ABSTRACT
Purpose:
Uveal melanoma (UM) is the most common intraocular malignant tumor in adults. Extracellular vesicles (EVs) have been extensively studied as a biomarker to monitor disease in patients. The study of new biomarkers in melanoma patients could prevent metastasis by earlier diagnosis. In this study, we determined the proteomic profile of EVs isolated from aqueous humor (AH), vitreous humor (VH), and plasma from UM patients in comparison with cancer-free control patients.
Methods:
AH, VH and plasma were collected from seven patients with UM after enucleation; AH and plasma were collected from seven cancer-free patients with cataract (CAT; control group). EVs were isolated using the membrane-based affinity binding column method. Nanoparticle tracking analysis (NTA) was performed to determine the size and concentration of EVs. EV markers, CD63 and TSG101, were assessed by immunoblotting, and the EV proteome was characterized by mass spectrometry.
Results:
Mean EV concentration was higher in all analytes of UM patients compared to those in the CAT group. In the UM cohort, the mean concentration of EVs was significantly lower in AH and plasma than in VH. In contrast, the mean size and size distribution of EVs was invariably identical in all analyzed analytes and in both studied groups (UM vs. CAT). Mass spectrometry analyses from the different analytes from UM patients showed the presence of EV markers.
Conclusion:
EVs isolated from AH, VH, and plasma from patients with UM showed consistent profiles and support the use of blood to monitor UM patients as a noninvasive liquid biopsy.
Keywords: Aqueous humor, Extracellular vesicles, Liquid biopsy, Plasma, Proteomic analysis, Uveal melanoma, Vitreous humor
Introduction
Uveal melanoma (UM) is a primary intraocular tumor in adults that accounts for less than 5% of all melanoma cases (1,2). The incidence of UM has remained stable at ~5.1 per million since the 1970s with subtle differences depending on geographic location, as well as environmental and occupational factors (1). Despite excellent control of local disease, prognosis remains poor due to metastatic progression affecting ~50% of patients (2-4). Mortality rates for UM are unchanged over the past decades (1).
Extracellular vesicles (EVs) have emerged as biomarkers in various cancers and provide valuable clinical information (5,6). Their use as a biomarker assay has gained interest as a new tool for monitoring of cancer patients; however, standardization and validation of EVs as a biomarker are needed (4,7).
EVs are small lipid bilayer particles released from all types of cells and found in different body fluids, most commonly the blood, but have also been detected in aqueous humor (AH) (8,9). EVs are classified mainly into exosomes (50-100 nm), microvesicles (100-1000 nm), and apoptotic bodies (50 nm-2 µm) based on their biogenesis, number, size, distinct biological functions and markers (10-16). Exosomes are a constitutive and abundant component of the vitreous (17).
EVs are involved in the transfer of biological macromolecules to recipient cells, and modulating various physiological and pathological processes, such as pathogen dissemination and regulation of the host immune system (18-20). Recent studies have shown that tumor cells release large amounts of EVs that can be uptaken by malignant and stromal cells, inducing tumor progression (21,22). They have been shown to play a major role in mediating metastasis, ranging from oncogenic reprogramming of malignant cells to the formation of pre-metastatic niches (23-25). Furthermore, our group and others have shown that cancer-derived exosomes can transfer bioactive molecules such as proteins, DNA, mRNAs, and miRNAs to recipient cells, thereby changing their function (26-29).
In ocular and cutaneous melanoma, the concentration of EVs and proteins is increased in patients compared to healthy individuals and has been shown to correlate with disease progression (30,31). Moreover, the profile of circulating EV-derived miRNAs is often altered in human cancers, and EVs from UM patients have been shown to contain miR-146, a potential circulating marker in UM (32). Recently, we have reported that the number of EVs produced and the profile of tumor-associated proteins vary between normal melanocytes and UM cell lines, and also between primary and metastatic UM cell lines (33). EVs released by metastatic melanoma cells were enriched in proteins (9,10,23) involved in the pre-metastatic niche formation (25), suggesting their role in preparing the environment for colonization by circulating tumor cells (CTCs).
There is a lack of detailed characterization of EVs in this disease as well as in nonblood-based liquid biopsy. In this study, our aim was to determine the proteomic profile of EVs isolated from AH, vitreous humor (VH), and plasma from patients with UM and to compare with cancer-free control patients.
Materials and methods
Patients
A total of 14 participants were enrolled for this study: 7 patients diagnosed with primary UM, and 7 healthy controls undergoing cataract surgery at the Department of Ophthalmology, Federal University of São Paulo (UNIFESP/EPM), Brazil. The patients were recruited from July 2019 to December 2019 at the Department of Ophthalmology of the UNIFESP/EPM. The clinical characteristics of the study population are described in Table I.
TABLE I -.
Clinical features of patients enrolled in this study
| Patients | Sex | Age (years) | Cell Types | Size | TNM |
|---|---|---|---|---|---|
| CAT1 | Male | 70 | N/A | N/A | N/A |
| CAT2 | Female | 77 | N/A | N/A | N/A |
| CAT3 | Female | 82 | N/A | N/A | N/A |
| CAT4 | Female | 77 | N/A | N/A | N/A |
| CAT5 | Female | 75 | N/A | N/A | N/A |
| CAT6 | Male | 76 | N/A | N/A | N/A |
| CAT7 | Female | 63 | N/A | N/A | N/A |
| UM1 | Male | 72 | Mixed UM, predominance of spindle cells affecting the ciliary body and choroid | 1.9 × 0.6 | pT4E |
| UM2 | Female | 86 | Epithelioid choroidal melanoma | 1.2 × 1.1 | pT3B |
| UM3 | Female | 53 | Mixed choroidal melanoma, predominance of spindle cells | 1.3 × 1.0 | pT3A |
| UM5 | Male | 63 | Mixed UM, predominance of spindle cells infiltrating the choroid and ciliary body | 1.2 × 1.5 | pT3B |
| UM6 | Female | 61 | Mixed UM, predominance of epithelioid cells | 1.0 × 0.8 | pT2 |
| UM8 | Female | 65 | Mixed UM, predominance of spindle cells infiltrating the choroid and ciliary body | 2.8 × 0.7 | pT4B |
| UM9 | Female | 39 | Mixed choroidal melanoma, predominance of epithelioid cells | 1.5 × 1.2 | pT3A |
Size refers to tumor size (base diameter × thickness [cm × cm]).
CAT = cataract, control group; N/A = not applicable; TNM = tumor, node, metastasis; UM = uveal melanoma.
This study was approved by the ethics committee investigational review board (CEP number 2198149) and adhered to the principles of the Declaration of Helsinki and Resolution 196/96 of the Ministry of Health, Brazil. Informed consent was obtained from all participants.
Sample collection
AH and plasma samples were collected from UM patients and controls. Additionally, VH samples were collected from UM patients. Peripheral blood (10 mL) was collected in ethylenediaminetetraacetic acid (EDTA) tubes. The tubes were centrifuged for 10 minutes at 1,900 × g), and plasma were collected. VH and AH samples from UM patients were collected from the enucleated eyes after the surgery with a syringe and fine needle. In the control group, AH samples were collected during cataract surgery. All routine surgical procedures were followed. All collected samples were kept at −80°C until the experimental procedure.
EV purification and characterization
The protocol for EV isolation was performed according to the guidelines of the International Society for Extracellular Vesicles (ISEV) (10). Samples were centrifuged at 16,000 × g for 10 minutes at 4°C to eliminate cellular debris. Then, EV isolation was performed using the exoEasy Maxi Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions (10,11,34-36). Isolated EVs were diluted 100 × in phosphate-buffered saline (PBS) and analyzed by nanoparticle tracking analysis (NTA) using the NanoSight NS300 instrument (Malvern Analytical, UK). PBS was used as a diluent. Samples and diluent were read in triplicates for 30 seconds at 20 frames per second. The NTA 3.2 software was used to estimate the concentration and size of the particles.
Immunoblotting
EVs isolated from patients and controls were lysed in RIPA buffer containing complete mini protease inhibitors (Sigma) at 4°C for 30 minutes. Samples were sonicated for 2 seconds (three times), and spun at 13,000 × g for 30 minutes at 4°C. Protein concentrations were quantified by the BCA assay (Thermo Fisher Scientific). Protein samples were processed for immunoblotting and mass spectrometry (MS).
EV-derived proteins (20 µg) were separated using 12% Mini-PROTEAN® precast polyacrylamide gel (Bio-Rad). Proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Membranes were blocked for 1 hour at room temperature with 5% nonfat dry milk in 1X Tris-buffer saline with 0.05% Tween 20 (TBST). Membranes were probed with anti-TSG101 (Abcam; 1:1,000) and anti-CD63 (Abcam; 1:1,000), anti-Alix (ThermoFisher Scientific 1:1,000), anti-β-actin (Sigma 1:1,000), anti-tenascin C (abcam 1:1,000), anti-vimentin (abcam 1:500) primary antibodies, followed by horseradish peroxidase (HRP)-conjugated goat anti-rabbit (Sigma 1:1,000) and goat anti-mouse (Sigma 1:3,000) secondary antibodies. Membranes were washed five times for 10 minutes each time after each incubation and developed using ECL prime Western blot detection (GE Healthcare). Protein signals were visualized using the ChemiDoc XRS + System.
MS analysis
MS analysis was performed in nine samples [AH (n = 3), plasma (n = 3), and VH (n = 3)] from UM-5, UM-6, and UM-8 patients; 20 µg of EV proteins from each sample was loaded onto a single stacking gel band to remove contaminants such as lipids, detergents, and salts. Each sample was run in duplicate.
The gel band was reduced with DTT (dithiothreitol), alkylated with iodoacetic acid, and digested with trypsin. Extracted peptides were resolubilized in 0.1% aqueous formic acid and loaded onto a Thermo Scientific Acclaim PepMap (75 μm inner diameter × 2 cm, C18 3 μm particle size) precolumn and then onto an Acclaim PepMap EASY-Spray (75 μm inner diameter × 15 cm with 2 μm C18, 2 µm beads) analytical column separation using a Dionex UltiMate 3000 uHPLC at 250 nL/min with a gradient of 2-35% organic (0.1% formic acid in acetonitrile) over 3 hours. Peptides were analyzed using a Thermo Orbitrap Fusion MS operating at 120,000 resolution (full width at half maximum in MS1) with Higher energy Collisional Dissociation (HCD) sequencing (15,000 resolution) at top speed for all peptides with a charge of 2+ or greater. The MS raw data were converted into *.mgf format (Mascot generic format) for searching using the Mascot 2.6.2 search engine (Matrix Science) against human protein sequences (Uniprot 2019). The database search results were loaded onto Scaffold Q+ Scaffold_4.10.0 (Proteome Sciences) for spectral counting, statistical treatment, data visualization, and quantification. Protein threshold >99%, peptide threshold >95%, and two of a minimum number of unique peptides were applied in Scaffold Q+ to increase the confidence level of identified proteins. Additional filters such as p-value cut-off of 0.05 and a fold-value change of ≥2 were used to identify the differential expression of proteins. The identified protein list in Scaffold was exported to Microsoft Excel and uploaded into the DAVID Bioinformatics database (version 6.8) for gene ontology (GO) analyses (i.e., biological process, cellular component, and KEGG pathway). In addition, bioinformatic analysis and Vesiclepedia database (37) search were performed using the FunRich software (version 3.1.3) (37-39).
Statistical analysis
Statistical analysis was performed using the GraphPad software (Prism, version 5.00 for Windows; GraphPad, San Diego, CA). The Mann-Whitney test was used to determine the statistical difference between respective groups. The results are expressed as mean ± standard deviation (SD). A p-value < 0.05 was considered significant.
Results
Characterization and isolation of EVs from plasma, AH, and VH
EVs were isolated from the plasma, AH, and VH of UM patients, and AH and plasma of cataract patients. Immunoblotting analysis showed the expression of EV markers CD63, TSG101, and Alix with different expression levels depending on the analyzed samples (Fig. 1A, B, and Supplementary Figure A). The expression of CD63 and Alix was higher in UM EVs than in CAT EVs (Fig. 1A, and Supplementary Figure A). Moreover, the expression of both CD63 and TSG101 was higher in EVs isolated from VH and plasma than in EVs isolated from AH (Fig. 1A, B). NTA from all samples showed that EVs ranged from 80 to 442 nm in size, with similar 10 percentile mean (D10) size (133 nm, 135 nm, and 139 nm) in plasma, AH, and VH, respectively (Fig. 1C, D). When analyzing sizes of isolated UM EVs, no difference was observed in all samples: 219 ± 26 nm (range: 168-241) in plasma, 211 ± 37 nm (range: 173-265) in AH, and 216 ± 71 nm (range: 110-314) in VH (Fig. 1D). Also, no difference was observed in the average size of EVs from AH and plasma between the UM and CAT groups (Fig. 1D).
Fig. 1 -.
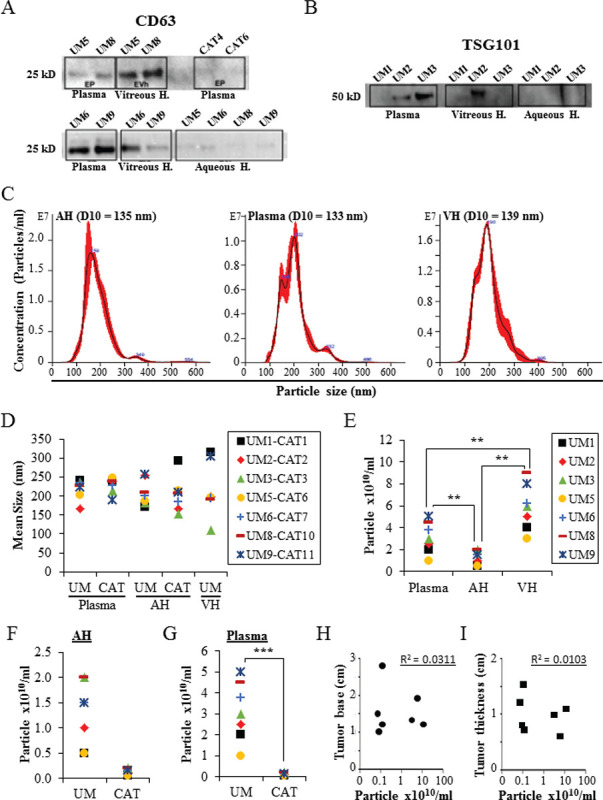
Characterization of EVs derived from AH, VH, and plasma. A,B) Proteins isolated from the different assay EVs (seven UM samples and two CAT samples) were analyzed by Western blot for the expression of specific EV markers (i.e., CD63 and TSG101). C) Nanosight analyses of EVs. Representative size distribution histograms showing data of EVs from AH, plasma, and VH. Note that mean EV sizes are similar. Histograms are displayed as averaged EV concentration (black line) and the variation between four repeated measurements indicating ±1 standard error of the mean (red outline). D) Mean size of EVs isolated from AH and plasma of UM (n = 7) and cataract-suffering (CAT, n = 7) patients, and from VH of UM patients (n = 7). E) Concentrations of EVs isolated from different analytes of seven UM patients. **p ˂ 0.01. F,G) Concentrations of EVs isolated from AH (F) and plasma (G) of UM (n = 7) and cataract-suffering (CAT, n = 7) patients. ***p ˂ 0.001. H and I) The concentrations of EVs isolated from the plasma of UM patients (n = 7) were plotted against ocular tumor size (base diameter (H) and thickness (I)). No correlation was found as shown by the correlation coefficient (R). Legend close to graph D applies to graphs D, F, and G. AH = aqueous humor; CAT = cataract; EV = extracellular vesicle; VH = vitreous humor; UM = uveal melanoma.
In the UM cohort, the concentration of EVs ranged from 2.6 × 109 to 9 × 1010 particles/mL in AH, VH, and plasma samples (Fig. 1E). The mean concentration of EVs in VH (6.6 × 1010 particles/mL) was significantly higher when compared to AH (1010 particles/mL, p < 0.01) and plasma (2.7 × 1010 particles/mL, p < 0.01) (Fig. 1E). No difference was observed in the concentration of AH-derived EVs between the UM and CAT groups. In contrast, the concentration of plasma-derived EVs was significantly higher in UM patients than in the CAT control group (p < 0.001) (Figs. 1F, G). Notably, we did not find any correlation between the concentrations of EVs isolated from UM patients and ocular tumor size (Fig. 1H, I).
EV protein cargo from plasma AH and VH
To gain an in-depth understanding of the protein cargo in EVs isolated from the different analytes, we performed whole proteomic analysis by MS. For this purpose, we focused our analysis on EVs isolated from three UM patients (UM-5, UM-6, and UM-8). Our goal from this analysis was to determine whether these EVs carried common protein cargo and also the nature of those proteins. We identified 542 proteins of which 498 (92%) overlap with EV proteins previously reported in the Vesiclepedia database (Supplementary Table A, List of EV-contained proteins identified by MS screening) (Fig. 2A) (37). As a readout for the purity of isolated EVs, we detected proteins that are specific to the tissue of origin (i.e., complement and coagulation factors in EVs from the plasma, melanocyte protein PMEL and HTRA1 in the VH, and beta- and gamma-crystallin in the AH) (Tab. II). In addition, protein cargo detected in isolated EVs included typical EV protein signatures such as ESCRT components CD81, CD63, CD9, HLA, annexins and syntenin (Supplementary Table A, List of EV-contained proteins identified by MS screening). Moreover, herein, we report the presence of 44 novel proteins not previously reported in the Vesiclepedia database (37); 2 are present in all EVs, 4 are present in EVs from plasma and VH, 4 are present in EVs from VH and AH, and the rest are unique to EVs from a single analyte (Tab. III).
Fig. 2 -.
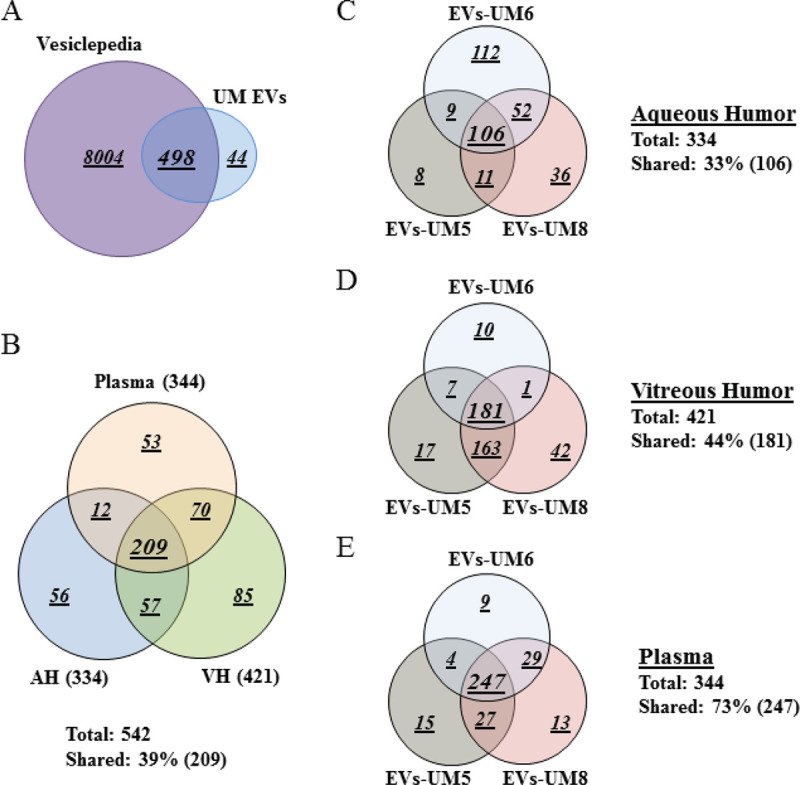
Plasma-derived EV protein cargo mirrored that of EVs isolated from AH and VH of UM patients. Venn diagram analyses. A) The majority of proteins isolated from EVs derived from the different analytes were shared with data published in Vesiclepedia database. B) EVs isolated from the three analytes shared 209 proteins (39%). C-E) Analyses of EV protein cargo in the same analytes from different donors. Note that these EVs shared 106 proteins (33%, C) in the aqueous humor, 181 proteins (44%, D) in the vitreous humor and 247 proteins (73%, E), which is in the same range of those shared between EVs from the three analytes (39%, see B). Data were collected from three UM patient analytes repeated twice each (UM5-1, UM5-2, UM6-1, UM6-2, UM8-1, and UM8-2). AH = aqueous humor; EV = extracellular vesicle; VH = vitreous humor; UM = uveal melanoma.
TABLE II -.
Protein readout for the purity of isolated EVs
| Spectrum Count | |||||
|---|---|---|---|---|---|
| Identified Proteins | ID | AH | VH | P | |
| Plasma | Coagulation factor V | FA5 | 2 | 0 | 146 |
| C4b-binding protein alpha chain | C4BPA | 1 | 7 | 127 | |
| Coagulation factor IX | FA9 | 1 | 16 | 79 | |
| von Willebrand factor | VWF | 0 | 0 | 72 | |
| Coagulation factor X | FA10 | 2 | 11 | 70 | |
| Multimerin-1 | MMRN1 | 0 | 0 | 33 | |
| Platelet glycoprotein Ib alpha chain | GP1BA | 0 | 2 | 22 | |
| C4b-binding protein beta chain | C4BPB | 0 | 0 | 14 | |
| Serum amyloid P-component | SAMP | 0 | 7 | 14 | |
| C-reactive protein | CRP | 1 | 1 | 13 | |
| Sushi, von Willebrand factor type A, EGF and pentraxin domain-containing protein 1 | SVEP1 | 0 | 0 | 2 | |
| Serum amyloid A-1 protein | SAA1 | 0 | 0 | 1 | |
| VH | Pigment epithelium-derived factor | PEDF | 37 | 111 | 13 |
| Retinol-binding protein 3 | RET3 | 3 | 111 | 0 | |
| Melanocyte protein PMEL | PMEL | 0 | 27 | 1 | |
| Serine protease HTRA1 | HTRA1 | 0 | 8 | 0 | |
| Retinaldehyde-binding protein 1 | RLBP1 | 0 | 2 | 0 | |
| Retinoschisin | XLRS1 | 0 | 2 | 0 | |
| Interphotoreceptor matrix proteoglycan 1 | IMPG1 | 0 | 1 | 0 | |
| AH and VH | Opticin | OPT | 12 | 12 | 0 |
| AH | Beta-crystallin B1 | CRBB1 | 179 | 2 | 1 |
| Alpha-crystallin A2 chain | CRYA2 | 146 | 0 | 0 | |
| Alpha-crystallin B chain | CRYAB | 139 | 7 | 0 | |
| Gamma-crystallin S | CRYGS | 97 | 1 | 0 | |
| Beta-crystallin A3 | CRBA1 | 76 | 0 | 0 | |
| Beta-crystallin A4 | CRBA4 | 52 | 0 | 0 | |
| Gamma-crystallin C | CRGC | 44 | 0 | 0 | |
| Gamma-crystallin D | CRGD | 42 | 0 | 0 | |
| Retinal dehydrogenase 1 | AL1A1 | 41 | 0 | 0 | |
| Filensin | BFSP1 | 2 | 0 | 0 | |
| Phakinin | BFSP2 | 2 | 0 | 0 | |
Data are derived from three patients (UM-5, UM-6, and UM-8) and samples were run in duplicates.
AH = aqueous humor; EV = extracellular vesicle; ID = alternative name; P = plasma; VH = vitreous humor.
TABLE III -.
Newly characterized protein from EVs isolated from plasma, aqueous humor, or vitreous humor
| Identified Proteins | ID | P | VH | AH |
|---|---|---|---|---|
| Complement C4-B | CO4B | Y | Y | Y |
| Beta-crystallin B1 | CRBB1 | Y | Y | Y |
| Vitamin K-dependent protein C | PROC | Y | Y | N |
| Immunoglobulin J chain | IGJ | Y | Y | N |
| L-selectin | LYAM1 | Y | Y | N |
| Neuropilin-2 | NRP2 | Y | Y | N |
| Soluble scavenger receptor cysteine-rich domain-containing protein SSC5D | SRCRL | Y | N | N |
| Extracellular matrix protein 2 | ECM2 | Y | N | N |
| Plexin domain-containing protein 1 | PLDX1 | Y | N | N |
| Retinol-binding protein 3 | RET3 | N | Y | Y |
| Opticin | OPT | N | Y | Y |
| Beta-1,4-glucuronyltransferase 1 | B4GA1 | N | Y | Y |
| Wnt inhibitory factor 1 | WIF1 | N | Y | Y |
| Beta-Ala-His dipeptidase | CNDP1 | N | Y | N |
| Receptor-type tyrosine-protein phosphatase zeta | PTPRZ | N | Y | N |
| Macrophage colony-stimulating factor 1 receptor | CSF1R | N | Y | N |
| Serpin E3 | SERP3 | N | Y | N |
| Cadherin-related family member 1 | CDHR1 | N | Y | N |
| Clusterin-like protein 1 | CLUL1 | N | Y | N |
| Retinaldehyde-binding protein 1 | RLBP1 | N | Y | N |
| Retinoschisin | XLRS1 | N | Y | N |
| Adipocyte plasma membrane-associated protein | APMAP | N | Y | N |
| Left-right determination factor 2 | LFTY2 | N | Y | N |
| Neuronal cell adhesion molecule | NRCAM | N | Y | N |
| Interphotoreceptor matrix proteoglycan 1 | IMPG1 | N | Y | N |
| Triggering receptor expressed on myeloid cells 2 | TREM2 | N | Y | N |
| Cathepsin L1 | CATL1 | N | Y | N |
| Endothelial lipase | LIPE | N | Y | N |
| BPI fold-containing family B member 4 | BPIB4 | N | Y | N |
| Semaphorin-3B | SEM3B | N | Y | N |
| Zinc transporter ZIP12 | S39AC | N | Y | N |
| Tsukushin | TSK | N | Y | N |
| Beta-crystallin A3 | CRBA1 | N | N | Y |
| Beta-crystallin A4 | CRBA4 | N | N | Y |
| Gamma-crystallin C | CRGC | N | N | Y |
| Gamma-crystallin D | CRGD | N | N | Y |
| Gamma-crystallin B | CRGB | N | N | Y |
| Beta-crystallin B3 | CRBB3 | N | N | Y |
| Filensin | BFSP1 | N | N | Y |
| Protein S100-B | S100B | N | N | Y |
| Secreted frizzled-related protein 3 | SFRP3 | N | N | Y |
| Phakinin | BFSP2 | N | N | Y |
| DNA polymerase theta | DPOLQ | N | N | Y |
| Protein kinase C-binding protein NELL2 | NELL2 | N | N | Y |
Data are derived from three patients (UM-5, UM-6, and UM-8) and samples were run in duplicates.
AH = aqueous humor; ID = alternative name; N = absent; P = plasma; VH = vitreous humor; Y = present.
Interestingly, 209 (39%) of the identified proteins were shared between EVs from the three assays (Fig. 2B). In addition, when we analyzed each analyte separately, we observed that EVs from the three samples shared 106 (33%) proteins in AH, 181 (44%) in VH, and 247 (73%) in plasma (Fig. 2C-E).
Proteins by GO analysis in specific biological processes
Of the proteins found in our proteomic analyses data from UM patients, 344 proteins were detected from plasma EVs, 334 in EVs from AH, and 421 in EVs from VH (Fig. 2B). To identify the physiological processes to which these proteins were associated, clustering was conducted into GO categories using the DAVID bioinformatics platform (Fig. 3). Characterization by biological process highlighted categories related to retina homeostasis, regulation of apoptosis, cell growth, and the activation of pathways involved in cancer cell biology (i.e., MAPK/ERK cascades). In addition, of the highly expressed proteins, several clustered in the categories of cell-cell adhesion and movement of cell or subcellular component (Fig. 3A). When clustering the proteins based on cellular component, we found they grouped into EV categories (i.e., vesicles) (Fig. 3B). Molecular functions clustering using KEGG pathway analysis revealed that isolated EVs were enriched for proteins related to immune escape from cancer, such as those involved in complement and coagulation cascades, and proteins involved in cell metabolic activities and interaction with extracellular matrix (ECM). Particularly, a panel of proteins clustered in the PI3k-Akt signaling pathway and the proteoglycan group were exclusively present in plasma-isolated EVs (Fig. 3C).
Fig. 3 -.
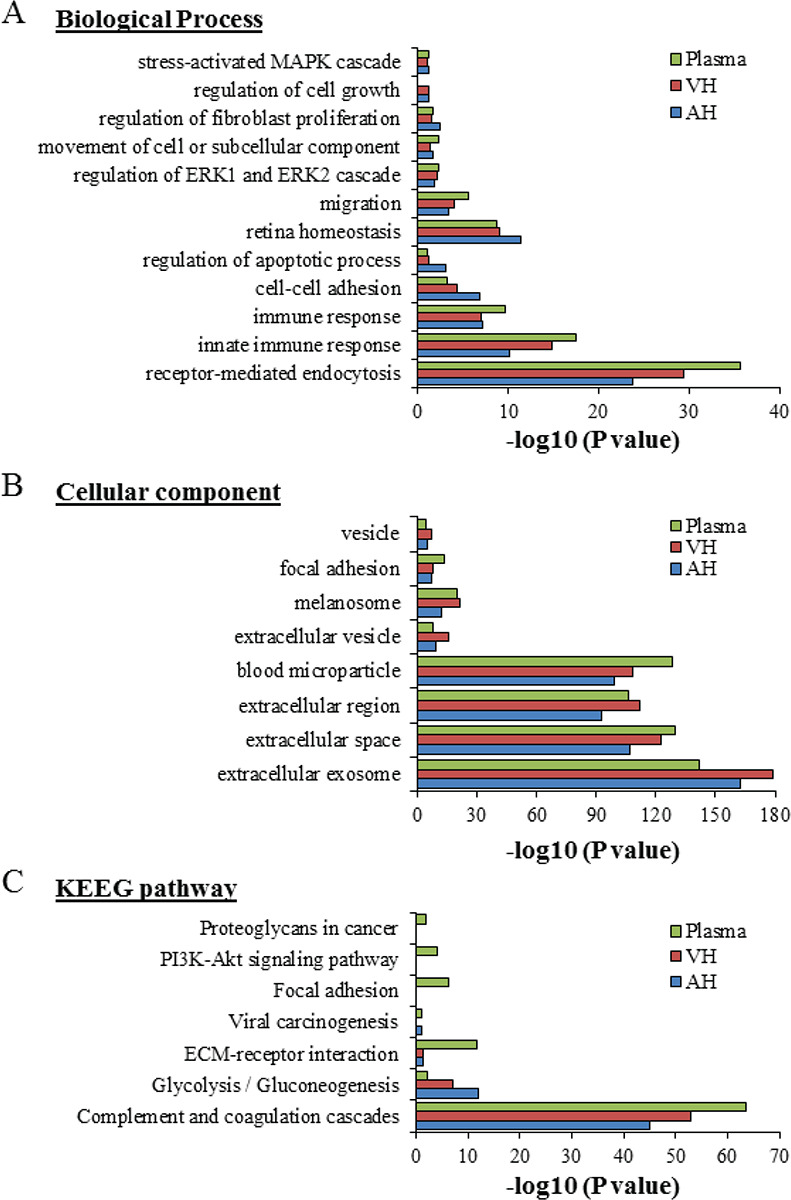
Gene ontology classification of EV protein cargo. The most enriched categories in biological process (A), cellular component (B), and molecular function (C) are shown. Data were collected from three UM patient analytes repeated twice (UM5-1, UM5-2, UM6-1, UM6-2, UM8-1, and UM8-2). EV = extracellular vesicle; UM = uveal melanoma.
UM arises from melanocytes of the uveal tract (25,34). EVs isolated from the AH and VH may contain proteins reflective of UM cells. We pooled our data from intraocular-derived EVs by focusing on proteins that regulate tumor growth and oncogenesis (Tab. IV). This identified a panel of proteins that are mainly involved in protecting cells against apoptosis, controlling cell growth, promoting angiogenesis, and inducing cell spreading (i.e., clusterin, alpha-enolase, fibulin-1, cathepsin, HSP, ECM1, MET, and GAS6). Moreover, vimentin (an intermediate filament protein that is overexpressed in epithelial tumors such as UMs) was detected in VH-derived EVs (Tab. IV) (36,38-40).
TABLE IV -.
Protein cargo from aqueous humor- or vitreous humor-derived EVs involved in cell proliferation, survival, and invasion
| Spectrum Count | |||
|---|---|---|---|
| Identified Proteins | Alternative Name | AH | VH |
| Clusterin | CLUS | 50 | 175 |
| Pigment epithelium-derived factor | PEDF | 37 | 111 |
| Alpha-enolase | ENOA | 30 | 7 |
| Vitronectin | VTNC | 29 | 100 |
| Gamma-enolase | ENOG | 7 | 2 |
| Cathepsin D | CATD | 6 | 64 |
| Fibulin-1 | FBLN1 | 6 | 4 |
| Myocilin | MYOC | 6 | |
| Heat shock protein HSP 90-alpha | HS90A | 5 | 10 |
| Galectin-1 | LEG1 | 3 | |
| Heat shock protein HSP 90-beta | HS90B | 3 | 4 |
| Extracellular matrix protein 1 | ECM1 | 2 | 5 |
| Growth arrest-specific protein 6 | GAS6 | 2 | |
| CD44 antigen | CD44 | 2 | 5 |
| C-reactive protein | CRP | 1 | 1 |
| Plexin domain-containing protein 2 | PXDC2 | 3 | |
| Ras-related protein Rab-1A | RAB1A | 1 | |
| Vimentin | VIME | 14 | |
| Cathepsin B | CATB | 8 | |
| Hepatocyte growth factor receptor | MET | 6 | |
| Cadherin-related family member 1 | CDHR1 | 6 | |
| Fibronectin | FINC | 38 | |
| Periostin | POSTN | 4 | |
| Legumain | LGMN | 3 | |
| Cathepsin F | CATF | 3 | |
AH = aqueous humor; EV = extracellular vesicle; VH = vitreous humor.
Data are derived from three patients (UM-5, UM-6, and UM-8) and samples were run in duplicates.
Plasma-isolated EVs were also enriched in proteins involved in the regulation of cell proliferation (i.e., SPARC, tenascin, plexin) and cell survival (i.e., clusterin), and the metastatic process such as metastatic niche organization (i.e., ECM1, ECM2, emilin, C-reactive protein [CRP], oncoprotein-induced transcript 3 [OIT3], and integrins) (Tab. V, and Supplementary Figure A) (41-44). These data suggest that, while AH- and VH-isolated proteins govern in situ UM growth and motility, those contained in the plasma-derived EVs are more involved in UM cell metastatic organotropism and the maintenance of the metastatic niche.
TABLE V -.
Protein cargo from UM plasma-derived EVs involved in cell proliferation and survival, and metastatic niche organization
| Identified Proteins | Alternative Name | Spectrum Count |
|---|---|---|
| Fibronectin | FINC | 130 |
| Vitronectin | VTNC | 109 |
| Clusterin | CLUS | 57 |
| Integrin alpha-IIb | ITA2B | 30 |
| Endoplasmin | ENPL | 28 |
| Integrin beta-3 | ITB3 | 22 |
| SPARC | SPRC | 22 |
| Nidogen-1 | NID1 | 16 |
| Vinculin | VINC | 14 |
| Tenascin | TENA | 13 |
| Pigment epithelium-derived factor | PEDF | 13 |
| C-reactive protein | CRP | 13 |
| Heat shock protein HSP 90-alpha | HS90A | 10 |
| Fibulin-1 | FBLN1 | 9 |
| Heat shock protein HSP 90-beta | HS90B | 7 |
| Endoplasmic reticulum chaperone BiP | BIP | 7 |
| CD44 antigen OS = Homo sapiens | CD44 | 6 |
| Heat shock cognate 71 kDa protein | HSP7C | 4 |
| Extracellular matrix protein 1 | ECM1 | 3 |
| Plexin domain-containing protein 2 | PXDC2 | 2 |
| Extracellular matrix protein 2 | ECM2 | 2 |
| Beta-parvin | PARVB | 2 |
| Caveolae-associated protein 2 | CAVN2 | 2 |
| Ras-related protein Rab-1A | RAB1A | 1 |
| Hepatocyte growth factor activator | HGFA | 1 |
| Oncoprotein-induced transcript 3 protein | OIT3 | 1 |
| EMILIN-1 | EMIL1 | 1 |
| Vascular endothelial growth factor receptor 3 | VGFR3 | 1 |
| Plexin-B1 | PLXB1 | 1 |
| Integrin beta-1 | ITB1 | 1 |
| Alpha-enolase | ENOA | 1 |
| Protein S100-A8 | S10A8 | 1 |
| Protein S100-A9 | S10A9 | 1 |
Data are derived from three patients (UM-5, UM-6, and UM-8) and samples were run in duplicates.
EV = extracellular vesicle; UM = uveal melanoma.
Discussion
EVs have been reported to regulate many aspects of physiological and pathological processes such as cancer. They carry substances that mirror the content of their cell of origin and have the capability to exhibit different biological functions on recipient cells via trafficking of different factors, that is, nucleic acids, proteins, lipids (10,21,44-52). EVs released from tumor cells promote cell proliferation, migration, invasion, angiogenesis, and metastases (54,57-63). EV cargo could be used as circulating biomarkers in liquid biopsy, mainly in the context of cancer. In the present study, we determined the proteomic profile of EVs isolated from AH, VH, and plasma from patients with UM in comparison with cancer-free control patients.
The size and distribution of EVs detected in the three samples were consistent with exosomes (10). In the blood samples, a significantly higher concentration of EVs was found in UM patients compared to the control group. This is in agreement with our recent observations that UM cell lines shed more EVs than normal choroidal melanocytes (33). Another study suggests that EV has potential roles in cancer progression and invasion (11). Interestingly, the mean concentration of EVs in VH from UM patients was higher when compared to plasma and AH, which seems normal as UM takes place in the posterior segment of the eye.
We showed that EVs derived from AH, VH, and plasma were positive for CD63 and TSG101 markers. Besides, the expression of CD63 was higher in UM EVs in comparison with EVs isolated from samples of control group. Our data corroborate with a study that demonstrated high levels of CD63 in exosomes isolated from plasma of melanoma patients (53). Also a study showed exosomal marker TSG101 was detected in plasma-derived exosome from ovarian cancer patients (21).
We observed that the plasma EV proteomic cargo resembles that of EVs obtained from AH and VH. Although we found that only 209 proteins (39%) were shared between EVs from the three samples (a value that reached 221 proteins [49%] and 279 proteins [57%] when taking into account only AH vs. plasma and VH vs. plasma, respectively), this is not surprising as the plasma is the common carrier of EVs from different tissues.
Moreover, proteomic mining of isolated EVs from UM group identified a set of proteins involved in oncogenesis (i.e., regulation of cell proliferation and survival, promotion of angiogenesis, and cell invasion) and metastasis (i.e., cell spreading and metastatic niche organization) (36,38-43,54). For example, SPARC abrogation has been reported to reduce cell proliferation in UM (41). Cathepsin, a lysosomal acid proteinase, was reported to be involved in different cancer types, especially in regulating UM invasion potential (36,40). Galectin has been shown to facilitate cell migration, to promote metastasis, and to be a hallmark for cancer aggressiveness (55,56). OIT3 is involved in the development and function of the liver, which is the primary site for UM metastasis (54). In addition, several integrins were detected in the isolated EVs from the UM group. These proteins are involved in adhesion to extracellular matrix components and specific organotropism of metastasizing cancer cells (43,64). The integrins present in the EV preparations demonstrate an upregulation of various signal transduction molecules such as S100-A. It has been shown that exosome-derived integrins are internalized by target cells and activate SRC phosphorylation and proinflammatory S100 gene expression (64). Furthermore, EVs from melanoma were found to upregulate S100 proteins in recipient target cells, resulting in vascular leakiness and promotion of metastasis (31,65).
Other proteins found in the datasets such as heat shock proteins and CRP are indicators of worse prognosis in UM (38,42). In addition, melanocyte-specific type I transmembrane glycoprotein (PMEL) was enriched in EVs from VH and less in EVs from plasma. This protein is released by proteolytic ectodomain shedding and may be used as a melanoma-specific blood marker (5,6,67,68).
Interestingly, the recovered protein cargo contained factors involved in cell proliferation, cell survival, oncogenesis, cell invasion, and metastatic niche organization. Together, these data suggest that plasma from UM patients could be used as liquid biopsy platform for patient diagnosis and noninvasive monitoring.
Using clustering analysis based on GO biological process, categories consistent with retinal homeostasis and activation of intracellular pathways involved in cancer cell biology were identified (i.e., MAPK/ERK cascades). Almost all UMs are characterized by mutations in one of GNAQ, GNA11, PLCB4, or CYSLTR2 genes, and these are upstream activators of the MAPK/ERK cascade (66).
One limitation of this study is the low number of analyzed samples for the proteomic characterization (three UM samples). However, the consistency of the data between the analyzed samples makes the conclusions valuable. Unfortunately, due to the lack of material, performing differential protein expression analysis is not possible at this stage. Studies including more samples are in progress to address this weakness.
Liquid biopsy is already distinguishing cancer-free individuals from non-small cell lung cancer patients and pancreatic ductal adenocarcinoma by the quantitative analysis of exosomal miR-21 and miR-10b, respectively (67). Intra-EV metabolites from prostate cancer patients before and after prostatectomy revealed novel biomarkers (68). One must remember that not only tumor cells release exosomal RNA to affect biological functions but also many normal cells will secrete the same exosomal RNA physiologically (69). As mentioned before, exosomal integrins could be used to predict organ-specific metastasis (64). Therefore, therapy supported by liquid biopsy could be driven in a premature way in case of early metastasis diagnosis or even somehow by targeting and blocking cancer pre-metastatic EV development. Certainly, this promising new tool has to be used with caution, and further studies are needed.
In conclusion, it has been observed that VH is significantly enriched in EVs when compared to AH and plasma in UM patients. EV concentrations in plasma and AH from UM patients was higher when compared to those in the cataract group. Proteomic analysis demonstrated that EVs from the different samples shared a panel of proteins, suggesting that circulating UM EVs mirrored the in situ shed of EVs (i.e., AH and VH). EVs isolated from AH, VH, and plasma from patients with UM showed consistent profiles and support the use of blood to monitor UM patients as a noninvasive liquid biopsy.
Acknowledgments
The authors would like to acknowledge the technical expertise and scientific support of the Proteomics facility of the MUHC-RI, especially Lorne Taylor and Amy Wong.
This article includes supplementary material.
Disclosures
Financial support: This work was supported by CNPq process No. 429571/2018-6, FAPESP, and CAPES. RBJ is an investigator for CNPq Brazil.
Conflict of interest: The authors report no conflict of interest.
Data Availability: All data generated and analyzed during this study are included in this manuscript.
References
- 1.Ortega MA, Fraile-Martínez O, García-Honduvilla N et al. Update on uveal melanoma: translational research from biology to clinical practice. [Review]. Int J Oncol. 2020;57(6):1262–1279. doi: 10.3892/ijo.2020.5140. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abildgaard SK, Vorum H. Proteomics of uveal melanoma: a minireview. J Oncol. 2013;2013:820953. doi: 10.1155/2013/820953. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuidervaart W, Hensbergen PJ, Wong MC et al. Proteomic analysis of uveal melanoma reveals novel potential markers involved in tumor progression. Invest Ophthalmol Vis Sci. 2006;47(3):786–793. doi: 10.1167/iovs.05-0314. PubMed [DOI] [PubMed] [Google Scholar]
- 4.Ramasamy P, Murphy CC, Clynes M et al. Proteomics in uveal melanoma. Exp Eye Res. 2014;118:1–12. doi: 10.1016/j.exer.2013.09.005. PubMed [DOI] [PubMed] [Google Scholar]
- 5.Surman M, Hoja-Łukowicz D, Szwed S et al. An insight into the proteome of uveal melanoma-derived ectosomes reveals the presence of potentially useful biomarkers. Int J Mol Sci. 2019;20(15):E3789. doi: 10.3390/ijms20153789. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Surman M, Stępień E, Przybyło M. Melanoma-derived extracellular vesicles: focus on their proteome. Proteomes. 2019;7(2):21.. doi: 10.3390/proteomes7020021. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Contreras-Naranjo JC, Wu HJ, Ugaz VM. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab Chip. 2017;17(21):3558–3577. doi: 10.1039/C7LC00592J. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pessuti CL, Costa DF, Ribeiro KS et al. Extracellular vesicles from the aqueous humor of patients with uveitis. Pan-Am J Ophthalmol. 2019;(1):1–3. doi: 10.4103/PAJO.PAJO_9_19. [DOI] [Google Scholar]
- 9.Perkumas KM, Hoffman EA, McKay BS, Allingham RR, Stamer WD. Myocilin-associated exosomes in human ocular samples. Exp Eye Res. 2007;84(1):209–212. doi: 10.1016/j.exer.2006.09.020. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Théry C, Witwer KW, Aikawa E et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Freitas D, Kim HS et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018;20(3):332–343. doi: 10.1038/s41556-018-0040-4. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramirez MI, Amorim MG, Gadelha C et al. Technical challenges of working with extracellular vesicles. Nanoscale. 2018;10(3):881–906. doi: 10.1039/C7NR08360B. PubMed [DOI] [PubMed] [Google Scholar]
- 13.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6(3):267–283. doi: 10.1586/epr.09.17. PubMed [DOI] [PubMed] [Google Scholar]
- 14.Devhare PB, Ray RB. Extracellular vesicles: novel mediator for cell to cell communications in liver pathogenesis. Mol Aspects Med. 2018;60:115–122. doi: 10.1016/j.mam.2017.11.001. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yáñez-Mó M, Siljander PR, Andreu Z et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4(1):27066. doi: 10.3402/jev.v4.27066. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao CY, Papoutsakis ET. Extracellular vesicles: exosomes, microparticles, their parts, and their targets to enable their biomanufacturing and clinical applications. Curr Opin Biotechnol. 2019;60:89–98. doi: 10.1016/j.copbio.2019.01.005. PubMed [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Weber SR, Lease J et al. Liquid biopsy of vitreous reveals an abundant vesicle population consistent with the size and morphology of exosomes. Transl Vis Sci Technol. 2018;7(3):6. doi: 10.1167/tvst.7.3.6. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiklander OPB, Brennan MÁ, Lötvall J, Breakefield XO, El Andaloussi S. Advances in therapeutic applications of extracellular vesicles. Sci Transl Med. 2019 May 15;11(492):eaav8521. doi: 10.1126/scitranslmed.aav8521. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campos JH, Soares RP, Ribeiro K, Andrade AC, Batista WL, Torrecilhas AC. Extracellular vesicles: role in inflammatory responses and potential uses in vaccination in cancer and infectious diseases. J Immunol Res. 2015;2015:832057. doi: 10.1155/2015/832057. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcilla A, Martin-Jaular L, Trelis M et al. Extracellular vesicles in parasitic diseases. J Extracell Vesicles. 2014;3(1):25040. doi: 10.3402/jev.v3.25040. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang B, Peng P, Chen S et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J Proteomics. 2013;80:171–182. doi: 10.1016/j.jprot.2012.12.029. PubMed [DOI] [PubMed] [Google Scholar]
- 22.Andrade LNS, Otake AH, Cardim SGB et al. Extracellular vesicles shedding promotes melanoma growth in response to chemotherapy. Sci Rep. 2019;9(1):14482. doi: 10.1038/s41598-019-50848-z. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angi M, Kalirai H, Prendergast S et al. In-depth proteomic profiling of the uveal melanoma secretome. Oncotarget. 2016;7(31):49623–49635. doi: 10.18632/oncotarget.10418. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazar I, Clement E, Ducoux-Petit M et al. Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Pigment Cell Melanoma Res. 2015;28(4):464–475. doi: 10.1111/pcmr.12380. PubMed [DOI] [PubMed] [Google Scholar]
- 25.Guo Y, Ji X, Liu J et al. Effects of exosomes on pre-metastatic niche formation in tumors. Mol Cancer. 2019;18(1):39. doi: 10.1186/s12943-019-0995-1. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plebanek MP, Angeloni NL, Vinokour E et al. Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nat Commun. 2017;8(1):1319. doi: 10.1038/s41467-017-01433-3. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015;219:278–294. doi: 10.1016/j.jconrel.2015.06.029. PubMed [DOI] [PubMed] [Google Scholar]
- 28.Chennakrishnaiah S, Tsering T, Aprikian S, Rak J. Leukobiopsy—a possible new liquid biopsy platform for detecting oncogenic mutations. Front Pharmacol. 2020;10:1608. doi: 10.3389/fphar.2019.01608. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdouh M, Floris M, Gao ZH, Arena V, Arena M, Arena GO. Colorectal cancer-derived extracellular vesicles induce transformation of fibroblasts into colon carcinoma cells. J Exp Clin Cancer Res. 2019;38(1):257. doi: 10.1186/s13046-019-1248-2. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eldh M, Olofsson Bagge R, Lässer C et al. MicroRNA in exosomes isolated directly from the liver circulation in patients with metastatic uveal melanoma. BMC Cancer. 2014;14(1):962. doi: 10.1186/1471-2407-14-962. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peinado H, Alečković M, Lavotshkin S et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ragusa M, Barbagallo C, Statello L et al. miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: pathological and diagnostic implications. Cancer Biol Ther. 2015;16(9):1387–1396. doi: 10.1080/15384047.2015.1046021. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsering T, Laskaris A, Abdouh M et al. Uveal melanoma-derived extracellular vesicles display transforming potential and carry protein cargo involved in metastatic niche preparation. Cancers (Basel). 2020;12(10):E2923. doi: 10.3390/cancers12102923. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding M, Wang C, Lu X et al. Comparison of commercial exosome isolation kits for circulating exosomal microRNA profiling. Anal Bioanal Chem. 2018;410(16):3805–3814. doi: 10.1007/s00216-018-1052-4. PubMed [DOI] [PubMed] [Google Scholar]
- 35.Enderle D, Spiel A, Coticchia CM et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS One. 2015;10(8):E0136133. doi: 10.1371/journal.pone.0136133. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu L, Wada M, Usagawa Y et al. Overexpression of cathepsin D in malignant melanoma. Fukuoka Igaku Zasshi. 2013;104(10):370–375. PubMed [PubMed] [Google Scholar]
- 37.Kalra H, Simpson RJ, Ji H et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10(12):E1001450. doi: 10.1371/journal.pbio.1001450. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faingold D, Marshall JC, Antecka E et al. Immune expression and inhibition of heat shock protein 90 in uveal melanoma. Clin Cancer Res. 2008;14(3):847–855. doi: 10.1158/1078-0432.CCR-07-0926. PubMed [DOI] [PubMed] [Google Scholar]
- 39.Wu X, Zhou J, Rogers AM et al. c-Met, epidermal growth factor receptor, and insulin-like growth factor-1 receptor are important for growth in uveal melanoma and independently contribute to migration and metastatic potential. Melanoma Res. 2012;22(2):123–132. doi: 10.1097/CMR.0b013e3283507ffd. PubMed [DOI] [PubMed] [Google Scholar]
- 40.Pardo M, García A, Antrobus R, Blanco MJ, Dwek RA, Zitzmann N. Biomarker discovery from uveal melanoma secretomes: identification of gp100 and cathepsin D in patient serum. J Proteome Res. 2007;6(7):2802–2811. doi: 10.1021/pr070021t. PubMed [DOI] [PubMed] [Google Scholar]
- 41.Maloney SC, Marshall JC, Antecka E et al. SPARC is expressed in human uveal melanoma and its abrogation reduces tumor cell proliferation. Anticancer Res. 2009;29(8):3059–3064. PubMed [PubMed] [Google Scholar]
- 42.Schicher N, Edelhauser G, Harmankaya K et al. Pretherapeutic laboratory findings, extent of metastasis and choice of treatment as prognostic markers in ocular melanoma—a single centre experience. J Eur Acad Dermatol Venereol. 2013;27(3):e394–e399. doi: 10.1111/jdv.12006. PubMed [DOI] [PubMed] [Google Scholar]
- 43.Janik ME, Lityńska A, Przybyło M. Studies on primary uveal and cutaneous melanoma cell interaction with vitronectin. Cell Biol Int. 2014;38(8):942–952. doi: 10.1002/cbin.10280. PubMed [DOI] [PubMed] [Google Scholar]
- 44.Lykke-Andersen S, Brodersen DE, Jensen TH. Origins and activities of the eukaryotic exosome. J Cell Sci. 2009;122(Pt 10):1487–1494. doi: 10.1242/jcs.047399. PubMed [DOI] [PubMed] [Google Scholar]
- 45.Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006;20(9):1487–1495. doi: 10.1038/sj.leu.2404296. PubMed [DOI] [PubMed] [Google Scholar]
- 46.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40(21):10937–10949. doi: 10.1093/nar/gks832. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Di Vizio D, Morello M, Dudley AC et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am J Pathol. 2012;181(5):1573–1584. doi: 10.1016/j.ajpath.2012.07.030. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kahlert C, Melo SA, Protopopov A et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289(7):3869–3875. doi: 10.1074/jbc.C113.532267. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Théry C, Boussac M, Véron P et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166(12):7309–7318. doi: 10.4049/jimmunol.166.12.7309. PubMed [DOI] [PubMed] [Google Scholar]
- 50.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125. PubMed [DOI] [PubMed] [Google Scholar]
- 51.Zaborowski MP, Balaj L, Breakefield XO, Lai CP. Extracellular vesicles: composition, biological relevance, and methods of study. Bioscience. 2015;65(8):783–797. doi: 10.1093/biosci/biv084. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inamdar S, Nitiyanandan R, Rege K. Emerging applications of exosomes in cancer therapeutics and diagnostics. Bioeng Transl Med. 2017;2(1):70–80. doi: 10.1002/btm2.10059. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Logozzi M, De Milito A, Lugini L et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4(4):E5219. doi: 10.1371/journal.pone.0005219. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu ZG, Du JJ, Zhang X et al. A novel liver-specific zona pellucida domain containing protein that is expressed rarely in hepatocellular carcinoma. Hepatology. 2003;38(3):735–744. doi: 10.1053/jhep.2003.50340. PubMed [DOI] [PubMed] [Google Scholar]
- 55.Sindrewicz P, Lian LY, Yu LG. Interaction of the oncofetal Thomsen-Friedenreich antigen with galectins in cancer progression and metastasis. Front Oncol. 2016;6:79. doi: 10.3389/fonc.2016.00079. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmed H, AlSadek DM. Galectin-3 as a potential target to prevent cancer metastasis. Clin Med Insights Oncol. 2015;9:113–121. doi: 10.4137/CMO.S29462. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al-Nedawi K, Meehan B, Micallef J et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–624. doi: 10.1038/ncb1725. PubMed [DOI] [PubMed] [Google Scholar]
- 58.Aga M, Bentz GL, Raffa S et al. Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene. 2014;33(37):4613–4622. doi: 10.1038/onc.2014.66. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdouh M, Hamam D, Gao ZH, Arena V, Arena M, Arena GO. Exosomes isolated from cancer patients’ sera transfer malignant traits and confer the same phenotype of primary tumors to oncosuppressor-mutated cells. J Exp Clin Cancer Res. 2017;36(1):113. doi: 10.1186/s13046-017-0587-0. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu J, Liao K, Zhou W. Exosomes regulate the transformation of cancer cells in cancer stem cell homeostasis. Stem Cells Int. 2018;2018:4837370. doi: 10.1155/2018/4837370. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen M, Ren X. New insights into the biological impacts of immune cell-derived exosomes within the tumor environment. Cancer Lett. 2018;431:115–122. doi: 10.1016/j.canlet.2018.05.040. PubMed [DOI] [PubMed] [Google Scholar]
- 62.Dos Anjos Pultz B, Andrés Cordero da Luz F, Socorro Faria S et al. The multifaceted role of extracellular vesicles in metastasis: priming the soil for seeding. Int J Cancer. 2017;140(11):2397–2407. doi: 10.1002/ijc.30595. PubMed [DOI] [PubMed] [Google Scholar]
- 63.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30(6):836–848. doi: 10.1016/j.ccell.2016.10.009. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoshino A, Costa-Silva B, Shen TL et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi: 10.1038/nature15756. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Che P, Yang Y, Han X et al. S100A4 promotes pancreatic cancer progression through a dual signaling pathway mediated by Src and focal adhesion kinase. Sci Rep. 2015;5(1):8453. doi: 10.1038/srep08453. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amaro A, Gangemi R, Piaggio F et al. The biology of uveal melanoma. Cancer Metastasis Rev. 2017;36(1):109–140. doi: 10.1007/s10555-017-9663-3. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li S, Yi M, Dong B, Tan X, Luo S, Wu K. The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. Int J Cancer. 2021;148(11):2640–2651. doi: 10.1002/ijc.33386. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puhka M, Takatalo M, Nordberg ME et al. Metabolomic profiling of extracellular vesicles and alternative normalization methods reveal enriched metabolites and strategies to study prostate cancer-related changes. Theranostics. 2017;7(16):3824–3841. doi: 10.7150/thno.19890. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vafaei S, Fattahi F, Ebrahimi M, Janani L, Shariftabrizi A, Madjd Z. Common molecular markers between circulating tumor cells and blood exosomes in colorectal cancer: a systematic and analytical review. Cancer Manag Res. 2019;11:8669–8698. doi: 10.2147/CMAR.S219699. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This article includes supplementary material.


