Abstract
Archaea are traditionally thought of as “extremophiles,” but recent studies have shown that marine planktonic Archaea make up a surprisingly large percentage of ocean midwater microbial communities, up to 60% of the total prokaryotes. However, the basic physiology and contribution of Archaea to community microbial activity remain unknown. We have studied Archaea from 200-m depths of the northwest Mediterranean Sea and the Pacific Ocean near California, measuring the archaeal activity under simulated natural conditions (8 to 17°C, dark and anaerobic) by means of a method called substrate tracking autoradiography fluorescence in situ hybridization (STARFISH) that simultaneously detects specific cell types by 16S rRNA probe binding and activity by microautoradiography. In the 200-m-deep Mediterranean and Pacific samples, cells binding the archaeal probes made up about 43 and 14% of the total countable cells, respectively. Our results showed that the Archaea are active in the uptake of dissolved amino acids from natural concentrations (nanomolar) with about 60% of the individuals in the archaeal communities showing measurable uptake. Bacteria showed a similar proportion of active cells. We concluded that a portion of these Archaea is heterotrophic and also appears to coexist successfully with Bacteria in the same water.
Archaea are normally regarded as organisms that thrive in high-temperature, high-salt, or extreme anaerobic environments, and they have generally been thought to be unable to compete against other microorganisms for limiting resources under “nonextreme” conditions (24, 30). However, ribosomal RNA genes and rRNA found in plankton of temperate (5 to 18°C) (7, 13, 14, 19, 28) and polar (−1.5°C) (22) aerobic seawater, usually from depths below the euphotic zone (>100 m deep), have revealed the widespread presence of Archaea, mostly close relatives of the Crenarchaeota, outside environments frequently associated with extreme conditions. Direct counts through fluorescent in situ hybridization (FISH) have provided the first look at these unique microorganisms, showing that they can constitute up to 60% of the cells in the prokaryotic community (15) but typically more like a few percent in the surface mixed layer and up to 30% at depths of a few hundred meters or more (8, 15, 22). Curiously, Crenarchaeota is a group originally thought to be made up only of thermophilic Archaea (7, 14, 30), and to date, no marine Crenarchaeota have been isolated in pure culture, and almost nothing is known about their activity and metabolic properties in situ (16, 24), including whether they are heterotrophs or autotrophs (e.g., ammonium or nitrite oxidizers). Their apparent widespread occurrence and persistence in the plankton alongside Bacteria in deep water remain a mystery.
We can postulate three possible explanations for this persistence: (i) the Archaea grow as plankton on dissolved substances (such as dissolved organic matter or inorganic nutrients), similar to the way most planktonic bacteria are thought to grow (2, 10), (ii) the Archaea grow in some specialized microzones within the planktonic habitat, such as sinking particles or anaerobic zones in animal guts, and are released into the plankton where they grow slowly or not at all, or (iii) the Archaea originated at some remote source and were dispersed into the plankton but are dormant (17, 33), neither growing nor being removed at appreciable rates, but with sufficient cellular rRNA content to be detected through various molecular methods.
To choose among these possibilities and generally examine archaeal activity, we used a triple labeling technique, which we have termed substrate tracking autoradiography fluorescence in situ hybridization (STARFISH) (26), that allows us to characterize the picoplankton community diversity by phylogenetic groups and simultaneously determine metabolic activity of each group in situ (25, 26). The technique applies a general fluorescent dye 4′,6-diamidino-2-phenylindole (DAPI) for total cell counts, fluorescently labeled oligonucleotide probes for whole-cell in situ hybridization of specific target groups, and a tritiated nutrient for detection of cell function by autoradiography. For this study, we chose dissolved amino acids as substrates, for two reasons: (i) they are ubiquitous and known to be an important source of C, N, and energy for marine microheterotrophs (11, 31), and (ii) their availability as a mixture of 15 highly specific activity-tritiated compounds permits sensitive measurement of single-cell uptake autoradiographically, even by small, slowly growing cells (26).
Here, we present evidence, based on the STARFISH method, suggesting that free-living planktonic marine Archaea from 200-m depths in the Mediterranean and off the California coast are actively involved in the heterotrophic uptake of dissolved amino acids from aerobic seawater, with activities apparently comparable to those of the Bacteria (based on the extent of clustering of silver grains around cells in microautoradiography). Hence, Archaea appear to be successfully vying with other heterotrophic microorganisms for dissolved amino acids.
MATERIALS AND METHODS
Sample preparation.
Two samples were collected outside the Villefranche-sur-Mer Bay, an oligotrophic environment in the French Mediterranean Sea (43°40.79′N, 7°17.88′W) on 21 July 1998, one sample at the surface (22.4°C) and one sample at 200 m (17.0°C) deep (Table 1), where we had previously detected nearly 60% of the total cells as Archaea (15). Similarly, two samples were collected in Monterey Bay, Northern California, at 36°45.49′N and 122°01.04′W on 14 July 1999, one sample at the surface (14.0°C) and one sample at 200 m deep (8.7°C) (Table 1). The chlorophyll maximum was detected at a 28-m depth in the Villefranche Bay and at 20 m deep in Monterey Bay. All samples were used unfiltered and divided into two live 100-ml subsamples in sterile culture flasks wrapped in aluminum foil and incubated with a mixture of 15 tritiated amino acids (premixed by Amersham, catalog no. TRK440, with an average specific activity of about 50 Ci/mmol) at trace levels of 5 nM at in situ temperatures. Surface samples from both sites were incubated for 13 h, while the 200-m-deep samples from Villefranche were incubated for 19 h, and the samples from Monterey Bay were incubated for 13 h. Controls consisted of two 100-ml subsamples from each depth killed in 10% formalin for 1 h prior to addition of 3H-labeled amino acids at 5 nM concentrations. Nutrient uptake was monitored over time for all of the live and killed subsamples by withdrawing 2-ml aliquots with replicates from each subsample, filtering each aliquot onto a 0.2-μm-pore-size Nucleopore polycarbonate filter, rinsing the filter four times with about 2 ml of 1× phosphate-buffered saline (PBS) and measuring the radioactivity with a scintillation counter by using EcosintA scintillation cocktail. In addition, total cell count (cells per milliliter) was determined with DAPI, and the radioactivity level in units of disintegration per minute (DPM) was normalized to the cell concentration (DPM/cell in Table 1). Once cells reached saturation (no further increase with time) levels, 3H-labeled-amino-acid uptake was halted by addition of formalin at a 10% concentration. Optimization of fluorescence was achieved by use of four archaeal probes (Table 2), each labeled at both 3′ and 5′ ends with the fluorochrome Cy3, and visualization was aided by image-intensified video microscopy (15). The 15 3H-labeled-amino-acid mixture used at trace levels (nanomolar concentration) served as an unspecialized substrate to maximize label uptake and detection of heterotrophic cells, without adding appreciable nutrients to these samples (26, 27). Other substrates such as sugars, lipids, and polymers, however, could be used to identify more specialized metabolic activities.
TABLE 1.
General characteristics of the Villefranche Bay and the Monterey Bay samples
| Site/date (maximum depth [m]) | Depth (m) | Temp (°C) | Salinity (ppt) | Oxygen (mg/liter) | Cells/ml (105) | Status (DPM/cella) |
|---|---|---|---|---|---|---|
| Villefranche/21 July 1998 (300) | 0 | 22.4 | 38.0 | 6.8 | 3.73 | Killed (4.78 × 10−5) |
| Live (3.32 × 10−2) | ||||||
| 200 | 17.0 | N/Ab | N/A | 1.82 | Killed (1.49 × 10−4) | |
| Live (5.07 × 10−2) | ||||||
| Monterey/14 July 1999 (886) | 0 | 14.0 | 33.75 | 9.64 | 21.3 | Killed (8.61 × 10−5) |
| Live (3.65 × 10−2) | ||||||
| 200 | 8.7 | 34.05 | 2.84 | 3.10 | Killed (2.62 × 10−4) | |
| Live (2.19 × 10−2) |
DPM/cell, radioactivity level in units of disintegrations per minute normalized to the cell concentration.
N/A, not applicable.
TABLE 2.
Oligonucleotide probe sequences, Escherichia coli positions, length, and references
| Name/target | Sequence (5′-3′)b | Position/length | Reference |
|---|---|---|---|
| Controla | CCTAGTGACGCCGTCGAC | N/Ac/18 | 26 |
| Bacteria | ACCGCTTGTGCGGGCCC | 927/17 | 5 |
| Archaea | GCGCCTGSTGCSCCCCGTAGGGCC | 338/24 | 5 |
| Marine GI Crenarchaeota-1 | CTCCTGACCACTTGAGGT | 541/18 | 15 |
| Marine GI Crenarchaeota-2 | TTAGGCCCAATAATCMTCCT | 554/20 | 19 |
| Marine GII Euryarchaeota | TTAGGCCCAATAAAAKCGAC | 554/20 | 19 |
Not expected to bind to any organism.
K = G or T, M = A or C, S = C or G.
N/A, not applicable.
Slide preparation.
In STARFISH, cells were transferred from a membrane filter onto Teflon glass slides (Cel Line Assoc.) with ten 7-mm-diameter wells that had previously been coated with a thin layer of Kodak photographic emulsion type NTB2. All procedures included precautions, such as gloves, to minimize RNase contamination. First, the emulsion was melted in a water bath at 43°C in a darkroom for 1 h. This emulsion was mixed with a gelatin solution (0.2% gelatin–0.02% chromium potassium sulfate) also at 43°C at a 50:50 (vol/vol) ratio. Each well on the slide was coated separately in a darkroom by pipetting about 20 μl and immediately withdrawing as much as possible of the emulsion-gelatin solution, leaving a thin layer of emulsion on the well. To see under extremely dim light conditions, a night vision scope with a close-focus lens (ITT Nightcam 310) was used in combination with a safe light (Testrite model 5 1/2) placed at least 2 m from the working area. The emulsion-gelatin on the slides was air dried for about 30 min in total darkness before cells were transferred.
Cell DAPI staining, filtration, and transfer.
In ordinary room light, 4 ml of formalin-fixed subsamples from the surface and 10 ml from the 200-m-deep samples were stained with 70 μl of a 0.1-μg/μl DAPI solution for 10 min (covered to protect it from light) and then filtered through a 0.2-μm-pore-size, 25-mm Nucleopore polycarbonate filter placed over a 0.8-μm-pore-size Millipore filter type AA. Staining with DAPI for less than 10 min led to a low DAPI fluorescence signal by the end of the STARFISH protocol. All of the filters were then rinsed four times with about 2 ml of 0.2-μm-filtered 1× PBS to remove unincorporated radioactive amino acids. With clean forceps, the Nucleopore and the Millipore filters were placed together with the cell side facing up on a 10-μl drop of 1× PBS in a petri dish (to prevent the filters from drying out), cut into eight equal pieces with a new, clean razor blade, and carried to the darkroom.
In the darkroom, cells were transferred from the filters onto Teflon slides by placing each one-eighth piece of the Nucleopore filter only (peeled off the Millipore filter) cell face down over a single emulsion-coated well, again with the aid of the night vision scope and a safe light positioned 2 m away.
Emulsion exposure and development.
The slides were left to dry in total darkness for 30 min before they were placed in a light-proof box and stored at 4°C for 3 days for emulsion exposure. The emulsion was developed to Kodak specifications as follows: 2 min in Dektol developer, a 10-s stop in deionized water, and 5 min in fixer. The slides were washed in deionized water for 2 min, the pieces of the Nucleopore filters were peeled off, and slides were air dried and prepared for whole-cell hybridization. At this point, the slides could be stored at −80°C for several weeks without noticeable changes in results.
In situ hybridization.
In situ hybridization took place in 10 μl of hybridization solution per well, containing 5× SET (1× SET is 150 mM NaCl–20 mM Tris-Cl [pH 7.8]–1 mM EDTA), 0.2% bovine serum albumin (acetylated; Sigma), 10% dextran sulfate (molecular weight, 500,000; Pharmacia), 0.01% polyadenylic acid (Sigma), and 0.1% sodium dodecyl sulfate (SDS) as described by DeLong et al. (9) and Braun-Howland et al. (3), and 50 ng of a 5-ng/μl concentration of Cy3-labeled oligonucleotide probe (Table 2). The slides were incubated at 43°C for 3 h, briefly washed with ∼5 ml of 0.2× SET at 43°C, and finally immersed three times in 0.2× SET at 43°C for 10 min each time. After air drying, the slides were mounted in glycerol–10× SET (50:50 [vol/vol]) solution under a 24-by-50-mm cover glass and kept at −20°C for at least 1 h before they were observed by fluorescence and transmitted light microscopy.
STARFISH cell count.
For each microscopic field observed, the following four types of counts were recorded: (i) total DAPI cell counts with UV excitation (Olympus type U-MWU UV filter; excitation wavelength, 330 to 385 nm; emission wavelength, >420 nm), (ii) probe fluorescence cell counts, which included autofluorescent and probe-labeled cell counts with a green-light excitation filter (Chroma Cy3, #U-M41007), (iii) microautoradiography counts (cells with associated silver grains) under transmitted light, and (iv) counts for cells labeled with a fluorescent probe and simultaneously labeled autoradiographically (overlap between green excitation and transmitted light). Based on these four counts, we quantified the abundance of cells labeled with the fluorescent probes as a percentage of the total DAPI-stained cells (the number of labeled cells with the probe divided by the number of DAPI-stained cells [count ii divided by i above]). Nearly 600 of the DAPI-stained cells were included in the computation of the percentages. Likewise, the abundance of all cells labeled autoradiographically with the tritiated amino acids (count iii divided by i above) and that of cells labeled both with the probe and the tritiated amino acids (count iv divided by i above) are presented.
Counts were acquired on an Olympus BMax epifluorescence microscope equipped with a UPlanApo objective lens (100×), a type HBO 100-Hg vapor lamp, and the filters previously described. The images were captured and intensified by using a microchannel plate image intensifier (COHU Intensified CCD camera model 5515-2001/0000), and the background was reduced by image averaging with a model DSP-2000 image processor (Dage-MTI, Inc., Michigan City, Ind.). The images were visualized with a Sony Trinitron color video monitor (model PVM-1353MD). This video system could display cells in real time with fluorescence considerably less than the fluorescence directly detectable by eye. Photomicrographs and microautoradiographs were captured with a Scion LG3 capture card and NIH Image software version 1.61.
RESULTS
Levels of prokaryotic cell-specific activity (DPM per cell) at the end of the incubation with the 3H-labeled-amino-acid mixture in the Villefranche Bay and the Monterey Bay samples were within the same order of magnitude (Table 1). Killed controls were ≤1% as radioactive as their live sample counterparts. Live surface prokaryotic-cell-specific activity levels between the two sites were <10% apart, but the Villefranche Bay 200-m-deep prokaryotic cells had a specific activity 2.4-fold greater than 200-m-deep prokaryotes in Monterey Bay (Table 1). Autoradiography for both samples confirmed that cells in the killed control did not take up the 3H-labeled amino acids at detectable levels (Table 1).
For the in situ hybridization counts, the negative-control counts were not subtracted from the probe counts unless specified. In all samples, the background level determined by the “no-probe” and the “control probe” counts consisted mostly of autofluorescent cells. These autofluorescent cells constituted <5% of the total counts on average, and ≤10% of those cells were detected taking up dissolved amino acids.
Villefranche-sur-Mer.
In the surface water, Archaea-labeled cell counts were low and statistically indistinguishable from the control probe counts (Table 3). Cells in the “no probe” counts were primarily autofluorescent cells such as cyanobacteria, whose pigment fluorescence is normally indistinguishable from the Cy3-probe fluorescence, and these did not generally take up the 3H-labeled amino acids (Table 3).
TABLE 3.
Percentage of total DAPI cells that are either autofluorescent (NP = no probe), labeled with fluorescent probes (CON = control probe; ARC = Archaea probe mix; BAC = Bacteria probe), labeled by microautoradiography (MAR) with 3H-labeled amino acids, or simultaneously labeled with both (probe and 3H-labeled amino acids) for surface and 200-m-deep Mediterranean Sea samplesa
| Depth (m) | NP | MAR | NP and MAR | CON | MAR | CON and MAR | ARC | MAR | ARC and MAR | BAC | MAR | BAC and MAR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 8.0 ± 1.2 | 30.5 ± 3.7 | 0.7 ± 0.4 | 5.0 ± 0.7 | 29.0 ± 1.7 | 0.7 ± 0.3 | 4.2 ± 1.1 | 29.3 ± 1.8 | 1.0 ± 0.6 | 56.2 ± 1.9 | 32.2 ± 1.8 | 27.3 ± 1.9 |
| 200 | 2.3 ± 0.7 | 33.3 ± 1.7 | 0.0 ± 0.0 | 0.83 ± 0.4 | 31.0 ± 3.8 | 0.0 ± 0.0 | 42.8 ± 5.5 | 39.5 ± 2.6 | 24.2 ± 4.0 | 46.3 ± 2.1 | 36.0 ± 2.5 | 30.8 ± 1.9 |
Each set of three columns is from a pair of duplicates from each depth.
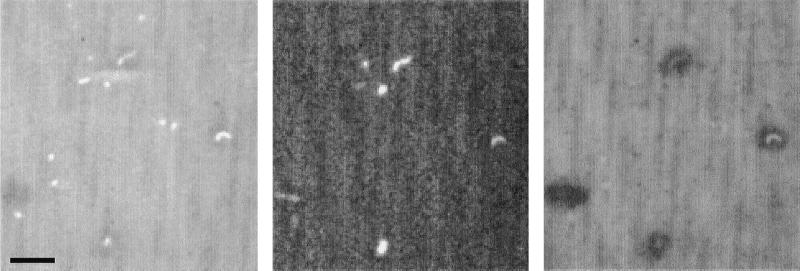
In the 200-m-deep water, however, cells labeled with the archaeal probe comprised about 43% (7.83 × 104 cells/ml) of the prokaryotic community (Fig. 1 and Table 3). These presumed Archaea were mostly curved rods (0.6 to 1.8 μm in length) or spheres (∼0.6 μm in diameter), and 57% of all such Archaea were also detected taking up 3H-labeled amino acids (Fig. 1 and Table 3). Control counts for both “no probe” and “control probe” were less than 3% of the total DAPI-detected cells at this deep-water site (Table 3).
FIG. 1.
Triple labeling of cells in situ. Live sample from a 200-m-deep French Mediterranean Sea site showing labels from DAPI, Cy3-fluorescent oligonucleotide probes specific for the Archaea domain, and autoradiography with a trace (5 nM total) mixture of 15 3H-labeled amino acids. All three panels are from the same microscope field. The left panel shows DAPI-stained cells, middle panel shows Archaea oligonucleotide probe-labeled cells, and right panel shows dark silver grains surrounding cells with 3H-labeled amino acids and faint Archaea probe fluorescence. The Archaea oligonucleotide probe sequences are shown in Table 2. Bar, 4 μm.
In addition to the archaeal probes, a Cy3-labeled probe targeting the Bacteria domain (5) was used in the same experiments. In the surface sample, 56% (2.09 × 105 cells/ml) of the DAPI cells were labeled with the bacterial probe, and half of those Bacteria-probe-labeled cells were simultaneously detected in the autoradiography taking up 3H-labeled amino acids (Table 3). At 200 m deep, almost half of the cells were labeled with the bacterial probe and two-thirds of those cells were also detected taking up 3H-labeled amino acids (Table 3). When we add the Archaea probe cell count to the Bacteria probe cell count, the total in surface waters is 56% (all bacteria), and at 200 m, the total is statistically indistinguishable from 100% of the DAPI-countable cells. The reason for the lower percentage of the total in the bacterium-dominated surface is unknown but may reflect an underestimate from the single bacterial probe compared to the multiple archaeal probes.
Monterey Bay.
The overall average percentage of autoradiography-positive counts in the Monterey Bay samples was nearly twice as high as that of the Villefranche Bay samples, with 60% of the total cells at the surface and 62% at 200 m deep being detected autoradiographically (Table 4). Archaea-probe counts (3%) were indistinguishable from the control counts at the surface. In the 200-m-deep sample from the Monterey Bay, the percentage of probe-labeled cells for all probes used was lower than those respective counts at the Monterey surface sample. Bacteria probe counts made up 44% (1.36 × 105 cells/ml) of the cells while Archaea counts increased to 14% (4.34 × 104 cells/ml) compared to the surface. About 64% of the Bacteria probe-labeled cells and 60% of the Archaea probe-labeled cells (number of probe-labeled cells and cells simultaneously recorded in autoradiography divided by the number of probe-labeled cells alone) were detected in autoradiography at this 200-m-deep site (Table 4).
TABLE 4.
Percentage of total DAPI cells that are either autofluorescent (NP = no probe), labeled with fluorescent probes (CON = control probe, ARC = Archaea probe mix, BAC = Bacteria probe) or are labeled by microautoradiography (MAR) with 3H-labeled amino acids or are simultaneously labeled with both (probe and 3H-labeled amino acids) for surface and 200-m deep Monterey Bay samplesa
| Depth (m) | NP | MAR | NP and MAR | CON | MAR | CON and MAR | ARC | MAR | ARC and MAR | BAC | MAR | BAC and MAR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2.8 ± 1.0 | 61.4 ± 3.0 | 0.4 ± 0.2 | 4.0 ± 1.3 | 60.3 ± 3.3 | 0.7 ± 0.7 | 3.0 ± 1.3 | 62.3 ± 3.3 | 0.7 ± 0.4 | NAb | NA | NA |
| 200 | 2.0 ± 0.8 | 64.3 ± 2.7 | 0.3 ± 0.1 | 2.6 ± 0.6 | 65.1 ± 2.3 | 0.6 ± 0.3 | 14.3 ± 4.3 | 65.1 ± 3.2 | 8.4 ± 2.6 | 43.6 ± 1.7 | 61.1 ± 2.8 | 27.8 ± 2.6 |
Each set of three columns is from a pair of duplicates from each depth.
NA, Not applicable.
DISCUSSION
This investigation used FISH in combination with autoradiography to assess both the abundance and activity of Archaea in a mixed natural marine prokaryotic community.
The results confirmed that Archaea can constitute a large fraction of the picoplankton. In addition, these results showing archaeal abundance greater at the subsurface depth and declining to nearly 40% of the total prokaryotic community at 200 m in the Mediterranean are similar to FISH results reported by Fuhrman and Ouverney (15). The lower percentages at 200 m deep off the California coast are in the range of Fuhrman and Ouverney's (15) counts from San Pedro Channel as well as a report from DeLong et al. (8), who found approximately 20% of Archaea of the total cell count at 100 m deep, offshore from Moss Landing, Calif., using polyribonucleotide probes with whole-cell in situ hybridization. Similar trends in archaeal abundance, but using dot blot hybridization methods, have been reported in California (19) and Antarctica (20, 21, 23). We did not find a significant number of archaeal cells near the surface. Previously, Massana et al. (19) and DeLong et al. (8) detected a relatively small amount of Archaea, mostly members of group II, near the surface off the coast of California by dot blot hybridization.
Results from microautoradiography applied in combination with FISH showed Archaea taking up dissolved organic nutrients at nanomolar concentrations under oxygenic and apparent nonextreme conditions. Measurement of in situ nutrient uptake by heterotrophic bacteria via autoradiography was first suggested in the mid 1960s by Brock and Brock (4) to better understand the nutritional requirements of microorganisms as well as estimate prokaryotic cell growth rate and production. Brock and Brock (4) also noticed that autoradiography alone was limited in assigning function to specific prokaryotic groups due to the lack of distinct morphological features among these microorganisms. STARFISH seems to help break the bacterioplankton “black box” not only in function (through autoradiography) but also in type (through FISH). Similar protocols have been proposed and applied in activated-sludge microbial communities (18) and other marine Bacteria (6).
Members of both the Archaea and the Bacteria domains were shown to coexist at the 200-m depth of the Mediterranean and California sites and apparently successfully compete for the same dissolved amino acid pool. Although this kind of microautoradiography is not yet an accurate way to measure the amount of uptake, there was not a qualitative difference between the labeling of the Archaea and Bacteria. These findings contradict the former conventional belief that Archaea are restricted to extreme environments due to lack of competitiveness against other microorganisms (24, 30). They also show that a significant portion of the archaeal community is using dissolved organic matter. Thus, these organisms are living at least partly heterotrophically, and this gives us a useful physiological clue as to their role in the ecosystem.
The percentage of total cells detected taking up tritiated amino acids varied among the Mediterranean and the California sites and with water depth within each site. The results show a higher proportion of autoradiographically active cells near the surface of Monterey Bay (60% of total cells, 12.8 × 105 cells/ml) than near the surface of the Villefranche Bay (30% of total cells, 1.1 × 105 cells/ml). Although the causes of such variations are not currently known, a higher percent of active cells may be attributed in part to higher productivity levels normally associated with zones of upwelling, such as the Monterey Bay, compared to oligotrophic waters, such as the Mediterranean Sea.
In comparison to previous results from similar environments, we are not aware of previous autoradiography experiments in Monterey, although near-shore samples from Southern California showed a wide range of percentages active, generally 35 to 85% (12, 17). We are unaware of previous published autoradiography results for samples from the Mediterranean for comparison, although our own unpublished experiments with samples from within Villefranche Bay (July 1996, about 500 m closer to shore and considerably shallower than the samples from this study) showed that 73 to 86% of the cells were active autoradiographically with 5 nM added amino acids (method described by Tabor and Neihof [32]). We do not know enough about these systems at this point to explain these differences, and they may have methodological as well as ecological causes.
The limits of sensitivity of STARFISH have not yet been determined, but they are compounded by the previously described limitations of both FISH (27) and autoradiography (29). The portion of probe-labeled archaeal cells that were not detected in the autoradiography, for example, probably represents a group of cells growing at a rate below our detection level and possibly not growing at all (26).
Previous work looking at the activity of whole-cell prokaryotic organisms by culture-independent methods required that an organism be recognized by its unique morphology through direct microscopic observations. Examples of such organisms include the aquatic filamentous Leucothrix mucor (4), as well as Microthrix parvicella and Nostocoida limicola found in activated sludge (1). However, measuring nutrient uptake by organisms with unique morphology in a mixed community is possible only once the classification system is determined, which normally requires pure cultures, and “morphospecies” may contain large genetic variations. Since the majority of microorganisms fall within a few morphotypes, alternative methods such as STARFISH (6, 18, 26) currently provide the only techniques for collecting information on in situ activity by specific microorganisms with nondistinct morphologies, which includes most of the bacterioplankton of natural systems.
Overall, the results reported here shed new light on microbial activity in ocean mid-depth waters. They suggest that Archaea, given their high abundance and activity, probably play a significant role in the organic and possibly inorganic biogeochemical cycles in the world's oceans, especially at greater depths.
ACKNOWLEDGMENTS
We greatly appreciate the support from many staff members of the Villefranche Marine Laboratory, especially Maria-Luiza Pedrotti and Sophie Beauvais. Liam Carr helped with the counts, and Bess Ward provided the water samples and auxiliary data from Monterey Bay.
This work was funded by the National Science Foundation grants DEB 9705523 and OCE 9906989, the University of Southern California Wrigley Institute for Environmental Studies, and a University of Southern California Sea grant.
REFERENCES
- 1.Andreasen K, Nielsen P H. Application of microautoradiography to the study of substrate uptake by filamentous microorganisms in activated sludge. Appl Environ Microbiol. 1997;63:3662–3668. doi: 10.1128/aem.63.9.3662-3668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azam F, Fenchel T, Field J G, Gray J S, Meyer-Reil L A, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- 3.Braun-Howland E B, Danielsen S A, Nierzwicki-Bauer S A. Development of a rapid method for detecting bacterial cells in situ using 16S rRNA-targeted probes. BioTechniques. 1992;13:928–933. [PubMed] [Google Scholar]
- 4.Brock T D, Brock M L. Autoradiography as a tool in microbial ecology. Nature. 1966;209:734–736. doi: 10.1038/209734a0. [DOI] [PubMed] [Google Scholar]
- 5.Brunk C F, Avaniss-Aghajani E, Brunk C A. A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl Environ Microbiol. 1996;62:872–879. doi: 10.1128/aem.62.3.872-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottrell M T, Kirchman D L. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–1697. doi: 10.1128/aem.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong E F, Taylor L T, Marsh T L, Preston C M. Visualization and enumeration of marine planktonic Archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol. 1999;65:5554–5563. doi: 10.1128/aem.65.12.5554-5563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 10.Fuhrman J A. Bacterioplankton roles in cycling of organic matter: the microbial food web. In: Falkowski P G, Woodhead A D, editors. Primary productivity and biogeochemical cycles in the sea. New York, N.Y: Plenum Press; 1992. pp. 361–383. [Google Scholar]
- 11.Fuhrman J A. Close coupling between release and uptake of dissolved free amino acids in seawater studied by an isotope dilution approach. Mar Ecol. 1987;37:45–52. [Google Scholar]
- 12.Fuhrman J A, Azam F. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar Biol. 1982;66:109–120. [Google Scholar]
- 13.Fuhrman J A, Davis A A. Widespread Archaea and novel bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar Ecol Prog Ser. 1997;150:275–285. [Google Scholar]
- 14.Fuhrman J A, McCallum K, Davis A A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 15.Fuhrman J A, Ouverney C C. Marine microbial diversity studied via 16S rRNA sequences: coastal cloning results and counting of native archaea with fluorescent single cell probes. Aquatic Ecol. 1998;32:3–15. [Google Scholar]
- 16.Hershberger K L, Barns S M, Reysenbach A-L, Dawson S, Pace N R. Wide diversity of Crenarchaeota. Nature. 1996;384:420. doi: 10.1038/384420a0. [DOI] [PubMed] [Google Scholar]
- 17.Karner M, Furhman J A. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee N, Nielsen P H, Andreasen K H, Juretschko S, Nielsen J L, Schleifer K-H, Wagner M. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massana R, Murray A E, Preston C M, DeLong E F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massana R, Taylor L T, Murrays A E, Wu K Y, Jeffrey W H, DeLong E F. Vertical distribution and temporal variation of marine planktonic archaea in the Gerlache Strait, Antartica, during early spring. Limnol Oceanogr. 1998;43:607–617. [Google Scholar]
- 21.Murray A E, Blakis A, Massana R, Strawzewski S, Passow U, Alldredge A, DeLong E F. A time series assessment of planktonic archaeal variability in the Santa Barbara Channel. Aquatic Microb Ecol. 1999;20:129–145. [Google Scholar]
- 22.Murray A E, Preston C M, Massana R, Taylor L T, Blakis A, Wu K, DeLong E F. Seasonal and spatial variability of bacterial and Archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray A E, Wu K Y, Moyer C L, Karl D M, DeLong E F. Evidence for circumpolar distribution of planktonic Archaea in the Southern Ocean. Aquatic Microb Ecol. 1999;18:263–273. [Google Scholar]
- 24.Olsen G J. Archaea, Archaea, everywhere. Nature. 1994;371:657–658. doi: 10.1038/371657a0. [DOI] [PubMed] [Google Scholar]
- 25.Ouverney C C. Ph.D. thesis. Los Angeles: University of Southern California; 1999. [Google Scholar]
- 26.Ouverney C C, Fuhrman J A. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl Environ Microbiol. 1999;65:1746–1752. doi: 10.1128/aem.65.4.1746-1752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouverney C C, Fuhrman J A. Increase in fluorescence intensity of 16S rRNA in situ hybridization in natural samples treated with chloramphenicol. Appl Environ Microbiol. 1997;63:2735–2740. doi: 10.1128/aem.63.7.2735-2740.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preston C M, Wu K Y, Molinski T F, DeLong E F. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers A W. Techniques of autoradiography. New York, N.Y: Elsevier/North-Holland Biomedical Press; 1979. [Google Scholar]
- 30.Stein J L, Simon M I. Archaeal ubiquity. Proc Natl Acad Sci USA. 1996;93:6228–6230. doi: 10.1073/pnas.93.13.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suttle C A, Chan A M, Fuhrman J A. Dissolved free amino acids in the Sargasso Sea: uptake and respiration rates, turnover times, and concentrations. Mar Ecol Prog Ser. 1991;70:189–199. [Google Scholar]
- 32.Tabor P S, Neihof R A. Improved microautoradiographic method to determine individual microorganisms active in substrate uptake in natural waters. Appl Environ Microbiol. 1982;44:945–953. doi: 10.1128/aem.44.4.945-953.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams S C, Hong Y, Danavall D C A, Howard-Jones M H, Gibson D, Frischer M E, Verity P G. Distinguishing between living and nonliving bacteria: evaluation of the vital stain propidium iodide and the combined use with molecular probes in aquatic samples. J Microbiol Methods. 1998;32:225–236. [Google Scholar]