Abstract
Background
The enterosalivary nitrate–nitrite–nitric oxide (NO3–NO2–NO) pathway generates NO following oral microbiota‐mediated production of salivary nitrite, potentially linking the oral microbiota to reduced cardiometabolic risk. Nitrite depletion by oral bacteria may also be important for determining the net nitrite available systemically. We examine if higher abundance of oral microbial genes favoring increased oral nitrite generation and decreased nitrite depletion is associated with a better cardiometabolic profile cross‐sectionally.
Methods and Results
This study includes 764 adults (mean [SD] age 32 [9] years, 71% women) enrolled in ORIGINS (Oral Infections, Glucose Intolerance, and Insulin Resistance Study). Microbial DNA from subgingival dental plaques underwent 16S rRNA gene sequencing; PICRUSt2 was used to estimate functional gene profiles. To represent the different components and pathways of nitrogen metabolism in bacteria, predicted gene abundances were operationalized to create summary scores by (1) bacterial nitrogen metabolic pathway or (2) biochemical product (NO2, NO, or ammonia [NH3]) formed by the action of the bacterial reductases encoded. Finally, nitrite generation‐to‐depletion ratios of gene abundances were created from the above summary scores. A composite cardiometabolic Z score was created from cardiometabolic risk variables, with higher scores associated with worse cardiometabolic health. We performed multivariable linear regression analysis with cardiometabolic Z score as the outcome and the gene abundance summary scores and ratios as predictor variables, adjusting for sex, age, race, and ethnicity in the simple adjusted model. A 1 SD higher NO versus NH3 summary ratio was inversely associated with a −0.10 (false discovery rate q=0.003) lower composite cardiometabolic Z score in simple adjusted models. Higher NH3 summary score (suggestive of nitrite depletion) was associated with higher cardiometabolic risk, with a 0.06 (false discovery rate q=0.04) higher composite cardiometabolic Z score.
Conclusions
Increased net capacity for nitrite generation versus depletion by oral bacteria, assessed through a metagenome estimation approach, is associated with lower levels of cardiometabolic risk.
Keywords: 16S rNA sequencing, blood pressure, epidemiology, insulin resistance, metagenomics, nitric oxide, oral microbiome
Subject Categories: Epidemiology; Diabetes, Type 2; High Blood Pressure; Risk Factors; Endothelium/Vascular Type/Nitric Oxide
Nonstandard Abbreviations and Acronyms
- ANR
assimilatory nitrate reduction
- CMZ
composite cardiometabolic Z score
- DNRA
dissimilatory nitrate reduction to ammonia
- FDR
false discovery rate
- KO
Kyoto Encyclopedia of Genes and Genomes Orthologs
- RD
respiratory denitrification
Clinical Perspective
What Is New?
Previous investigations have observed that nitrate‐reducing oral bacteria and the nitrite‐generating capacity of the oral microbiome are associated with reduced cardiometabolic disease risk through the enterosalivary NO3–NO2–NO pathway of NO generation.
This study uses predicted metagenomic content from 16S rRNA sequencing to operationalize different components of the NO3–NO2–NO pathway to investigate whether the different pathways of bacterial nitrate metabolism, levels of a specific reductase gene, or the overall balance/ratio between nitrite generation and nitrite depletion are of greatest relevance to cardiometabolic outcomes.
Our results suggest that nitrite depletion by oral bacteria can influence the NO3–NO2–NO pathway, and gene scores that conceptually capture the net NO‐generating potential of the oral microbiome were most strongly and consistently associated with cardiometabolic outcomes.
What Are the Clinical Implications?
This knowledge will aid in clinical trial designs that aim to enrich oral nitrate reducers by identifying individuals with low levels of net NO‐producing capacity and characterizing an intermediate biomarker of efficacy.
Furthermore, a high net nitrite‐generating capacity could be a characteristic used to identify potential candidate bacteria species for probiotics to enhance enterosalivary NO generation.
Nitric oxide (NO) is an important signaling molecule for cardiometabolic health, and is involved in vasodilation, glucose metabolism, and other physiological functions. 1 , 2 , 3 The oral microbiome contributes to NO generation through the enterosalivary nitrate–nitrite–NO (NO3–NO2–NO) pathway. 3 Oral bacteria reduce salivary nitrate (NO3) derived from dietary nitrate or metabolism of endogenously produced NO to nitrite (NO2), which is swallowed and made systemically available for further reduction into NO in the blood vessels and tissues. 3 The potential importance of the oral microbiome in this pathway and the beneficial effect of this NO pathway on blood pressure and metabolism has been demonstrated through several experimental studies. 1 , 3 , 4 Decreases in salivary and plasma nitrite, 5 , 6 , 7 , 8 , 9 , 10 , 11 and corresponding increases in blood pressure and plasma glucose levels, 6 , 7 , 8 , 12 following antibacterial mouthwash use and presumed lower oral bacteria levels, have been observed. Likewise, associations between bacterial taxa and genes coding for bacterial enzymes along the NO3–NO2–NO pathway, and blood pressure levels have been found, 13 , 14 further emphasizing the importance of the oral microbiome.
Because nitrite is thought to act as a storage pool for bioavailable NO, most studies have focused on the nitrate‐reducing capacity (ie, the nitrite‐generating capacity of the oral microbiome). 7 , 9 , 10 , 13 However, some oral bacteria can also further reduce salivary nitrite to NO or ammonia (NH3), affecting the amount of salivary nitrite swallowed and absorbed into the circulation for systemic NO generation. Figure 1 illustrates the 3 main pathways of bacterial nitrogen metabolism. 3 , 15 Previous authors have suggested that an oral microbiome that allows nitrite accumulation is beneficial, and that the overall balance between nitrite‐generating versus nitrite‐depleting capacity may be more important than the absolute nitrate‐reducing capacity of the oral microbiome. 16 In an experimental study, no association was observed between nitrate reductase genes and blood pressure, but higher levels of NO‐forming nitrite reductase genes and lower levels of NH3‐forming nitrite reductase genes were observed to be associated with lower systolic blood pressure. 14 This suggests that salivary nitrite depletion by oral bacteria can influence the NO3–NO2–NO pathway and may be associated with blood pressure as well, and that the association may vary dependent on the product (NO or NH3) of that nitrite depletion. Larger population‐based studies examining how in addition to nitrite generation (ie, nitrate reduction), the nitrite depletion by oral bacteria may also be associated with cardiometabolic risk, can help validate findings from smaller experimental studies highlighting the importance of oral bacteria in cardiometabolic health.
Figure 1. Representation of the 3 major bacterial pathways for nitrate reduction to nitrite.
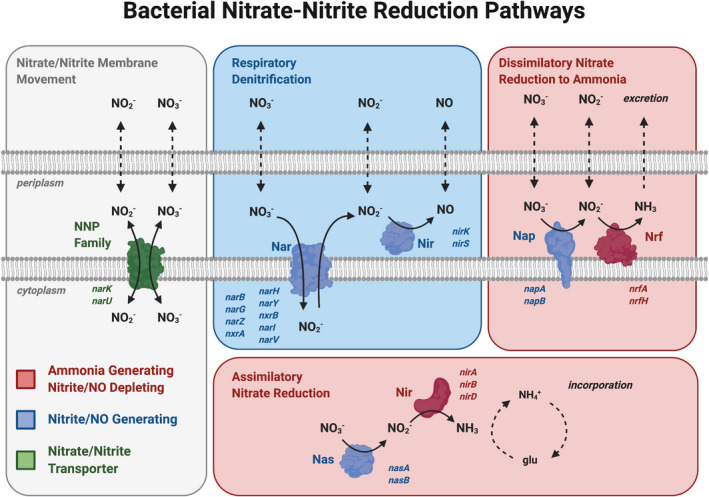
(Adapted from Koch et al 3 and Sparacino‐Watkins et al 15 and created with BioRender.com). Pathways, proteins and genes are color coded. Blue genes and proteins indicate NO2 or NO‐generating enzymes, whereas red indicates nitrite depletion to NH3. Nitrite and nitrate move through the plasma membrane through NNP (nitrate/nitrite transporter ) (in green), coded by the gene narK. The blue box contains the RD pathway that increases nitrite and NO availability through the nitrate reductase Nar (coded by genes narB, narG, narZ, nxrA, narH, narY, nxrB, narI, narV) and nitrite reductase Nir (coded by genes nirK and nirS), respectively, and thus characterized as a nitrite/NO‐generating pathway. Red boxes contain the DNRA and ANR pathways characterized as nitrite‐depleting pathways. Following nitrite generation by nitrate reductases Nap (coded by genes napA and napB) and Nas (coded by genes nasA and nasB), nitrite is depleted by the NH3‐producing enzyme proteins Nir (coded by the genes nirA, nirB, and nirD), and Nrf (coded by the genes nrfA and nrfB). Dotted arrows represent the diffusion of the nitrate, nitrite, nitric oxide, and ammonia in and out of the cell. ANR indicates assimilatory nitrate reduction; DNRA, dissimilatory nitrate reduction to ammonia; glu, glutamine; Nar, Nap, Nas, nitrate reductase; NH3, ammonia; NH4+, ammonium; Nir, Nrf, nitrite reductase; NO2, nitrite; NO3, nitrate; NNP, nitrate/nitrite transporter; and RD, respiratory denitrification.
Therefore, this study aimed for a more nuanced interrogation of the role of the oral microbiome in the enterosalivary pathway on cardiometabolic health by operationalizing different components of the NO3–NO2–NO pathway. Prior attempts simply focused on the relative abundance of nitrate‐reducing bacterial taxa, but it is unknown whether the different pathways of bacterial nitrate metabolism, levels of a specific reductase gene, or the overall balance between nitrite generation and depletion is of greatest relevance to cardiometabolic risk. Recent tools predicting gene abundances from 16S rRNA sequencing data show reasonable performance for human data sets in particular, with correlations of 0.79 to 0.88 with shotgun metagenomics results. 17 , 18 , 19 Therefore, using predicted gene abundances from 16S rRNA sequencing data, we aimed to examine the association of the nitrite‐generating and nitrite‐depleting capacity of the oral microbiome with cardiometabolic risk. We hypothesize that a higher abundance of oral microbial genes favoring increased oral nitrite generation and decreased nitrite depletion are associated with a better cardiometabolic profile.
Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Study Population
ORIGINS (Oral Infections, Glucose Intolerance, and Insulin Resistance Study) is a prospective cohort study investigating the relationship between the subgingival microbial community composition and impaired glucose metabolism. 20 , 21 From February 2011 to May 2013, 300 men and women were enrolled in wave 1 of the study, and from January 2016 to January 2020 an additional 800 individuals were enrolled during wave 2 of the study. Inclusion criteria were (1) age 20 to 55 years; (2) no diabetes (type 1 or type 2) based on participant self‐report of no previously diagnosed disease, hemoglobin A1c (HbA1c) values <6.5%, and fasting plasma glucose <126 mg/dL; (3) no history of myocardial infarction, congestive heart failure, stroke, or chronic inflammatory conditions based on participant self‐report. The Columbia University and University of Minnesota institutional review boards approved the protocol. All participants provided informed consent. Only wave 2 participants who had 16S rRNA microbial data from subgingival plaque samples and were not missing important baseline risk factors or outcome data were included in this study.
Subgingival Plaque Collection and Bacterial Assessments
Subgingival plaque samples (6–8 teeth per participant) were collected from predefined index teeth using sterile curettes after removal of the supragingival plaque as previously described. 20 The samples were pooled by shallow (probing depth of <4 mm) versus deep (probing depth ≥4 mm) collection sites, and suspended in 300 µL of Tris‐EDTA buffer (50 mmol/L Tris, 1 mmol/L EDTA; pH 7.6). Microbial DNA was extracted using the MasterPure Gram Positive DNA Positive Purification kit (Lucigen).
Only pooled samples from shallow sites (n=764) were used for the main analyses, because deeper probing depths are often associated with periodontal disease, and microbiome from these sites could reflect local inflammatory disease processes and environmental pressures that may alter the composition of the oral microbiome. Sensitivity analyses using a weighted average of within‐mouth gene abundances across pooled samples from both deep and shallow sites showed similar results to our main analyses and are presented in Figure S1.
16S rRNA Sequencing and Prediction of Gene Abundances Using PICRUSt2
We used 50 ng of DNA in polymerase chain reaction amplification targeting the V3 to V4 region of the 16S rRNA gene (using 341F/806R universal primers), and polymerase chain reaction products were purified using AMPure beads. 22 , 23 For each library, 100 ng were pooled, gel purified, and quantified using a bioanalyzer, and 12 pM of the library mixture was spiked with 20% PhiX and run on a MiSeq (Illumina) platform. Raw reads were analyzed with QIIME2 24 (the data curation pipeline is outlined in Data S1). Overall, 44 776 283 sequences were generated for final analysis (median of 37 067 sequences per sample).
We then used the bioinformatics tool PICRUSt2 18 through the qiime2 plugin, which uses the amplicon sequence variant relative abundance table, together with a reference database of known microbial genomes, to predict metagenomic content from 16S rRNA marker gene sequencing. The output is in the form of a matrix of Kyoto Encyclopedia of Genes and Genomes Orthologs (KO), functional orthologs containing a group of bacterial genes coding for that molecular‐level function, and the predicted count of each KO in each sample. Relative gene abundance was then calculated for each KO as the proportion of counts for that individual KO over the total counts of all KOs in the sample.
Identification of Functional Gene Orthologs on the Enterosalivary Pathway
KOs containing bacterial genes of interest on the NO3–NO2–NO pathway were identified from the Kyoto Encyclopedia of Genes and Genomes Pathways bacterial nitrogen metabolism pathway. 25 , 26 These KOs correspond to nitrate reductase enzymes (ie, nitrite‐generating) such as Nar, Nap, Nas, and the nitrite reductase enzymes (ie, nitrite‐depleting) Nir and Nrf in the 3 bacterial nitrate reduction pathways illustrated in Figure 1. Because nitrate transport is thought to be critical for nitrate reduction, we examined the KO corresponding to NNP, the nitrate/nitrite transporter responsible for majority of nitrate transport as well. 3 Therefore, a total of 16 KOs representing 21 genes of interest across 3 bacterial pathways were identified for our exposure (Table S1).
Operationalization of the Gene Abundance Exposure
Bacterial nitrate reduction to nitrite occurs along 3 major metabolic pathways: (1) respiratory denitrification (RD), (2) dissimilatory nitrate reduction to ammonia (DNRA), and (3) assimilatory nitrate reduction (ANR) (Figure 1). Each pathway has a different function, location, and nitrate and nitrite reductase enzymes involved. Following nitrite generation, nitrite depletion occurs differently for each pathway as well, leading to NO in the RD pathway and NH3 in the DNRA and ANR pathways. Based on these bacterial nitrate reduction pathways, 2 main approaches were used to operationalize the nitrite‐generating and nitrite‐depleting gene abundances, by pathway or biochemical product created.
Exposure Operationalization by Pathway
Microbial gene abundances were summed according to their pathway, creating 3 pathway summary scores: an RD pathway score, ANR pathway score, and DNRA pathway score. For example, the RD score includes only nar and nir relative gene abundances (Table S1). This enables an examination of which, if any, bacterial nitrogen metabolic pathways were associated with cardiometabolic risk.
Exposure Operationalization by Biochemical Product
Microbial gene abundances scores were also summed by the biochemical product, NO2, NO, or NH3, formed from the action of the bacterial reductase enzymes (ie, nitrate reductase or nitrite reductase) along the NO3–NO2–NO pathway of NO generation. Therefore, 3 summary scores were created: a summary score of microbial genes that generated nitrite from nitrate (NO2 score), depleted nitrite to form NO (NO score), or depleted nitrite to NH3 (NH3 score). These product summary scores are distinct from the pathway summary scores, because they can include genes from >1 pathway. For example, the NO2 summary score includes nitrate reductases genes (nar, nap, nas) from across the 3 pathways, representing the total nitrate‐reducing capacity of the oral microbiome. The NH3 summary score includes both nrf and nir gene abundances from the DNRA and ANR pathway, which reduce nitrite to NH3, whereas the NO summary score includes only nir gene abundances from the RD pathway. Table S1 details the KOs and corresponding bacterial genes used for each pathway and product summary score.
Ratios of Nitrite Generation to Nitrite Depletion
As the overall balance between nitrite‐generating versus nitrite‐depleting capacity of the oral microbiome is of interest, nitrite generation‐to‐depletion ratios of gene abundances were created. First, a ratio of NO2 versus NO+NH3 was created. NO formation in the mouth has been suggested to have systemic effects as well 14 ; therefore, we also created a ratio comparing NO2 versus NH3, and another ratio, NO2+NO versus NH3, which considered both NO2 and NO generation as contributing to the overall effect of the enterosalivary pathway and NH3 as depleting from this pathway. Finally, to assess the importance of nitrite depletion end products, we examined if the ratio of depletion of nitrite into NO, directly beneficial to cardiometabolic health, or NH3 (ie, a NO versus NH3 ratio) was associated with cardiometabolic risk.
Pathways were characterized as nitrite/nitric oxide generating (RD pathway) or nitrite depleting (DNR and ANR pathways), and ratios of nitrite generation‐to‐depletion pathways were created as well as RD versus ANR, RD versus DNRA, and RD versus ANR+DNRA.
Cardiometabolic Risk
Plasma glucose, HbA1c, and insulin were measured from blood collected following an overnight fast with standard methods. 20 , 27 Insulin resistance was measured using the Homeostasis Model Assessment for Insulin Resistance (HOMA‐IR) values calculated from fasting insulin and glucose levels. 28 Seated resting systolic and diastolic blood pressures were measured in triplicate, and the last 2 measurements averaged.
The primary outcome of interest, overall cardiometabolic risk, was calculated as the average of the summed Z scores of the following variables: HbA1c, fasting plasma glucose, insulin, HOMA‐IR, and systolic and diastolic blood pressure. Z scores were calculated for each cardiometabolic risk variable by subtracting the study population mean value, divided by the population standard deviation. Insulin resistance and plasma insulin were natural log‐transformed to address their skewed distributions before standardization into Z scores. A higher composite cardiometabolic Z score (CMZ) represents worse cardiometabolic health.
Risk Factor Assessment
Cardiometabolic risk factors were measured by trained research assistants as previously described. 20 Questionnaires assessed information on age, sex, race and ethnicity (non‐Hispanic Black, non‐Hispanic White, Hispanic, other (including American Indian or Alaska Native, Native Hawaiian or Pacific Islander, and Asian), educational level (less than a bachelor’s degree, bachelor’s degree, higher than a bachelor’s degree), and cigarette smoking (current, former, or never smoking). Body mass index was calculated as weight (in kilograms)/height (meters2) from in‐person physical assessments. From clinical oral examinations, periodontal measures were obtained, and the periodontal status of the participants classified according to the Centers for Disease Control and Prevention/American Academy of Periodontology diagnosis classification (none/mild versus moderate/severe). 29
Statistical Analysis
Individual relative gene abundances were first summed into pathway and biochemical product summary scores as described above, and then arcsine square root transformed to stabilize the variance and create a more normally distributed continuous summary score variable for use in linear regression analyses. 30 Ratios of nitrite generation‐to‐depletion (described above) underwent natural log‐transformation before statistical analyses.
We performed multivariable linear regression analyses with composite and individual cardiometabolic risk variable Z‐scores as the outcome, and the arcsine square root transformed relative gene abundance summary scores and log‐transformed ratios as the predictor variables, adjusting for the potential confounders (sex, age, and race and ethnicity) in the simple adjusted model. We additionally adjusted for education, smoking status, body mass index, and periodontal disease status in the fully adjusted model. These adjustments were made either a priori based on previous studies or on their association with nitrite generation‐to‐depletion ratio NO versus NH3 and the primary outcome CMZ score at an α=0.20 threshold of significance (Table S2). The false discovery rate (FDR) q value calculated according to the Benjamini‐Hochberg method was used to account for multiple‐hypothesis testing. 31 All results are presented as the change in the dependent variable for a 1 SD change in the independent variable. The key results are also graphically presented as exposure quartiles, with the cardiometabolic risk biomarkers in their original scale for interpretability. Exploratory analyses of the relationship between the 16 individual natural log‐transformed relative gene abundances and cardiometabolic risk were also conducted, but no clear patterns were observed, and the results are presented in Figure S2.
Results
Participant Characteristics and Predicted Gene Abundance Counts
Of the 764 participants (mean [SD] age=32 [9] years, 71% women), 71% of participants had none or mild periodontitis as defined per the Centers for Disease Control and Prevention/American Academy of Periodontology guidelines, and a majority were never smokers (82%) (Table). The mean relative gene abundances for the bacterial nitrate and nitrite reductases are presented in Table S1. Overall, the bacterial gene with the highest relative gene abundance was nar, with a 0.08% total mean relative gene abundance, and the lowest was the nas gene, representing 0.001% of all genes in the sample (Table S1). Of the total bacterial metagenomic content, 0.098% was nitrate‐reducing (nar, nap, nas). Almost all of the bacterial genes of interest were detected in 100% of participants. Only genes nirS, narB, nasB, and nasA were not prevalent in all participants, represented in only 94%, 62%, 21%, and 2% of participants, respectively.
Table 1.
Characteristics of the 764 Diabetes‐Free Adults in the ORIGINS Study
| Characteristic | Mean (SD) or n (%) | ≤ Median CMZ | > Median CMZ | P value |
|---|---|---|---|---|
| Age, y | 31.66 (9.36) | 29.30 (7.96) | 34.03 (10.06) | <0.0001 |
| Sex | <0.0001 | |||
| Women | 541 (70.81%) | 305 (79.84%) | 236 (61.78%) | |
| Men | 223 (29.19%) | 77 (20.16%) | 146 (38.22%) | |
| Race and ethnicity | <0.0001 | |||
| Hispanic | 217 (28.40%) | 146 (38.22%) | 71 (18.59%) | |
| Non‐Hispanic White | 111 (14.53%) | 30 (7.85%) | 81 (21.20%) | |
| Black | 212 (27.75%) | 89 (23.30%) | 123 (32.20%) | |
| Other (including American Indian/Alaska Native, Native Hawaiian/Pacific Islander, and Asian) | 218 (28.53%) | 113 (29.58%) | 105 (27.49%) | |
| Education | <0.0001 | |||
| < Bachelor’s degree | 171 (22.38%) | 52 (13.61%) | 119 (31.15%) | |
| Bachelor’s degree | 365 (47.77%) | 204 (53.40%) | 161 (42.15%) | |
| > Bachelor’s degree | 204 (26.70%) | 115 (30.10%) | 89 (23.30%) | |
| Smoking status | 0.82 | |||
| Never | 630 (82.46%) | 315 (82.46%) | 315 (82.46%) | |
| Former | 43 (5.63%) | 23 (6.02%) | 20 (5.24%) | |
| Current | 49 (6.41%) | 23 (6.02%) | 26 (6.81%) | |
| BMI, kg/m2 | 25.52 (5.81) | 22.91 (3.77) | 28.14 (6.29) | <0.0001 |
| Periodontal disease | <0.0001 | |||
| None/mild | 540 (70.68%) | 294 (76.96%) | 246 (64.40%) | |
| Moderate/severe | 217 (28.40%) | 87 (22.77%) | 130 (34.04%) | |
| Bacterial gene abundance summary scores | ||||
| RD pathway* | 0.033 (0.008) | 0.034 (0.008) | 0.033 (0.008) | 0.01 |
| ANR pathway* | 0.017 (0.005) | 0.017 (0.005) | 0.017 (0.005) | 0.54 |
| DNRA pathway* | 0.020 (0.005) | 0.020 (0.005) | 0.020 (0.005) | 0.37 |
| NO2 product* | 0.031 (0.006) | 0.031 (0.006) | 0.030 (0.007) | 0.02 |
| NO product* | 0.018 (0.004) | 0.018 (0.004) | 0.017 (0.005) | 0.01 |
| NH3 product* | 0.025 (0.004) | 0.025 (0.004) | 0.025 (0.004) | 0.78 |
| Ratios of nitrite generation‐to‐depletion gene abundances | ||||
| RD vs ANR ratio † | 4.18 (2.19) | 4.37 (2.35) | 4.00 (2.01) | 0.02 |
| RD vs DNRA ratio † | 3.99 (5.11) | 4.26 (5.83) | 3.72 (4.26) | 0.14 |
| RD vs ANR+DNRA ratio † | 1.65 (0.87) | 1.72 (0.89) | 1.58 (0.85) | 0.02 |
| NO2 vs NH3 ratio † | 1.66 (0.72) | 1.72 (0.74) | 1.61 (0.70) | 0.03 |
| NO vs NH3 ratio † | 0.57 (0.30) | 0.6 (0.30) | 0.55 (0.30) | 0.01 |
| NO+NO2 vs NH3 ratio † | 2.23 (0.99) | 2.32 (1.01) | 2.15 (0.96) | 0.02 |
| NO2 vs NO+NH3 ratio † | 1.02 (0.27) | 1.04 (0.27) | 1.01 (0.28) | 0.06 |
| Cardiometabolic risk biomarkers | ||||
| Insulin | 7.68 (4.62) | 5.10 (1.96) | 10.26 (5.06) | <0.0001 |
| HOMA‐IR | 1.64 (1.09) | 1.02 (0.40) | 2.26 (1.19) | <0.0001 |
| HbA1C | 5.26 (0.33) | 5.12 (0.27) | 5.40 (0.32) | <0.0001 |
| Fasting plasma glucose | 84.78 (8.22) | 80.70 (5.45) | 88.87 (8.48) | <0.0001 |
| Diastolic blood pressure | 72.29 (9.05) | 67.20 (6.54) | 77.39 (8.33) | <0.0001 |
| Systolic blood pressure | 116.70 (12.28) | 110.18 (8.90) | 123.23 (11.71) | <0.0001 |
Values are presented as mean (SD) or n (%). P values are for ANOVA F statistics or χ2 tests for differences in level of covariates between participants with less than median CMZ score and more than median CMZ score. Missing values: n=6 for race and ethnicity, n=24 for education, n=42 for smoking status, n=11 for BMI, n=7 for periodontal status. ANR indicates assimilatory nitrate reduction; BMI, body mass index; CMZ, composite cardiometabolic score that averages the Z scores for the individual cardiometabolic risk variables; DNRA, dissimilatory nitrate reduction to ammonia; HbA1c, hemoglobin A1c; HOMA‐IR, Homeostatic Model Assessment of Insulin Resistance; NH3, ammonia; NO, nitric oxide; NO2, nitrite; ORIGINS, Oral Infections, Glucose Intolerance, and Insulin Resistance Study and RD, respiratory denitrification.
Arcsine square root transformed summary scores of relative gene abundance.
Ratios of nitrite generation‐to‐depletion proportions of pathway and product relative gene abundances before log‐transformation.
The mean (SD) gene summary scores are presented in the Table. RD gene abundance pathway and NO2 and NO product summary scores were higher in those with CMZ below the median. Similarly, ratios of nitrite generation to depletion were appreciably larger in those with CMZ scores below the median CMZ, and mostly indicated a net nitrite or NO generating versus depleting capacity.
Association of Pathway‐Specific Gene Abundances Summary Scores With Cardiometabolic Risk
In unadjusted analyses, the RD pathway was overall inversely associated with several cardiometabolic risk biomarkers (Figure 2A, upper panels). Upon adjustment for age, sex and race/ethnicity in the simple adjusted model, these associations were attenuated. Higher combined nitrite‐depleting pathways summary score (ANR+DNRA) was overall associated with higher cardiometabolic risk, though this was statistically significant only for HbA1c (q=0.03) (Figure 2A, middle panels). In the fully adjusted models, these associations no longer met the FDR threshold for significance, but remained P<0.05.
Figure 2. Association between various measures of nitrogen metabolism and cardiometabolic risk Z scores, unadjusted, adjusted for sex, age, race and ethnicity in the simple adjusted model, and additionally for education, smoking status, body mass index, and periodontal disease in the fully adjusted model. Results from n=764 participants in the ORIGINS (Oral Infections, Glucose Intolerance, and Insulin Resistance Study).
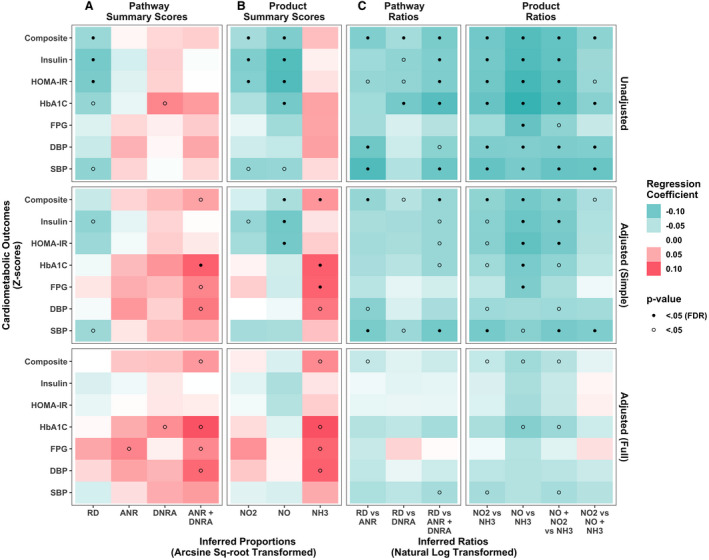
This heatmap represents the β coefficients (change in cardiometabolic risk Z score per 1 SD increase in metagenomic variable) from (1) unadjusted linear regression models (upper panels), (2) simple adjusted multivariable linear regression models (middle panels), and (3) fully adjusted multivariable linear regression models (lower panels) regressing cardiometabolic risk Z scores on the following measures of nitrogen metabolism. A, Abundance of microbial genes in the following 3 pathways were considered: respiratory denitrification (RD), dissimilatory nitrate reduction to ammonia (DNRA), and assimilatory nitrate reduction (ANR). B, Abundance of microbial genes for reductase enzymes that produce the following biochemical products: NO2, NO, and NH3. C, Nitrite generation vs depletion ratios of microbial gene abundances for pathways (RD vs ANR, RD vs DNRA, RD vs ANR+DNRA) or products (NO2 vs NH3, NO vs NH3, NO+NO2 vs NH3, NO2 vs NO+NH3). Green represents inverse associations, and red represents positive associations. Darker colors represent a larger effect estimate. The values from the analyses used to create this heatmap are presented in Table S3. DBP indicates diastolic blood pressure; FDR, false discovery rate; FPG, fasting plasma glucose; HbA1c, hemoglobin A1c; HOMA‐IR, Homeostatic Model Assessment of Insulin Resistance; NH3, ammonia; NO2, nitrite; and SBP, systolic blood pressure.
Association of Biochemical Product‐Specific Gene Abundances Summary Scores With Cardiometabolic Risk
Higher NO2 and NO summary scores were strongly associated with lower levels of cardiometabolic biomarkers in unadjusted analyses (Figure 2B). In the simple adjustment model, the inverse associations of NO summary score with overall CMZ, insulin, and HOMA‐IR met the FDR threshold for significance, whereas statistically significant positive associations emerged between NH3 summary score (reflective of nitrite depletion away from the enterosalivary pathway) and higher overall CMZ, HbA1c, and fasting plasma glucose (Figure 2B). One SD higher NH3 summary score was associated with a 0.06 (q=0.04) higher CMZ (Table S3). In the fully adjusted models, only NH3 summary score was associated with higher cardiometabolic risk at the P<0.05 threshold. Figure 3A illustrates that overall cardiometabolic risk worsened across increasing quartiles of NH3 summary score, demonstrating a significant linear trend for HbA1c, glucose, and diastolic blood pressure. Pairwise comparisons between the highest versus lowest quartile of NH3 summary score were statistically significant for HbA1c and glucose.
Figure 3. Plasma insulin, Homeostatic Model Assessment of Insulin Resistance (HOMA‐IR), hemoglobin A1c (HbA1c), fasting plasma glucose (FPG), diastolic blood pressure (DBP), and systolic blood pressure (SBP) values across increasing quartiles of (A) NH3 product summary score and (B) NO vs NH3 nitrite generation to depletion predicted gene abundance ratio.
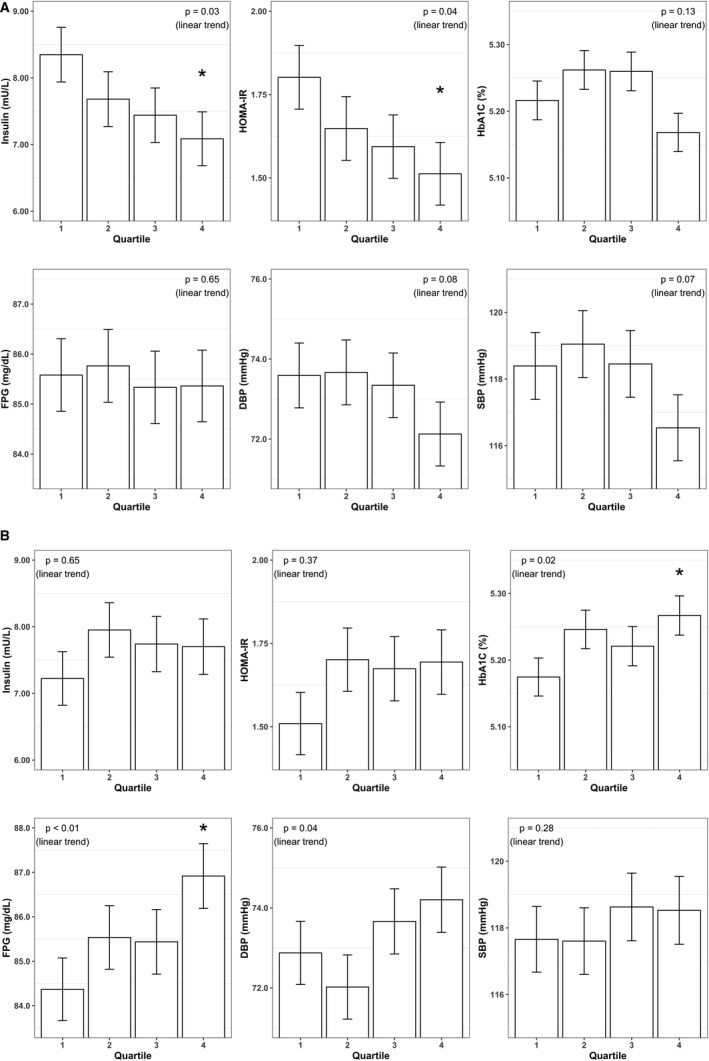
In fully adjusted models controlling for sex, age, race and ethnicity, education, smoking status, body mass index, and periodontal disease status. * P<0.05 for pairwise comparisons between the first and fourth quartile. NH3 indicates ammonia.
Association of Ratios of Nitrite Generation‐to‐Depletion Gene Abundance With Cardiometabolic Risk
The nitrite generation‐to‐depletion ratios all showed inverse associations with cardiometabolic risk in unadjusted analyses (Figure 2C), indicating that a net nitrite‐generating gene abundance was associated with better cardiometabolic risk. Focusing on the biochemical product ratios as their summary scores had stronger associations with cardiometabolic risk, the NO versus NH3 ratio remained statistically significant (FDR q<0.05) with all the cardiometabolic risk biomarkers in simple adjusted models except blood pressure (Figure 2C, middle panels). One SD higher NO versus NH3 summary ratio was associated with a −0.10 (q=0.03) lower CMZ (Table S3). In addition, ratios of NO2+NO versus NH3 and NO2 versus NH3, were significantly associated with CMZ and systolic blood pressure (q<0.05). Alternatively, the ratio capturing nitrate reductase genes to nitrite reductase genes (NO2 versus NO+NH3) was associated only with systolic blood pressure (q=0.03) and not overall cardiometabolic risk. In fully adjusted models, the ratios of NO2+NO versus NH3, NO2 versus NH3, and NO versus NH3, remained inversely associated with composite CMZ at the P<0.05 threshold only (Figure 2C, lower panels).
The means of cardiometabolic risk biomarkers across quartiles of the ratio of NO versus NH3 generation are presented in Figure 3B. The geometric means of HOMA‐IR values and insulin observed in the highest versus lowest quartile of NO versus NH3 across increasing quartiles of NO versus NH3 ratio were 1.80 versus 1.51, and 8.4 versus 7.1 mU/L, respectively (P < 0.05).
Discussion
In this cross‐sectional study using predicted metagenomic content from 16S rRNA sequencing, we found gene function scores representing enhanced nitrite generation‐to‐depletion (eg, NO versus NH3) associated with lower values of a composite cardiometabolic Z score (ie, lower cardiometabolic risk). Conversely, nitrite‐generating gene abundance alone (NO2 product summary score) was not associated with any markers of cardiometabolic health, whereas the NH3 product summary score (suggestive of nitrite depletion) was strongly associated with a higher cardiometabolic risk.
These findings extend our previous work demonstrating an inverse association between higher levels of a priori identified nitrate‐reducing taxa and cardiometabolic health. 13 The results show that more precise scores using hypothesis‐driven metagenomic inference approaches can improve prediction of cardiometabolic risk. The impact of nitrite depletion on the NO generating capacity of the enterosalivary pathway has been suggested by several authors, but few have explicitly tested the association of nitrite depletion with cardiometabolic risk or attempted to operationalize nitrite/NO generation‐versus‐depletion capacity of the oral microbiome. Our findings suggest that in addition to nitrite generation (ie, nitrate‐reducing capacity), nitrite depletion by oral bacteria is of relevance to the enterosalivary pathway of NO generation as well. In this population of healthy diabetes‐free individuals, the net balance of generation versus consumption of nitrite by bacteria in the mouth may have a greater association with cardiometabolic risk. Furthermore, the net balance of the product generated by nitrite depletion, NO versus NH3, may also be a meaningful predictor of cardiometabolic health.
The lack of association between higher levels of bacterial nitrate reductase genes or nitrate‐reducing bacteria and blood pressure has been observed in other studies. 14 , 32 For example, Burleigh et al report that greater abundance of nitrate‐reducing oral bacteria and correspondingly greater salivary nitrate to nitrite reduction was not related to plasma nitrite and blood pressure. The authors hypothesized that as the mean relative abundance of bacteria containing at least 1 nitrate reductase gene was already high (mean [SD], 41% [6%], with a lowest abundance of 29%), a saturation threshold for circulating nitrite may have been reached, and any additional bacterial nitrate reduction did not influence systemic NO levels. 33 It may be possible that for many people there is a substantial abundance of nitrate‐reducing oral bacteria, beyond which no additional effects of nitrite generation are observed. Additionally, regardless of nitrate‐reducing capacity, most nitrate‐reducing organisms also have functional potential for depleting nitrite. Thus, it is possible that with sufficient nitrate‐reducing capacity of the oral microbiome, nitrite depletion and its steady‐state becomes an important factor determining nitrite accumulation and NO production potential. Our results support this notion by showing that gene ratios and gene product scores that conceptually capture the net NO‐generating potential of the oral microbiome were most strongly and consistently associated with cardiometabolic risk.
Our results indicate that oral nitrite depletion can influence the enterosalivary NO pathway, and its effects may vary by the biochemical product formed (NO or NH3). NO produced in the mouth may have direct interaction with the circulation through the vascular tissues of the mouth. 34 Oral NO may also have systemic effects affecting tissues (eg, gut and liver) distal to its production. 35 , 36 While NO has a short half‐life of <1 second, NO metabolites formed including nitrite and S‐nitrosothiols have a longer half‐life of minutes and can contribute to signaling activities outside the mouth. 4 , 37 Our findings of an association with composite cardiometabolic Z score for NO product summary score and the NO versus NH3 ratio in simple adjusted models, reinforce the idea that NO formed orally contributes to the beneficial systemic effects of vasodilation and metabolism‐modulation mediated through the enterosalivary NO3–NO2–NO pathway. We also observed significant associations between higher levels of NH3‐forming nitrite reductase genes and higher cardiometabolic risk, which is consistent with previous studies. Tribble et al likewise found an association between higher levels of NH3‐forming nitrite reductase genes and higher systolic blood pressure. 14 The role of NH3 in the NO3–NO2–NO pathway is not clear, but an increase in NH3 production may lead to an increase in pH, reducing environmental factors conducive for NO2‐to‐NO reduction for some species, 38 or create selective pressures on the oral microbiome toward other species. Additionally, more NH3 generation also portends depleted nitrite stores.
In a recent study aimed at identifying bacterial species for potential nitrate‐reducing probiotics, it was noted that bacterial isolates with the best nitrate‐reducing capacity (Rothia aeria, Rothia dentocariosa, and Rothia mucilaginosa) also encoded genes for both NO2‐to‐NO and NO2‐to‐NH3 nitrite reductase enzymes, as well as the nitrate transporter gene, narK. 38 This suggests that an optimal taxa contributing to the NO pathway is one that has the ability to reduce nitrite to NO or NH3 depending on the different environmental pressures, and further supports the relevance of bacterial nitrite reductase genes to the enterosalivary NO pathway. Although in previous analyses we did not find any consistent associations with cardiometabolic risk for individual a priori identified taxa, 13 correlations between these taxa and the gene abundance summary scores and ratios in this study (Data S2, Figure S3) support the finding of Rothia dentocariosa as an important bacterial species for net NO generation, and identifies other potential candidate bacterial species that may help achieve a net nitrite generating capacity such as Actinomyces naeslundii, Corynebacterium durum, and Corynebacterium matruchotti.
Our study has several strengths. First, ORIGINS collected a robust set of risk factor data allowing for control of confounding variables. The metagenomic analyses extend previous findings using a summary score of a priori selected bacterial taxa found to be most associated with nitrate‐reducing capacity. 13 Using predicted metagenomic content enables us to further tease apart oral bacterial nitrogen metabolism and its influence on systemic NO bioavailability, testing several hypotheses about the relative importance of specific bacterial gene abundances and pathways. Both approaches are of importance. The nitrate‐reducing taxa summary score optimizes the exposure measurement, using only the presence of a small number of bacterial species most associated with nitrate‐reducing capacity, and has use as a clinical biomarker of oral microbiome‐mediated enterosalivary NO generation. On the other hand, unlike for nitrate reduction, the key bacterial species most associated with nitrite reduction have yet to be identified. This is further complicated by the fact that nitrite can be reduced to NO or NH3, and the different products may influence the enterosalivary pathway differently. 39 Using the gene abundances of all taxa present incorporates a community picture of the microbiome functional activity and its complex interactions, which may be more informative than the characterization of only a handful of species. 16 The knowledge gained from using the predicted gene abundances of nitrite generation‐to‐depletion ratios is important for future research that tries to maximize the salivary nitrite swallowed, such as in the development of probiotics and trial protocols that try to manipulate the enterosalivary pathway of NO generation. Future methodological work could combine both approaches by identifying taxa from the taxa summary score most strongly associated with actual gene abundances shown to be the best predictors of cardiometabolic risk (eg, NO versus NH3 ratios or NH3 biochemical product summary score). Such information can be used to further refine and optimize the taxa summary score, and improve the specificity of this measure as a predictor of cardiometabolic risk. Doing so will aid in clinical trial designs that aim to enrich oral nitrate reducers by identifying individuals with low levels of NO producing capacity and characterizing an intermediate biomarker of efficacy.
Finally, there is increased attention on the bias of comparisons using the relative abundance of microbial data 40 . The use of compositional differential abundance or reference frames is advocated for circumventing some of these biases, 41 and the results of the nitrite generation‐to‐depletion ratios may be more robust than that of the biochemical product and pathway gene summary scores. Furthermore, as individual gene abundances on the same pathway are highly correlated, the NO versus NH3 ratio that compares genes from different pathways may have the largest power for teasing out associations with cardiometabolic risk.
Limitations
This study has some limitations. Associations observed between nitrite generation‐to‐depletion gene abundance and cardiometabolic risk met the FDR threshold for statistical significance in simple adjusted models, whereas in the fully adjusted models they only reached a P<0.05 threshold, limiting how conclusive these findings are and highlighting the need for future replication. Our study used cross‐sectional data, and reverse causation may explain our findings. It should be noted that the nitrite‐generating and nitrite‐depleting functional capacity in our population was inferred from 16S rRNA gene sequences and known bacterial genomes in available databases. Therefore, the findings still rely on taxonomic identified 16S rRNA marker gene sequences and do not reflect the true functional gene sequences from the microbiome. 7 , 10 The 16S rRNA sequencing does not enable resolution for strain‐level variation within species (eg, for nitrate‐reduction capacity between strains) or account for horizontal gene transfer between bacteria. Future studies with metagenomic shotgun sequencing and metatranscriptomics capturing actual genomic content and variable gene expression and metabolic activity of the oral microbiome would be useful in addressing potential misclassification.
The predicted gene abundance summary scores also assumed that nitrate or nitrite produced by one pathway and/or in a certain location in the bacterial cell (Figure 1) can be acted on by reductases from another pathway, or transported interchangeably through the cell, and excreted to be swallowed for systemic bioavailability. We could not account for the local environmental factors, including oral pH, and oxygen and nitrate availability, which may influence the expression and activity of nitrate and nitrite reductase, and thus the amount of NO available systemically. 3 , 15 , 34 For example, nitrite can be chemically reduced to NO under acidic conditions in the mouth. 34 Thus, the ratio of NO versus NH3 observed to be associated with cardiometabolic risk in our study could reflect environmental local factors that influence oral microbiome composition, such as pH that determines whether nitrite is further reduced to NO or NH3, 42 rather than the systemic effects of bacterial nitrite reduction. Although the exact mechanisms of how oral nitrite reductase genes are associated with the enterosalivary NO generation are unclear, our findings warrant further investigations into how these processes are linked. Furthermore, although we did not observe stark differences in the associations of the predicted gene abundances with cardiometabolic risk in analyses using shallow samples only (Figure 2) compared with weighted averaged gene abundances across samples from shallow and deep sites (Figure S1), oral disease status may influence nitrite generation to depletion. Future in‐depth examinations of the interactions between oral disease and the enterosalivary pathway can move toward a better understanding of the effects of local environmental factors. Similarly, the interplay of conditions in the gut (eg, pH) and the gut microbiome on the enterosalivary pathway could not be accounted for. NO produced by gut bacteria such as Lactobacilli can affect mucus generation and blood flow, 43 potentially influencing the circulatory uptake of swallowed nitrate and nitrite, and modifying nitrite and NO bioavailability. 44 Likewise, acid‐reducing proton pump inhibitors have been shown to blunt the effect of nitrate supplementation on blood pressure reductions. 45
Finally, although the tongue microbiome is believed to be the main site of oral nitrate reduction, 46 we were only able to use the subgingival plaque microbiome. A recent study of subgingival plaque from 739 participants observed Rothia dentocariosa as consistently one of the most abundant species regardless of periodontal health status, 47 suggesting that subgingival plaque is at least abundant in key bacterial species for the enterosalivary pathway. Nevertheless, the correlation of the nitrite generation‐to‐depletion capacity of the tongue and subgingival microbiome is unknown and of methodological interest for future studies.
Conclusions
We observed gene abundances representing higher nitrite generating‐to‐depleting capacity to be associated with lower cardiometabolic risk. Our findings suggest that nitrite depletion plays an important role in the enterosalivary pathway and provides support for future studies examining the interplay of nitrite generation and depletion and cardiometabolic health.
Sources of Funding
This research was supported by National Institutes of Health (grants R00 DE018739, R21 DE022422, and R01 DK102932) to Dr Demmer. This publication was also supported by the National Center for Advancing Translational Sciences, National Institutes of Health through grant number UL1TR001873. R. Molinsky was supported by institutional training grant number T32HL007779 from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosures
None.
Supporting information
Data S1–S2
Tables S1–S3
Figures S1–S3
Acknowledgments
The authors thank the following individuals for their invaluable contributions to this research: C. Mclaughin, B. Batista, and V. Rivera for their assistance with recruitment; R. Celenti for her efforts in performing phlebotomy and processing and analyzing plaque samples; A. Zuk and G. Zulaika for their leadership and excellent program coordination; and Drs Arora, Pokherel, Silfa, and Spinell for their skilled examinations and essential participant engagement. The authors are also profoundly grateful to the ORIGINS participants for their participation in this research.
Supplemental Material for this article is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.121.023038
For Sources of Funding and Disclosures, see page 12.
References
- 1. Lundberg JO, Weitzberg E, Gladwin MT. The nitrate‐nitrite‐nitric oxide pathway in physiology and therapeutics. Nat Rev Drug Discov. 2008;7:156–167. doi: 10.1038/nrd2466 [DOI] [PubMed] [Google Scholar]
- 2. Sansbury BE, Hill BG. Regulation of obesity and insulin resistance by nitric oxide. Free Radic Biol Med. 2014;73:383–399. doi: 10.1016/j.freeradbiomed.2014.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koch CD, Gladwin MT, Freeman BA, Lundberg JO, Weitzberg E, Morris A. Enterosalivary nitrate metabolism and the microbiome: intersection of microbial metabolism, nitric oxide and diet in cardiac and pulmonary vascular health. Free Radic Biol Med. 2017;105:48–67. doi: 10.1016/j.freeradbiomed.2016.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lundberg JO, Carlstrom M, Weitzberg E. Metabolic effects of dietary nitrate in health and disease. Cell Metab. 2018;28:9–22. doi: 10.1016/j.cmet.2018.06.007 [DOI] [PubMed] [Google Scholar]
- 5. Govoni M, Jansson EA, Weitzberg E, Lundberg JO. The increase in plasma nitrite after a dietary nitrate load is markedly attenuated by an antibacterial mouthwash. Nitric Oxide. 2008;19:333–337. doi: 10.1016/j.niox.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 6. Woessner M, Smoliga JM, Tarzia B, Stabler T, Van Bruggen M, Allen JD. A stepwise reduction in plasma and salivary nitrite with increasing strengths of mouthwash following a dietary nitrate load. Nitric Oxide. 2016;54:1–7. doi: 10.1016/j.niox.2016.01.002 [DOI] [PubMed] [Google Scholar]
- 7. Kapil V, Haydar SM, Pearl V, Lundberg JO, Weitzberg E, Ahluwalia A. Physiological role for nitrate‐reducing oral bacteria in blood pressure control. Free Radic Biol Med. 2013;55:93–100. doi: 10.1016/j.freeradbiomed.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bondonno CP, Liu AH, Croft KD, Considine MJ, Puddey IB, Woodman RJ, Hodgson JM. Antibacterial mouthwash blunts oral nitrate reduction and increases blood pressure in treated hypertensive men and women. Am J Hypertens. 2015;28:572–575. doi: 10.1093/ajh/hpu192 [DOI] [PubMed] [Google Scholar]
- 9. Ashworth A, Cutler C, Farnham G, Liddle L, Burleigh M, Rodiles A, Sillitti C, Kiernan M, Moore M, Hickson M, et al. Dietary intake of inorganic nitrate in vegetarians and omnivores and its impact on blood pressure, resting metabolic rate and the oral microbiome. Free Radic Biol Med. 2019;138:63–72. doi: 10.1016/j.freeradbiomed.2019.05.010 [DOI] [PubMed] [Google Scholar]
- 10. Bescos R, Ashworth A, Cutler C, Brookes ZL, Belfield L, Rodiles A, Casas‐Agustench P, Farnham G, Liddle L, Burleigh M, et al. Effects of chlorhexidine mouthwash on the oral microbiome. Sci Rep. 2020;10:5254. doi: 10.1038/s41598-020-61912-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhurakivska K, Troiano G, Caponio VCA, Dioguardi M, Laino L, Maffione AB, Lo Muzio L. Do changes in oral microbiota correlate with plasma nitrite response? A systematic review. Front Physiol. 2019;10. doi: 10.3389/fphys.2019.01029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beals JW, Binns SE, Davis JL, Giordano GR, Klochak AL, Paris HL, Schweder MM, Peltonen GL, Scalzo RL, Bell C. Concurrent beet juice and carbohydrate ingestion: influence on glucose tolerance in obese and nonobese adults. J Nutr Metab. 2017;2017:6436783. doi: 10.1155/2017/6436783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goh CE, Trinh P, Colombo PC, Genkinger JM, Mathema B, Uhlemann A‐C, LeDuc C, Leibel R, Rosenbaum M, Paster BJ, et al. Association between nitrate‐reducing oral bacteria and cardiometabolic outcomes: results from origins. J Am Heart Assoc. 2019;8:e013324. doi: 10.1161/JAHA.119.013324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tribble GD, Angelov N, Weltman R, Wang B‐Y, Eswaran SV, Gay IC, Parthasarathy K, Dao D‐H, Richardson KN, Ismail NM, et al. Frequency of tongue cleaning impacts the human tongue microbiome composition and enterosalivary circulation of nitrate. Front Cell Infect Microbiol. 2019;9:39. doi: 10.3389/fcimb.2019.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sparacino‐Watkins C, Stolz JF, Basu P. Nitrate and periplasmic nitrate reductases. Chem Soc Rev. 2014;43:676–706. doi: 10.1039/C3CS60249D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hyde ER, Andrade F, Vaksman Z, Parthasarathy K, Jiang H, Parthasarathy DK, Torregrossa AC, Tribble G, Kaplan HB, Petrosino JF, et al. Metagenomic analysis of nitrate‐reducing bacteria in the oral cavity: implications for nitric oxide homeostasis. PLoS One. 2014;9:e88645. doi: 10.1371/journal.pone.0088645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. PICRUST2 for prediction of metagenome functions. Nat Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Douglas GM, Maffei VJ, Zaneveld J, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. PICRUST2: an improved and extensible approach for metagenome inference. bioRxiv. 2019:672295. [Google Scholar]
- 19. Sun S, Jones RB, Fodor AA. Inference‐based accuracy of metagenome prediction tools varies across sample types and functional categories. Microbiome. 2020;8:46. doi: 10.1186/s40168-020-00815-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Demmer RT, Jacobs DR Jr, Singh R, Zuk A, Rosenbaum M, Papapanou PN, Desvarieux M. Periodontal bacteria and prediabetes prevalence in origins: the oral infections, glucose intolerance, and insulin resistance study. J Dent Res. 2015;94:201s–211s. doi: 10.1177/0022034515590369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Demmer RT, Breskin A, Rosenbaum M, Zuk A, LeDuc C, Leibel R, Paster B, Desvarieux M, Jacobs DR Jr, Papapanou PN. The subgingival microbiome, systemic inflammation and insulin resistance: the oral infections, glucose intolerance and insulin resistance study (origins). J Clin Periodontol. 2017;44:255–265. doi: 10.1111/jcpe.12664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. The Forsyth Institute . Human Oral Microbe Identification using Next Generation Sequencing (HOMINGS). Sample Preparation. https://homings.forsyth.org/sampleprep.html. Accessed 29/12/2017
- 23. Gomes BP, Berber VB, Kokaras AS, Chen T, Paster BJ. Microbiomes of endodontic‐periodontal lesions before and after chemomechanical preparation. J Endod. 2015;41:1975–1984. doi: 10.1016/j.joen.2015.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al‐Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2016;45:D353–D361. doi: 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanehisa Laboratories . Kegg nitrogen metabolism‐reference pathway map00910. KEGG Pathway Database. https://www.genome.jp/kegg‐bin/show_pathway?map00910 Updated 9/1/2020. Accessed 31/1/2020
- 27. Demmer RT, Breskin A, Rosenbaum M, Zuk A, LeDuc C, Leibel R, Paster B, Desvarieux M, Jacobs DR Jr, Papapanou PN. The subgingival microbiome, systemic inflammation and insulin resistance: the oral infections, glucose intolerance and insulin resistance study (origins). J Clin Periodontol. 2017;44:255–265. doi: 10.1111/jcpe.12664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta‐cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883 [DOI] [PubMed] [Google Scholar]
- 29. Eke PI, Page RC, Wei L, Thornton‐Evans G, Genco RJ. Update of the case definitions for population‐based surveillance of periodontitis. J Periodontol. 2012;83:1449–1454. doi: 10.1902/jop.2012.110664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morgan XC, Kabakchiev B, Waldron L, Tyler AD, Tickle TL, Milgrom R, Stempak JM, Gevers D, Xavier RJ, Silverberg MS, et al. Associations between host gene expression, the mucosal microbiome, and clinical outcome in the pelvic pouch of patients with inflammatory bowel disease. Genome Biol. 2015;16:67. doi: 10.1186/s13059-015-0637-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B (Methodol). 1995;289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 32. Burleigh M, Liddle L, Muggeridge DJ, Monaghan C, Sculthorpe N, Butcher J, Henriquez F, Easton C. Dietary nitrate supplementation alters the oral microbiome but does not improve the vascular responses to an acute nitrate dose. Nitric Oxide. 2019;89:54–63. doi: 10.1016/j.niox.2019.04.010 [DOI] [PubMed] [Google Scholar]
- 33. Burleigh MC, Liddle L, Monaghan C, Muggeridge DJ, Sculthorpe N, Butcher JP, Henriquez FL, Allen JD, Easton C. Salivary nitrite production is elevated in individuals with a higher abundance of oral nitrate‐reducing bacteria. Free Radic Biol Med. 2018;120:80–88. doi: 10.1016/j.freeradbiomed.2018.03.023 [DOI] [PubMed] [Google Scholar]
- 34. Schreiber F, Stief P, Gieseke A, Heisterkamp IM, Verstraete W, de Beer D, Stoodley P. Denitrification in human dental plaque. BMC Biol. 2010;8:24. doi: 10.1186/1741-7007-8-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elrod JW, Calvert JW, Gundewar S, Bryan NS, Lefer DJ. Nitric oxide promotes distant organ protection: evidence for an endocrine role of nitric oxide. Proc Natl Acad Sci. 2008;105:11430–11435. doi: 10.1073/pnas.0800700105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duncan C, Dougall H, Johnston P, Green S, Brogan R, Leifert C, Smith L, Golden M, Benjamin N. Chemical generation of nitric oxide in the mouth from the enterosalivary circulation of dietary nitrate. Nat Med. 1995;1:546–551. doi: 10.1038/nm0695-546 [DOI] [PubMed] [Google Scholar]
- 37. Kelm M. Nitric oxide metabolism and breakdown. Biochim Biophys Acta. 1999;1411:273–289. doi: 10.1016/S0005-2728(99)00020-1 [DOI] [PubMed] [Google Scholar]
- 38. Rosier BT, Moya‐Gonzalvez EM, Corell‐Escuin P, Mira A. Isolation and characterization of nitrate‐reducing bacteria as potential probiotics for oral and systemic health. Front Microbiol. 2020;11. doi: 10.3389/fmicb.2020.555465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Doel JJ, Hector MP, Amirtham CV, Al‐Anzan LA, Benjamin N, Allaker RP. Protective effect of salivary nitrate and microbial nitrate reductase activity against caries. Eur J Oral Sci. 2004;112:424–428. doi: 10.1111/j.1600-0722.2004.00153.x [DOI] [PubMed] [Google Scholar]
- 40. McLaren MR, Willis AD, Callahan BJ. Consistent and correctable bias in metagenomic sequencing experiments. Elife. 2019;8:e46923. doi: 10.7554/eLife.46923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, Zengler K, Knight R. Establishing microbial composition measurement standards with reference frames. Nat Commun. 2019;10:2719. doi: 10.1038/s41467-019-10656-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kraft B, Tegetmeyer HE, Sharma R, Klotz MG, Ferdelman TG, Hettich RL, Geelhoed JS, Strous M. The environmental controls that govern the end product of bacterial nitrate respiration. Science. 2014;345:676–679. doi: 10.1126/science.1254070 [DOI] [PubMed] [Google Scholar]
- 43. Tiso M, Schechter AN. Nitrate reduction to nitrite, nitric oxide and ammonia by gut bacteria under physiological conditions. PLoS One. 2015;10:e0119712. doi: 10.1371/journal.pone.0119712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Petersson J, Carlström M, Schreiber O, Phillipson M, Christoffersson G, Jägare A, Roos S, Jansson EÅ, Persson AEG, Lundberg JO, et al. Gastroprotective and blood pressure lowering effects of dietary nitrate are abolished by an antiseptic mouthwash. Free Radic Biol Med. 2009;46:1068–1075. doi: 10.1016/j.freeradbiomed.2009.01.011 [DOI] [PubMed] [Google Scholar]
- 45. Montenegro MF, Sundqvist ML, Larsen FJ, Zhuge Z, Carlstrom M, Weitzberg E, Lundberg JO. Blood pressure‐lowering effect of orally ingested nitrite is abolished by a proton pump inhibitor. Hypertension. 2017;69:23–31. doi: 10.1161/HYPERTENSIONAHA.116.08081 [DOI] [PubMed] [Google Scholar]
- 46. Doel JJ, Benjamin N, Hector MP, Rogers M, Allaker RP. Evaluation of bacterial nitrate reduction in the human oral cavity. Eur J Oral Sci. 2005;113:14–19. doi: 10.1111/j.1600-0722.2004.00184.x [DOI] [PubMed] [Google Scholar]
- 47. Papapanou PN, Park H, Cheng B, Kokaras A, Paster B, Burkett S, Watson CWM, Annavajhala MK, Uhlemann AC, Noble JM. Subgingival microbiome and clinical periodontal status in an elderly cohort: the WHICAP ancillary study of oral health. J Periodontol. 2020;91:S56–S67. doi: 10.1002/JPER.20-0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Amir A, McDonald D, Navas‐Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, et al. Deblur rapidly resolves single‐nucleotide community sequence patterns. mSystems. 2017;2:e00191–e00116. doi: 10.1128/mSystems.00191-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Scikit‐learn DV. Machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 50. Mougeot JL, Stevens CB, Cotton SL, Morton DS, Krishnan K, Brennan MT, Lockhart PB, Paster BJ, Bahrani Mougeot FK. Concordance of homim and homings technologies in the microbiome analysis of clinical samples. J Oral Microbiol. 2016;8:30379. doi: 10.3402/jom.v8.30379 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1–S2
Tables S1–S3
Figures S1–S3


