spectrometry.
Oxidation and partial inactivation of pediocin PA-1 during storage.
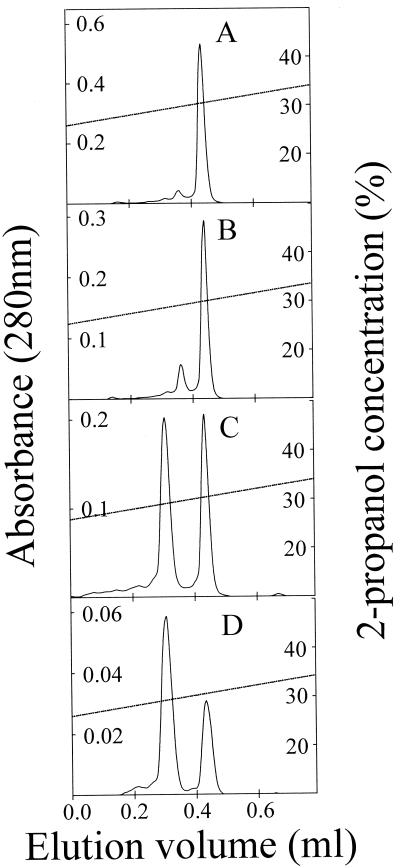
During storage pediocin-like bacteriocins that contain a methionine residue change to a less active form (10, 26; see below), apparently due to oxidation of the methionine sulfur atom to sulfoxide. This oxidation increases the molecular mass by 16 Da. For pediocin PA-1, the kinetics of this conversion was determined by separating the unoxidized and oxidized forms by reverse-phase chromatography after the bacteriocin was stored for various lengths of time under various conditions (Fig. 2). The relative amounts of the two forms were determined from the reverse-phase chromatography absorbance profiles. The first of the two peptide forms to elute from the reverse-phase column had the molecular mass (as determined by mass spectrometry) expected for pediocin PA-1 containing an oxidized methionine (4,640 Da), whereas the second form to elute had the molecular mass expected for the active unoxidized form of pediocin PA-1 (4,624 Da). The specific activity of the latter was about 100-fold greater than that of the former.
FIG. 2.
Reverse-phase chromatography of purified pediocin PA-1 after storage. Pediocin PA-1 was purified and stored in 100% 2-propanol–0.1% TFA at room temperature for 1 (A), 7 (B), 14 (C), and 28 (D) days, and aliquots of stored samples were analyzed by reverse-phase chromatography in order to detect relative amounts of oxidized and unoxidized pediocin PA-1. Mass spectrometry showed that the first of the two peaks contains a peptide whose mass is 16 Da greater than the mass of the peptide in the second peak (see text).
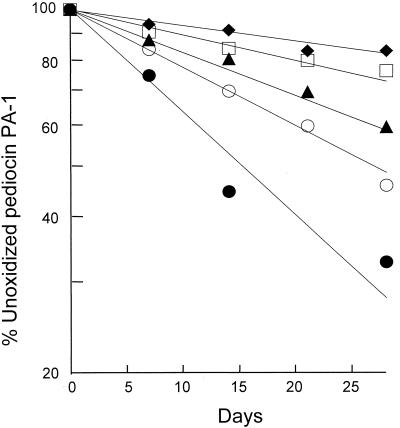
The conversion of active unoxidized pediocin PA-1 to the less active oxidized form followed first-order kinetics with half-lives of 15, 27, 42, 65, and 100 days when preparations were stored at room temperature in 100, 50, 25, 10, and 0% propanol respectively, containing 0.1% (vol/vol) trifluoracetic acid (TFA) (Fig. 3). Freezing the bacteriocin protected it from oxidation. After 55 days (in 20 mM phosphate buffer, pH 7), no oxidation was detected when preparations were stored at −20°C, in contrast to the 20 to 30% oxidation that occurred when preparations were stored at either 4°C or room temperature.
FIG. 3.
Kinetics of oxidation of pediocin PA-1. The peptide was stored in 100% (●), 50% (○), 25% (▴), 10% (□), or 0% (⧫) isopropanol containing 0.1% TFA. The degree of oxidation was determined by reverse-phase chromatography (see text).
Mutational effects on bacteriocin activity.
The susceptibility of pediocin PA-1 to inactivation, apparently because of oxidation of its methionine residue, prompted us to construct methionine-free mutant pediocin PA-1 molecules. Seven indicator strains were used to test the effect that the mutations had on potency and target cell specificity. Four of these strains (L. sake NCDO 2714, L. coryneformis subsp. torquens NCDO 2740, E. faecalis NCDO 581, and C. piscicola UI49) were sensitive to both pediocin PA-1 and sakacin P, whereas three of the strains (P. pentosaceus FBB63B, P. acidilactici NCDO 1859, and L. mesenteroides subsp. dextranicum NCDO 529) were about 100 to 1,000 times more sensitive to pediocin PA-1 than to sakacin P (Table 2). The latter three strains were, consequently, useful for analyzing whether replacement of the methionine residue (which is absent in sakacin P [Fig. 1]) significantly alters the target cell specificity of pediocin PA-1.
TABLE 2.
Activities of pediocin PA-1, sakacin P, and mutant pediocin PA-1 molecules against various indicator strainsa
| Bacterium | MIC (nM)b
|
|||||
|---|---|---|---|---|---|---|
| Pediocin PA-1 | ped[M31A] | ped[M31I] | ped[M31L] | ped[M31D] | Sakacin P | |
| L. sake | 0.2 | 0.1 | 0.1 | 0.1 | 20 | 0.3 |
| L. coryneformis | 0.1 | 0.1 | 0.1 | 0.1 | 10 | 0.4 |
| E. faecalis | 0.2 | 0.2 | 0.2 | 0.2 | 30 | 0.2 |
| C. piscicola | 0.5 | 0.5 | 0.5 | 0.6 | 60 | 0.2 |
| P. pentosaceus | 0.1 | 0.4 | 0.5 | 0.2 | NDc | 40 |
| P. acidilactici | 2 | 10 | 10 | 6 | ND | >400 |
| L. mesenteroides | 4 | 4 | 10 | 10 | ND | 250 |
Bacteriocin activity was measured as described in Materials and Methods. The indicator strains used in the bacteriocin assay were L. sake NCDO 2714 (type strain), L. coryneformis subsp. torquens NCDO 2740, E. faecalis NCDO 581, C. piscicola UI49, P. pentosaceus FBB63B, P. acidilactici NCDO 1859, and L. mesenteroides subsp. dextranicum NCDO 529.
The MIC is the concentration of bacteriocin that inhibited growth of the indicator strain by 50%. The values are the results of at least three independent measurements, and the standard deviations were less than 50%.
ND, not determined.
The mutant pediocin PA-1 molecules in which the methionine residue was replaced by a hydrophobic amino acid residue (alanine, isoleucine, or leucine) were as active as pediocin PA-1 and sakacin P against the four strains that were sensitive to both pediocin PA-1 and sakacin P (Table 2). These mutant molecules were only slightly less potent (two to five times less potent) than pediocin PA-1 against the three strains that were sensitive to pediocin PA-1 but relatively resistant to sakacin P, but they were more potent than sakacin P (Table 2). Replacing the methionine residue with a negatively charged hydrophilic amino acid residue (aspartate) rather than with a hydrophobic residue resulted in a 100-fold reduction in the bacteriocin activity (Table 2).
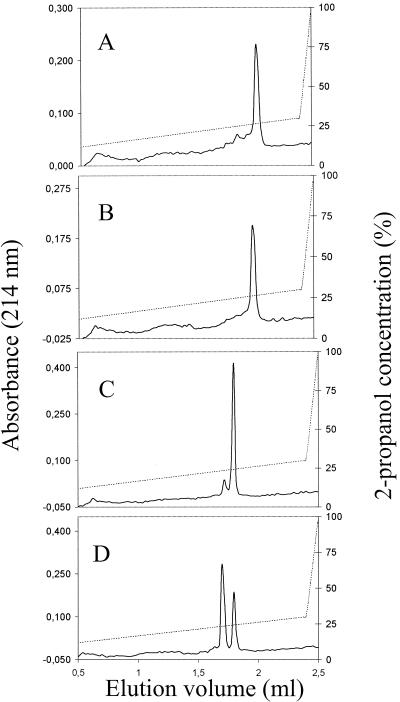
A more stable variant of pediocin PA-1 was clearly obtained by replacing the methionine residue with either alanine, leucine, or isoleucine. In contrast to pediocin PA-1, the methionine-free mutant pediocin PA-1 molecules retained activity, and no oxidation was detected even after 4 weeks of storage at room temperature in the presence of 80% propanol (Fig. 4). Methionine-free mutant molecules in 25% propanol–0.1% TFA in fact remained active without detectable oxidation for more than 7 months at 4°C or for 70 days at room temperature.
FIG. 4.
Reverse-phase chromatography of purified ped[M31L] and pediocin PA-1 after storage. ped[M31L] (A and B) and pediocin PA-1 (C and D) were purified and stored in 80% 2-propanol–0.1% TFA at −80°C (A and C) or room temperature (B and D) for 28 days, and aliquots of stored samples were then analyzed by reverse-phase chromatography. The mass expected for ped[M31L] was obtained upon analysis of the peak fractions in panels A and B by mass spectrometry, and the mass expected for pediocin PA-1 was obtained upon analysis of the peak fraction in panel C and the second of the two peak fractions in panel D. The first of the two peak fractions in panel D contained a peptide (pediocin PA-1 with oxidized methionine) whose mass was 16 Da greater than the mass of pediocin PA-1. Results similar to those shown in panels A and B were obtained with ped[M31A] and ped[M31I].
DISCUSSION
Pediocin PA-1 readily changed to a less active form, which had a molecular mass that was 16 Da greater and which was somewhat more polar (as judged by reverse-phase chromatography), as one would expect upon oxidation of the methionine sulfur atom to sulfoxide. The change followed first-order kinetics, with the half-life varying between a few weeks and several months, apparently depending on the amount oxygen present. Oxygen, a nonpolar molecule, is expected to have higher solubility in organic solvents with low polarities than in water, and this might be the reason why the conversion rate in water-propanol mixtures increased with increasing propanol concentration. Pediocin PA-1 was clearly much more stable when the methionine residue was replaced by either an alanine, isoleucine, or leucine residue, indicating that methionine was indeed the destabilizing residue in pediocin PA-1.
When a bacteriocin contained four cysteine residues, as is the case in pediocin PA-1, our expression system produced about equal amounts of three variants, each displaying one of the three possible patterns of disulfide formation (9). In contrast, the wild-type producer of pediocin PA-1 yielded basically one variant with correct disulfide bridges (9). A protein present in the wild-type producer of pediocin PA-1, but not in our expression system, thus apparently helps generate the correct disulfide bridges (9). The formation of incorrect disulfide bridges may explain why more complex absorbance profiles were obtained in the last reverse-phase chromatography step when we purified the mutant bacteriocins and pediocin PA-1 produced by our expression system, compared to the simple absorbance peak which was obtained when we purified pediocin PA-1 produced by the wild-type producer.
Despite similarities in their primary structures, the pediocin-like bacteriocins have different target cell specificities (8). Their hydrophobic-amphiphilic C-terminal halves appear to be important in determining their specificities, since hybrid bacteriocins containing N- and C-terminal regions from different pediocin-like bacteriocins have antimicrobial spectra similar to those of the bacteriocins from which the C-terminal halves are derived (10). The fact that 15-mer fragments from the C-terminal half of pediocin PA-1, but not fragments from the N-terminal half, inhibit pediocin PA-1 to a greater extent than they inhibit other closely related pediocin-like bacteriocins also suggests that the C-terminal half contains important specificity determinants (11). The disulfide bridge present in the C-terminal half of some pediocin-like bacteriocins is clearly one such specificity determinant. Introducing this bridge in sakacin P (which naturally lacks the bridge) by inserting two cysteine residues made the target cell specificity of sakacin P more similar to that of pediocin PA-1 (which naturally contains the bridge), whereas removing the bridge in pediocin PA-1 by replacing cysteine with serine residues made the specificity of pediocin PA-1 more similar to that of sakacin P (9). Other residues in the C-terminal half may also influence the target cell specificity. Replacement of the methionine residue in the C-terminal half of pediocin PA-1 appeared, however, to have only a minor effect on the target cell specificity, since ped[M31A], ped[M31L], and ped[M31I] were as potent as pediocin PA-1 against the four strains that were sensitive to both pediocin PA-1 and sakacin P and were only slightly less potent than pediocin PA-1, but much more potent than sakacin P, against the three strains that were sensitive to pediocin PA-1 but relatively resistant to sakacin P. Replacing the methionine residue with a hydrophilic negatively charged residue (aspartate) instead of a hydrophobic residue resulted in a marked decrease in potency against all strains tested, which is consistent with the proposal that the region interacts with the hydrophobic part of target cell membranes (10, 18). Similarly, replacement of methionine with a hydrophilic but uncharged threonine residue reduces the activity (18). Among the pediocin-like bacteriocins, pediocin PA-1 is perhaps the molecule which is considered to be the most promising antimicrobial additive. Making pediocin PA-1 more stable by replacing the methionine with another hydrophobic residue and retaining the bacteriocin activity is an important step in developing pediocin PA-1 into a useful antimicrobial additive.
ACKNOWLEDGMENT
This work was supported by a grant from the Norwegian Research Council.
REFERENCES
- 1.Aukrust T W, Brurberg M B, Nes I F. Transformation of Lactobacillus by electroporation. Methods Mol Biol. 1995;47:201–208. doi: 10.1385/0-89603-310-4:201. [DOI] [PubMed] [Google Scholar]
- 2.Axelsson L, Holck A. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J Bacteriol. 1995;177:2125–2137. doi: 10.1128/jb.177.8.2125-2137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Axelsson L, Katla T, Bjørnslett M, Eijsink V G H, Holck A. A system for heterologus expression of bacteriocins in Lactobacillus sake. FEMS Microbiol Lett. 1998;168:137–143. doi: 10.1111/j.1574-6968.1998.tb13266.x. [DOI] [PubMed] [Google Scholar]
- 4.Biswas S R, Ray P, Johnson M C, Ray B. Influence of growth conditions on the production of a bacteriocin, pediocin AcH, by Pediococcus acidilactici H. Appl Environ Microbiol. 1991;57:1265–1267. doi: 10.1128/aem.57.4.1265-1267.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Ludescher R D, Montville T J. Electrostatic interactions, but not the YGNGV consensus motif, govern the binding of pediocin PA-1 and its fragments to phospholipid vesicles. Appl Environ Microbiol. 1997;63:4770–4777. doi: 10.1128/aem.63.12.4770-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Shapira R, Eisenstein M, Montville T J. Functional characterization of pediocin PA-1 binding to liposomes in the absence of a protein receptor and its relationship to a predicted tertiary structure. Appl Environ Microbiol. 1997;63:524–531. doi: 10.1128/aem.63.2.524-531.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chikindas M L, Garcia-Garcera M J, Driessen A J M, Ledeboer A M, Nissen-Meyer J, Nes I F, Abee T, Konings W N, Venema G. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl Environ Microbiol. 1993;59:3577–3584. doi: 10.1128/aem.59.11.3577-3584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eijsink V G H, Skeie M, Middelhoven P H, Brurberg M B, Nes I F. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl Environ Microbiol. 1998;64:3275–3281. doi: 10.1128/aem.64.9.3275-3281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fimland G, Johnsen L, Axelsson L, Brurberg M B, Nes I F, Eijsink V G H, Nissen-Meyer J. A C-terminal disulfide bridge in pediocin-like bacteriocins renders bacteriocin activity less temperature dependent and is a major determinant of the antimicrobial spectrum. J Bacteriol. 2000;182:2643–2648. doi: 10.1128/jb.182.9.2643-2648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fimland G, Blingsmo O R, Sletten K, Jung G, Nes I F, Nissen-Meyer J. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl Environ Microbiol. 1996;62:3313–3318. doi: 10.1128/aem.62.9.3313-3318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fimland G, Jack R, Jung G, Nes I F, Nissen-Meyer J. The bactericidal activity of pediocin PA-1 is specifically inhibited by a 15-mer fragment that spans the bacteriocin from the center toward the C terminus. Appl Environ Microbiol. 1998;64:5057–5060. doi: 10.1128/aem.64.12.5057-5060.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fregeau Gallagher N L, Sailer M, Niemczura W P, Nakashima T T, Stiles M E, Vederas J C. Three-dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine miscelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry. 1997;36:15062–15072. doi: 10.1021/bi971263h. [DOI] [PubMed] [Google Scholar]
- 13.Hastings J W, Sailer M, Johnson K, Roy K L, Vederas J C, Stiles M. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidium. J Bacteriol. 1991;173:7491–7500. doi: 10.1128/jb.173.23.7491-7500.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Héchard Y, Dérijard B, Letellier F, Cenatiempo Y. Characterization and purification of mesentericin Y105, an anti-Listeria bacteriocin from Leuconostoc mesenteroides. J Gen Microbiol. 1992;138:2725–2731. doi: 10.1099/00221287-138-12-2725. [DOI] [PubMed] [Google Scholar]
- 15.Henderson J T, Chopko A L, van Wassenaar D. Purification and primary structure of pediocin PA-1 produced by Pediococcus acidilactici PAC-1.0. Arch Biochem Biophys. 1992;295:5–12. doi: 10.1016/0003-9861(92)90480-k. [DOI] [PubMed] [Google Scholar]
- 16.Holck A, Axelsson L, Birkeland S E, Aukrust T, Blom H. Purification and amino acid sequence of sakacin A, a bacteriocin from Lactobacillus sake Lb706. J Gen Microbiol. 1992;138:2715–2720. doi: 10.1099/00221287-138-12-2715. [DOI] [PubMed] [Google Scholar]
- 17.Marugg J D, Gonzalez C F, Kunka B S, Ledeboer A M, Pucci M J, Toonen M Y, Walker S A, Zoetmulder L C M, Vandenbergh P A. Cloning, expression and nucleotide sequence of genes involved in the production of pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1992;58:2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller K W, Schamber R, Osmanagaoglu O, Ray B. Isolation and characterization of pediocin AcH chimeric protein mutants with altered bactericidal activity. Appl Environ Microbiol. 1998;64:1997–2005. doi: 10.1128/aem.64.6.1997-2005.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nes I F, Diep D B, Håvarstein L S, Brurberg M B, Eijsink V, Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 20.Nes I F, Holo H, Fimland G, Hauge H H, Nissen-Meyer J. Unmodified peptide-bacteriocins (class II) produced by lactic acid bacteria. In: Dutton C J, Haxell M A, McArthur H A I, Wax R G, editors. Peptide antibiotics: discovery, modes of action and application, section B. Distribution of antimicrobial peptides, in press. New York, N.Y: Marcel Dekker, Inc.; 2000. [Google Scholar]
- 21.Nes I F, Eijsink V G H. Regulation of group II peptide bacteriocin synthesis by quorum-sensing mechanisms. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 175–192. [Google Scholar]
- 22.Nieto Lozano J C, Nissen-Meyer J, Sletten K, Peláz C, Nes I F. Purification and amino acid sequences of a bacteriocin produced by Pediococcus acidilactici. J Gen Microbiol. 1992;138:1985–1990. doi: 10.1099/00221287-138-9-1985. [DOI] [PubMed] [Google Scholar]
- 23.Nissen-Meyer J, Hauge H H, Fimland G, Eijsink V G H, Nes I F. Ribosomally synthesized antimicrobial peptides produced by lactic acid bacteria: their function, structure, biogenesis, and their mechanism of action. Recent Res Dev Microbiol. 1997;1:141–154. [Google Scholar]
- 24.Nissen-Meyer J, Holo H, Håvarstein L S, Sletten K, Nes I F. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol. 1992;174:5686–5692. doi: 10.1128/jb.174.17.5686-5692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissen-Meyer J, Nes I F. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch Microbiol. 1997;167:67–77. [PubMed] [Google Scholar]
- 26.Quadri L E N, Sailer M, Roy K L, Vederas J C, Stiles M E. Chemical and genetic characterization of bacteriocins produced by Carnobacterium piscicola LV17B. J Biol Chem. 1994;269:12204–12211. [PubMed] [Google Scholar]
- 27.Stoffels G, Nissen-Meyer J, Gudmundsdottir A, Sletten K, Holo H, Nes I F. Purification and characterization of a new bacteriocin isolated from a Carnobacterium sp. Appl Environ Microbiol. 1992;58:1417–1422. doi: 10.1128/aem.58.5.1417-1422.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tichaczek P S, Nissen-Meyer J, Nes I F, Vogel R F, Hammes W P. Characterization of the bacteriocins curvacin A from Lactobacillus curvatus LTH1174 and sakacin P from L. sake LTH673. Syst Appl Microbiol. 1992;15:460–468. [Google Scholar]
- 29.von Wright A, Tynkkynen S, Suominen M. Cloning of a Streptococcus lactis subsp. lactis chromosomal fragment associated with the ability to grow in milk. Appl Environ Microbiol. 1987;53:1584–1588. doi: 10.1128/aem.53.7.1584-1588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Henz M E, Fregeau Gallagher N L, Chai S, Gibbs A C, Yan L Z, Stiles M E, Wishart D S, Vederas J C. Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria. Biochemistry. 1999;38:15438–15447. doi: 10.1021/bi991351x. [DOI] [PubMed] [Google Scholar]