ABSTRACT
Bacterial species in the polymicrobial community evolve interspecific interaction relationships to adapt to the survival stresses imposed by neighbors or environmental cues. Pseudomonas aeruginosa and Staphylococcus aureus are two common bacterial pathogens frequently coisolated from patients with burns and respiratory disease. Whether the application of commonly used antibiotics influences the interaction dynamics of the two species still remains largely unexplored. By performing a series of on-plate competition assays and RNA sequencing-based transcriptional profiling, we showed that the presence of the cephalosporin antibiotic cefotaxime or the quinolone antibiotic levofloxacin at subinhibitory concentration contributes to selecting P. aeruginosa from the coculture with S. aureus by modulating the quorum-sensing (QS) system of P. aeruginosa. Specifically, a subinhibitory concentration of cefotaxime promotes the growth suppression of S. aureus by P. aeruginosa in coculture. This process may be related to the increased production of the antistaphylococcal molecule pyocyanin and the expression of lasR, which is the central regulatory gene of the P. aeruginosa QS hierarchy. On the other hand, subinhibitory concentrations of levofloxacin decrease the competitive advantage of P. aeruginosa over S. aureus by inhibiting the growth and the las QS system of P. aeruginosa. However, pqs signaling of P. aeruginosa can be activated instead to overcome S. aureus. Therefore, this study contributes to understanding the interaction dynamics of P. aeruginosa and S. aureus during antibiotic treatment and provides an important basis for studying the pathogenesis of polymicrobial infections.
IMPORTANCE Increasing evidence has demonstrated the polymicrobial characteristics of most chronic infections, and the frequent communications among bacterial pathogens result in many difficulties for clinical therapy. Exploring bacterial interspecific interaction during antibiotic treatment is an emerging endeavor that may facilitate the understanding of polymicrobial infections and the optimization of clinical therapies. Here, we investigated the interaction of cocultured P. aeruginosa and S. aureus with the intervention of commonly used antibiotics in clinic. We found that the application of subinhibitory concentrations of cefotaxime and levofloxacin can select P. aeruginosa in coculture with S. aureus by modulating P. aeruginosa QS regulation to enhance the production of antistaphylococcal metabolites in different ways. This study emphasizes the role of the QS system in the interaction of P. aeruginosa with other bacterial species and provides an explanation for the persistence and enrichment of P. aeruginosa in patients after antibiotic treatment and a reference for further clinical therapy.
KEYWORDS: Pseudomonas aeruginosa, bacterial interaction, antibiotics, quorum sensing, competition, RNA sequencing
INTRODUCTION
Increasing clinical evidence has revealed the polymicrobial characteristics of the majority of chronic infections (1–3). Cocolonization of bacterial pathogens in a common ecosystem or infection nidus frequently results in a variety of physical and chemical interactions among species. In addition to biofilms, which encapsulate different bacterial pathogens in a dense matrix and protect the microcommunity from host immunity, antibiotic clearance, and other environmental stresses, bacteria also evolve capacities to resolve survival competition with their neighbors or coexist with them in the same ecological niche (4–9).
Pseudomonas aeruginosa and Staphylococcus aureus are the two most relevant bacterial pathogens in patients with chronic respiratory infections, especially the genetic disease cystic fibrosis (10–15). It is recognized that P. aeruginosa has a significant growth advantage over S. aureus because it secretes a series of antistaphylococcal metabolites that are controlled by the quorum-sensing (QS) system. However, S. aureus usually coexists with P. aeruginosa by producing small-colony variants (16–19). Remarkably, the frequent emergence and prevalence of QS-deficient P. aeruginosa isolates (typically lasR mutants), which lose the ability to kill S. aureus, enable persistent cocolonization by the two species in host lung or even facilitate the formation of mutually beneficial interactions (20). Furthermore, both the virulence and resistance of P. aeruginosa and S. aureus can be enhanced by the each other’s presence and thus worsen the outcomes of diseases (7, 14, 18, 21–24).
Antibiotic therapy is a routine method of treating bacterial infection. However, the heterogeneous diffusion of antibiotics in host tissues or the obstruction created by biofilms may reduce the functional concentration of antibiotics and enhance the tolerance of bacterial cells. In this case, the phenotype and intracellular transcription of coexisting bacterial species will also be changed by antibiotics at subinhibitory concentrations (7, 25–27). It is reported that although P. aeruginosa has an innate growth advantage over S. aureus in coculture, repeated antibiotic therapy frequently selects methicillin-resistant S. aureus (MRSA) in patients and causes the shift of the dominant species from P. aeruginosa to S. aureus (10, 19, 22, 28). Our prior work (29) further identified that the application of subinhibitory aminoglycoside antibiotics induced a large scale of transcriptional changes in P. aeruginosa and cocultured MRSA. Specifically, subinhibitory streptomycin inhibited the QS regulation of P. aeruginosa but increased the iron acquisition capacity of MRSA. As a consequence, P. aeruginosa and MRSA could coexist by forming a periodical change in frequencies along with the intermittent application of aminoglycosides (29).
We have also noticed that the growth of P. aeruginosa treated with subinhibitory cefotaxime (CTX) concentrations on a MRSA lawn produced a remarkably larger inhibitory zone than normal coculture (29). A previous study reported that later-generation cephalosporins, including cefotaxime, appear to enhance the QS-controlled virulence of P. aeruginosa (30). These observations indicated that the cephalosporin antibiotic cefotaxime may enhance the growth inhibition ability of P. aeruginosa on MRSA by promoting the expression of the QS system. Hence, in this study, we further explored the roles of CTX and another quinolone antibiotic commonly used against respiratory infections in the clinic, levofloxacin (LVX), on the interaction of coisolated P. aeruginosa and MRSA by using a series of phenotypic identification and RNA sequencing methods. We found that both CTX and LVX could select P. aeruginosa from the coculture with MRSA, although the functional mechanisms were different.
RESULTS
Influences of CTX and LVX on P. aeruginosa and MRSA each grown on a lawn of the other.
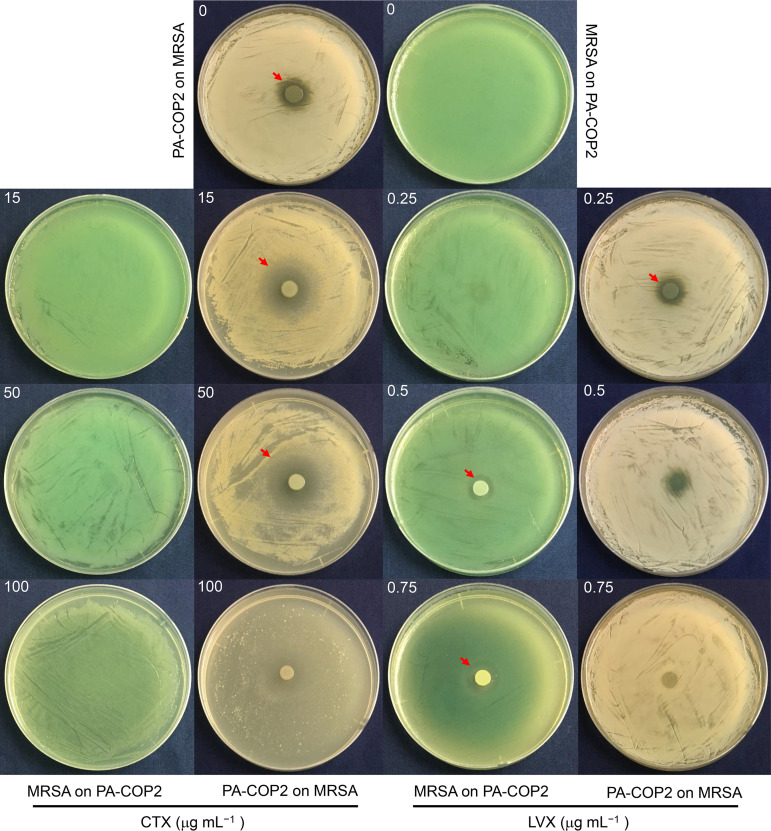
The influences of CTX and LVX on the interactions of P. aeruginosa and MRSA were first studied by coculturing each organism on a lawn of the other. P. aeruginosa PA-COP2 grew normally on an S. aureus MRSA-COP112 lawn on a blank lysogeny broth (LB) plate and produced a small inhibitory zone, while MRSA-COP112 could hardly grow on a PA-COP2 lawn (Fig. 1). Our prior work determined that the MICs of CTX on PA-COP2 and MRSA-COP112 were around 200 and 50 μg mL−1, and the MICs of LVX on these strains were 4 and >8 μg mL−1, respectively (29, 31). Here, we showed that PA-COP2 produced distinctly larger inhibitory zone on a MRSA-COP112 lawn on LB-CTX plates than on blank LB plates, while MRSA-COP112 failed to grow on a PA-COP2 lawn on plates containing any tested concentrations of CTX (Fig. 1). On the other hand, MRSA-COP112 began to grow on a PA-COP2 lawn when the concentration of LVX was 0.5 μg mL−1 and produced an inhibitory zone around the colony. Growth of PA-COP2 on a MRSA-COP112 lawn was gradually inhibited on the plates with increasing concentrations of LVX but left a distinct green dot or blank trace (Fig. 1). Moreover, although the resistance of P. aeruginosa reference strain PAO1 to CTX and LVX was weaker than that of PA-COP2 (31), outcomes of interaction between PAO1 and MRSA-COP112 were similar to those between PA-COP2 and MRSA-COP112 (see Fig. S1 in the supplemental material). These results preliminarily suggested that the intervention of subinhibitory CTX and LVX might have different effects on the interactions of P. aeruginosa and MRSA.
FIG 1.
Growth of PA-COP2 and MRSA-COP112 on lawns of the other on LB plates supplemented with different concentrations of cefotaxime (CTX) and levofloxacin (LVX). Equal amounts of PA-COP2 and MRSA-COP112 were each spotted on lawns of the other and cultured for 24 h. Images are representative of three independent replicates. Dishes were 9 cm in diameter. Red arrows indicate the edges of inhibitory zones.
Extracellular product-mediated interactions of P. aeruginosa and MRSA with CTX or LVX intervention.
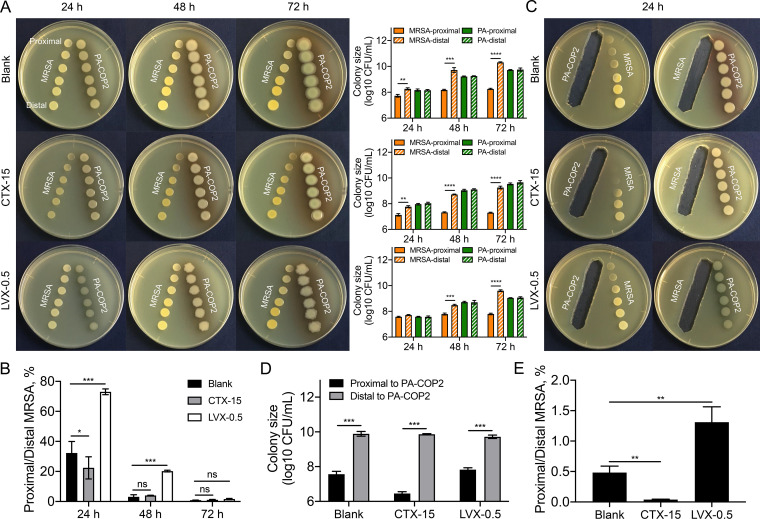
Previous studies have confirmed that the growth suppression of S. aureus by P. aeruginosa is mainly attributed to the extracellular metabolites of P. aeruginosa with antistaphylococcal activities (18, 19). In this study, we performed a series of pairwise proximity assays to investigate the extracellular-product-mediated interactions of P. aeruginosa and MRSA with the intervention of subinhibitory concentrations of CTX (15 μg mL−1) or LVX (0.5 μg mL−1). The growth of MRSA-COP112 was slowed by treatment with 15 μg mL−1 of CTX but was similar to that of the blank control on the plate containing 0.5 μg mL−1 of LVX (Fig. S2A). In contrast, the growth of PA-COP2 was slightly inhibited by 15 μg mL−1 of CTX or 0.5 μg mL−1 of LVX (Fig. S2B). The results of pairwise growth assays showed that compared to the distal colonies, the growth of proximal MRSA-COP112 on blank LB or LB-CTX plates was significantly suppressed by neighboring PA-COP2 after 24 h. In contrast, the growth of proximal MRSA-COP112 on LB-LVX plate was significantly inhibited by PA-COP2 after 48 h (Fig. 2A). To eliminate the killing effects of antibiotics on bacterial cells, the colony sizes of MRSA-COP2 at the proximal ends were normalized to those at the distal ends on the same plates. We found that CTX could significantly promote the growth suppression of MRSA-COP2 by PA-COP2 at 24 h, while LVX protected MRSA-COP112 from growth inhibition by PA-COP2 (Fig. 2B).
FIG 2.
Pairwise proximity assay of PA-COP2 and MRSA-COP112. (A) Pairwise growth of PA-COP2 and MRSA-COP112 on LB plates containing subinhibitory CTX (15 μg mL−1) or LVX (0.5 μg mL−1) concentrations for different time durations. The proximal and distal colonies of the two species were harvested for CFU enumeration. (B) Proximity of PA-COP2 and MRSA-COP112 to each other’s extracellular products on LB plates containing subinhibitory CTX (15 μg mL−1) or LVX (0.5 μg mL−1) concentrations for 24 h. Images are representative of three independent replicates. Dishes were 9 cm in diameter. (C) The colony sizes of MRSA-COP2 at the proximal ends were normalized (proximal CFU/distal CFU × 100) to those at the distal ends on the same plates (corresponding to panel A). (D) Colony sizes of MRSA-COP112 proximal and distal to the extracellular products of PA-COP2 on blank LB plates and LB plates containing 15 μg mL−1 of CTX or 0.5 μg mL−1 of LVX (corresponding to panel B). (E) The colony sizes of MRSA-COP2 at the proximal ends were normalized (proximal CFU/distal CFU × 100) to those at the distal ends on the same plates (corresponding to panel B). Data are means and standard deviations (SD) for three independent replicates and were compared by using two-tailed paired (A and D) or unpaired (C and E) t tests. *, P < 0.05; P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
We further verified the influences of CTX and LVX on the interaction of P. aeruginosa and MRSA by growing each on the extracellular products of the other. The results showed that the suppression of MRSA-COP112 growth by PA-COP2 extracellular products was significantly enhanced by the presence of CTX but alleviated by LVX (Fig. 2C to E). Similar outcomes were obtained for the interactions of MRSA-COP112 and PAO1 on LB plates containing 15 μg mL−1 of CTX or 0.25 μg mL−1 of LVX (Fig. S3 and S4). Therefore, these results demonstrated that the intervention of subinhibitory CTX might promote the selection of P. aeruginosa in the coculture with MRSA, while this process might be delayed by subinhibitory LVX concentrations.
Interaction dynamics of P. aeruginosa and MRSA with CTX or LVX intervention.
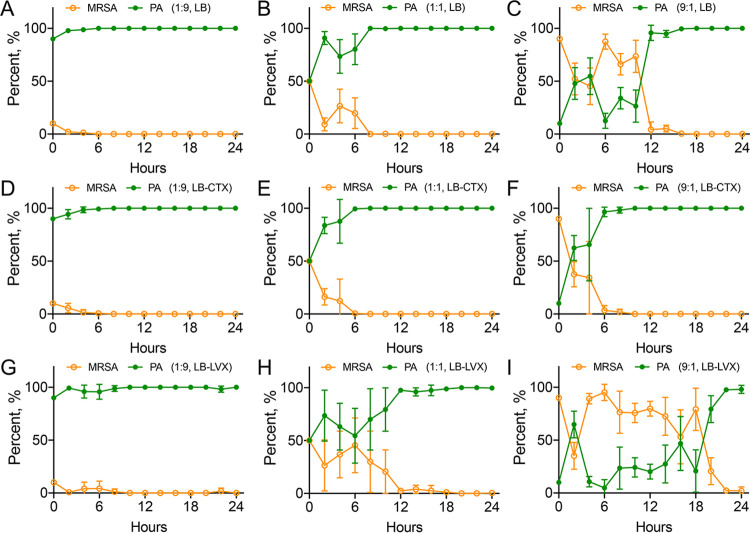
We then investigated the interaction dynamics of PA-COP2 and MRSA-COP112 by using the on-plate competition assay developed in our prior work (29). Bacterial cells of the two species were mixed in different ratios and cocultured on blank LB and plates containing 15 μg mL−1 of CTX or 0.5 μg mL−1 of LVX. On blank LB plates, PA-COP2 always defeated MRSA-COP112 and became the dominating species in cocultures that started with different ratios of them (Fig. 3A to C; Fig. S5A to C). As expected, the presence of subinhibitory CTX accelerated the decline of MRSA-COP112 population compared to those on blank LB plates (Fig. 3D to F; Fig. S5D to F). Specifically, it took PA-COP2 approximately 12 to 16 h to dominate the coculture started from a low population proportion (ratio of MRSA-COP112 to PA-COP2 = 9:1) on blank LB plates, while it took only 4 to 8 h on CTX plates (Fig. 3C and F; Fig. S5C and F). On the other hand, subinhibitory LVX prolonged the interacting time of PA-COP2 and MRSA-COP112 compared to blank LB plates (Fig. 3G to I; Fig. S5G to I), especially in the coculture started from low population proportion of PA-COP2 (ratio of MRSA-COP112 to PA-COP2 = 9:1). However, PA-COP2 was still the dominating species at the end of the competition. These results suggested that the presence of subinhibitory concentrations of either CTX or LVX would select P. aeruginosa in the mixed coculture with MRSA. However, this process could be accelerated by CTX but delayed by LVX.
FIG 3.
Interaction dynamics of cocultured PA-COP2 and MRSA-COP112. On-plate competition of PA-COP2 and MRSA-COP112 on blank LB plates (A to C) and LB plates containing subinhibitory concentrations of CTX (D to F) and LVX (G to I). Equal amounts of PA-COP2 and MRSA-COP112 were mixed at ratios of 9:1 (A, D, and G), 1:1 (B, E, and H), and 1:9 (C, F, and I) and cocultured for different times as described elsewhere (29). Bacterial cells from each time point were harvested for CFU enumeration and cell discrimination on blank LB and King’s B plates. Data are means and SD for six independent replicates.
CTX and LVX stimulate pyocyanin production in P. aeruginosa.
In the on-lawn competition experiments described above (Fig. 1), we noticed that PA-COP2 produced more pyocyanin pigment on CTX or LVX plates, even on the plates containing higher concentrations of antibiotics, and fewer bacterial cells. We verified this phenomenon by smearing pure PA-COP2 on blank LB plates and the plates containing CTX (15 μg mL−1) or LVX (0.5 μg mL−1). As shown in Fig. 4A, the CTX and LVX plates were greener than the blank. We then quantified the production of pyocyanin by culturing PA-COP2 in LB broth containing different concentrations of antibiotics. Although increasing the concentration of antibiotics significantly decreased the cell densities, PA-COP2 produced significantly larger amounts of pyocyanin than on the blank control, especially in LB-CTX medium (Fig. 4B and C). Similar results were obtained when the reference strain PAO1 was cultured in LB broth containing different concentrations of CTX or LVX (Fig. 4D and E). These results confirmed that the presence of subinhibitory CTX or LVX would promote the production of pyocyanin in P. aeruginosa.
FIG 4.
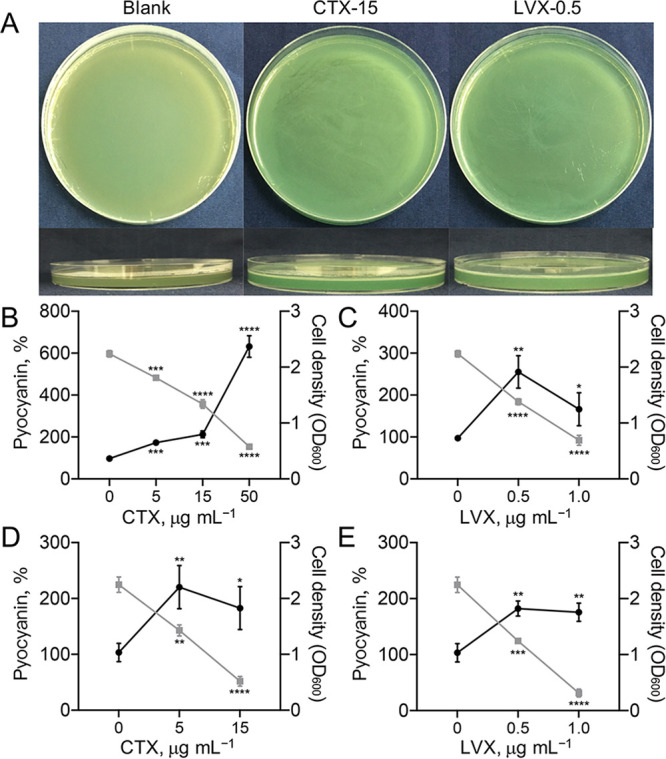
Subinhibitory CTX and LVX concentrations promote the production of pyocyanin in Pseudomonas aeruginosa. (A) Colors of PA-COP2 on blank LB, LB-CTX (15 μg mL−1), and LB-LVX (0.5 μg mL−1) plates. Images are representative of three independent replicates. (B to E) Percent pyocyanin levels of PA-COP2 (B and C) and P. aeruginosa PAO1 (D and E) in LB broth containing different concentrations of CTX (B and D) and LVX (C and E). Data are means and SD for three independent replicates and were compared by using two-tailed unpaired t tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Black dots indicate pyocyanin level as a percentage of the blank control value; gray squares indicate cell density.
Influence of CTX or LVX on P. aeruginosa transcriptional profile.
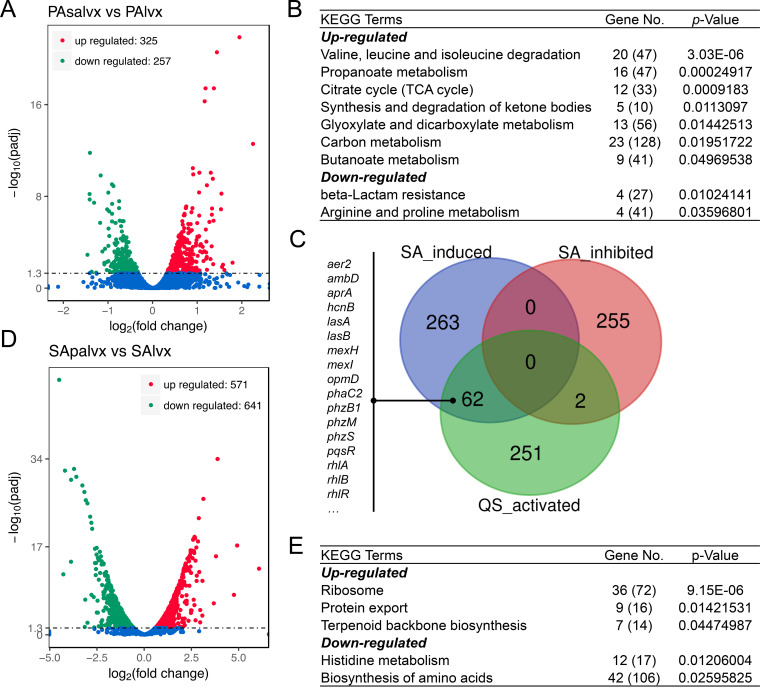
We then profiled the transcriptional change of PA-COP2 with CTX or LVX intervention by using RNA sequencing (RNA-seq). The results showed that compared to the control, a subinhibitory CTX concentration (15 μg mL−1) upregulated the expression of 275 genes (enriched in ribosome, citrate cycle, starch, and sucrose metabolism, carbon metabolism, and fructose and mannose metabolism) and downregulated the expression of 115 genes (enriched in sulfur metabolism, oxidative phosphorylation, beta-lactam resistance, ribosome, and ABC transporters) of PA-COP2 (Fig. S6A and B and Data Set S1). In contrast, subinhibitory LVX (0.5 μg mL−1) caused more extensive transcriptional changes in PA-COP2. Specifically, the 1,199 upregulated genes were significantly enriched in sulfur metabolism, purine metabolism, riboflavin metabolism, and RNA degradation, while the 1,288 downregulated genes were enriched in valine, leucine, and isoleucine degradation, bacterial chemotaxis, starch and sucrose metabolism, biosynthesis of siderophore group nonribosomal peptides, propanoate metabolism, synthesis and degradation of ketone bodies, and tryptophan metabolism (Fig. S6C and D and Data Set S2).
Influence of CTX or LVX on P. aeruginosa QS regulation.
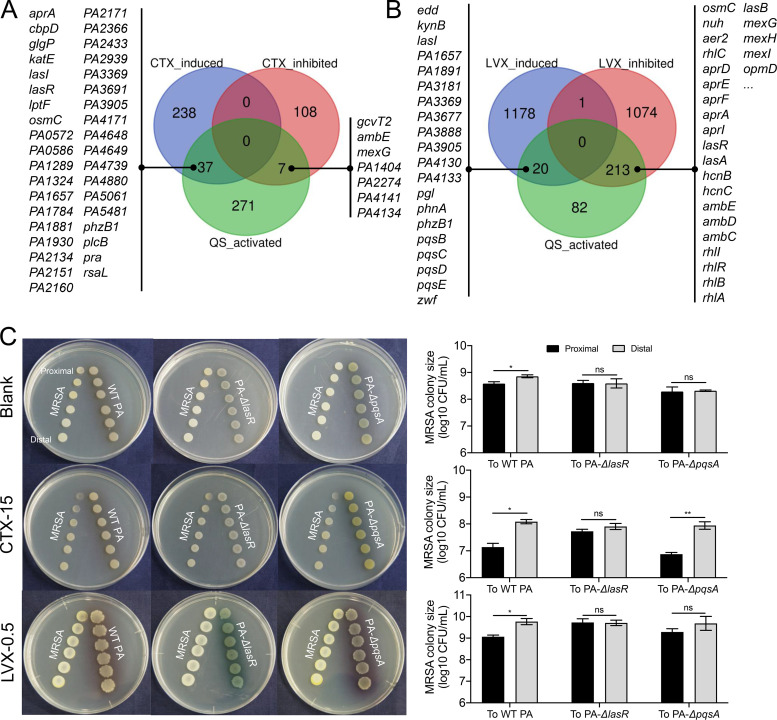
The production of pyocyanin is a complex process positively controlled by P. aeruginosa QS circuitry (32). To further explore the effect of subinhibitory CTX or LVX on the QS regulation of PA-COP2, the significantly differentially expressed genes identified by RNA-seq were mapped to the list of QS-activated genes in P. aeruginosa reported by Schuster et al. (33). The result showed that subinhibitory CTX upregulated the expression of 37 genes (including lasR and phzB1) and downregulated 7 genes activated by the QS system (Fig. 5A; Fig. S7). In PA-COP2 treated with subinhibitory LVX concentrations, 20 QS-activated genes, including the pqs operon and phzB1, were identified among the upregulated genes, while 213 QS-activated genes (315 in total), including the core regulatory genes lasR and rhlR and the majority of the typical downstream functional genes, were all downregulated (Fig. 5B; Fig. S7). These data suggested that subinhibitory LVX could decrease the expression of lasR and rhlR and their downstream genes but activate pqs signaling in P. aeruginosa.
FIG 5.
Influence of subinhibitory CTX and LVX concentrations on QS regulation in PA-COP2. (A and B) Effect of subinhibitory concentrations of CTX (A) and LVX (B) on the expression of QS-regulated genes in PA-COP2. The genes that were significantly differentially expressed in PA-COP2 treated with CTX (15 μg mL−1) and LVX (0.5 μg mL−1) were mapped to the list of QS-activated genes (n = 315) released by Schuster et al. (33). (C) Pairwise growth of MRSA-COP112 with PA-COP2, PA-ΔlasR, or PA-ΔpqsA on blank LB and LB-CTX plates for 24 h and on LB-LVX plates for 48 h. Images are representative of three independent replicates. Data are means and SD for three independent replicates and were compared by using a two-tailed paired t test. *, P < 0.05; **, P < 0.01; ns, not significant.
We then verified the roles of las and pqs QS systems in the interaction of PA-COP2 with MRSA-COP112 by performing a batch of pairwise proximity assays. The results showed that knockout of lasR or pqsA abolished the growth inhibition ability of PA-COP2 on MRSA-COP112 on blank LB plates (Fig. 5C). The presence of subinhibitory CTX failed to promote the growth inhibition of PA-ΔlasR on MRSA-COP112 but restored the ability of PA-ΔpqsA to inhibit the growth of MRSA-COP112 (Fig. 5C). This suggested that lasR might be the target of CTX in promoting the growth inhibition of PA-COP2 on MRSA-COP112. On the other side, MRSA-COP112 grew normally in proximity to wild-type (WT) PA-COP2, PA-ΔlasR, or PA-ΔpqsA on LB-LVX plates after 24 h but was significantly inhibited only by WT PA-COP2 after 48 h (Fig. 5C; Fig. S8). We further tested the influence of LVX (0.25 μg mL−1) on the QS regulation of PAO1 by using RNA-seq. We found that subinhibitory LVX significantly upregulated the expression of 475 genes and downregulated 294 genes in PAO1 (Fig. S9A and Data Set S3). After these differentially expressed genes were mapped to the list of QS activated genes, 8 upregulated genes (including pqsD in the pqs operon) and 74 downregulated genes (including lasR and rhlR and their typical downstream genes), were identified (Fig. S9B). Therefore, these results collectively indicated that the las and pqs QS systems played an important role in the competition of P. aeruginosa and MRSA with CTX or LVX intervention. Subinhibitory LVX inhibited the lasR regulon but might activate pqs signaling in P. aeruginosa to defeat MRSA during further culture.
Influence of LVX on the transcriptional patterns of interacting P. aeruginosa and MRSA.
RNA-seq was then conducted to profile the transcriptional changes of PA-COP2 and MRSA-COP112 during interspecific interaction. In the colony of PA-COP2 grown on LB-LVX plate (Fig. 1), the presence of a MRSA-COP112 lawn upregulated the expression of 325 genes and downregulated 257 genes in PA-COP2 (Fig. 6A; Data Set S4). The upregulated genes of PA-COP2 were mainly enriched in valine, leucine, and isoleucine degradation, propanoate metabolism, citrate cycle, synthesis and degradation of ketone bodies, etc. The downregulated genes were mainly enriched in beta-lactam resistance and arginine and proline metabolism (Fig. 6B). Intriguingly, when the differentially expressed genes of PA-COP2 on LB-LVX plates with a MRSA-COP112 lawn were mapped to a list of QS-activated genes, we found that 62 QS-activated genes, including the pyocyanin synthesis genes phzM, phzS, and phzB1, the regulatory genes rhlR and pqsR, and the typical QS-controlled downstream genes in PA-COP2, were upregulated by the presence of MRSA-COP112 (Fig. 6C). These results revealed that although subinhibitory LVX suppressed the las-QS regulation of P. aeruginosa (Fig. 5B), the presence of MRSA might still induce P. aeruginosa to produce QS-controlled extracellular products.
FIG 6.
Influence of subinhibitory LVX concentrations on the transcriptional pattern of interacting PA-COP2 and MRSA-COP112. Cells of PA-COP2 and MRSA-COP112 each cultured on lawns of the other on LVX plates (0.5 μg mL−1) were harvested at 24 h and subjected to RNA-seq. (A to C) Volcano plots (A), significantly enriched KEGG terms (B), and expression of QS-activated genes (C) for the differentially expressed genes in PA-COP2 cultured on MRSA-COP112 lawns on LVX plates compared to LVX-treated PA-COP2. (D and E) Volcano plots (D) and significantly enriched KEGG terms (E) for the differentially expressed genes in MRSA-COP112 cultured on PA-COP2 lawns on LVX plates compared to LVX-treated MRSA-COP112. In panels B and E, gene number is the number of differentially expressed genes relating to the pathway, while the number in parentheses is the total number of genes relating to the pathway.
On the other hand, compared to the low impact (3 upregulated genes and 1 downregulated) of LVX (0.5 μg mL−1) on the transcription of MRSA-COP112 (Fig. S10), the presence of PA-COP2 lawn upregulated 571 genes and downregulated 641 genes in MRSA-COP112 on LB-LVX plates (Fig. 6D; Data Set S5). The upregulated genes of MRSA-COP112 were mainly enriched in ribosome, protein export, and terpenoid backbone biosynthesis. The downregulated genes were mainly enriched in histidine metabolism and biosynthesis of amino acids (Fig. 6E). Therefore, these results confirmed that P. aeruginosa and MRSA could remarkably influence each other’s transcriptional network in an LVX background and emphasized the central role of the P. aeruginosa QS system in the complicated interactions of the two species.
DISCUSSION
It is well known that P. aeruginosa has an innate competitive advantage over S. aureus by virtue of its QS system, which dominates the production of various antistaphylococcal metabolites (17–19). The emergence and enrichment of P. aeruginosa lasR mutants contribute to the coexistence of P. aeruginosa with S. aureus or other bacterial pathogens (7, 19, 20). In consideration of the nonuniform distribution of antibiotics in host tissues or bacterial community, we focused on studying the effects of commonly used antibiotics on bacterial interspecific interactions at subinhibitory concentrations. In this study, we identified the influences of CTX and LVX on the interactions of P. aeruginosa and S. aureus and found that subinhibitory CTX or LVX influences the P. aeruginosa QS system and selects P. aeruginosa from the coculture with S. aureus.
A previous study by Okada et al. (30) tested the impact of cephalosporin antibiotics from each generation on P. aeruginosa QS regulation by using several indicator genes positively controlled by QS. They found that the expression levels of QS-activated genes increased with the later generations of cephalosporins (30). Our present study reports that the third-generation cephalosporin CTX could promote pyocyanin production and upregulate a portion of the genes controlled by the las QS system of P. aeruginosa (Fig. 4 and 5A). This might be plausibly explained by the result of molecular docking, which predicts that CTX can correctly bind only to LasR with lower Gibbs free energy but fewer active sites than the native signal 3-oxo-C12-homoserin lactone (HSL) (Fig. S11 to S13). In any case, our present study demonstrates that subinhibitory CTX could significantly promote the growth suppression of MRSA by P. aeruginosa with an intact lasR regulon (Fig. 2 and 5C) and thus emphasizes the important role of the las QS system in mediating the accelerated selection of P. aeruginosa from the coculture with MRSA.
Our prior work revealed that the aminoglycoside antibiotic streptomycin could suppress the QS system and reduce the production of pyocyanin in P. aeruginosa (29). In the present study, we found that the quinolone antibiotic LVX could also downregulate the las and rhl QS systems of P. aeruginosa. However, the pqs QS system and pyocyanin production of P. aeruginosa were promoted by subinhibitory LVX (Fig. 4 and 5B; Fig. S7). This might be explained by the fixed binding of LVX to two active sites of LasR in a form different from that of the native signal (Fig. S11 to S13), which thus antagonistically inhibited the expression of the las and rhl QS system, since LasR sits on top of RhlR in the QS hierarchy (34). The increased pqs signaling might be related to fact that the binding of LVX to PqsR is similar to that of the native Pseudomonas quinolone signal (PQS), which can activate the expression of partial genes controlled by the las and rhl QS systems (32). Correspondingly, although subinhibitory LVX concentrations delayed the growth suppression of MRSA-COP112 by PA-COP2 and prolonged the competition process compared to blank LB plates, PA-COP2 was always the final dominating species in cocultures started with any species ratio (Fig. 2 and 3G to I). These results are also supported by the observation that the presence of LVX failed to restore the growth inhibition ability of PA-COP2 pqsA mutant on MRSA-COP112 (Fig. 5C). In general, the pqs QS system is considered a backup for the las QS system and contributes to the extended survival of P. aeruginosa population in host tissues (32, 35). Clinical evidence showed that although P. aeruginosa isolates with spontaneous loss-of-function mutations in lasR were frequently detected in sputum samples, hardly any pqsR mutants have been reported (36–38). Some environmental cues, such as iron level, can also activate pqs signaling in a lasR-independent pathway (39, 40). Therefore, our present study further expands the functional aspect of P. aeruginosa pqs signaling to bacterial interspecific interaction by using antibiotics as environmental cues.
Moreover, compared to the inhibited growth of MRSA-COP112 on a PA-COP2 lawn on the plates containing low concentrations of LVX (≤0.25 μg mL−1), MRSA-COP112 could grow on a PA-COP2 lawn and produce an inhibitory zone around the colony when the LVX concentration was higher than 0.5 μg mL−1 (Fig. 1). However, growth suppression of PA-COP2 by MRSA-COP112 was not observed in the extracellular-product-mediated pairwise proximity assay (Fig. 2B). After checking the in situ transcriptions of interacting PA-COP2 and MRSA-COP112 colonies, we did not identify any specific genes which might contribute to the suppression of PA-COP2 by MRSA-COP112 (Fig. 6; Data Sets S4 and S5). These results indicated that the mutual inhibition of PA-COP2 and MRSA-COP112 determined by LVX concentration in the on-lawn coculture assays is different from that on LB-streptomycin plates. Our previous study showed that PA-COP2 and MRSA-COP112 were capable of inhibiting each other on LB plates containing the same concentration of streptomycin, and the inhibition of MRSA-COP112 on PA-COP2 was associated with their differentiated capacities in iron acquisition (29). On the other hand, we found that in comparison to the significantly downregulated las QS system of PA-COP2 by subinhibitory LVX (Fig. 5B), the presence of a MRSA-COP112 lawn induced higher expression levels of QS-controlled genes in PA-COP2 than in that treated solely with LVX (Fig. 6C). This is in agreement with previous findings that the presence of S. aureus could enhance the QS-related virulence of P. aeruginosa (24). These results collectively revealed the complicated interaction of P. aeruginosa and MRSA with LVX intervention, and the interaction process might be associated with the bactericidal effect of LVX and the influences of LVX on the intracellular transcription of the two species. It is believed that further genetic characterizations of P. aeruginosa and S. aureus will help identify the detailed mechanism of their interaction.
In conclusion, the present study characterized the dynamics of P. aeruginosa and S. aureus interactions in the presence of the commonly used antibiotics CTX and LVX. Both antibiotics are beneficial to the fitness of P. aeruginosa in coculture with S. aureus, although the underlying functional mechanisms are different. The enhanced ability of P. aeruginosa to inhibit growth of S. aureus on CTX plates is partially related to the induction of the las QS system in P. aeruginosa. In contrast, LVX inhibits the las and rhl QS systems of P. aeruginosa but activates pqs signaling to overcome S. aureus. Therefore, this study provides an explanation for the enrichment of P. aeruginosa in the lungs of patients with polymicrobial colonization after antibiotic therapy and lays an important basis for further studying the complicated interactions and pathogenesis of bacterial individuals in the community.
MATERIALS AND METHODS
Bacterial strains and medium.
The clinical P. aeruginosa strain COP2 (PA-COP2), the isogenic lasR mutant (PA-ΔlasR), the pqsA mutant (PA-ΔpqsA), the coisolated S. aureus strain MRSA-COP112 from the sputum of a patient with chronic obstructive pulmonary disease (COPD), and the P. aeruginosa reference strain PAO1 were preserved in the laboratory and are described elsewhere (19, 29, 31). Single colonies of all the strains were cultured in LB broth overnight (16 to 18 h) at 37°C. Bacterial cells were then harvested and adjusted to an optical density at 600 nm (OD600) of 1.0 in 1.0 mL of sterile saline for use.
Culture conditions.
For the on-plate inhibition assay, 100 μL of P. aeruginosa or S. aureus was smeared on LB plates containing different concentrations of cefotaxime (CTX) or levofloxacin (LVX) (Sigma-Aldrich). After the plate surface of one species was naturally dried, 5 μL of the other species was spotted on the center of the plate and statically cultured for 24 h at 37°C.
For the pairwise proximity assay, equal amounts (5 μL) of P. aeruginosa and S. aureus were successively spotted on the two sides of blank LB plates and plates containing 15 μg mL−1 of CTX or 0.5 μg mL−1 of LVX. The distance between spots of the same species was set as 0.5 cm, while the distance between different species ranged from 0.5 to 3.0 cm. After static culture for 24 h, 48 h, and 72 h at 37°C, the colony sizes at the proximal and distal ends were determined by enumerating the CFU.
For the extracellular-product-mediated interspecific interaction, inoculation was similar to that for the pairwise proximity assay. However, one species (5 μL) was first spotted on one side of the plates and statically cultured for 24 h at 37°C. Subsequently, all the colonies were removed with a sterile blade, and then the other species was successively spotted on the other side at increasing distances from the spaces occupied by the removed species. All the experiments described above were independently repeated three times.
On-plate competition assay.
The influence of subinhibitory concentrations of CTX or LVX on the interaction of PA-COP2 and MRSA-COP112 was studied by using on-plate competition assay as described previously (29). Briefly, sterile plastic tubes (1.0 cm in diameter, 1.0 cm in length) were inserted into the center of blank LB plates and the plates containing 15 μg mL−1 of CTX or 0.5 μg mL−1 of LVX during plate preparation. Mixtures (1:9, 1:1, and 9:1) of PA-COP2 and MRSA-COP112 were spotted (5 μL) on the inner region of tubes and statically cultured at 37°C for different times. In this way, the growth of a PA-MRSA mixed colony will not spread around on the plate and the interaction of the two species can be limited in a fixed region. The colony sizes and proportions of PA-COP2 and MRSA-COP112 were monitored by enumerating the CFU on blank LB and King’s B plates. All the experiments were independently repeated six times.
Pyocyanin production assay.
The influence of subinhibitory CTX or LVX concentrations on the production of pyocyanin by PA-COP2 was determined by using the method described by Essar et al. (41). Briefly, 10 μL of PA-COP2 was inoculated into 2.0 mL of LB broth containing different concentrations of CTX or LVX and statically cultured at 37°C for 24 h. Pyocyanin was extracted from the supernatants by using chloroform and 0.2 N HCl, followed by measuring the OD520.
Transcriptomic analysis.
PA-COP2 and MRSA-COP112 were spotted (5 μL) on CTX (15 μg mL−1) and LVX (0.5 μg mL−1) plates, respectively. PA-COP2 and MRSA-COP112 were each spotted (5 μL) on lawns of the other on CTX and LVX plates as described for the on-plate inhibition assay above. All the bacterial colonies from each experiment were harvested after 24 h and subjected to total RNA isolation using the Cell Total RNA isolation kit with genomic DNA (gDNA) removal (Forgene, Chengdu, China). RNA samples from two parallel experiments with at least three independent biological replicates were used for library construction. Prokaryotic strand-specific RNA-seq was performed on a HiSeq 2500 platform by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). Bowtie2-2.2.3 (42) and HTSeq v0.6.1 (43) were used to map the read data to the genomes of P. aeruginosa PAO1 (NCBI accession number AE004091.2) and S. aureus USA300 (NCBI accession number CP000255.1) and assemble the transcriptome, respectively. The values of differential gene expression were calculated by DESeq 2 (44) using expected fragments per kilobase of transcript per million fragments, and an adjusted P value (Padj) of <0.05 was set as the criterion. Enrichment of KEGG and GO terms among the differentially expressed genes were performed by using clusterProfiler 4.0 (45). Finally, Venn Diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to mapped the differentially expressed genes of P. aeruginosa to the list of QS-activated genes established by Schuster et al. (33).
Molecular docking.
The software AutoDock 4.2.6 (https://autodock.scripps.edu/download-autodock4/) and AutoDockTools (https://ccsb.scripps.edu/mgltools/) were used to predict the interactions of CTX and LVX to the three main QS regulatory proteins of P. aeruginosa. The pdb files containing the crystal structures of CTX, LVX, LasR, RhlR, and PqsR were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov). The docking of ligands and proteins was performed according to the manufacturer’s guidelines.
Statistical analyses.
Data analysis and statistical tests were performed by using GraphPad Prism version 9.0 (San Diego, CA, USA). Mean values and standard deviations were compared by using a two-tailed paired or unpaired t test or one-way analysis of variance (ANOVA).
Data availability.
The data from this study are presented in the article and supplemental material. The raw RNA sequencing data are available from the NCBI database under accession number PRJNA600167.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31970131, 81922042, and 82172285), the Sichuan Science and Technology Program (2021JDJQ0042), and the 1·3·5 project of excellent development of discipline of West China Hospital of Sichuan University (ZYYC21001). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no conflict of interest exists.
K.Z., X.Z., and Y.C. designed research; K.Z., J.L., X.Y., Q.Z., and Y.W. performed research; W.L., H.Z., B.P., X.W., and X.Z. contributed new reagents/analytic tools; K.Z., J.L., and Y.C. analyzed data; and K.Z. wrote the paper. All authors contributed to the article and approved the submitted version.
Footnotes
Supplemental material is available online only.
Contributor Information
Xikun Zhou, Email: xikunzhou@scu.edu.cn.
Yiwen Chu, Email: chuyiwen@cdu.edu.cn.
Maia Kivisaar, University of Tartu.
REFERENCES
- 1.Brogden KA, Guthmiller JM, Taylor CE. 2005. Human polymicrobial infections. Lancet 365:253–255. 10.1016/S0140-6736(05)17745-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team . 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pragman AA, Berger JP, Williams BJ. 2016. Understanding persistent bacterial lung infections: clinical implications informed by the biology of the microbiota and biofilms. Clin Pulm Med 23:57–66. 10.1097/CPM.0000000000000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Little AE, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. 2008. Rules of engagement: interspecies interactions that regulate. microbial communities. Annu Rev Microbiol 62:375–401. 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 5.Cornforth DM, Foster KR. 2013. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol 11:285–293. 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 6.Short FL, Murdoch SL, Ryan RP. 2014. Polybacterial human disease: the ills of social networking. Trends Microbiol 22:508–516. 10.1016/j.tim.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orazi G, O’Toole GA. 2019. '“It takes a village”: mechanisms underlying antimicrobial recalcitrance of polymicrobial biofilms. J Bacteriol 202:e00530-19. 10.1128/JB.00530-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buch PJ, Chai Y, Goluch ED. 2019. Treating polymicrobial infections in chronic diabetic wounds. Clin Microbiol Rev 32:e00091-18. 10.1128/CMR.00091-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vandeplassche E, Tavernier S, Coenye T, Crabbé A. 2019. Influence of the lung microbiome on antibiotic susceptibility of cystic fibrosis pathogens. Eur Respir Rev 28:190041. 10.1183/16000617.0041-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahlgren HG, Benedetti A, Landry JS, Bernier J, Matouk E, Radzioch D, Lands LC, Rousseau S, Nguyen D. 2015. Clinical outcomes associated with Staphylococcus aureus and Pseudomonas aeruginosa airway infections in adult cystic fibrosis patients. BMC Pulm Med 15:67. 10.1186/s12890-015-0062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yonker LM, Cigana C, Hurley BP, Bragonzi A. 2015. Host-pathogen interplay in the respiratory environment of cystic fibrosis. J Cyst Fibros 14:431–439. 10.1016/j.jcf.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maliniak ML, Stecenko AA, McCarty NA. 2016. A longitudinal analysis of chronic MRSA and Pseudomonas aeruginosa co-infection in cystic fibrosis: a single-center study. J Cyst Fibros 15:350–356. 10.1016/j.jcf.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Cystic Fibrosis Foundation. 2017. Patient registry 2016 annual data report. Cystic Fibrosis Foundation, Bethesda, MD. [Google Scholar]
- 14.Limoli DH, Hoffman LR. 2019. Help, hinder, hide and harm: what can we learn from the interactions between Pseudomonas aeruginosa and Staphylococcus aureus during respiratory infections? Thorax 74:684–692. 10.1136/thoraxjnl-2018-212616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Song W, Chen X, Gong P. 2019. Analysis of pathogens and drug resistance in patients with community acquired lower respiratory tract infection. Chinese Foreign Med Res 17:172–173. [Google Scholar]
- 16.Hoffman LR, Déziel E, D'Argenio DA, Lépine F, Emerson J, McNamara S, Gibson RL, Ramsey BW, Miller SI. 2006. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 103:19890–19895. 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biswas L, Biswas R, Schlag M, Bertram R, Götz F. 2009. Small-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa. Appl Environ Microbiol 75:6910–6912. 10.1128/AEM.01211-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hotterbeekx A, Kumar-Singh S, Goossens H, Malhotra-Kumar S. 2017. In vivo and in vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front Cell Infect Microbiol 7:106. 10.3389/fcimb.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao K, Du L, Lin J, Yuan Y, Wang X, Yue B, Wang X, Guo Y, Chu Y, Zhou Y. 2018. Pseudomonas aeruginosa quorum-sensing and type VI secretion system can direct interspecific coexistence during evolution. Front Microbiol 9:2287. 10.3389/fmicb.2018.02287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frydenlund Michelsen C, Hossein Khademi SM, Krogh Johansen H, Ingmer H, Dorrestein PC, Jelsbak L. 2016. Evolution of metabolic divergence in Pseudomonas aeruginosa during long-term infection facilitates a proto-cooperative interspecies interaction. ISME J 10:1323–1336. 10.1038/ismej.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirketerp-Møller K, Jensen PØ, Fazli M, Madsen KG, Pedersen J, Moser C, Tolker-Nielsen T, Høiby N, Givskov M, Bjarnsholt T. 2008. Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol 46:2717–2722. 10.1128/JCM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sagel SD, Gibson RL, Emerson J, McNamara S, Burns JL, Wagener JS, Ramsey BW, Inhaled Tobramycin in Young Children Study Group; Cystic Fibrosis Foundation Therapeutics Development Network . 2009. Impact of Pseudomonas and Staphylococcus infection on inflammation and clinical status in young children with cystic fibrosis. J Pediatr 154:183–188. 10.1016/j.jpeds.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seth AK, Geringer MR, Galiano RD, Leung KP, Mustoe TA, Hong SJ. 2012. Quantitative comparison and analysis of species-specific wound biofilm virulence using an in vivo, rabbit-ear model. J Am Coll Surg 215:388–399. 10.1016/j.jamcollsurg.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 24.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. 2013. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci USA 110:1059–1064. 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skindersoe ME, Alhede M, Phipps R, Yang L, Jensen PO, Rasmussen TB, Bjarnsholt T, Tolker-Nielsen T, Høiby N, Givskov M. 2008. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:3648–3663. 10.1128/AAC.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jo A, Ahn J. 2016. Phenotypic and genotypic characterisation of multiple antibiotic-resistant Staphylococcus aureus exposed to subinhibitory levels of oxacillin and levofloxacin. BMC Microbiol 16:170. 10.1186/s12866-016-0791-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, Zeng J, Wang L, Lan KES, Wang L, Xiao Q, Luo Q, Huang X, Huang B, Chen C. 2017. Antibiotics promote Escherichia coli-Pseudomonas aeruginosa conjugation through inhibiting quorum sensing. Antimicrob Agents Chemother 61:e01284-17. 10.1128/AAC.01284-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubert D, Réglier-Poupet H, Sermet-Gaudelus I, Ferroni A, Le Bourgeois M, Burgel P-R, Serreau R, Dusser D, Poyart C, Coste J. 2013. Association between Staphylococcus aureus alone or combined with Pseudomonas aeruginosa and the clinical condition of patients with cystic fibrosis. J Cyst Fibros 12:497–503. 10.1016/j.jcf.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Chen X, Lin J, Yuan Y, Huang T, Du L, Prithiviraj B, Zhang A, Wang X, Chu Y, Zhao K. 2021. Antibiotic intervention redisposes bacterial interspecific interacting dynamics in competitive environments. Environ Microbiol 23:7432–7444. 10.1111/1462-2920.15461. [DOI] [PubMed] [Google Scholar]
- 30.Okada BK, Li A, Seyedsayamdost MR. 2019. Identification of the hypertension drug guanfacine as an antivirulence agent in Pseudomonas aeruginosa. Chembiochem 20:2005–2011. 10.1002/cbic.201900129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao K, Yuan Y, Li J, Pan W, Yan C, Fu H, Lin J, Yue B, Wang X, Gou X, Chu Y, Zhou Y. 2019. Phenotypic and genetic characterization of Pseudomonas aeruginosa isolate COP2 from the lungs of COPD patients in China. Pathog Dis 77:ftz038. 10.1093/femspd/ftz038. [DOI] [PubMed] [Google Scholar]
- 32.Déziel E, Gopalan S, Tampakaki AP, Lépine F, Padfield KE, Saucier M, Xiao G, Rahme LG. 2005. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol 55:998–1014. 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- 33.Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185:2066–2079. 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balasubramanian D, Schneper L, Kumari H, Mathee K. 2013. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res 41:1–20. 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen R, Déziel E, Groleau M-C, Schaefer AL, Greenberg EP. 2019. Social cheating in a Pseudomonas aeruginosa quorum-sensing variant. Proc Natl Acad Sci USA 116:7021–7026. 10.1073/pnas.1819801116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA 103:8487–8492. 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L, Jelsbak L, Marvig RL, Damkiær S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Høiby N, Sommer MOA, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci USA 108:7481–7486. 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao K, Huang T, Lin J, Yan C, Du L, Song T, Li J, Guo Y, Chu Y, Deng J, Wang X, Liu C, Zhou Y. 2020. Genetic and functional diversity of Pseudomonas aeruginosa in patients with chronic obstructive pulmonary disease. Front Microbiol 11:598478. 10.3389/fmicb.2020.598478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diggle SP, Winzer K, Chhabra SR, Worrall KE, Cámara M, Williams P. 2003. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol 50:29–43. 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- 40.Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Cámara M, Williams P. 2007. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol 14:87–96. 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 41.Essar DW, Eberly L, Hadero A, Crawford IP. 1990. and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol 172:884–900. 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. 2021. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2:100141. 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download aem.00592-22-s0002.xlsx, XLSX file, 0.05 MB (47.2KB, xlsx)
Data Set S2. Download aem.00592-22-s0003.xlsx, XLSX file, 0.2 MB (235.1KB, xlsx)
Data Set S3. Download aem.00592-22-s0004.xlsx, XLSX file, 0.08 MB (80.4KB, xlsx)
Data Set S4. Download aem.00592-22-s0005.xlsx, XLSX file, 0.06 MB (64.7KB, xlsx)
Data Set S5. Download aem.00592-22-s0006.xlsx, XLSX file, 0.1 MB (123.5KB, xlsx)
Fig. S1 to S13 and descriptions of Data Sets S1 to S5. Download aem.00592-22-s0001.pdf, PDF file, 7.3 MB (7.3MB, pdf)
Data Availability Statement
The data from this study are presented in the article and supplemental material. The raw RNA sequencing data are available from the NCBI database under accession number PRJNA600167.