Abstract
Despite recent success in transforming various thermophilic gram-type-positive anaerobes with plasmid DNA, use of shuttle vectors for the expression of genes other than antibiotic resistance markers has not previously been described. We constructed new vectors in order to express heterologous hydrolytic enzymes in our model system, Thermoanaerobacterium saccharolyticum JW/SL-YS485. Transformed Thermoanaerobacterium expressed active enzyme, indicating that this system may function as an alternate expression host, especially for genes with a thermophilic origin. To develop further the genetic system for T. saccharolyticum JW/SL-YS485, two improved Escherichia coli-Thermoanaerobacterium shuttle vectors, pRKM1 and pRUKM, were constructed. Furthermore, the kanamycin resistance cassette alone and the kanamycin resistance cassette plus the cellobiohydrolase gene (cbhA) from Clostridium thermocellum JW20 were integrated into the xylanase gene (xynA) region of the Thermoanaerobacterium chromosome via homologous recombination using pUC-based suicide vectors pUXK and pUXKC.
Species of the genera Thermoanaerobacter and Thermoanaerobacterium (9, 24) are representatives of the recently extended group gram-type-positive (25) thermophilic anaerobic bacteria (Fermicutes). Their optimal growth temperatures are usually between 60 and 70°C, and some can grow at up to almost 80°C; they are of interest for a variety of applications (1, 15, 24, 26). Species of Thermoanaerobacterium are widely distributed and can efficiently utilize xylan, a potentially inexpensive substrate for industrial fermentation (15). Several enzymes from Thermoanaerobacterium saccharolyticum strain JW/SL-YS485 have been isolated and characterized, and various genes have been cloned and sequenced (10, 12–14). Due to lack of developed genetic systems for any member belonging to this group of microorganisms, T. saccharolyticum JW/SL-YS485 was chosen as a model organism for the development of genetic tools (16).
Until now, only a few vectors capable of transforming gram-positive anaerobic thermophiles (T. [basonym Clostridium] thermosaccharolyticum, Thermoanaerobacter [basonym Clostridium] thermohydrosulfuricus, and Thermoanaerobacter mathranii) have been described (8, 20; A. Clausen and B. Ahring, Abstr. Thermophile 98, poster abstr. G-P51, 1998). We have previously reported the development of the shuttle vector pIKM1 for transformation of Thermoanaerobacterium strain JW/SL-YS485 (16), which is a strain of T. saccharolyticum (F. Rainey and J. Wiegel, unpublished results). Briefly, the construct pIKM1 is based on the Escherichia coli-Clostridium acetobutylicum shuttle vector pIMP1 and contains the thermostable kanamycin cassette from Streptococcus faecalis plasmid pKD102 (21). After electrotransformation into T. saccharolyticum JW/SL-YS485, plasmid pIKM1 mediated in this strain resistance to up to 400 μg of kanamycin ml−1 at 48°C and 200 μg ml−1 at 60°C. We report here three aspects of extending our model system.
First, we describe the construction and utilization of pIKM3 to pIKM8 for the successful expression of a thermophilic mannanase and various cellulolytic enzymes in T. saccharolyticum JW/SL-YS485. Second, we constructed the new shuttle vectors pRKM1 and pRUKM1 for improved transformation, since the pIKM1-based shuttle vectors had some disadvantages, including low expression levels and low transformation efficiencies. Finally, we developed suicide vectors that facilitated chromosomal integration.
MATERIALS AND METHODS
Culture conditions:
Thermoanaerobacterium strain JW/SL-YS485 (DSM 8691), now identified as T. saccharolyticum JW/SL-YS485 (Rainey and Wiegel, unpublished), isolated in our laboratory, was grown either in prereduced mineral medium (24) or in the same medium supplemented with 50 to 800 μg of kanamycin/ml (selective medium). Wild-type JW/SL-YS485 is sensitive to 50 μg of kanamycin/ml but resistant to 100 μg of ampicillin/ml. E. coli TG1 (supE hsdΔ5 thi Δ[lac-proAB] [F′ traD36 proAB lacIq ZΔM15]) was grown in Luria-Bertani medium; selective medium was supplemented with either ampicillin (100 μg ml−1) or kanamycin (50 μg ml−1).
Plasmid construction.
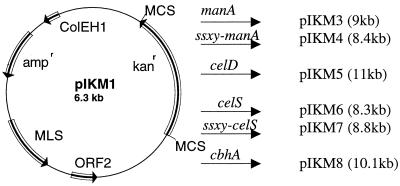
Plasmid pIKM3 (9 kb [Fig. 1]) was constructed as follows. Using standard subcloning procedures (2), plasmid pLAL602 was digested with EcoRI/BamHI, and the 2.7-kb band containing the manA gene from Caldicellululosiruptor strain Rt8B4 (4) was isolated from a 1% agarose gel after electrophoresis. The DNA was subsequently ligated into EcoRI/BamHI-digested pIKM1 (6.3 kb). Plasmids pIKM5 (11 kb) and pIKM6 (8.3 kb) were correspondingly constructed by ligating the cloned celD/celS from Clostridium thermocellum (7, 23) into EcoRI/KpnI- and XbaI/KpnI-cut pIKM1, respectively (Fig. 1). Plasmids pIKM4 (8.4 kb) and pIKM7 (8.8 kb) were constructed by ligating the signal sequence, amplified by PCR from the 5′ end of T. saccharolyticum JW/SL-YS485 xynA, in frame to manA and celS, respectively. Plasmid pIKM8 (10.1 kb) was constructed by inserting the PCR-amplified cbhA from C. thermocellum into XbaI/SmaI-cut pIKM1.
FIG. 1.
Construction of plasmids pIKM3, pIKM4, pIKM5, pIKM6, pIKM7, and pIKM8 from E. coli-Thermoanaerobacterium shuttle vector pIKM1. MCS, multiple cloning site; MLS, macrolide lincosamide streptogramicidin.
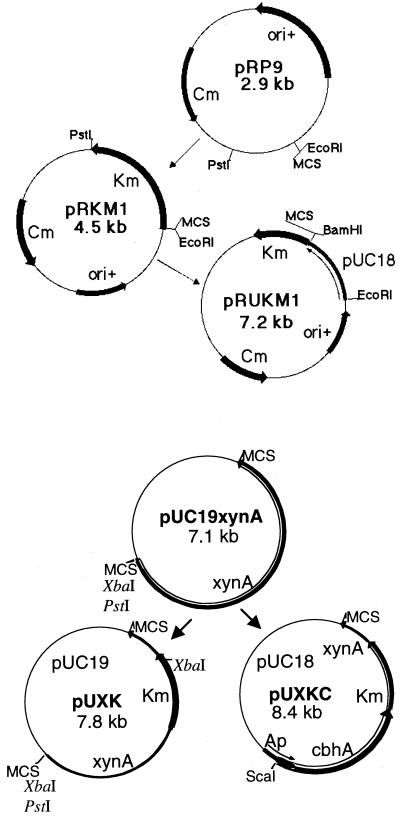
Plasmid pRKM1 was constructed as follows (Fig. 2). Using standard subcloning procedures (2), plasmid pKD102 (21) was digested with EcoRI/PstI, and the 1.6-kb band containing the kanamycin resistance cassette was isolated from a 1% agarose gel after electrophoresis. The DNA was subsequently ligated into EcoRI/PstI-digested pRP9 (2.9 kb), yielding plasmid pRKM1 (4.5 kb). Plasmid pRUKM1 was constructed by ligating pUC18 into pRKM1, both digested with EcoRI/BamHI.
FIG. 2.
Construction of shuttle vectors pRKM1 and pRUKM1 and suicide vectors pUXK and pUXKC. MCS, multiple cloning site.
The suicide vector pUXK was constructed by ligating the kanamycin resistance cassette from shuttle vector pIKM1 digested with SmaI/PstI into EcoRV/NsiI-digested pUC19xynA, which carries the cloned T. saccharolyticum xynA gene. Similarly, suicide vector pUXKC was constructed by fusing PCR-amplified cbhA from C. thermocellum to the kanamycin resistance cassette on pUXK (Fig. 2).
Transformation of T. saccharolyticum JW/SL-YS485.
Transformations were performed as described previously (16). Briefly, cells were grown in Hungate tubes with 10 ml of prereduced mineral medium at 60°C. Isonicotinic acid hydrazide (isoniacin) was added to cultures in early exponential phase at a final concentration of 4 μg ml−1 to weaken the cell wall (5). Cells were harvested and washed in sterile prereduced water or 270 mM sucrose (N2 flushed; 1 μmol of Na2S/ml added). Transformation was performed via a single electroporation pulse (1.25 kV, 400 Ω, 25 mF) using a Bio-Rad gene pulser.
Plasmid isolation.
Plasmid DNA from transformed E. coli TG1 was isolated using commercial miniprep kits (Qiagen and Promega). Plasmid DNA from transformed T. saccharolyticum JW/SL-YS485 was initially extracted via the modified isolation procedure of O'Sullivan and Klaenhammer (17) as described previously (16). In addition, after lysozyme-mediated cell lysis, a commercial miniprep kit (Qiagen) was successfully used.
PCR amplification.
PCR amplification was performed by use of a Perkin-Elmer thermal cycler according to the manufacturer's instructions. Taq polymerase and Tfl polymerase (Promega) were used in the reactions with the supplied buffers. Primers for PCR amplification were designed using the OLIGO software and contained restriction enzymes recognition sites at their 5′ ends for subsequent subcloning. PCR products were first cloned into pGM-T (Promega) before subcloning was performed. The primers used were as follows: primer 1 (signal sequence xynA amplification for ligation to manA and celS), 5′ ATCGTCTAGATAGTGTCAGTGGTT 3′; primer 2 (signal sequence xynA amplification for ligation to manA), 5′ TCATCCATGGTGCTTGTTCAGTAG 3′; primer 3 (signal sequence xynA amplification for ligation to celS), 5′ TCATCTGCAGTTGCTGCTTGTTCAGTA 3′; primer 1 (cbhA amplification), 5′ CCCGGGATTAACTGTTTGTGCGCTAT 3′; primer 2 (cbhA amplification), 5′ TCTAGAGCTCTTATCGATATGGCAATTC 3′.
Southern hybridization.
The probe against the 5′-end 0.4 kb of the T. saccharolyticum xynA gene was generated using a PCR-DIG (digoxigenin) labeling kit (Boehringer Mannheim) according to the manufacturer's instructions. The commercial nucleotide mix containing DIG-labeled dUTP was diluted one-half with nonlabeled nucleotide mix to increase product yield. The sequences of the two primers were 5′ ATCGTCTAGATAGTGTCAGTGGTT 3′ (primer 1) and 5′ TCATCCATGGTGCTTGTTCAGTAG 3′ (primer 2).
Enzyme assay.
Mannanase activity was determined at neutral pH using azo-carob galactomannan as a substrate. Uncut substrate was precipitated with 95% ethanol and centrifuged, and the amount of dye released into the supernatant was measured at 590 nm as instructed by the manufacturer (Megazyme, Sydney, New South Wales, Australia), using a standard curve generated for the specific lot. Endoglucanase, cellobiohydrolase, and exoglucanase activities were determined by measuring the amount of reducing sugars released from 7L CMC (carboxymethyl cellulose) (Hercules, Wilmington, Del.), acid-swollen cellulose, Avicel, or crystalline cellulose, employing the p-hydroxybenzoic acid hydrazide assay as described by Lever (11), using glucose for preparation of standard curves. Additionally, p-nitrophenol-β-d-cellobioside was used as a substrate for cellobiohydrolase, and the release of p-nitrophenol was determined at 405 nm as described previously (27). Enzyme activities were determined at neutral pH and are expressed in units defined as the formation of 1 μmol of product per min at 60°C. Specific activities are given in units per milligram of protein (determined by Bradford assay).
RESULTS AND DISCUSSION
Effects of the shuttle vector on growth of T. saccharolyticum JW/SL-YS485.
Plasmid pIKM1 (16) was used to insert a thermophilic mannanase from Caldicellulosiruptor (4) (yielding pIKM3 and pIKM4) and an endoglucanase (pIKM5), an exoglucanase (pIKM6/7), and a cellobiohydrolase (pIKM8) from C. thermocellum (7, 23, 27) as described above. All plasmids displayed relatively good stability in transformed T. saccharolyticum JW/YS485 as demonstrated by comparing the numbers of CFU on selective (50 μg of kanamycin/ml) and nonselective (no antibiotic) plates. Under the nonselective conditions of growth in the absence of kanamycin for 50 generations, at least 20% of the transformed Thermoanaerobacterium cells still retained the plasmid. All transformants displayed growth characteristics similar to those of the wild type, as judged by total cell protein patterns, microscopy of cell shapes, and growth rates. The growth rates and yields for the wild type and the transformants at 50 and 60°C were very similar (i.e., at 50°C, 141 min for the wild type versus 143 to 145 for the transformants; at 60°C, 52 min [wild type] versus 52/53 min [pIKM7/pIKM8], 55 min [pIKM3], and 57 min [pIKM5]).
Expression of mannanase.
Mannanase activity in T. saccharolyticum is highly regulated and repressed during growth on monosaccharides such as glucose (unpublished results). Thus, mannanase-encoding manA from Caldicellulosiruptor (4) was subcloned into the E. coli-Thermoanaerobacterium shuttle vector pIKM1 to yield plasmid pIKM3 (Fig. 1). Because the 5′ end of manA encodes a negatively charged residue atypical of a signal sequence, we inserted a gene construct that encodes a protein that has the signal sequence from the extracellular T. saccharolyticum JW/SL-YS485 xylanase (XynA) ligated to the activity-bearing carboxy-terminal domain of the mannanase into pIKM1, yielding pIKM4 (Fig. 1). Both constructs (plasmids pIKM3 and pIKM4) mediated mannanase activity in transformed E. coli (10 U/ml for both constructs) as well as T. saccharolyticum (4 U/ml for pIKM3 and 4.5 U/ml for pIKM4). However, activity in both hosts was primarily intracellular. Endogenous mannanase activity in T. saccharolyticum is repressed under growth on glucose, which results in an adaptation time of about 16 h for growth on mannan (locust bean gum). The expressed enzyme activities in transformed Thermoanaerobacterium, however, allowed for a shorter lag time (less than 3 h) for growth on locust bean gum.
Expression of endoglucanase.
The endoglucanase (CelD) from C. thermocellum degrades the artificial cellulose CMC as well as, albeit slowly, microcrystalline Avicel (6). The celD gene from C. thermocellum was subcloned into the E. coli-Thermoanaerobacterium shuttle vector pIKM1 to give plasmid pIKM5 (Fig. 1). In contrast to the recombinant protein in E. coli that accumulates in inclusion bodies (6), the enzyme was expressed in its active form in transformed T. saccharolyticum. Cell extracts of T. sacharolyticum transformed with plasmid pIKM5 exhibited an endoglucanase activity of 1.8U/mg on CMC and a lower activity on Avicel. Growth on CMC was slightly improved. Whereas the wild-type grows on the artificial celluloses LV CMC (Sigma) and 7L CMC (Hercules) to final optical densities of 0.12 and 0.15, transformed T. saccharolyticum (pIKM5) showed consistently improved growth up to optical densities of 0.15 (LV CMC) and 0.2 (7L CMC).
Expression of exoglucanase.
The exoglucanase (CelS) from C. thermocellum possesses activity against crystalline cellulose in connection with the cellulose-binding domain containing the structural backbone of the cellulosome, encoded by cipA (23). The celS clone is truncated at the 5′ end and lacks the promoter region as well as the region encoding the signal sequence. The celS gene was subcloned into the E. coli-Thermoanaerobacterium shuttle vector pIKM1 to give plasmid pIKM6 (Fig. 1). As expected, no cellulase activity in Thermoanaerobacterium transformed with plasmid pIKM6 was detected. The presumed promoter region containing the 0.4-kb 5′ end of the extracellular xylanase (XynA) from strain JW/SL-YS485 was ligated to the celS clone, resulting in a fusion protein which has the XynA signal sequence fused to the exoglucanase (CelS), producing plasmid pIKM7 (Fig. 1). Plasmid pIKM7 mediated exoglucanase activity in transformed Thermoanaerobacterium. Exoglucanase activity was low with CMC as a substrate, but a significantly higher activity of 4 U/mg of protein was observed with acid-swollen cellulose. This is in agreement with the activities described for the urea-renatured recombinant protein in E. coli (23). Again, in contrast to the recombinant protein in E. coli, where the enzyme accumulates in inclusion bodies, in transformed Thermoanaerobacterium the enzyme was expressed in its active form. Excretion mediated by the xynA signal sequence was only limited, strengthening the argument for the presence of more complex requirements for recognition and translocation of extracellular enzymes.
Expression of cellobiohydrolase.
The gene for the cellobiohydrolase (CbhA) from C. thermocellum strain F7 was previously cloned and sequenced (22, 27). This enzyme has been shown to contain intrinsic cellulose-binding domains and thus can act independently of the cellulosome complex. We designed PCR primers (see Materials and Methods) according to the sequence information and amplified the cellobiohydrolase from C. thermocellum JW20. The PCR-amplified cbhA was subcloned into pIKM1, and the resulting vector pIKM8 was used to transform Thermoanaerobacterium. Using CMC as the substrate, cell extracts of these transformants showed an activity of 8 U/mg of protein. Activity on the artificial substrate p-nitrophenol-β-d-cellobioside was detected above the background which was caused by an endogenous β-d-glucosidase. Apparently the signal sequence from C. thermocellum JW20 is not recognized or processed correctly in T. saccharolyticum, resulting in intracellular accumulation of the enzyme.
Construction of new shuttle vectors.
To obtain an alternative transformation system for our model system in T. saccharolyticum JW/SL-YS485 (16), new shuttle vectors were constructed and designated pRKM1 and pRUKM1. Plasmid pRKM1 (Fig. 2), based on plasmid pRP9 from Bacillus stearothermophilus (3), was constructed as described in Materials and Methods and used to transform E. coli TG-1. The plasmid construct was verified by restriction analysis of plasmid DNA extracted from cultures grown in Luria broth containing 50 μg of kanamycin/ml. Because plasmid pRKM1 does not contain a typical gram-negative origin of replication and thus transforms E. coli only at low frequencies, we generated plasmid pRUKM1 by fusion of plasmid pRKM1 with pUC18 (Fig. 2), leading to significantly improved transformation frequencies in E. coli. Transformation with pRUKM1 was performed as described previously (16); the observed transformation efficiency varied between 10 and 1,000 transformants per μg of plasmid DNA. This frequency is similar to the efficiencies achieved with the previously described shuttle vector pIKM1. Despite standardization of the procedure, the remaining variability in transformation efficiency is likely due to difficulties with the autoplast generation and/or recovery that is part of the transformation protocol. Plasmid DNA isolated from transformed T. saccharolyticum yielded the characteristic restriction pattern (data not shown). Thus, these plasmids did not undergo rearrangement in Thermoanaerobacterium and were relatively stable in transformants. The stability was the same as for the derivatives of pIKM1 described above. We observed only negligible differences in the doubling times of the transformants and the wild-type strain JW/SL-YS485.
Plasmid purifications from transformed JW/SL-YS485 consistently gave higher yields than previously observed with the shuttle vector pIKM1, which indicates a higher copy number of the new shuttle vectors. The newly developed shuttle vectors pRKM and pRUKM1 mediate kanamycin resistance to higher levels than previously observed with shuttle vector pIKM1. T. saccharolyticum JW/SL-YS485 is resistant to ampicillin at 100 μg ml−1 and chloramphenicol at 50 μg ml−1 but sensitive to kanamycin at an MIC of 25 μg ml−1 (60°C, pH 6.2). After transformation with plasmids pRKM1 and pRUKM1, the strain became resistant to 800 μg of kanamycin ml−1 at 48°C and 400 μg ml−1 at 60°C. This resistance is twice as high as that found with the shuttle vector pIKM1. In addition to allowing for an alternative plasmid replicon, the new vectors have the potential to increase expression levels of heterologous genes in Thermoanaerobacterium.
Chromosomal integration of the kanamycin cassette and the cbhA gene.
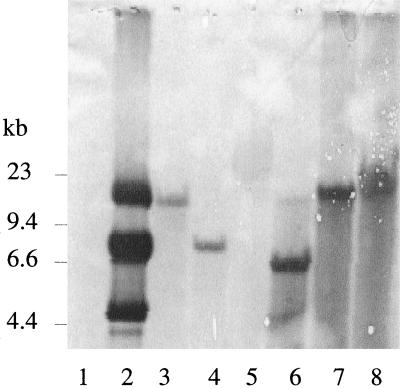
To grow transformed Thermoanaerobacterium over an extended period in the absence of selective pressure while maintaining the recombinant activity, we chose to obtain integration of foreign genes into the chromosome of T. saccharolyticum JW/SL-YS485. Integration was achieved via homologous recombination using a suicide vector containing the kanamycin resistance cassette and the xynA gene from strain JW/SL-YS485 as the site of integration. The xynA gene product is required only during growth on xylan; XynA has previously been sequenced, and the PCR-amplified 5′ end of this gene has been used to direct secretion of hydrolytic enzymes (V. Mai and J. Wiegel, Abstr. 98th Gen. Meet. Am. Soc. Microbiology, abstr. O-63, 1998). Suicide vectors pUXK (containing the kanamycin resistance cassette) and pUXKC (containing the kanamycin resistance cassette and cbhA gene from C. thermocellum) were constructed as described in Materials and Methods (Fig. 2). The vectors are based on pUC18. The new constructs do not contain a gram-type-positive thermostable replicon and thus function as suicide vectors in Thermoanaerobacterium. The plasmid constructs were verified by restriction analysis and used to transform JW/SL-YS485. The successful chromosomal integration of the suicide vector was verified by restriction analysis followed by Southern blot analysis (Fig. 3). Southern blot analysis of DNA isolated from T. saccharolyticum transformed with plasmid pUXK or the same DNA digested with enzymes which do not cut within the suicide vector (Fig. 3, lanes 7 and 8) showed no signs of plasmid bands. Digestion of DNA with enzymes cutting within the sequence of pUXK released the expected vector DNA from the chromosome (Fig. 3, lane 6), indicating insertion of the vector into the chromosome via a single-crossover mechanism. Chromosomal insertion by a similar mechanism mediated by suicide plasmids carrying antibiotic resistance has previously been observed in Methanococcus maripuladis (18) and in a thermophilic bacillus (19).
FIG. 3.
Southern blot analysis of DNA isolated from Thermoanaerobacterium strain YS485 wild type and YS485 transformed with suicide vector pUXK. The DIG-labeled probe against the 5′ end of xynA was generated as described in Materials and Methods. Plasmid pUXK contains neither NsiI nor NcoI but one PstI recognition site. The probe hybridized with the PstI-linearized pUXK. Lanes: 1, lambda HindIII (bands are not visible on Southern blots; however, the positions of the four largest bands on the agarose gel are marked on the side); 2, pUXK undigested; 3, YS485 wild type digested with PstI; 4, YS485 wild type digested with NsiI; 5, YS485 wild type digested with NcoI; 6, YS485/pUXK digested with PstI; 7, YS485/pUXK digested with NsiI; 8, YS485/pUXK digested with NcoI.
Expression of cbhA coding for the cellobiohydrolase after integration.
Integration of pUXKC into Thermoanaerobacterium resulted in the expression of cbhA from C. thermocellum, as judged by the presence of cellobiohydrolase activity. This cellobiohydrolase has been shown to contain intrinsic cellulose-binding domains and thus can act on cellulose independently from the cellulosome complex (22, 27). The observed cellobiohydrolase activity in Thermoanaerobacterium after the chromosomal integration of pUXKC was 6.5 U/mg, which compares to 8 U/mg in Thermoanaerobacterium transformed with the previously described shuttle vector pIKM8.
Chromosomal integration has been achieved for the first time in any gram-type-positive anaerobic thermophile. The activity levels in Thermoanaerobacterium after integration via suicide vector pUXKC are similar to the levels achieved with the shuttle vector pIKM8; however, selective pressure is no longer required after integration.
Conclusions.
The utilization of cellulolytic plant materials requires the synergistic action of hemicellulolytic as well as cellulolytic enzymes. In an initial inquiry into the feasibility of improving the utilization of such abundant and inexpensive substrates by thermophilic anaerobes, we successfully expressed four such enzymes in Thermoanaerobacterium by using shuttle vectors pIKM3-8, demonstrating that improvement of substrate utilization in thermophilic anaerobes can be achieved principally via expression of hydrolytic genes from shuttle vectors. However, a better secretion of extracellular enzymes and the integration of a whole suite of enzymes need to be achieved to successfully convert these bacteria into industrially useful strains. The improvements described above for a genetic system of Thermoanaerobacterium, our model system for gram-positive anaerobic thermophiles, show the general feasibility of genetic manipulations in this important group of microorganisms. The utilization of the improved shuttle vectors and especially the use of the herein demonstrated chromosomal integration should facilitate further genetic research of Thermoanaerobacterium and related gram-type-positive anaerobic bacteria such as the cellulolytic thermophile C. thermocellum or the ethanol-producing Thermoanaerobacter ethanolicus.
ACKNOWLEDGMENTS
We thank P. Bergquist, P. Beguin, and D. Wu for the gifts of cloned manA, celD, and celS, respectively, and N. E. Welker for the gift of plasmid pRP9. We also thank Cara Runsick Mitchell for editing the manuscript.
REFERENCES
- 1.Aguilar A. Extremophile research in the European Union: from fundamental aspects to industrial expectations. FEMS Microbiol Rev. 1996;18:89–92. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 3.De Rossi E, Brigidi P, Welker N E, Riccardi G, Matteuzzi D. New shuttle vector for cloning in Bacillus stearothermophilus. Res Microbiol. 1994;145:579–583. doi: 10.1016/0923-2508(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 4.Gibbs M D, Elinder A U, Reeves R A, Bergquist P L. Sequencing, cloning and expression of a β-1,4-mannanase gene, manA, from the extremely thermophilic anaerobic bacterium, Caldicellulosiruptor Rt8B4. FEMS Microbiol Lett. 1996;141:37–43. doi: 10.1111/j.1574-6968.1996.tb08360.x. [DOI] [PubMed] [Google Scholar]
- 5.Hermans J, Boschloo J G, de Bont J A M. Transformation of M. aurum by electroporation: the use of glycine, lysozyme and isonicotinic acid hydrazide in enhancing transformation efficiency. FEMS Microbiol Lett. 1990;72:221–224. [Google Scholar]
- 6.Joliff G, Beguin P, Juy M, Millet J, Ryter A, Poljak R, Aubert J P. Isolation, crystallization and properties of a new cellulase of Clostridium thermocellum overproduced in E. coli. Bio/Technology. 1986;4:896–900. [Google Scholar]
- 7.Joliff G, Beguin P, Aubert J P. Nucleotide sequence of the cellulase gene celD encoding endoglucanase of Clostridium thermocellum. Nucleic Acids Res. 1986;14:8605–8613. doi: 10.1093/nar/14.21.8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klapatch T R, Guerinot M L, Lynd L R. Electrotransformation of Clostridium thermosaccharolyticum. J Ind Microbiol. 1996;16:342–347. doi: 10.1007/BF01570112. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y E, Jain M K, Lee C, Lowe S E, Zeikus J G. Taxonomic distinction of saccharolytic thermophilic anaerobes: description of Thermoanaerobacterium xylanaolyticum gen. nov., sp. nov., and Thermoanaerobacterium saccharolyticum gen. nov., sp. nov; reclassification of Thermoanaerobium brockii, Clostridium thermosulfurogenes, and Clostridium thermohydrosulfuricum E100–69 as Thermoanaerobacter brockii comb. nov., Thermoanaerobacterium thermosulfurigenes comb. nov., and Thermoanaerobacter thermohydrosulfuricus comb. nov., respectively: and transfer of Clostridium thermohydrosulfuricum 39E to Thermoanaerobacter ethanolicus. Int J Syst Bacteriol. 1993;43:41–51. [Google Scholar]
- 10.Lee Y E, Ramesh M V, Zeikus J G. Cloning, sequencing and biochemical characterization of xylose isomerase from Thermoanaerobacterium saccharolyticum strain B6A-RI. J Gen Microbiol. 1993;139:1227–1234. doi: 10.1099/00221287-139-6-1227. [DOI] [PubMed] [Google Scholar]
- 11.Lever M. Colorimetric and fluorometric carbohydrate determination with p-hydroxybenzoic acid hydrazide. Biochem Med. 1973;7:274–281. doi: 10.1016/0006-2944(73)90083-5. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y-S, Gherardini F C, Matuschek M, Bahl H, Wiegel J. Cloning, sequencing, and expression of the gene encoding a large S-layer-associated endoxylanase from Thermoanaerobacterium sp. strain JW/SL-YS 485 in Escherichia coli. J Bacteriol. 1996;178:1539–1547. doi: 10.1128/jb.178.6.1539-1547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y-S, Gherardini F C, Matuschek M, Bahl H, Wiegel J. Purification and cloning of a thermostable xylose (glucose) isomerase with an acidic pH optimum from Thermoanaerobacterium strain JW/SL-YS 485. J Bacteriol. 1996;178:5938–5945. doi: 10.1128/jb.178.20.5938-5945.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorenz W W, Wiegel J. Isolation, analysis, and expression of two genes from Thermoanaerobacterium sp. strain JW/SL-YS485: a beta-xylosidase and a novel xylan esterase with cephalosporin C deacetylase activity. J Bacteriol. 1997;179:5436–5441. doi: 10.1128/jb.179.17.5436-5441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe S E, Jain M K, Zeikus J G. Biology, ecology, and biotechnological applications of anaerobic bacteria adapted to environmental stresses in temperature, pH, salinity, or substrates. Microbiol Rev. 1993;57:451–509. doi: 10.1128/mr.57.2.451-509.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mai V, Lorenz W W, Wiegel J. Transformation of Thermoanaerobacterium sp. strain JW/SL-YS485 with plasmid pIKM1 conferring kanamycin resistance. FEMS Microbiol Lett. 1997;148:163–167. [Google Scholar]
- 17.O'Sullivan D J, Klaenhammer T R. Rapid Mini-Prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus. Appl Environ Microbiol. 1993;59:2730–2734. doi: 10.1128/aem.59.8.2730-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandbeck K E, Leigh J A. Recovery of an integration shuttle vector from tandem repeats in Methanococcus maripuladis. Appl Environ Microbiol. 1991;57:2762–2763. doi: 10.1128/aem.57.9.2762-2763.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shenday A, Rao M. Chromosomal gene integration and enhanced xylanase production in an alkaliphilic thermophilic Bacillus sp. (NCIM 59) Biochem Biophys Res Commun. 1993;195:776–784. doi: 10.1006/bbrc.1993.2113. [DOI] [PubMed] [Google Scholar]
- 20.Soutschek-Bauer E, Hartl L, Staudenbauer W L. Transformation of Clostridium thermohydrosulfuricum DSM 568 with plasmid DNA. Biotechnol Lett. 1985;7:705–710. [Google Scholar]
- 21.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the S. faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 22.Tuka K, Zverlov V V, Bumazkin B K, Velikovorskaya G A, Strongin A Y. Cloning and expression of Clostridium thermocellum genes coding for thermostable exoglucanases (cellobiohydrolases) in E. coli cells. Biochem Biophys Res Commun. 1990;169:1055–1060. doi: 10.1016/0006-291x(90)92001-g. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, Kruus K, Wu J H D. Cloning and DNA sequence of the gene coding for Clostridium thermocellum cellulase SS (CelS) a major cellulosome component. J Bacteriol. 1993;175:1293–1302. doi: 10.1128/jb.175.5.1293-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiegel J. The anaerobic thermophilic bacteria. In: Kristjansson J K, editor. Thermophilic bacteria. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 105–184. [Google Scholar]
- 25.Wiegel J. Distinction between the Gram reaction and the Gram type of bacteria. Int J Syst Bacteriol. 1981;31:88. [Google Scholar]
- 26.Wiegel J, Ljungdahl L G. The importance of thermophilic bacteria in biotechnology. Crit Rev Biotechnol. 1986;3:39–107. [Google Scholar]
- 27.Zverlov V V, Velikodvorskaya G V, Schwarz W H, Bronnenmeier K, Kellermann J, Staudenbauer W L. Multidomain structure and cellulosomal localization of the Clostridium thermocellum cellobiohydrolase CbhA. J Bacteriol. 1998;180:3091–3099. doi: 10.1128/jb.180.12.3091-3099.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]