Abstract
Two hundred and twenty-six Salmonella enterica serotype Typhimurium isolates were examined for the presence of integron-associated gene cassettes. All but two of the non-DT104 isolates, together with DT104 isolates, contained gene cassettes. Amplicons of 1.5 kbp each were found in two non-DT104 isolates, encoding a dhfrI gene (trimethoprim resistance) linked to an aadA gene (streptomycin and spectinomycin resistance), by site-specific recombination. DT104 isolates of resistance (R) type ACSSuT possessed the recently described 1.0- and 1.2-kbp gene cassettes. Macrorestriction analysis with XbaI and DNA probing mapped ant(3")-1a, blaPSE-1, and dhfrI genes to large multiresistant gene clusters in a DT170a isolate and a DT193 isolate. In contrast, all DT104 isolates (R-type ACSSuT) possessed a conserved 10-kbp Xba1 DNA fragment. Our study highlights the occurrence of integrons (and antimicrobial resistance determinants) among serotype Typhimurium isolates other than DT104. Larger and previously unrecognized multiresistance gene clusters were identified in these isolates by DNA probing.
Salmonella enterica serotype Typhimurium is recognized as a significant human pathogen. It is currently estimated that, of the 40,000 Salmonella isolates reported annually to the Centers for Disease Control and Prevention, 8.5% are identified as serotype Typhimurium. These organisms are often resistant to five or more antimicrobial agents, including ampicillin (A), chloramphenicol (C), streptomycin (S), sulfonamides (Su), and tetracycline (T)—the characteristic resistance (R) type ACSSuT. Reporting of resistance to fluoroquinolones or extended-spectrum cephalosporins is also increasing annually (9). Serotype Typhimurium is frequently present in the gastrointestinal tracts of cattle, pigs, poultry, and other animal species and is transferred to humans via the food chain. In particular, Serotype Typhimurium definitive phage type 104 (DT104), R-type ACSSuT, has emerged as a global health problem. While most Salmonella infections are self-limiting, occasionally a more invasive illness can develop that requires effective antimicrobial agent-based therapy. Emerging (antimicrobial) multidrug resistance (MDR) is associated with the therapeutic and nontherapeutic uses of these agents in food animals.
Acquisition and dissemination of genetic determinants of antimicrobial resistance are accounted for in part by R plasmids and transposons (6, 20, 22, 25, 31). Often, the frequency with which resistance to sulfonamide is encountered suggests the involvement of a third mechanism linked to a novel group of naturally occurring mobile genetic elements, integrons. Integrons may be found as part of the Tn21 transposon family or located on broad-host-range plasmids (8, 13). Three integron classes, denoted I through III, have been described. Class I integrons are significant in clinical isolates. The fundamental class I integron structure consists of a 5′-conserved segment (5′CS) and a 3′-conserved segment (3′CS). The former encodes an integrase gene, intI (23), containing the attI recombination site, necessary for integrating gene cassettes. The 3′CS contains two well characterized open reading frames (ORFs); qacEΔI, conferring resistance to quaternary ammonium compounds; and the sulI encoding sulfonamide resistance determinant (18). Sulfonamide resistance is therefore a useful marker for the presence of class I integrons. Located between these conserved regions are gene cassettes that vary in length and molecular organization. Several gene cassettes have been characterized, the majority of which contain antimicrobial resistance-encoding genes (20, 21). Some cassettes may have two antimicrobial resistance genes fused by site-specific recombination, via the conserved 59-base element, located downstream of the ORF. These structures have recently been mapped to the chromosome of serotype Typhimurium (30). Transduction experiments with a host-specific P-22-like phage facilitated the horizontal transfer of these chromosomal located multiresistance genes (26) to recipient strains.
The emergence of MDR among bacterial species is causing concerns among medical, veterinary, and public health professionals (3). Epidemiological data show that the most common source of MDR DT104 and other MDR serotype Typhimurium infections in humans is the consumption of contaminated food of animal origin and direct contact with livestock (31). Although cattle are recognized as a major reservoir for serotype Typhimurium DT104, increasing numbers are being reported in other animals, including porcine and avian populations (22). To extend these observations and determine their relevance in this geographical region we studied 226 randomly collected serotype Typhimurium isolates (183 DT104 types and 34 non-DT104 phage types together with 7 nontypeables) for the presence of class I integrons and mapped these genes in a subset of the isolates. The significance of our findings, together with implications for the continuing and uniformed usage of antimicrobial agents, are discussed.
MATERIALS AND METHODS
Bacterial strains.
A total of 226 S. enterica serotype Typhimurium isolates were investigated in this study. A number of isolates are referred to in the text; a complete listing of all isolates and their relevant characteristics may be found at the following web site address: www.cit.ie/courses/courses.htm. These isolates were collected between the end of 1997 and 1999 from animal (clinical), human, and food sources. Cork University Hospital, Waterford Regional Hospital, the Cork Regional Veterinary Laboratory, and the Cork County Council Food Laboratory each submitted isolates to the Molecular Diagnostics Unit (MDU) for study. All isolates were identified as S. enterica serotype Typhimurium based on colony morphology and serotyping. In addition, all isolates were phage typed, tested for their susceptibility to a range of antimicrobial agents, and finally stored and maintained as previously outlined by Daly et al. (6).
Gene cassette amplification and analysis.
Inserted gene cassettes were amplified using Int1F and Int1B primers as described by Lévesque et al. (13). Amplification conditions were identical to those reported previously (6). Each amplicon was individually excised from an agarose gel and purified using the QIAquick gel extraction kit (Qiagen, West Sussex, United Kingdom) according to the manufacturer's instructions. Gel slice-extracted DNA was then reamplified using the same reaction conditions outlined above to assess the quality of the gel excision step. Following this step the PCR product was directly ligated to pCR2.1 (Invitrogen, BV, Amsterdam, The Netherlands) and cloned according to the manufacturer's recommendations. All gene cassettes were cloned in this manner, and the corresponding constructs were carefully screened for the correct insert before purification using the Wizard Plus SV Minipreps DNA Purification system (Promega, Madison, Wis.). Inserts were sequenced using the Int1F and Int1B primers and dye terminator chemistry protocols with cycle sequencing (Beckman Coulter).
PFGE.
Representative Salmonella serotype Typhimurium isolates were prepared for pulsed-field gel electrophoresis (PFGE) as described by Maslow et al. (16) with some minor modifications. Briefly, cells were grown overnight in tryptone soy broth (Oxoid, Hampshire, United Kingdom), washed in PIV buffer (containing 10 mM Tris-HCl [pH 7.6] and 1 M NaCl), and mixed with an equal volume of 2% (wt/vol) InCert agarose (FMC Bioproducts, Rockland, Maine). The culture-agarose mixture was placed in 1-ml syringe barrels (Becton Dickinson, Dublin, Ireland) and allowed to solidify on ice for 30 min. Cell lysis was achieved using a mixture containing Lysozyme (Sigma, Poole, United Kingdom) at a final concentration of 0.1 mg/ml and RNase (Sigma) at a final concentration of 20 μg/ml in lysis buffer (6 mM Tris [pH 7.6], 1 M NaCl, 100 mM EDTA, 0.5% Brij-58 [polyoxyethylene-20-cetyl-ether], 0.2% sodium deoxycholate, 0.5% sodium lauroyl sarcosine). Plugs were washed three times in ESP solution (0.5 M EDTA [pH 8], 10% [wt/vol] lauroylsarcosine, and proteinase K [Sigma] at a final concentration of 100 mg/liter). The first two washes were at 50°C for 2 h, followed by a third wash overnight at the same temperature. Inactivation of proteinase K (Sigma) was performed in one 4-h wash, followed by one overnight wash in 1× TE-PMSF (10 mM Tris-HCl [pH 7.6], 0.1 mM EDTA, 1.5 mM phenylmethylsulfonyl fluoride), followed by two 2-h washes in 1× TE.
Restriction endonuclease digestion was carried out on 1- to 2-mm slices of each prechilled gel mold by adding 5 U of XbaI in multicore buffer (final concentration, 1×), bovine serum albumin (final concentration, 0.1 mg/ml; Promega) to a final volume of 100 μl with sterile distilled water. Plugs were allowed to digest for 3 h at 37°C. Samples were electrophoresed in a 1% (wt/vol) agarose gel (SeaKem Gold; FMC Bioproducts) in 0.5× TBE (45 mM Tris, 45 mM borate [pH 8.3], 1 mM EDTA) for 20 h at 10°C. Electrophoresis was performed at 200 V using a Gene Navigator system with a hexagonal electrode array (Pharmacia LKB Biotechnology AB, Uppsala, Sweden) in the interpolation mode pulsing from 1 through 40 s. Molecular weight markers included mid-range PFG Markers (New England BioLabs, Hertfordshire, United Kingdom), and digoxigenin-labeled DNA molecular weight marker grade II (Roche Diagnostics, Lewes, East Sussex, United Kingdom). Gels were stained with 0.5 mg of ethidium bromide per liter in distilled water before being photographed over a UV transilluminator.
Southern blotting.
After PFGE, DNA fragments were transferred to 15-by-20-cm Nytran Nylon Membranes (Schleicher & Schuell, Dassel, Germany) essentially using the method previously described by Southern (27). DNA was allowed to transfer overnight, after which the membrane was air dried and baked at 80°C for 1 to 2 h. Probing and detection of DNA fragments was performed using a DIG-DNA labeling and detection kit (Roche Diagnostics). All protocols, buffers, and reagents recommended by the kit manufacturer were used throughout. Briefly, all membranes were prehybridized for 1 h at 55°C, followed by hybridization overnight with the corresponding DIG-labeled DNA probe at 55°C. Stringency washes of the membrane were performed at 68°C.
Generation of nonradiolabeled DNA probes.
Probe generation was performed by PCR on 1 μl (ca. 500 ng) of each plasmid construct containing a previously characterized gene cassette (as described below). Reaction conditions were identical to those described previously (6) except that 1 μl of DIG-dUTP (1 nmol/μl) (Roche Diagnostics) was included in the deoxynucleoside triphosphate mix. This step ensures simultaneous amplification and labeling from the corresponding DNA template. Except for the dhfrI gene probe, all other probes were prepared in this way. Since the dhfrI gene was located proximal and within the same 1.5-kbp gene cassette containing a gene coding for an aminoglycoside modifying enzyme aadA, it was necessary to design a primer set to selectively generate the dhfrI probe. These primers were dhfrI F (5′-GTG AAA CTA TCA CTA ATG GTA GCT-3′ [24-mer]) and dhfrI R (5′-ACC CTT TTG CCA GAT TTG GTA ACT-3′ [24-mer]). Amplification was performed in a MiniCycler (MJ Research, Inc., Watertown, Mass.) using the following temperature profile: predenaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 1 min, 65°C for 1 min, 72°C for 1 min and a final extension step at 72°C for 5 min. This reaction yielded the expected DIG-labeled 473-bp dhfrI-specific DNA product which was then used as a probe.
Nucleotide sequence accession numbers.
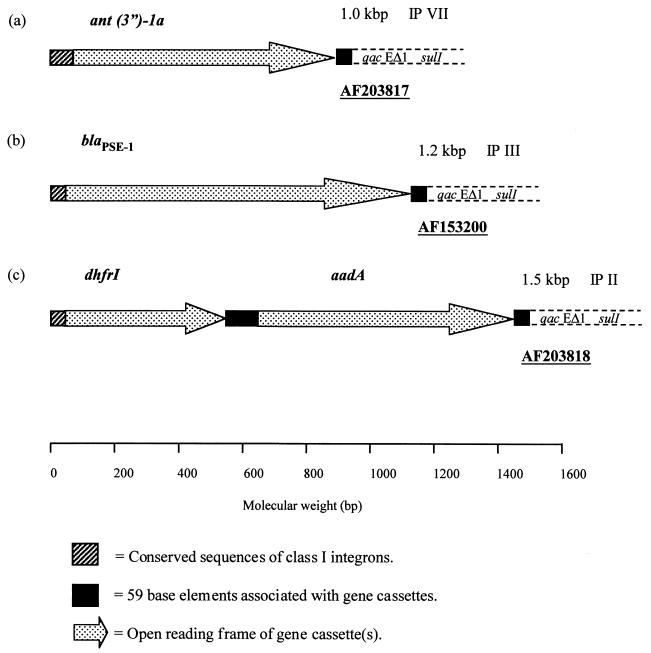
The complete coding sequence of the ant(3")-1a gene has been assigned GenBank accession no. AF203817. The complete coding sequence of the blaPSE-1 gene was assigned GenBank accession no. AF153200, and the 1.5-kbp amplicon containing the coding sequences for dhfrI followed by aadA was assigned GenBank accession no. AF203818.
RESULTS
PCR based detection of integron-associated antimicrobial resistance gene cassettes.
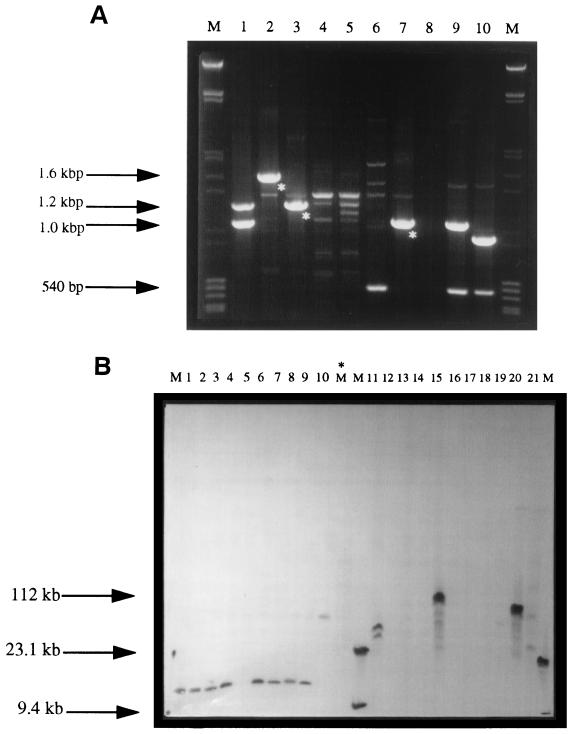
A random collection of 226 serotype Typhimurium isolates (Table 1), cultured from human (133 isolates), veterinary (clinical) (including 32 bovine, 18 porcine, 5 ovine, 2 poultry, 4 equine, 6 canine, and 3 additional isolates), and food sources (23 isolates) were tested for gene cassettes by PCR. Class I integron-associated gene cassettes were amplified with primers described previously (13). All DT104 isolates and related isolates (including DT104b and PT U302), constituting 81% (183 of 226 isolates) of the study collection (Table 1), produced amplified products (Fig. 1A). Within this group, 174 of the 183 DT104 complex isolates (95%) consisted of two copies of a class I integron containing 1.0- and 1.2-kbp gene cassettes (Fig. 1A, lane 1). This DNA fragment pattern was denoted as integron pattern (IP) type I (Table 1) and was also demonstrated in other, unrelated, DT104 isolates cultured previously (6, 17, 22, 25) from human, veterinary, and food sources. All IP type I isolates were R-type ACSSuT (Table 1). Eight of the remaining DT104 isolates contained the 1.0-kbp amplicon alone (IP-VIII; Fig 1A, lane 7) with the R-type SSu. The occurrence of the latter IP profile is rare among DT104, and CIT-F44 (DT104) contained a single 1.2-kbp amplified gene cassette (IP-III; Fig 1A, lane 3) of R-pattern ASu (see also Table 2).
TABLE 1.
Summary of 226 Salmonella serotype Typhimurium isolates analyzed by the MDU
| Phage type | R typea | IP type | Source
|
No. of isolates (subtotal) | ||
|---|---|---|---|---|---|---|
| H | V | F | ||||
| 104 | ACSSuT | I | 107 | 47 | 8 | 162 |
| 104 | ACSSuTTp | I | 2 | 2 | 0 | 4 |
| 104 | ACSSuTN | I | 1 | 0 | 0 | 1 |
| 104b | ACSSuTK | I | 0 | 0 | 3 | 3 |
| 104b | ACSSuTTp | I | 1 | 2 | 0 | 3 |
| 104 | SSu | VII | 8 | 0 | 0 | 8 |
| 104 | ASu | III | 0 | 0 | 1 | 1 |
| U302 | ACSSuTTp | I | 0 | 1 | 0 | 1 (183) |
| 193 | ASSuTTp | II | 0 | 0 | 1 | 1 |
| 193 | ASSuTN | IV | 0 | 0 | 1 | 1 |
| 193 | ASSuT | IV | 1 | 3 | 2 | 6 |
| 193 | SuTTp | IV | 0 | 1 | 0 | 1 |
| 193 | ASuTTp | IV | 0 | 1 | 0 | 1 |
| 193 | SSuT | IV | 0 | 1 | 0 | 1 (11) |
| 195 | T | IV | 5 | 0 | 0 | 5 |
| 195 | SuTTp | IV | 0 | 1 | 0 | 1 |
| 195 | STp | IV | 0 | 0 | 1 | 1 |
| 195 | SuTp | IV | 0 | 0 | 1 | 1 |
| 195 | ASSuT | VI | 1 | 0 | 0 | 1 |
| 195 | ASSuT | IV | 2 | 0 | 0 | 2 |
| 195 | Sensitive | IV | 0 | 1 | 0 | 1 (12) |
| 204a | ASSuT | none | 0 | 0 | 2 | 2 (2) |
| 208 | T | IV | 0 | 3 | 0 | 3 |
| 208 | SuTTp | IV | 0 | 2 | 0 | 2 (5) |
| 170a | SuTTp | IV | 0 | 3 | 0 | 3 |
| 170a | ASSuTTp | II | 0 | 1 | 0 | 1 (4) |
| 15a | SuTTp | IV | 0 | 0 | 1 | 1 |
| 15a | T | IV | 1 | 0 | 0 | 1 (2) |
| NT | SuTp | V | 0 | 0 | 1 | 1 |
| NT | SuTp | IV | 1 | 0 | 0 | 1 |
| NT | ACSSuT | I | 2 | 0 | 0 | 2 |
| NT | Sensitive | IV | 1 | 0 | 0 | 1 |
| NT | Sensitive | V | 0 | 0 | 1 | 1 |
| NT | S | V | 0 | 1 | 0 | 1 (7) |
R type, antimicrobial resistance pattern; A, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfonamide; T, tetracycline; Tp, trimethoprim; N, nalidixic acid; K, kanamycin.
FIG. 1.
(A) Amplified gene cassettes and corresponding IP groups of representative strains of S. enterica serotype Typhimurium. After PCR, 10 μl of the amplified reaction mixture was loaded onto a 1% agarose gel in 1× Tris-EDTA-acetate (TAE) buffer containing 0.1 μg of ethidium bromide per ml. Samples were horizontally electrophoresed at 100 V for 90 min. The lanes marked M contain grade III molecular weight markers ranging in size from 0.56 to 21.2 kb and grade V molecular weight markers ranging in size from 8 to 597 bp (Roche Diagnostics). The asterisks represent the gene cassettes selected for sequencing. Lanes (IP type): 1, CIT-F45 (I); 2, CIT-F41 (II); 3, CIT-F44 (III); 4, CIT-V75 (IV); 5, CIT-V111 (V); 6, CIT-H11 (VI); 7, CIT-H183 (VII); 8, CIT-F46 (none detected); 9, E. coli R100.1 (control); 10, E. coli R751 (control). (B) Following PFGE, the XbaI macrorestricted DNA fragments were transferred to nylon membranes. Individual gene cassettes were then used to probe the membranes. The Southern blot using the blaPSE-1 gene as a probe is shown. The lanes marked M contain DIG-labeled DNA molecular weight marker grade II (Roche Diagnostics). The unlabeled mid-range PFG Markers (New England BioLabs) in lane M* were included for fragment sizing before Southern transfer. Lanes (IP type): 1, CIT-V38 (I); 2, CIT-F45 (I); 3, CIT-H164 (I); 4, CIT-H176 (I); 5, CIT-H183 (VII); 6, CIT-V115 (I); 7, CIT-F107 (I); 8, CIT-H144 (I); 9, CIT-V37 (I); 10, CIT-F34 (IV); 11, CIT-F41 (II); 12, CIT-F44 (III); 13, CIT-V60 (IV); 14, CIT-V75 (IV); 15, CIT-V127 (II); 16, CIT-V129 (IV); 17, CIT-F40 (V); 18, CIT-F105 (V); 19, CIT-H195 (IV); 20, E. coli R100.1; 21, E. coli R751.
TABLE 2.
Southern blotting of representative Salmonella serotype Typhimurium isolates and probe hybridization using selected gene cassettesa
| Phage type | R type | Presence (+) or absence (−) of 10-kb XbaI fragment | IP type | Probe hybridization
|
||
|---|---|---|---|---|---|---|
| ant(3")-1a | pseI | dhfrI | ||||
| DT104 | ||||||
| CIT-V38 | ACSSuT | + | I | + | + | − |
| CIT-F45 | ACSSuT | + | I | + | + | − |
| CIT-H164 | ACSSuT | + | I | + | + | − |
| CIT-H176 | ACSSuTTp | + | I | + | + | − |
| CIT-H183 | SSu | − | VII | − | − | − |
| CIT-V115 | ACSSuT | + | I | + | + | − |
| CIT-F44 | ASu | − | III | − | − | − |
| DT104 related | ||||||
| CIT-F107 (104b) | ACSSuTK | + | I | + | + | − |
| CIT-H144 (104b) | ACSSuTTp | + | I | + | + | + |
| CIT-V37 (U302) | ACSSuTTp | + | I | + | + | − |
| DT193 | ||||||
| CIT-F34 | ASSuTN | − | IV | + | + | + |
| CIT-F41 | ASSuTTp | − | II | + | + | + |
| DT195, CIT-V60 | SuTTp | − | IV | − | − | − |
| DT208, CIT-V75 | T | − | IV | − | − | − |
| DT170a | ||||||
| CIT-V127 | ASSuTTp | − | II | + | + | + |
| CIT-V129 | SuTTp | − | IV | − | − | − |
| NT | ||||||
| CIT-F40 | Sensitive | − | V | − | − | − |
| CIT-F105 | SuTp | − | V | − | − | − |
| CIT-H195 | SuTp | − | IV | + | + | − |
| Controls | ||||||
| E. coli R100.1 | − | a | + | + | + | |
| E. coli R751 | − | b | + | + | − | |
ant(3")-1a, aminoglycoside-modifying enzyme coding gene; pseI, β-lactamase gene; dhfrI, dihydrofolate reductase gene. R type, antimicrobial resistance pattern. IP types a and b were previously defined (6). A, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfonamides; T, tetracycline; N, nalidixic acid; K, kanamycin. In the phage types, the letters V, H, and F refer to veterinary, human, and food origins, respectively.
Gene cassettes were also identified in non-DT104 isolates, including DT193 (n = 11), −195 (n = 12), −208 (n = 5), −170a (n = 4), and −15a (n = 2), and non-phage-typeable (NT) (n = 7) isolates (Table 1). Class I integron structures were not detected in two DT204a isolates (Table 1). Two of the non-typeable isolates (CIT-H143 and CIT-H201 [Table 1]) displayed the same IP type I and R types (ACSSuT) found in DT104 isolates. When gene cassettes were amplified in the remaining non-DT104 isolates, the numbers and/or the size of the DNA products was distinct from the predominant IP type I, DT104 pattern. In one example, a large 1.5-kbp amplicon was identified in both CIT-F41 (DT193, IP type II) and CIT-V127 (DT170a, IP type II) (Fig. 1A, lane 2).
DNA sequencing.
Gene cassettes denoted by the asterisks in Fig. 1A were completely characterized, including the 1.5-kbp amplicon from CIT-F41 (DT-193; IP type II), the 1.2-kbp DNA fragment in CIT-F44 (DR104; IP type III), and the single 1.0-kbp band from CIT-H183 (DR104; IP type VII) (Fig. 1A, lanes 2, 3, and 7; Fig. 2). IP types IV, V, and VI (Fig. 1A, lanes 4, 5, and 6) contained weakly amplified DNA products. These were not always reproducible and were not considered further at this time.
FIG. 2.
Schematic representation of the molecular organization of antimicrobial agent resistance-encoding ORFs sequenced from selected gene cassettes (see Fig. 1A). The direction of transcription is given by the arrowheads in the figure. Also included are the identities of each ORF, the corresponding GenBank accession numbers, the IP profile types, and the sizes of the amplified gene cassettes. Conserved integron-associated features are also shown. An approximate molecular weight scale is included at the bottom of the figure.
Sequence analysis (including BLAST searches of the current databases) of the 1.0-kbp amplicon (R-type SSu and Fig. 1A, lane 7) identified an ORF of 789 bp encoding an ant(3")-1a gene (Fig. 2a and Table 2). Similarly, the single 1.2-kbp amplicon (R-type ASu and Fig. 1A, lane 3), contained an ORF of 993 bp (Fig. 2b) and was identical to a previously characterized blaPSE-1 gene (22, 25). When the 1.5-kbp amplified cassette (Table 2; Fig. 1A, lane 2) was analyzed, two site-specific recombined ORFs of 471 and 789 bp were noted (FIG. 2C). BLAST analysis identified these ORFs as a dhfrI gene conferring resistance to trimethoprim followed by aadA coding for an aminoglycoside-modifying enzyme.
Along with the ORFs outlined above, further comparisons of these sequences identified the inverted repeat elements (Fig. 2a, b, and c), consisting of the GTT triplet insertion point (4, 5, 10, 15) immediately adjacent to the 5′ end of the integrated cassette(s) and the 59 base elements at the cassette 3′ end of the gene (5, 10, 14, 28).
Mapping gene cassettes on the Salmonella serotype Typhimurium chromosome.
Antimicrobial agent resistance-encoding genes derived from integrons were previously mapped to the chromosome of the host (1–3, 22, 25). To determine whether this situation extends to Irish serotype Typhimurium and to directly compare non-DT104 and DT104 isolates, nineteen isolates (Table 2) were chosen for investigation. Two Escherichia coli K-12 isolates were included for comparison (25). Genomic DNA was prepared from all of these isolates and subjected to macrorestriction with XbaI. After PFGE and Southern blotting, the gels were sequentially probed with the DNA probes derived from amplified gene cassettes (Table 2; Fig. 2a through c) outlined above.
Analysis of the Southern blots identified a XbaI DNA fragment of approximately 10 kbp in all DT104 and related isolates of R-type ACSSuT (see Fig. 1B, lanes 1 to 4 and 6 to 9). Only the ant(3")-1a and blaPSE-1 probes mapped to this fragment, identifying a multiresistance gene cluster or “resistance island” in the genome of serotype Typhimurium (1–3, 17, 22). In other, more-drug-sensitive, DT104 isolates (e.g., CIT-H183 R-type SSu [Fig. 1B, lane 5] and CIT-F44, R-type ASu [Fig. 1B, lane 12]) this 10-kbp XbaI “resistance island” was not detected after probing.
Previously unrecognized and apparently larger multiresistance gene clusters were observed in non-DT104 isolates, including CIT-F34 (DT193), CIT-F41 (DT193), and CIT-V127 (DT170a) (Table 2 and Fig. 1B, lanes 10, 11, and 15, respectively). The hybridizing XbaI DNA fragment in CIT-F34 (DT193) was located at approximately 82 kbp (Fig. 1B, lane 10), containing the ant(3")-1a and blaPSE-1 genes. Two positive signals were also observed in CIT-F41, of approximately 48.5 and 63.5 kbp, to which all probes mapped. The 1.5-kbp dhfrI-aadA gene cassette (IP type II) was amplified from the latter isolate. However, the largest XbaI DNA fragment hybridizing to all DNA probes was found in CIT-V127 (DT170a), located at approximately 112 kbp. The latter isolate had an IP type II profile containing the recombined dhfrI and aadA genes on a 1.5-kbp gene cassette with the R-type ASSuTTp. Non-DT104 serotype Typhimurium containing a gene cassette-encoding dhfrI gene mapped to these very large multiresistance DNA fragments. In addition, two weakly hybridizing signals in CIT-H144 (DT104b, IP type I, R-type ACSSuTTp, and Table 2) were detected with the dhfrI DNA probe alone (data not shown). These XbaI DNA fragments were approximately 6.5 and 170 kbp in size.
DISCUSSION
Rapid dissemination of antimicrobial resistance through bacterial populations has long been attributed to the presence of plasmids and transposons. Over the last decade transposon-like structures have been identified which were capable of integrating into bacterial genomes, presenting a third mechanism contributing to the spread of drug resistance (6, 14, 20, 22). These integron structures are responsible for a substantial proportion of antimicrobial resistance to the more commonly used antibiotics and, in many cases, are often plasmid-borne (11). Recently, integrons have been recognized in the chromosome of serotype Typhimurium (30). There is concern that the integration of resistance determinants into the chromosome may contribute to enhanced persistence of antimicrobial resistance even in the absence of antimicrobials.
Use of antimicrobials (for both therapeutic and nontherapeutic purposes) in food animals is the dominant factor contributing to the increase in the reporting of drug-resistant isolates (7, 9). In particular, the spread of the penta-resistant serotype Typhimurium DT104 is causing global concern with potentially significant impact on public health. It is now acknowledged that these organisms are transmitted from animals to humans via the food chain (31, 32). An understanding of molecular mechanisms of resistance will provide useful markers to assist future studies investigating the evolution of MDR (1, 3, 7).
Two hundred and twenty-six serotype Typhimurium isolates were randomly collected over a 2-year period. The majority of these were DT104 and DT104-related isolates (77%) of R-type ACSSuT. With the exception of DT204a, all phage types had at least one class I integron, as determined by amplification of a gene cassette(s). Seven IP profiles denoted I through VII were detected, suggesting that considerable drug resistance encoding potential may exist in these organisms. Ninety-five percent of the DT104 and DT104-related isolates had two integrons, both of which were previously characterized. The latter pattern, denoted IP type I, consisted of two amplified cassettes of 1.0 kbp encoding the ant(3")-1a gene and a larger 1.2-kbp amplicon containing the blaPSE-1 gene. In earlier reports, DNA probe experiments, long-range PCR, and DNA sequencing strategies identified a 10-kbp chromosomal fragment in DT104 isolates (3, 22) containing the latter integrons. Recombined between these structures was an R plasmid expressing chloramphenicol and tetracycline resistance (1, 3). Based on the mapping data reported here, it is reasonable to suggest that this multiresistant gene cluster, or resistance island, is conserved among Irish DT104 strains of R-type ACSSuT. Importantly, chloramphenicol and more recently florfenicol (1, 2) resistance in DT104 and DT104-related strains correlated with the presence of the 10-kbp XbaI chromosomal DNA band. Chloramphenicol resistance is recognized as a good marker for DT104 (9). This marker may be useful in prioritizing the investigation of an outbreak cluster (1, 12).
Although our isolates are a random collection and are not truly representative of isolates in Ireland generally, the non-DT104 isolates provided the opportunity to directly compare their integron structures and corresponding mapping locations with those of MDR-DT104. Forty-three isolates (19%) in this collection were non-DT104 serotype Typhimurium representing four phage types, including DT193, DT195, DT170a, and DT208 and three non-phage-typeable isolates. Ninety-five percent (41 of 43) produced amplified gene cassettes of IP types II, IV, V, and VI. CIT-F41 (DT193) and CIT-V127 (DT170a) both contained a 1.5-kbp gene cassette encoding two site-specific recombined ORFs encoding resistance to trimethoprim and spectinomycin-streptomycin, respecitvely. A similar arrangement was recently reported for a transmissible plasmid in E. coli (24) and in Klebsiella pneumoniae, Serratia marcescens, and Psuedomonas aeruginosa (11, 23, 29), highlighting the potential for gene transfer to other bacteria, including unrelated species. Interestingly, IP-II types containing the dhfrI gene mapped the latter to apparently larger multiresistance gene clusters in the isolates CIT-F41 (DT193) and CIT-V127 (DT170a) compared with the conserved 10-kbp XbaI DNA fragment associated with MDR-DT104 of R-type ACSSuT. In DT104b isolates additionally resistant to trimethoprim (e.g., CIT-H144), the dhfrI gene was probably plasmid encoded, as this was the only DNA probe to map to large DNA fragments, greater than 10 kbp in size, in this case. Molecular organization of the MDR-DT104 10-kbp XbaI DNA fragment may represent a minimal “resistance island,” which has the potential to act as a “molecular sink” into which R-plasmids and other mobile genetic elements, including integrons, are recombined. Indiscriminate use of antimicrobial agents (including those for human therapeutic use) could speed the evolution of these structures and contribute to the spread of drug resistance (6, 7, 9, 14). However, confirmation of this hypothesis must await a more complete characterization of these putative “resistance islands.”
Definition of the (molecular) epidemiology of MDR-DT104 may be valuable in controlling the spread of these pathogens. These organisms were detected initially almost simultaneously in different geographical regions, with increasing evidence suggesting that DT104 has a clonal origin (6, 7, 19) independent of its source. All serotype Typhimurium isolates from human, veterinary, and food sources analyzed to date in this laboratory by DNA amplification (DAF) produced several DNA banding profiles (reference 6 and unpublished data), with the majority (>80%) being accounted for within the DAF-I group (6). Although the current strain collection is not truly representative of all Irish isolates, these data provide some clues as to the reservoir of MDR-DT104 and non-DT104 in Ireland. Furthermore, PFGE analysis of one-third of this collection of strains failed to distinguish between the DT104 isolates from any of the source groups, further supporting the clonal nature of this organism (unpublished data). This feature presents a significant technical challenge to the future tracking of MDR-DT104 by molecular methods. If these organisms are to be ultimately controlled it is essential that, in addition to understanding the epidemiology of DT104, the association between drug usage and the continued emergence of MDR-DT104 also be addressed. Molecular analysis of antimicrobial resistance indicated that the same gene cassettes account for MDR in isolates from diverse geographical regions (3, 6, 22, 25), with MDR-DT104 emerging as a result of antimicrobial selective pressure in humans and in food animals. Resistance to antimicrobial agents increases with the addition of new drugs (9). Currently, it is unknown whether any virulence determinants map to these multiresistance clusters and if increased virulence can be favored by antimicrobial selection (3). Finally, the stability of the latter clusters after drug removal needs to be assessed, in addition to advocating a more limited use of antimicrobial agents, in relation to growth promotion and routine animal prophylaxis.
ACKNOWLEDGMENTS
We thank Helen O'Shea, Emma Fanning, Patrick Wall, and Martin Cormican for critical comments on the manuscript, and we also thank all those who submitted isolates for study. We thank John Murphy and Dolores Crowley for technical assistance.
M.D. is the recipient of a scholarship from Cork Institute of Technology and laterally a postdoctoral fellowship awarded by the Irish government Science Agency, Enterprise Ireland (PD/2000/010). We are grateful for the financial support provided by Cork County Council and the Food Safety Authority of Ireland which partly funded this study.
REFERENCES
- 1.Arcangioli M A, Leroy-Setrin S, Martel J L, Chaslus-Dancla E. Evolution of chloramphenicol resistance, with emergence of cross resistance to florfenicol, in bovine Salmonella Typhimurium strains implicates definitive phage type (DT) 104. J Med Microbiol. 2000;49:103–110. doi: 10.1099/0022-1317-49-1-103. [DOI] [PubMed] [Google Scholar]
- 2.Arcangioli M A, Leroy-Setrin S, Martel J L, Chaslus-Dancla E. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella Typhimurium DT-104. FEMS Microbiol Lett. 1999;15:327–332. doi: 10.1111/j.1574-6968.1999.tb13586.x. [DOI] [PubMed] [Google Scholar]
- 3.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella Typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collis C M, Hall R M. Site specific deletion and rearrangement of integron insert genes catalysed by the integron DNA integrase. J Bacteriol. 1992;174:1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collis C M, Hall R M. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol Microbiol. 1992;6:2875–2885. doi: 10.1111/j.1365-2958.1992.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 6.Daly M, Buckley J, Power E, O'Hare C, Cormican M, Cryan B, Wall P G, Fanning S. Molecular characterization of Irish Salmonella enterica serotype Typhimurium: detection of class I integrons and assessment of genetic relationships by DNA amplification fingerprinting. Appl Environ Microbiol. 2000;66:614–619. doi: 10.1128/aem.66.2.614-619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis M A, Hancock D D, Besser T E, Rice D H, Gay J M, Gay C, Gearhart L, DiGiacomo R. Changes in antimicrobial resistance among Salmonella enterica serovar Typhimurium isolates from humans and cattle in the Northwestern United States, 1982–1997. Emerg Infect Dis. 1999;5:802–805. doi: 10.3201/eid0506.990610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falbo V, Carattoli A, Tosini F, Pezzella C, Dionisi A M, Luzzi I. Antibiotic resistance conferred by a conjugative plasmid and a class I integron in Vibrio cholerae O1 El Tor strains isolated in Albania and Italy. Antimicrob Agents Chemother. 1999;43:693–696. doi: 10.1128/aac.43.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glynn M K, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo F J. Emergence of multidrug resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–1338. doi: 10.1056/NEJM199805073381901. [DOI] [PubMed] [Google Scholar]
- 10.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination crossover point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 11.Kazama H, Kizu K, Iwasaki M, Hamashima H, Sasatsu M, Arai T. Isolation and structure of a new integron that includes a streptomycin resistance gene from the R plasmid of Pseudomonas aeruginosa. FEMS Microbiol Lett. 1995;134:137–141. doi: 10.1111/j.1574-6968.1995.tb07927.x. [DOI] [PubMed] [Google Scholar]
- 12.Khan A A, Nawaz M S, Khan S A, Cerniglia C E. Detection of multidrug-resistant Salmonella typhimurium DT104 by multiplex polymerase chain reaction. FEMS Microbiol Lett. 2000;182:355–360. doi: 10.1111/j.1574-6968.2000.tb08921.x. [DOI] [PubMed] [Google Scholar]
- 13.Lévesque C, Piche L, Larose C, Roy P H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucey B, Crowley D, Moloney P, Cryan B, Daly M, O'Halloran F, Threlfall E J, Fanning S. Integron like structures in Campylobacter spp. of human and animal origin. Emerg Infect Dis. 2000;6:50–55. doi: 10.3201/eid0601.000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez E, de la Cruz F. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 1990;9:1275–1281. doi: 10.1002/j.1460-2075.1990.tb08236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maslow J N, Slutsky A M, Arbeit R D. Application of pulsed field gel electrophoresis to molecular epidemiology. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and application. Washington, D.C.: American Society for Microbiology; 1993. pp. 563–572. [Google Scholar]
- 17.Ng L K, Mulvey M R, Martin I, Peters G A, Johnson W. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT 104. Antimicrob Agents Chemother. 1999;43:3018–3021. doi: 10.1128/aac.43.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulsen I T, Littlejohn T G, Radström P, Sundstrom L, Sköld O, Swedberg G, Skurray R A. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajashekara G, Haverly E, Halverson D A, Ferris K E, Lauer D C, Nagaraja K V. Multidrug resistant Salmonella Typhimurium DT104 in poultry. J Food Prot. 2000;63:155–161. doi: 10.4315/0362-028x-63.2.155. [DOI] [PubMed] [Google Scholar]
- 20.Recchia G D, Hall R M. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 1997;5:389–394. doi: 10.1016/S0966-842X(97)01123-2. [DOI] [PubMed] [Google Scholar]
- 21.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 22.Ridley A, Threlfall E J. Molecular epidemiology of antibiotic resistant genes in multiresistant epidemic Salmonella typhimurium DT 104. Microb Drug Resist. 1998;4:113–118. doi: 10.1089/mdr.1998.4.113. [DOI] [PubMed] [Google Scholar]
- 23.Sallen B, Rajoharison A, Desvarenne S, Malibat C. Molecular epidemiology of integron associated antibiotic resistance genes in clinical isolates of Enterobactericeae. Microb Drug Resist. 1995;1:195–202. doi: 10.1089/mdr.1995.1.195. [DOI] [PubMed] [Google Scholar]
- 24.Sandvang D. Novel streptomycin and spectinomycin resistance genes as a gene cassette within a class 1 integron isolated from Escherichia coli. Antimicrob Agents Chemother. 1999;43:3036–3038. doi: 10.1128/aac.43.12.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandvang D, Aarestrup F M, Jensen L B. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica typhimurium DT104. FEMS Microbiol Lett. 1998;160:37–41. doi: 10.1111/j.1574-6968.1998.tb12887.x. [DOI] [PubMed] [Google Scholar]
- 26.Schmieger H, Schickmaier P. Transduction of multiple drug resistance of Salmonella enterica serovar typhimurium DT104. FEMS Microbiol Lett. 1999;170:251–256. doi: 10.1111/j.1574-6968.1999.tb13381.x. [DOI] [PubMed] [Google Scholar]
- 27.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 28.Stokes H W, Ogorman D B, Recchia G D, Parsekhian M, Hall R M. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 29.Sundström L, Sköld O. The dhfrI trimethoprim resistance gene of Tn7 can be found at specific sites in other genetic surroundings. Antimicrob Agents Chemother. 1990;34:642–650. doi: 10.1128/aac.34.4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Threlfall E J, Frost J A, Ward L R, Rowe B. Epidemic in cattle and humans of Salmonella typhimurium DT104 with chromosomally integrated multiple drug resistance. Vet Rec. 1994;134:577. doi: 10.1136/vr.134.22.577. [DOI] [PubMed] [Google Scholar]
- 31.Wall P G, Morgan D, Lamden K, Griffin M, Threlfall E J, Ward L R, Rowe B. Transmission of multi-resistant strains of Salmonella typhimurium from cattle to man. Vet Rec. 1995;136:591–592. doi: 10.1136/vr.136.23.591. [DOI] [PubMed] [Google Scholar]
- 32.Wall P G, Morgan D, Lamden K, Griffin M, Threlfall E J, Ward L R, Rowe B. A case control study of infection with an epidemic strain of multiresistant Salmonella typhimurium DT104 in England and Wales. Commun Dis Rep. 1994;4:R130–R135. [PubMed] [Google Scholar]