ABSTRACT
Staphylococcus hominis is frequently isolated from human skin, and we hypothesize that it may protect the cutaneous barrier from opportunistic pathogens. We determined that S. hominis makes six unique autoinducing peptide (AIP) signals that inhibit the major virulence factor accessory gene regulator (agr) quorum sensing system of Staphylococcus aureus. We solved and confirmed the structures of three novel AIP signals in conditioned medium by mass spectrometry and then validated synthetic AIP activity against all S. aureus agr classes. Synthetic AIPs also inhibited the conserved agr system in a related species, Staphylococcus epidermidis. We determined the distribution of S. hominis agr types on healthy human skin and found S. hominis agr-I and agr-II were highly represented across subjects. Further, synthetic AIP-II was protective in vivo against S. aureus-associated dermonecrotic or epicutaneous injury. Together, these findings demonstrate that a ubiquitous colonizer of human skin has a fundamentally protective role against opportunistic damage.
KEYWORDS: Staphylococcus aureus, coagulase-negative staphylococci, quorum sensing, skin microbiota
INTRODUCTION
The skin is a hostile environment to many microorganisms due to its acidic pH, fatty acids, antimicrobial peptides, dryness, and constant exposure to UV radiation and other environmental challenges (1). Still, healthy skin is colonized by a diverse flora of bacteria, archaea, fungi, and viruses that thrive in this environment (1, 2). Coagulase-negative staphylococci (CoNS) are dominant bacterial skin colonizers and active participants in the cutaneous microenvironment (1, 3). CoNS inhabit distinct niches in and on skin appendages, i.e., hair follicles, glands, and epidermal and dermal tissues, and prevent opportunistic pathogens from infecting healthy skin by mechanisms collectively termed colonization resistance (1, 4–7). While the full depth and breadth of CoNS colonization resistance mechanisms remain to be determined, CoNS can drive epidermal barrier development (8, 9), educate or tune the cutaneous immune response (10), and produce a variety of antimicrobial molecules (11, 12) to promote barrier function and integrity.
The most frequently isolated and best characterized CoNS from human skin is Staphylococcus epidermidis (1, 13). There are now many examples of how S. epidermidis mediates skin colonization resistance, including cross talk with keratinocytes (14), synergy between S. epidermidis small molecules and host antimicrobial peptides (15–17), and active secretion of bactericidal molecules (18, 19). Given its relative abundance on the skin, it is unsurprising that S. epidermidis has historically been used for studies assumed to be representative of all CoNS. However, recent evidence suggests that S. epidermidis leads a far more dichotomous lifestyle as both symbiont and opportunistic pathogen (20, 21). S. epidermidis is the leading cause of medical implant-associated infections, which often develop into antibiotic-resistant biofilms requiring implant removal and replacement and placing a tremendous burden on the health care system (22–24). In addition, certain S. epidermidis strains, akin to pathogenic Staphylococcus aureus, can expand and exacerbate barrier degradation in atopic dermatitis (AD) and Netherton syndrome (NS) lesions (25–27). Taken together, S. epidermidis remains an important skin colonizer with protective benefits but also represents a significant challenge to homeostasis depending on the context of the interaction.
There is emerging evidence that other commensal CoNS may play a more protective role on skin than previously appreciated. Staphylococcus hominis is the second most frequently isolated CoNS from healthy human skin (3, 28). Unlike S. aureus or S. epidermidis, S. hominis does not expand in AD lesions, and skin colonization with S. hominis in infancy is correlated with a reduced likelihood of developing AD later in life (2, 29, 30). S. hominis may also play an active role in skin protection; several groups have described bactericidal molecules made by strains of S. hominis that selectively kill S. aureus (11, 12, 31, 32). Further, one lantibiotic-producing S. hominis strain even showed promise as an anti-S. aureus topical treatment for patients with AD in a phase 1 randomized clinical trial (12).
Aside from the antibacterial activity described above, we and others have begun to describe the importance of interspecies competition in maintaining skin integrity. There are now many examples of CoNS competing with S. aureus and protecting host skin from damage through the conserved two-component quorum sensing (QS) accessory gene regulator (agr) system (33–40). S. aureus is the most common etiological agent of skin and soft tissue infections in the United States, and productive S. aureus skin infection requires agr signaling (41–43). Thus, targeting agr to dampen S. aureus virulence factor production has been proposed as a potential alternative to antibiotic therapy (44, 45).
In all staphylococci, agr (agrBDCA) signals via its cognate autoinducing peptide (AIP) in a cell density-dependent manner (Fig. 1A) (46, 47). At sufficient external concentration, AIP binds to the membrane-bound histidine kinase receptor (AgrC) and induces receptor dimerization, followed by phosphorylation of the response regulator, AgrA. AgrA then binds between chromosomal promoters P2 and P3 to induce transcription of the agrBDCA operon and the major effector transcript, RNAIII. In S. aureus, the RNAIII molecule posttranscriptionally regulates the expression of a suite of virulence factors including toxins such as the canonical alpha-toxin, exoenzymes including several serine and cysteine proteases, and immune evasion factors (47). In S. epidermidis, the agr regulon is diminished in size because CoNS do not possess as many virulence factors as S. aureus (48, 49). However, the cysteine protease EcpA, which is necessary for S. epidermidis barrier degradation in AD and NS lesions, is under agr control (25, 26, 48). In S. hominis, the precise agr regulon and RNAIII effector molecule have not been determined, but most CoNS agr regulons are hypothesized to contain phenol-soluble modulins (PSMs) and a small number of proteases and lipases similar to those of S. epidermidis.
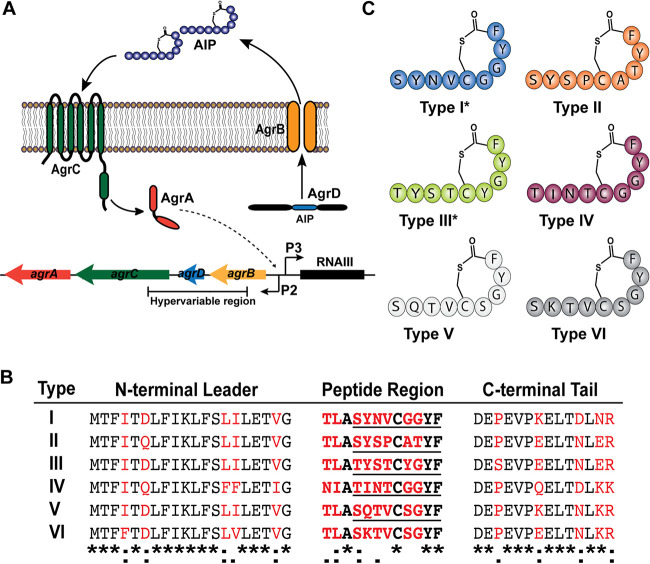
FIG 1.
S. hominis makes six AIP variants. (A) Schematic of the conserved staphylococcal accessory gene regulator quorum sensing system. (B) Predicted agrD sequences for each S. hominis agr type identified by PCR. Nonconserved residues are marked in red. The Clustal Omega sequence alignment between all strains is shown at the bottom, with asterisks representing fully conserved residues, colons representing residues with strongly similar properties, and periods representing residues with weakly similar properties. The confirmed peptide sequence of each AIP is underlined. (C) Representative images of predicted AIP structures for each agr type. Previously confirmed structures are marked with an asterisk.
There are four allelic variants of S. aureus and S. epidermidis agr determined by a hypervariable region spanning agrBDC (46, 47, 50). Every staphylococcal strain possesses a single agr type, which senses and responds to its cognate AIP signal. All described AIPs are 7 to 12 amino acids in length, with a conserved thiolactone or lactone ring constraining the last five residues in the C terminus (47). Only a limited number of agr types have been identified for most CoNS, and the functional or fitness advantages of agr heterogeneity remain unclear (47, 50). Intriguingly, some but not all CoNS AIPs inhibit S. aureus agr signaling via interspecies interference, including two recently described AIPs from S. hominis isolates (38, 51). One of these AIPs fully abrogated S. aureus skin damage in a mouse model of atopic injury (38), while the other showed inhibitory activity against S. aureus agr types I to III with in vitro reporters (51). Screening a collection of S. hominis isolates, we discovered more S. hominis agr allelic variation than previously reported.
Given the abundance and ubiquity of S. hominis on healthy skin and the potential of S. hominis to actively protect the host from infection, we sought to definitively characterize novel S. hominis AIP signals and the potential implications of S. hominis agr allelic variation for human skin health. Our data highlight the remarkable strain-level diversity of S. hominis skin isolates and suggest a significant role for S. hominis interspecies cross talk in preventing opportunistic skin infection and epidermal damage.
RESULTS
Identification of six S. hominis agr types.
Previous work identified at least three classes of S. hominis agr allelic variation by sequence (here referred to as AIP-II) (38) or confirmed AIP structure (here referred to as AIP-I [38] and AIP-III [51]). We screened our collection of S. hominis skin isolates (see Table S1 in the supplemental material) by PCR to determine if any other allelic variants might be present. We confirmed previously published agrD sequences for AH5009 (type I) (38), AH5006 (type II) (38), and AH4545 (type III) (51) and identified three novel AIP sequences (types IV to VI) (Fig. 1B). All classes of S. hominis AIPs contained a conserved alanine as the third residue of the peptide region, a conserved cysteine as the eighth residue of the peptide region, and a conserved tyrosine and phenylalanine as the eleventh and twelfth peptide region residues, respectively (Fig. 1B) (38, 51). Like other published sequences, the S. hominis AIP leader, peptide, and tail regions varied outside of the four aforementioned conserved residues in the peptide region (36, 38, 47, 48, 51). Based on previously published structures of S. hominis AIP-I and -III, we illustrated predicted structures of each S. hominis AIP (Fig. 1C).
List of strains and plasmids used in this study. Download Table S1, DOCX file, 0.02 MB (16.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
CM from S. hominis isolates inhibits MRSA agr signaling.
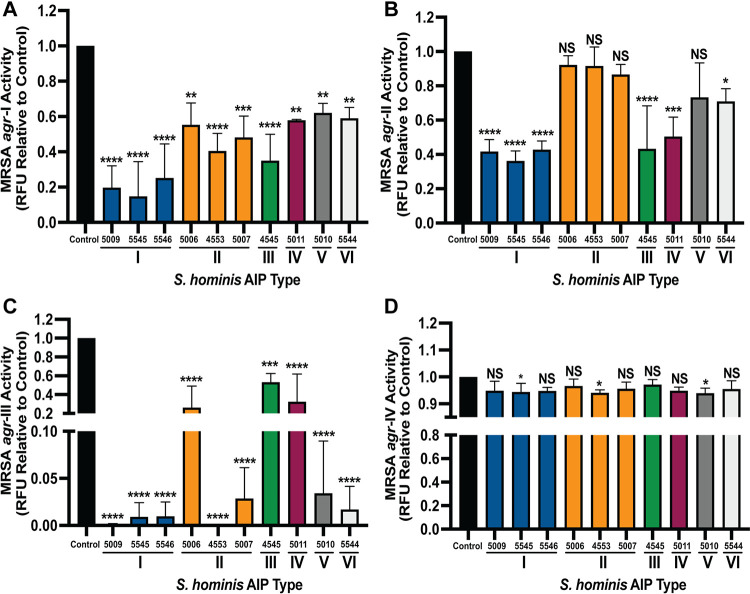
There is a rapidly growing body of work on CoNS AIPs that inhibit S. aureus agr signaling via interspecies cross talk (33–40). To determine if all S. hominis AIP types inhibit S. aureus agr signaling, we incubated methicillin-resistant S. aureus (MRSA) with the agr reporter plasmid P3::YFP (agr-I to -IV) (where YFP is yellow fluorescent protein) with 10% (vol/vol) cell-free conditioned medium (CM) from S. hominis isolates. We confirmed previously published findings that S. hominis AIP-I is a potent inhibitor of MRSA agr-I, -II, and -III with no effect on growth (Fig. 2A; see Fig. S1A in the supplemental material) (38). Each of the other S. hominis AIP classes also inhibited MRSA agr-I and reduced fluorescent signal to 40 to 60% of that of the reporter-only control. MRSA agr-I growth was not impacted in the presence of CM from any S. hominis strain (Fig. S1A). MRSA agr-II was inhibited by S. hominis AIP-I, -III, -IV, and -VI strains but not by any AIP-II-producing strain or by an AIP-V strain (Fig. 2B). No effect on reporter growth was observed (Fig. S1B). MRSA agr-III was highly susceptible to inhibition by all classes of S. hominis AIP, with the greatest inhibition by AIP-I, -V, and -VI (Fig. 2C). S. hominis AIP-II was also a strong inhibitor of MRSA agr-III, but the level of inhibition appeared to be strain dependent. It was previously reported that the level of AIP production can be strain dependent even within the same agr type (36), thus leading to differing levels of cross-inhibition. While CM from most S. hominis isolates did not impact the growth of MRSA agr-III, 10% CM from strain AH5011 (AIP-IV) attenuated reporter growth (Fig. S1C) during the first 8 h of incubation. Reporter growth in AH5011 CM ultimately recovered to the same optical density at 600 nm (OD600) as the reporter-only control by 24 h (Fig. S1C). For MRSA agr-IV signaling, only S. hominis strains producing AIP-I, -II, or -V inhibited fluorescent signal (Fig. 2D), and no significant effect on type IV reporter growth was observed with any type of S. hominis CM (Fig. S1D). Our results are consistent with previous literature which suggests that MRSA agr-IV is more resistant to interspecies cross talk than other MRSA agr types (33, 36).
FIG 2.
S. hominis conditioned medium (CM) inhibits MRSA agr signaling. MRSA agr-I P3::YFP (A), agr-II P3::YFP (B), agr-III P3::YFP (C), and agr-IV P3::YFP (D) reporter strains were incubated with 10% cell-free CM from S. hominis strains for 24 h. The 24-h fluorescent point (relative fluorescence units [RFU]) is shown relative to the reporter-only control. Data were pooled from three independent experiments. Significance was determined by ordinary one-way ANOVA with Dunnett’s multiple-comparison test (degree of freedom [df] = 32, F = 12.00 [A], 12.56 [B], 21.65 [C], 1.78 [D]). Mean values ± SD are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. NS, not significant.
MRSA growth in the presence of S. hominis conditioned medium. (A) Growth of MRSA agr-I P3::YFP over 24 h in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains. Growth is reported as the optical density at 600 nm. (B) Growth of MRSA agr-II P3::YFP over 24 h in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains. (C) Growth of MRSA agr-III P3::YFP over 24 h in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains. (D) Growth of MRSA agr-IV P3::YFP over 24 h in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains. Results are pooled from three independent experiments. Mean values ± SD are shown. Download FIG S1, DOCX file, 0.3 MB (273.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
LC-MS identification and validation of S. hominis AIPs.
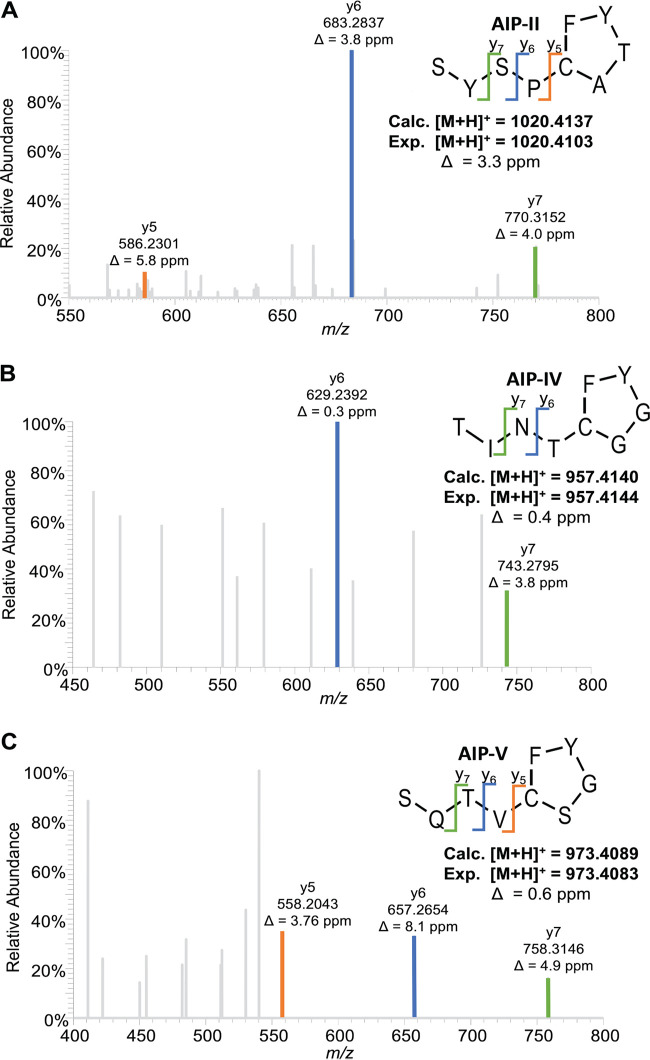
Since previous studies identified and validated the structures of S. hominis AIP-I (38) and AIP-III (51), we next performed mass spectrometric analysis of CM from representative isolates of S. hominis AIP-II, AIP-IV, and AIP-V to identify the structure of these novel AIPs. For AIP-II, a nine-residue AIP (SYSPc[CATYF]) was detected (Fig. 3A) with calculated and measured m/z values for the [M + H]+ ion of 1,020.4137 and 1,020.4103 (Δ = 3.3 ppm), respectively. For AIP-IV, a nine-residue AIP (TINTc[CGGYF]) was detected (Fig. 3B) with calculated and measured m/z values for the [M + H]+ ion of 957.4140 and 957.4144 (Δ = 0.4 ppm), respectively. For AIP-V, a nine-residue AIP (SQTVc[CSGYF]) was detected (Fig. 3C) with calculated and measured m/z values for the [M + H]+ ion of 973.4089 and 973.4083 (Δ = 0.6 ppm), respectively. We further validated the AIP-II, -IV, and -V structures by analyzing synthetic AIP. Liquid chromatography-mass spectrometry (LC-MS) analysis of each synthetic AIP demonstrated matching retention time, accurate mass, and fragmentation patterns between each native structure identified in the CM and the corresponding synthetic standard (Fig. S2).
FIG 3.
Identification and validation of three novel S. hominis AIPs. The amino acid sequence and thiolactone structure of S. hominis AIP-II (A), AIP-IV (B), and AIP-V (C) were solved and confirmed using MS-MS analysis. Characteristic y ions are listed for each AIP. The calculated mass of the protonated peptide (Calc.) is shown beneath each structure in comparison to the experimental mass (Exp.) and mass error.
Synthetic AIP validation. (A) Selected-ion chromatograms (SIC) for the calculated AIP-II mass (m/z = 1,020.4137) from strain AH4553 for both the spent medium (top) and the synthetic AIP in TSB (bottom). (B) MS-MS spectra for identified AIP-II ion in both the spent medium (top) and the synthetic AIP in TSB (bottom). (C) SIC for the calculated AIP-IV mass (m/z = 957.4137) from strain AH5011 for both the spent medium (top) and the synthetic AIP in TSB (bottom). (D) MS-MS spectra for identified AIP-IV ion in both the spent medium (top) and the synthetic AIP in TSB (bottom). (E) SIC for the calculated AIP-V mass (m/z = 973.4089) from strain AH5010 for both the spent medium (top) and the synthetic AIP in TSB (bottom). (F) MS-MS spectra for identified AIP-V ion in both the spent medium (top) and the synthetic AIP in TSB (bottom). Download FIG S2, DOCX file, 0.7 MB (774.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
An AIP was not detected in CM from the S. hominis type VI strain, and we were unable to identify any other type VI strains in our collection or in the National Center for Biotechnology Information (NCBI) database. The predicted AIP-VI structure was a single amino acid change from AIP-V (Q5K), and given its rarity in our collection and in published genomes, we excluded it from further analyses. Together, all five identified S. hominis AIPs were nine amino acids in length with a conserved thiolactone ring between the fifth-residue cysteine and ninth-residue phenylalanine. The AIPs diverged at the second and fourth tail residues, and AIP-II was the only S. hominis AIP with a threonine as the seventh residue rather than a glycine.
Synthetic S. hominis AIPs inhibit MRSA QS.
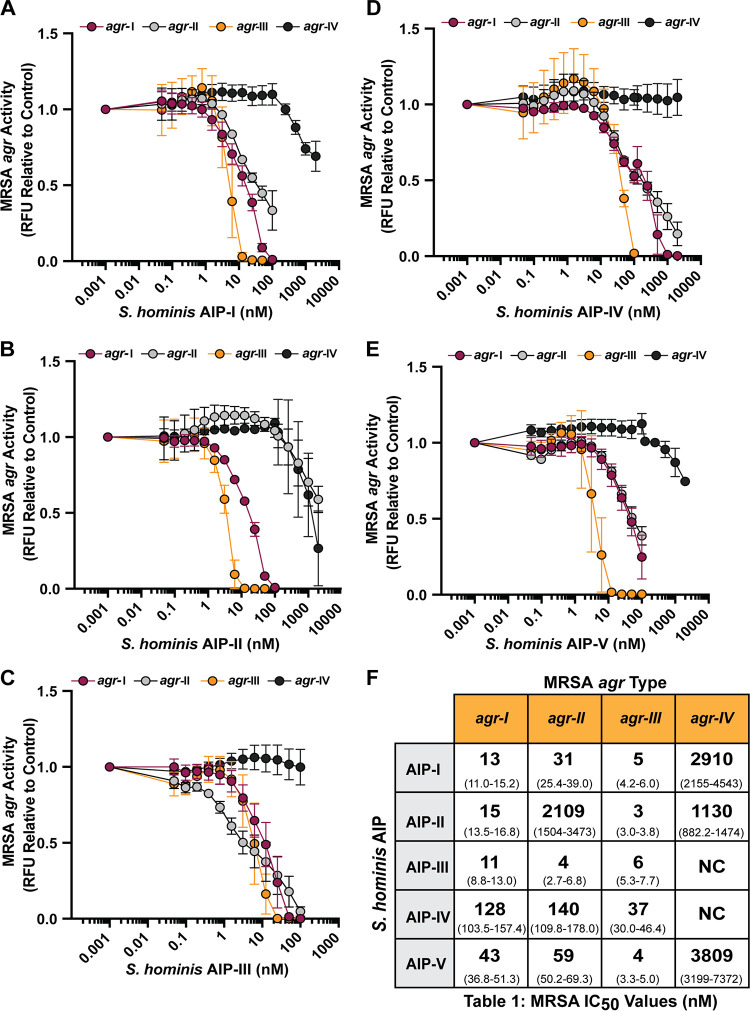
To profile the specific activity of each S. hominis AIP, MRSA agr P3::YFP reporters type I to IV were treated with a dose response of synthetic AIP-I (Fig. 4A), AIP-II (Fig. 4B), AIP-III (Fig. 4C), AIP-IV (Fig. 4D), or AIP-V (Fig. 4E). Previous studies demonstrated that many CoNS AIPs inhibit MRSA AgrC signaling with affinity constants in the low nanomolar range (36, 38, 47). Consistent with previous characterization and our initial findings with CM, S. hominis synthetic AIP-I potently inhibited MRSA agr-I (50% inhibitory concentration [IC50], 13 nM), agr-II (IC50, 31 nM), and agr-III (IC50, 5 nM) but was a poor inhibitor of agr-IV (IC50, 2,910 nM) (Fig. 4F) (38). Also consistent with our CM findings, synthetic S. hominis AIP-II was a poor inhibitor of MRSA agr-II (IC50, 2,109 nM) (Fig. 4F). MRSA agr-III was highly susceptible to all classes of synthetic S. hominis AIPs, with IC50 values in the low nanomolar range (Fig. 4F). Finally, MRSA agr-IV was poorly inhibited by synthetic AIP-I, -II, and -V and was not inhibited by AIP-III or -IV (Fig. 4F).
FIG 4.
Synthetic S. hominis AIPs inhibit MRSA agr signaling. (A to E) MRSA P3::YFP reporters were incubated with increasing doses of S. hominis synthetic AIP-I (A), AIP-II (B), AIP-III (C), AIP-IV (D), or AIP-V (E). Fluorescent intensity relative to the reporter only at the 24-h time point is shown. (F) IC50 values (nM) were calculated for each AIP-reporter pair from a four-parameter nonlinear regression curve. The 95% confidence interval for each IC50 value is reported in parentheses. Results were pooled from three independent experiments. Mean ± SD values are shown. NC, not calculated.
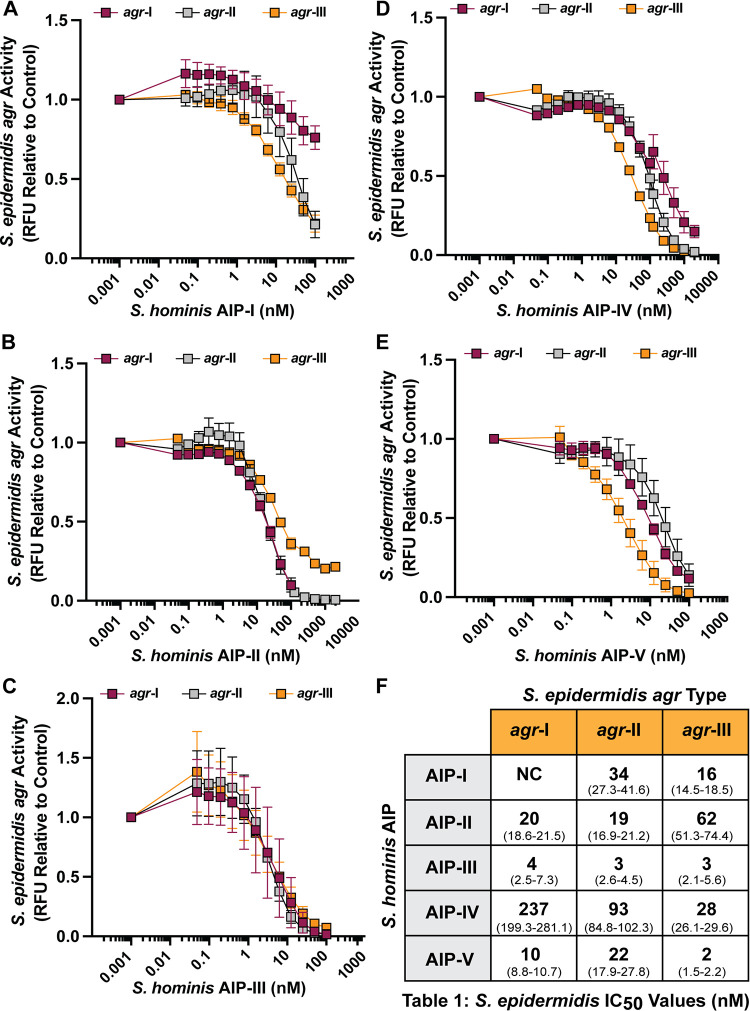
Synthetic S. hominis AIPs inhibit S. epidermidis QS.
S. epidermidis is an abundant CoNS on healthy human skin but is also an opportunistic or “accidental” pathogen that can worsen disease outcomes for patients with AD or NS through production of the agr-regulated cysteine protease EcpA (20, 25, 26). To determine if S. hominis AIPs could block S. epidermidis agr signaling, we incubated S. epidermidis agr P3::sGFP reporter types I to III (where sGFP is superfolder green fluorescent protein), which are the most common S. epidermidis agr types (50), with a dose response of each synthetic S. hominis AIP (Fig. 5A to E). Consistent with previous findings, S. hominis synthetic AIP-I was a poor inhibitor of S. epidermidis agr-I and even boosted the fluorescent signal in the first 8 h of incubation (Fig. S3) (25). However, AIP-I strongly inhibited S. epidermidis agr-II and -III signaling with low nanomolar potency (Fig. 5F). S. hominis synthetic AIP-II, -III, and -V were broadly inhibitory across all classes of S. epidermidis agr signaling (Fig. 5F), while AIP-IV was a relatively poor inhibitor of S. epidermidis agr-I (IC50, 237 nM) and agr-II (IC50, 93 nM) but more effective against agr-III (IC50, 28 nM) (Fig. 5F). CM (10%, vol/vol) from select S. hominis strains was also broadly inhibitory against all S. epidermidis agr types with no effect on growth (Fig. S4).
FIG 5.
S. hominis synthetic AIPs inhibit S. epidermidis agr signaling. (A to E) S. epidermidis P3::sGFP reporters were incubated with increasing doses of S. hominis synthetic AIP-I (A), AIP-II (B), AIP-III (C), AIP-IV (D), or AIP-V (E). Fluorescent intensity relative to the reporter only at the 24-h time point is shown. (F) IC50 values (nM) were calculated for each AIP-reporter pair from a four-parameter nonlinear regression curve. The 95% confidence interval for each IC50 value is reported in parentheses. Results are pooled from three independent experiments. Mean ± SD values are shown. NC, not calculated.
S. epidermidis agr-I early fluorescence is boosted by S. hominis AIP-I. Relative fluorescence (RFU) is shown compared to reporter-only control for S. epidermidis agr-I P3::sGFP incubated with increasing doses of S. hominis AIP-I over 24 h. Results were pooled from three independent experiments. Mean values ± SD are shown. Download FIG S3, DOCX file, 0.1 MB (94.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
S. hominis conditioned medium inhibits S. epidermidis agr quorum sensing. (A) S. epidermidis agr-I P3::sGFP incubated with 10% cell-free conditioned medium from S. hominis strains for 24 h. The 24-h fluorescent point is shown relative to reporter only. (B) S. epidermidis agr-I P3::sGFP growth reported as the OD600 in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains for 24 h. (C) S. epidermidis agr-II P3::sGFP 24-h fluorescent point relative to reporter only. (D) S. epidermidis agr-II P3I::sGFP growth (OD600) in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains for 24 h. (E) S. epidermidis agr-III P3::sGFP 24-h fluorescent point relative to reporter only. (F) S. epidermidis agr-III P3::sGFP growth (OD600) in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains for 24 h. Significance was determined by ordinary one-way ANOVA with Dunnett’s multiple-comparison test. Results were pooled from three independent experiments. Mean values ± SD are shown. *, P < 0.05; ****, P < 0.0001. Download FIG S4, DOCX file, 0.2 MB (236.9KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
S. hominis agr type distribution in a healthy skin cohort.
Healthy human skin is often dominated by a large proportion of S. epidermidis agr-I strains, mixed with smaller subpopulations of agr-II and -III strains (50). Given that CoNS agr heterogeneity could be important for mediating both inter- and intraspecies cross talk (20, 38, 50), we first determined the relative frequency of five representative S. hominis agrD sequences in all published S. hominis genomes in the NCBI database. S. hominis agr-I and -II were the most frequently deposited genomes (31 and 43 hits, respectively), agr-III and -IV were found less frequently but with similar numbers of hits (14 and 16, respectively), and agr-V was rarely found (4 hits) (Fig. 6A).
FIG 6.
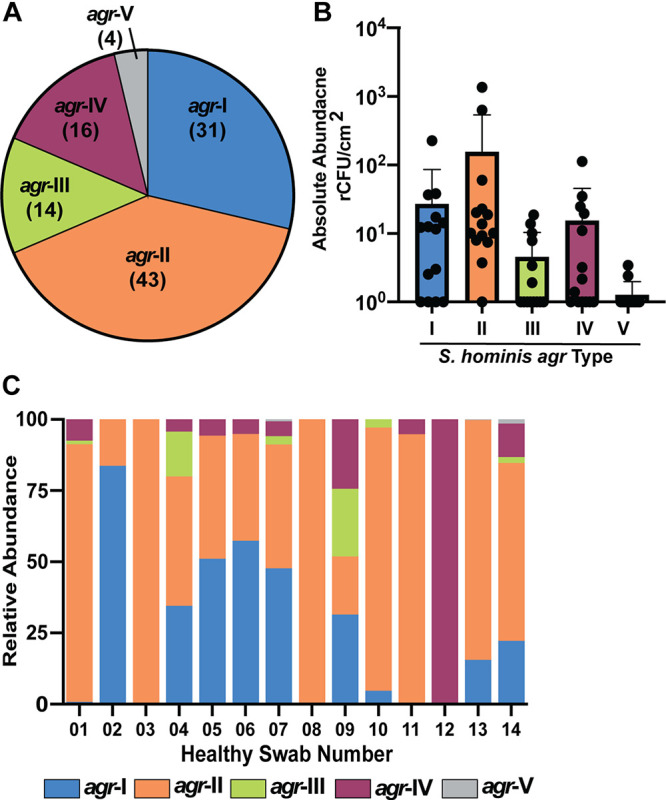
S. hominis agr-I and agr-II are the most abundant agr types on healthy human skin. (A) Relative frequency of S. hominis agr types identified in all published S. hominis genomes in the NCBI database. (B) Absolute abundance by quantitative PCR of S. hominis agr types in a 5-cm2 swab of the antecubital crease pooled from 14 healthy skin swabs. (C) Relative abundance of each S. hominis agr type stratified by patient swab.
Next, we analyzed S. hominis agr types by quantitative PCR from DNA extracted from skin swabs from the antecubital crease of 14 healthy skin donors (25). This analysis also found that S. hominis agr-I and -II were the most dominant agr types by absolute abundance (Fig. 6B; Fig. S5). S. hominis agr-III and -IV were less abundant, while agr-V was undetectable in most swab samples (Fig. 6B). When we stratified total S. hominis agr type abundance by individual donor, we found that many samples were unique in their relative distribution of S. hominis agr types (Fig. 6C). For example, some swabs were dominated by a single S. hominis agr type (i.e., HC01, -02, -03, -08, -11, and -12) while other samples contained a more diverse mix of agr types (i.e., HC04, -05, -06, -07, and -09) (Fig. 6C). Generally, most samples were dominated by relatively more S. hominis agr-I or S. hominis agr-II, with few samples containing equivalent distributions of these two agr types.
Validation of S. hominis agr type distribution in healthy skin swabs. (A) Table of qPCR primers used in this study. (B) qPCR primer specificity validation for each S. hominis agr type. S. epidermidis (SE) genomic DNA (gDNA) standards were also included to determine any off-target amplification. Download FIG S5, DOCX file, 0.2 MB (209.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
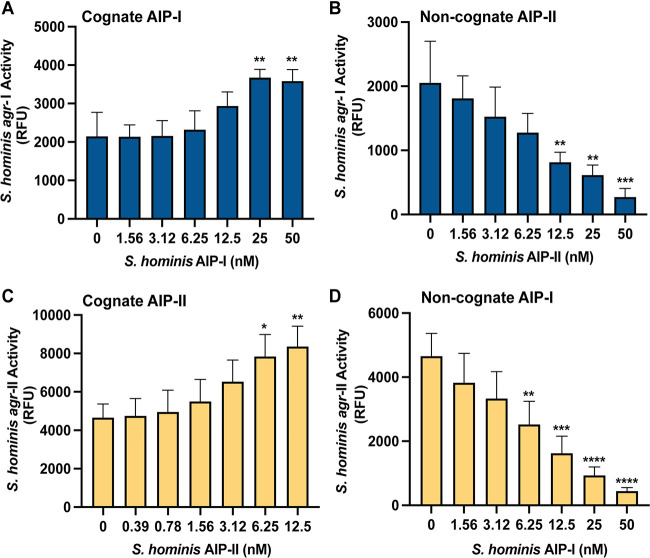
S. hominis agr-I and agr-II intraspecies competition.
Because S. hominis agr-I and agr-II were the most abundant agr classes in the NCBI database and on healthy skin, we sought to determine if they might compete with each other via intraspecies cross talk. Previous reports demonstrated that heterogeneous S. epidermidis agr groups can mediate intraspecies interference, potentially giving one agr type a competitive advantage over another (48, 50). We transformed representative S. hominis agr-I and -II strains with the previously characterized staphylococcal agr P3::sGFP reporter plasmid pCM41 (36) and treated each with a dose of cognate and noncognate synthetic AIP. A concentration of 25 nM or greater of synthetic AIP-I significantly boosted S. hominis agr-I fluorescent signal (Fig. 7A) with no effect on growth (Fig. S6A). Synthetic AIP-II inhibited agr-I signaling in a dose-dependent manner with low nanomolar IC50 (IC50, 9 nM; 95% confidence interval [CI] = 5.1 to 16.8) (Fig. 7B) and no effect on reporter growth (Fig. S6B). For S. hominis agr-II, 12.5 nM synthetic AIP-II was optimal to boost fluorescent signal with no effect on growth (Fig. 7C; Fig. S6C). Synthetic AIP-I similarly inhibited agr-II fluorescent signaling in a dose-dependent manner with a low nanomolar IC50 (IC50, 7 nM; 95% CI = 5 to 11) (Fig. 7D) with no impact on reporter growth (Fig. S6D).
FIG 7.
S. hominis agr-I and agr-II intraspecies cross talk. (A) Relative fluorescent values of S. hominis agr-I P3::YFP incubated with increasing doses of synthetic AIP-I at 8 h of incubation. (B) Relative fluorescent values of S. hominis agr-I P3::YFP incubated with increasing doses of synthetic AIP-II at 8 h of incubation. (C) Relative fluorescent values of S. hominis agr-II P3::YFP incubated with increasing doses of synthetic AIP-II at 8 h of incubation. (D) Relative fluorescent values of S. hominis agr-II P3::YFP incubated with increasing doses of synthetic AIP-I at 8 h of incubation. Results were pooled from three independent experiments. Significance was determined by ordinary one-way ANOVA with Dunnett’s multiple-comparison test (df = 20, F = 8.53 [A], 9.75 [B], 6.40 [C], 17.23 [D]). Mean values ± SD are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
S. hominis agr-I and agr-II intraspecies cross-talk with synthetic AIPs. (A) S. hominis agr-I growth (measured by OD600) with increasing concentrations of AIP-I. (B) S. hominis agr-I growth with increasing concentrations of AIP-II. (C) S. hominis agr-II growth with increasing concentrations of AIP-II. (D) S. hominis agr-II growth (OD600) with increasing concentrations of AIP-I. (E) S. hominis agr-I growth with increasing doses of AIP-III. (F) S. hominis agr-I fluorescence with increasing doses of AIP-III. (G) S. hominis agr-I growth with increasing doses of AIP-IV. (H) S. hominis agr-I fluorescence with increasing doses of AIP-IV. (I) S. hominis agr-I growth with increasing doses of AIP-V. (J) S. hominis agr-I fluorescence with increasing doses of AIP-V. (K) S. hominis agr-II growth with increasing doses of AIP-III. (L) S. hominis agr-II fluorescence with increasing doses of AIP-III. (M) S. hominis agr-II growth with increasing doses of AIP-IV. (N) S. hominis agr-II fluorescence with increasing doses of AIP-IV. (O) S. hominis agr-II growth with increasing doses of AIP-V. (P) S. hominis agr-II fluorescence with increasing doses of AIP-V. Growth curves are representative of three independent experiments. For fluorescence curves, results were pooled from three independent experiments. Mean values ± SD are shown. Download FIG S6, DOCX file, 0.6 MB (615.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We also investigated other relationships between S. hominis AIPs and agr-I or agr-II signaling. Synthetic AIP-III, -IV, or -V did not impact the growth of the S. hominis agr-I or -II reporters at any tested concentration (Fig. S6). Both agr-I and -II early fluorescent signals were boosted in a dose-dependent manner by the addition of synthetic AIP-III (Fig. S6F and S6L). We found weaker interactions with AIP-IV or -V. AIP-IV slightly boosted early agr-I signaling but had negligible effects on agr-II, while AIP-V had weak to negligible effects on agr-I and modestly inhibited agr-II (Fig. S6G to J and M to P) The strongest intraspecies interactions were observed between agr-I and agr-II (Fig. 7).
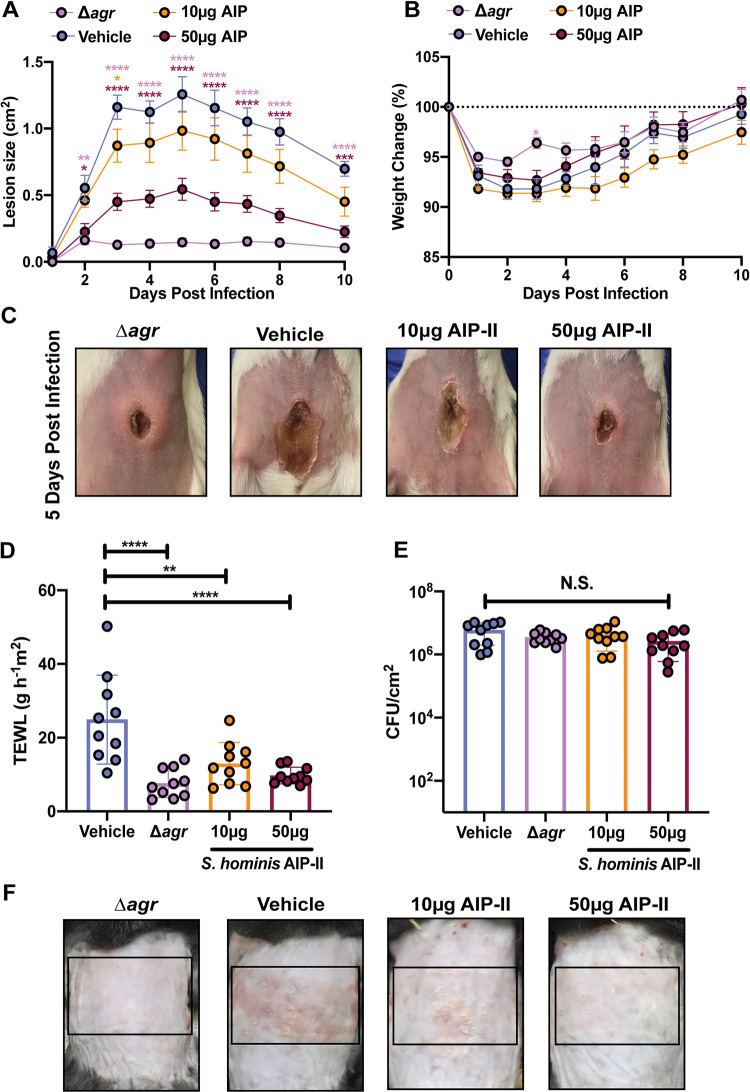
S. hominis AIP-II protects murine skin from MRSA QS-mediated injury.
S. aureus is the most common cause of skin and soft tissue infections in the United States and can also colonize and exacerbate disease in patients with AD and NS (26, 29, 52). Many virulence factors associated with S. aureus skin infection are under the agr regulon (42, 43, 47). It was previously reported that S. hominis AIP-I effectively mitigates MRSA-associated AD symptoms in a mouse model (38). However, since we also found that S. hominis agr-II was a dominant healthy human skin colonizer in most of our swab donors, we hypothesized that AIP-II may also be protective against S. aureus skin infections. Using an established model of USA300 MRSA dermonecrosis to assess synthetic AIP-II efficacy in an acute infection, we found that mice treated with synthetic S. hominis AIP-II developed significantly smaller lesions than did mice treated with vehicle treatment alone, and lesion size was dose dependent (Fig. 8A). Furthermore, mice administered 50 μg of S. hominis synthetic AIP-II tended to lose less weight than their vehicle counterparts, although this trend was not statistically significant (Fig. 8B). Lesion severity in mice administered a 50-μg dose of synthetic AIP-II closely resembled that of mice administered an agr mutant after 5 days of infection, suggesting potent inhibition of MRSA agr signaling in this model (Fig. 8C).
FIG 8.
S. hominis AIP-II protects murine skin from MRSA damage. (A) Dermonecrotic lesion size for mice administered a 10- or 50-μg dose of synthetic S. hominis AIP-II compared to that of mice administered wild type (WT) plus vehicle (DMSO) or Δagr mutant plus vehicle control. (B) Weight change for indicated groups. (C) Representative images of dermonecrotic lesion size 5 days postinfection. Data were pooled from two independent experiments (n = 10 per group). Mean ± SEM is shown. Significance was determined by two-way ANOVA with Dunnett’s multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Transepithelial water loss 72 h postinfection for mice administered a 10- or 50-μg dose of synthetic S. hominis AIP-II compared to that of mice administered WT plus vehicle (DMSO) or Δagr mutant plus vehicle control. (E) Total CFU burden in epicutaneous lesions at 72 h postinfection (hpi) for the indicated groups. (F) Representative images of epicutaneous damage at 72 hpi. Data were pooled from two independent experiments (n = 10 per group). Mean ± SD is shown. Significance was determined by ordinary one-way ANOVA with Dunnett’s multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. N.S., not significant.
We also investigated the ability of S. hominis AIP-II to protect the skin barrier from MRSA degradation with an established model of epicutaneous infection (akin to AD in humans) (38, 53). After 72 h of topical association with MRSA, mice that received 10 or 50 μg of synthetic S. hominis AIP-II retained more of their barrier function by transepithelial water loss (TEWL) measurement, similar to mice receiving agr-null MRSA, than those receiving the vehicle control (Fig. 8D). This was not due to MRSA loss or killing, as no differences in CFU between groups was observed (Fig. 8E). Gross morphology of the skin also revealed less redness, scaling, and erythema in the AIP-II-treated mice than in the vehicle control mice (Fig. 8F). We also found that coapplication of an S. hominis AIP-II-producing strain (AH4553) with MRSA resulted in a strong trend toward skin barrier protection as assessed by TEWL at 72 h postinfection (Fig. S7A). Mouse skin that received equivalent numbers of CFU of S. hominis and MRSA had significantly less redness and scaling than mouse skin that received MRSA application alone (Fig. S7B). S. hominis cochallenge was not as protective as administration of synthetic AIP-II, likely due to the amount of AIP-II produced on skin by this strain or the tropism of S. hominis for human rather than mouse skin. However, our results are consistent with previously published observations of the protective role of an S. hominis AIP-I-producing strain (38).
S. hominis in competition with MRSA protects skin from epicutaneous damage. (A) Transepithelial water loss 72 h postinfection for mice challenged with MRSA only, S. hominis only, or equivalent numbers of CFU of MRSA and S. hominis, or uninfected (UI; saline only). (B) Representative images of mouse back skin at 72 h postinfection with the indicated groups. Results were pooled from two independent experiments (n = 10 for all groups except UI [n = 9]). Mean ± SD is shown. Significance was determined by ordinary one-way ANOVA with the Holm-Sidak multiple-comparison test. Download FIG S7, DOCX file, 0.4 MB (426.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Transcriptional profiling of S. hominis AgrA-regulated genes.
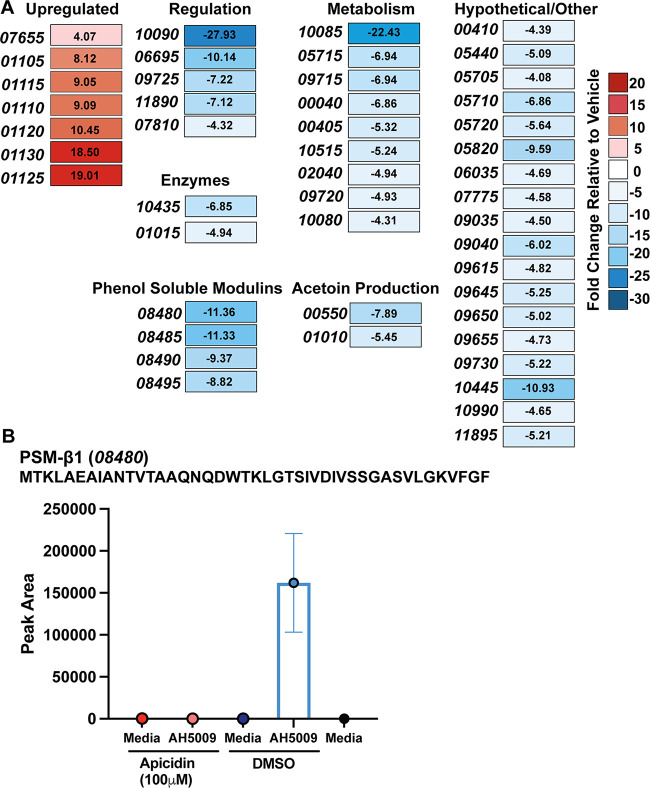
We profiled genes under transcriptional control of AgrA in our representative S. hominis agr-I strain (AH5009) to better understand fundamental molecular regulation in an understudied CoNS and to identify genes that could facilitate S. hominis skin colonization. CoNS are difficult to genetically manipulate, so we took an alternative approach to predict genes under agr control. Apicidin is a specific inhibitor of the agr response regulator AgrA, which has previously been characterized for its activity against MRSA (54). Using the S. hominis agr-I P3::sGFP reporter strain, we confirmed that apicidin inhibited S. hominis agr-I signaling with no effect on growth (Fig. S8A). To evaluate genes that could be under the transcriptional control of AgrA, we conducted transcriptome sequencing (RNA-seq) analysis on apicidin-treated S. hominis agr-I compared to a vehicle (dimethyl sulfoxide [DMSO])-treated control. Using a 4-fold cutoff for differential gene expression (false discovery rate P value, ≤0.05), we found that 40 genes were downregulated in the apicidin-treated samples compared to the control and that 7 genes were significantly upregulated (Fig. 9A; Table S2).
FIG 9.
Genes under transcriptional control of S. hominis AgrA. (A) RNA-seq heat map of genes with greater than a 4-fold change in expression and a false discovery rate P value of less than 0.05 in 100 μM apicidin-treated S. hominis compared to wild-type vehicle (DMSO)-treated cells. Fold changes are indicated in each box. Numbers in italics refer to the last five digits of the locus tag. (B) Mass spectrometric validation of the absence of PSM-β1 production in S. hominis treated with 100 μM apicidin. Results are averages from triplicate injections of biological triplicates (9 independent injections). The PSM-β1 predicted sequence and locus tag number are shown above the graph. The ion detected was at m/z 908.6785, which corresponds to the predicted mass (908.6819 Δm = 3.7 ppm) of PSM-β1with five protons and a +5 charge ([M + 5H]5+).
Apicidin inhibition of S. hominis agr-I signaling is AgrA dependent. (A) S. hominis agr-I P3::sGFP growth (OD600) and fluorescence (relative fluorescent units) in the presence of 100 μM apicidin or equivalent DMSO control over 24 h. Results were pooled from three independent experiments, and mean values ± SD are shown. (B) LC-MS selected ion chromatogram for the m/z range of 908.6774 to 908.6864 (chosen from the calculated m/z of the PSM-β1 peptide with five protons; [M + 5H]5+, 908.6819) for S. hominis AH5009 (C5). The data show PSM-β1 presence in vehicle-treated culture (DMSO) (top) and PSM-β1 absence in 100 μM apicidin-treated S. hominis AH5009 (bottom). Results are representative chromatograms from among those collected for biological replicates. (Average peak area for replicates is shown in Fig. 9 of the main text.) Download FIG S8, DOCX file, 0.4 MB (459.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
RNA-seq analysis of differentially expressed transcripts in apicidin-treated S. hominis agr-I. Download Table S2, DOCX file, 0.02 MB (16.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
As expected, several genes, including four beta class phenol-soluble modulins (PSMs) (MOV58_08480, down 11.36-fold; MOV58_08485, down 11.33-fold; MOV58_08490, down 9.37-fold; and MOV58_08495, down 8.82-fold) and a predicted alpha-beta hydrolase (putative lipase; MOV58_10435, down 6.85-fold) were significantly downregulated (Fig. 9A). PSMs are under direct transcriptional control of AgrA (47, 54); therefore, we functionally validated our RNA-seq findings by assessing the presence or absence of S. hominis PSMs in apicidin (100 μM)- or DMSO-treated cultures. Of the four predicted beta-type PSMs in S. hominis AH5009, we found that only PSM-β1 was consistently detected by mass spectrometry. The mass spectrometric analyses confirmed that this PSM was present only in S. hominis cultures treated with DMSO, not in those treated with apicidin (Fig. 9B; Fig. S8B).
Outside of our 4-fold cutoff but in support of agr inhibition, we found RNAIII (down 3.40-fold), agrD (down 3.08-fold), and agrB (down 2.89-fold) downregulated. We also found two genes involved in acetoin production (MOV58_00550, down 7.89-fold; MOV58_01010, down 5.45-fold) significantly downregulated as well as multiple transcriptional regulators (MOV89_10090, down 27.93-fold; MOV58_09725, down 7.22-fold; and MOV58_07810, down 4.32-fold) and a choloylglycine hydrolase family protein putatively involved in bile salt hydrolysis and/or penicillin hydrolysis (55) (MOV58_01015, down 4.94-fold) (Fig. 9A). Unlike the S. aureus or S. epidermidis agr regulon, we did not find any proteases significantly downregulated. Additionally, homologs of the S. epidermidis cysteine protease gene ecpA, the extracellular elastase gene sepA, or the serine protease gene esp were not identified by a BLAST (Basic Local Alignment Search Tool) (56) search of the S. hominis C5 (AH5009) genome.
DISCUSSION
S. hominis is the second most frequently isolated colonizer of human skin and is often found on moist, sebaceous, or foot sites, with a particular affinity for colonizing axillae and pubic regions (28, 57, 58). Here, we have identified that S. hominis has the most agr heterogeneity (6 unique types) of any CoNS species. We utilized LC-MS to confirm three novel AIP structures (AIP-II, -IV, and V). Together with previously published structures of AIP-I (38) and -III (51), all five identified AIPs are the same size (9 amino acids) but vary significantly in amino acid composition. We profiled CM and synthetic AIP activity against MRSA agr quorum sensing and found that S. hominis AIPs are variable but often potent agr inhibitors. Going further, we also found that S. hominis synthetic AIPs mediate distinct and often inhibitory patterns of interspecies cross talk with the three most common S. epidermidis agr types (50). We determined a potential role for S. hominis agr variability in protecting host skin from opportunistic staphylococcal infections, as we showed that S. hominis AIP-II was a potent MRSA quorum quencher in murine models of acute damage and topical degradation. We addressed S. hominis intraspecies competition for the first time and found that the two most abundant S. hominis agr classes (agr-I and agr-II) in our healthy human skin swab samples were also the most potent agr cross-inhibitors in vitro. Finally, we took a novel approach to predict genes regulated by S. hominis AgrA and found several gene candidates that could potentiate S. hominis skin colonization.
Our results bridge several recent studies which suggest that S. hominis is a protective human skin commensal with the capacity to make AIPs that inhibit noncognate quorum sensing systems (32, 38, 51). Here, we identified 6 S. hominis agr types, though potentially more may be revealed with greater sequencing depths in metagenomic studies or with typing of more skin isolates (Fig. 1). We validated that S. hominis AIP-I (38) and -III (51) inhibit MRSA agr and that S. hominis AIP-I is a poor inhibitor of S. epidermidis agr-I signaling (25). Intriguingly, we found that S. hominis AIP-II does not inhibit MRSA agr-II in either CM or synthetic AIP assays. This lack of activity was surprising considering that AIP-II was such a strong inhibitor of other MRSA agr systems but could be due to the unusual insertion of a threonine (rather than a glycine) in the seventh residue of the AIP (Fig. 1). In a previously published alignment of 32 CoNS agrD sequences, only Staphylococcus haemolyticus and Staphylococcus caprae agr-II had a threonine in the seventh AIP residue (47). Potentially, this threonine could hinder AIP binding in the MRSA type-II AgrC pocket, but further investigations of structure-function relationships between CoNS AIPs and MRSA AgrC receptors are warranted.
We also found that at least one S. hominis strain (AH5011) makes a molecule that is specifically bacteriostatic or bactericidal against the MRSA agr type-III reporter strain MW2, which is a community-acquired USA400 clone (see Fig. S1 in the supplemental material). USA400 (agr-III) strains are most often associated with toxic shock syndrome but have also been associated with necrotizing pneumonia and infective endocarditis (59). It is not unusual for CoNS to make bacteriocin or lantibiotic-type molecules with high specificity for bacterial species or strains (18). Such specificity is one reason why there is continued interest in microbiome-inspired therapeutic discovery, including the discovery and development of S. hominis-derived anti-S. aureus therapeutics targeted toward patients with AD or NS (11, 12). Our observation that S. hominis makes a molecule that is a potentially specific anti-USA400 agent suggests we may one day be able to achieve highly individualized, lineage-specific anti-MRSA treatments. In the emerging postantibiotic age, S. hominis- and CoNS-derived therapeutics represent an exciting and necessary new avenue for antimicrobial development (60).
Another unusual observation from our study was the slight boost in early S. epidermidis agr signaling with the addition of S. hominis synthetic AIP-I (Fig. S3). Most quorum sensing cross talk has been explored in the context of inhibition rather than activation (33, 36, 38, 40). Given the dissimilarities between S. hominis AIP-I (SYNVc[CATYF]) and S. epidermidis AIP-I (DSVc[CASYF]) (48), we did not expect early S. epidermidis signal boosting. Intriguingly, S. hominis AIP-III was previously shown to boost MRSA agr-IV signaling, as assessed by a β-lactamase activity assay (51). This was the first identification of interspecies activation and, in conjunction with our results, indicates that this may be a more common type of interaction than previously appreciated. Further investigations of the potentially promiscuous nature of CoNS AIPs are necessary and could significantly improve our understanding of global quorum sensing dynamics during homeostatic skin colonization versus infection. Moreover, it remains difficult to determine if CoNS AIPs are made on healthy human skin or in what quantity. Future studies may reveal whether AIPs can be detected on skin and if some AIP types are more prevalent than others.
In addition to understanding interspecies cross talk mediated by S. hominis, we also sought to investigate how the remarkably high level of S. hominis agr variability might impact skin colonization or intraspecies cross talk dynamics. To our knowledge, there have been no other reports of CoNS with as many as six agr allelic variants. There are four agr allelic variants of the most common skin CoNS, S. epidermidis (48, 50). Species lower in abundance like Staphylococcus warneri (51) and S. haemolyticus (51), each have one confirmed agr type. Another low-abundance commensal, Staphylococcus simulans, was recently shown to have three agr allelic variants, although only one of these (agr-I) was associated primarily with human skin isolates (36). When we analyzed the S. hominis agr type distribution in published genomes on NCBI, we found that S. hominis agr-I and -II were most highly represented (Fig. 6). Although our sample size was quite modest, given that there are only 160 genome annotation and assembly reports for S. hominis compared to 13,551 for S. aureus or 1,106 for S. epidermidis, we further validated our NCBI results by determining S. hominis agr type distribution on the antecubital crease in a cohort of 14 healthy volunteers (Fig. 6). Our human skin swab data combined with our observations of significant intraspecies cross talk between S. hominis agr-I and agr-II lend support to the current hypothesis that agr variability may be important for niche competition or kin selection (40). Previous work showed that certain MRSA or S. epidermidis agr types are more prevalent in some disease states than others. For example, S. aureus agr-IV is most often associated with scalded skin syndrome (61), while S. aureus agr-I is a predominant isolate in community and hospital-associated infections in the United States (62, 63). S. epidermidis agr-I is the most dominant skin colonizer (50) but also the most common S. epidermidis isolate in AD lesions (38). Potentially, S. hominis agr-I and -II actively compete on skin for certain niches or resources, although more targeted analysis of S. hominis agr type distribution across different body sites is necessary to better understand these interactions.
Our study was limited by the small number of volunteers (n = 14) and singular time point. Nevertheless, it is the first assessment of S. hominis agr type distribution on healthy skin and supports future investigation of this underappreciated skin commensal in colonization resistance. Future studies could expand the number of volunteers and time points, similar to a recent metagenomics investigation of S. epidermidis strain-level variation on healthy skin (50). More time points could also reveal if or how S. hominis populations change on skin over time or if perturbations to the skin microbiome may alter S. hominis population dynamics. Finally, given the relative difficulty of genetic manipulation in S. hominis, we did not characterize every interaction between every S. hominis agr type, and additional work is needed to better understand why certain S. hominis AIPs (AIP-I and -II) are intraspecies inhibitors while others including AIP-III act as intraspecies activators.
We and others have shown that agr signaling is necessary for S. epidermidis colonization of porcine skin (48) and for MRSA epicutaneous colonization and induction of inflammation in a mouse infection model (64). This is due to the fact that the agr regulon in either species controls production of a variety of multifunctional proteases, lipases, and phenol-soluble modulins that can mediate colonization as well as invasive infection (20, 47, 48). We found several expected genes under transcriptional control of AgrA in S. hominis agr-I, including four distinct beta-type PSMs and one putative lipase (Fig. 9). Outside of anticipated genes, we also found a variety of metabolically associated genes that were downregulated. Of particular interest were the genes associated with acetoin production (Fig. 9). l-Lactate is a major component of human sweat, and acetoin is a breakdown product of microbial l-lactate metabolism (65). Human body odor is significantly associated with the microbial breakdown of sweat components, and several studies have characterized potential deodorant agents to reduce S. epidermidis breakdown of these metabolites (65, 66). Aside from l-lactate metabolism, S. hominis is already known to contribute to human body odor through the clade-specific C-S lyase-mediated breakdown of thioalcohols (67). Our results indicate a potentially interesting quorum sensing-mediated body odor production pathway, but more targeted work with specific knockout strains is needed to fully understand this mechanism.
While we were primarily interested in the downregulated genes in the apicidin data set, there were also 7 genes that were significantly upregulated (Fig. 9). Intriguingly, 6 of the 7 genes were colocalized on the S. hominis AH5009 genome, ranging from locus tag MOV58_01105 to MOV58_01130. While the specific function of these genes is unclear given the few prior functional studies in any S. hominis strain, MOV58_01110 was predicted by BLASTx to be an arylamine N-acetyltransferase, MOV58_01120 was predicted to be an ATP binding protein, MOV58_01130 was predicted to be a helix-turn-helix transcriptional regulator, and MOV58_01125 was predicted to be a DUF3169 protein. In S. aureus, proteins in this domain of unknown function family are predicted to have 6 transmembrane loops, although their function has not been defined (68). These predicted functions suggest that a drug sensing and metabolizing system is upregulated in response to apicidin treatment. This putative drug response was not found in previous RNA-seq characterization of apicidin-treated MRSA, where the most highly upregulated gene was betB, encoding glycine betaine aldehyde dehydrogenase (54).
Taken together, our results imply that S. hominis is a ubiquitous commensal with potentially protective roles in maintaining human health and skin integrity. We show that S. hominis agr signaling and allelic variation may be important mechanisms of skin colonization resistance against opportunistic staphylococcal pathogens. Future work may continue to uncover even more beneficial roles of this underappreciated commensal. As we move away from traditional antibiotics and toward individualized medicine and bacteriotherapies (11, 12), it is imperative to understand the fundamental molecular mechanisms driving commensal-host relationships. There is mounting evidence that CoNS and other skin commensals can be used with great success to remediate skin barrier function during AD flares (12) or even to inhibit tumor proliferation (69), yet we still know little about mechanisms underpinning CoNS skin colonization, what virulence factors CoNS encode or could acquire, or how shifts in skin population dynamics or transfer of genes may impact clinical outcomes for patients administered these new therapies. S. hominis is a promising candidate for many new and exciting translational applications, and we will continue to benefit from deeper investigations of its fundamental roles in colonization resistance as well as its potential clinical applications.
MATERIALS AND METHODS
Ethics statement.
Seven-week-old female BALB/c mice or 8-week-old male and female C57BL/6J mice were purchased from Jackson Laboratories and housed in specific-pathogen-free facilities at the University of Colorado Anschutz Medical Center Animal Facility. Mice were allowed to acclimate for 1 week prior to experimentation. At experimental endpoints, mice were euthanized via CO2 inhalation followed by cervical dislocation. All animal work was approved by and performed in accordance with the Institutional Animal Care and Use Committee of the University of Colorado Anschutz Medical Campus under protocol numbers 00486 and 00941.
Human subjects and skin swab collection.
Bacterial DNA from adult human skin swabs was used from a previously published collection where swabs of surface microbiota from a 5-cm2 area of the antecubital fossa skin of both the left and right arms were collected from 14 healthy subjects and 13 patients with AD (25). Only healthy samples were used in this study. Swabs were collected according to protocols approved by the University of California, San Diego (UCSD), institutional review board (project no. 140144), and written informed consent was obtained from all subjects.
Collection of bacteria from human subjects.
Collection of bacterial isolates from human skin was carried out according to protocols approved by the UCSD Institutional Review Board (project no. 071032), and informed consent was obtained from all subjects. Some S. hominis isolates used in this study (see Table S1 in the supplemental material; Gallo strains) were collected from a previous study at UCSD under the aforementioned protocol number (32, 38).
Growth conditions and reagents.
Bacterial strains used in this study are listed in Table S1. S. hominis skin and nasal isolates labeled “this study” in Table S1 were confirmed to be S. hominis by matrix-assisted laser desorption/ionization (MALDI)–time of flight (TOF) mass spectrometry prior to experimentation. All staphylococcal strains were grown in tryptic soy broth (TSB) at 37°C with shaking at 220 rpm. Escherichia coli was grown in LB at 37°C with shaking at 220 rpm. For strains with pDB59 or pCM41, chloramphenicol was added to a final concentration of 10 μg/mL. For strains with pCM40, erythromycin was added to a final concentration of 10 μg/mL. S. hominis synthetic AIPs were custom synthesized by AnaSpec, Inc.
PCR identification of S. hominis agr types.
Genomic DNA was isolated from S. hominis strains using the Puregene yeast/bact kit (Qiagen) with a modified protocol: 4 mL of cells grown overnight (ON) in TSB were pelleted and resuspended in 0.5 mL phosphate-buffered saline (PBS). The suspension was homogenized in a bead beater with 1-mm zirconia/silica beads (BioSpec) for three 1-min intervals with 1 min on ice between the homogenization intervals. After homogenization, the manufacturer’s protocol was continued with the omission of the RNase A step and the addition of a 1-h incubation on ice after the addition of protein precipitation solution. Samples were submitted to the Barbara Davis Center Bioresource Core for Sanger sequencing. Oligonucleotides used to sequence S. hominis agrD were as follows: forward, 5′-GCATGAATTCAGTCAAGGAGAGTGGCACA-3′; reverse, 5′-CGAGGATCCAAACCATCCATATCATTTTCTCTT -3′.
LC-MS identification of S. hominis AIPs.
Identification of AIPs was achieved using LC-MS analysis by previously described methods (36). In brief, a single isolated colony of S. hominis strains containing each agr type was inoculated into 6 mL of TSB and incubated at 37°C with 250 rpm shaking for 24 h. Cultures were then diluted 1:200 and returned to the incubator for 18 to 20 h, until the OD600 indicated that cell growth had reached stationary phase. Each culture was centrifuged at 10,000 rpm and filtered through a 0.22-μm surfactant-free cellulose acetate (SFCA) membrane. Filtered medium was then subjected to solid-phase extraction using Strata-X-C strong-cation-exchange, reversed-phase columns, according to the manufacturer’s recommended protocol. Eluent was dried under a stream of nitrogen at room temperature and resuspended in 120 μL of 80:20 water-methanol.
Samples were analyzed on a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific, Waltham, MA) with a heated electrospray ionization source coupled to an Acquity ultrahigh-performance liquid chromatography (UPLC) system (Waters Corp., Milford, MA) by use of a previously described method (36). In brief, a 7-μL injection of spent medium was eluted from an Acquity BEH C18 column (1.7 μm, 2.1 by 50 mm; Waters) at 0.3 mL/min using a binary solvent system consisting of 0.1% formic acid (A) and acetonitrile (CH3CN) with 0.1% formic acid (B). The solvent gradient initiated with a 1.5-min isocratic hold at 20% B and was followed by a linear increase to 60% B over 5 min. The gradient was held isocratic from 6.5 min to 7.0 min and then increased to 100% B at 8.0 min. The column was washed at 100% B for 1 min and then returned to the starting conditions to allow reequilibration for 1.0 min prior to the next injection. The first 1.5 min of eluent was diverted to waste. Synthetic standards for each of the observed AIP structures were purchased from AnaSpec, EGT (Fremont, CA), and subjected to the same LC-MS analysis. The accurate mass, retention time, and fragmentation patterns for synthetic standards were compared to those of putative AIP ions in spent medium to confirm the structure.
Analysis of the frequency of unique S. hominis agr types.
Previously determined S. hominis agr type I to VI sequences were used as a database for determining the frequency of agr types across all current S. hominis genomes on the NCBI genome database. S. hominis strains were downloaded and annotated with Prokka (1.13.7) (70), and BLAST was used for AgrD protein (region containing unique autoinducing peptide sequences) alignment (56, 71, 72). Based upon the detectable S. hominis agr types confirmed in the NCBI database, primers were designed for quantifying S. hominis agr type absolute abundance by quantitative PCR (qPCR). For measurement of absolute abundance, standards for CFU/μg of genomic DNA were created from representative strains of S. hominis agr types I to V grown to mid-exponential phase (approximate OD600, 0.5), followed by both serial dilution live count plating (10−1 to 10−12; 5-μL droplets) on mannitol salt agar and genomic DNA isolation. For qPCR-based absolute abundance counts of S. hominis agr types I to V in human skin swabs, the representative strain standards of S. hominis agr types I to V were used to generate standard curves and the unknown quantities in healthy human skin swabs were assessed by use of the S. hominis agr type-specific primers. Human skin swab genomic DNA was from a previous study involving nonlesional and lesional bilateral antecubital crease samples from 14 healthy and 13 atopic dermatitis (AD) individuals (25).
S. hominis agr::P3 reporter electroporation.
S. hominis AH5009 (agr-I) and S. hominis AH4553 (agr-II) were made electrocompetent by following Staphylococcus epidermidis: Methods and Protocols (73). The previously described agr reporter plasmid pCM41 (36) was midi prepped from the restriction-modification-deficient E. coli DC10B host with the Invitrogen PureLink midi prep kit in accordance with the manufacturer’s guidelines (36). One to 5 μg of plasmid was used for each transformation.
Fluorescent agr reporter assays.
For conditioned medium (CM) assays, S. hominis strains and MRSA reporters were grown ON for 20 h as desribed in growth conditions and reagents. One milliliter of S. hominis culture was pelleted, and CM was filtered through a Costar Spin-X centrifuge tube (0.22 μm, cellulose acetate filter). Reporters were prepared by subculturing 1:500 in fresh TSB with chloramphenicol. CM was added at 20% (vol/vol) to a 96-well black culture plate (Corning) and 2-fold serially diluted to 0.15% (vol/vol). One hundred microliters of reporter was added to a final volume of 200 μL per well. Cultures were grown in a Stuart humidified incubator at 37°C with shaking at 1,000 rpm. At hourly time points up to 24 h, plates were measured on a Tecan Group Ltd. Infinite Pro plate reader to quantify growth (optical density at 600 nm) and YFP signal (excitation, 480 nm; emission, 515 nm). For synthetic AIP experiments, peptide was resuspended in neat DMSO or a DMSO control was added from stocks of 20 μM to the indicated concentrations shown in Figures 4 and 5. S. hominis reporter assays were conducted in the same manner as MRSA reporter assays.
Murine dermonecrosis model.
To prepare bacteria for the dermonecrosis model, USA300 MRSA (strain LAC) was grown ON in TSB and then subcultured 1:100 in fresh TSB and allowed to grow to early exponential phase (OD600, 0.5 to 0.7). Bacterial cells were washed and pelleted in phosphate-buffered saline (PBS) and resuspended in sterile saline to achieve an inoculum of 1 × 108 CFU in 50 μL. The inoculum concentration was verified by colony counting after 24 h of incubation at 37°C. One day prior to challenge, BALB/c mouse abdomens were shaved and residual hair was removed with a 30-s application of Nair (Church & Dwight Co., Inc.). Immediately prior to injection, abdomens were sanitized with alcohol wipes. Fifty-microliter inoculum suspensions containing 1 × 108 CFU MRSA and either S. hominis AIP-II (10 μg or 50 μg in neat DMSO) or DMSO alone were injected intradermally. MRSA and AIP were mixed immediately prior to injection. Body weights were measured before infection and every day thereafter for a period of 7 days. To determine lesion size, digital images were taken using a Canon PowerShot ELPH 180 camera and analyzed with ImageJ (NIH) software.
Murine epicutaneous infection.
To prepare bacteria for AIP experiments, strain LAC was grown ON in TSB and then subcultured 1:50 in fresh TSB and allowed to grow to an OD600 of 1. Bacterial cells were washed and pelleted in phosphate-buffered saline (PBS) and resuspended in sterile saline to achieve an inoculum of 1 × 108 CFU in 100 μL. LAC and synthetic AIP-II (10 or 50 μg) or vehicle (DMSO) were combined immediately prior to application on gauze. For competition experiments, LAC and S. hominis AH4553 (agr-II) were grown ON in TSB and then subcultured 1:50 in fresh TSB and allowed to grow to an OD600 of 1. Bacterial cells were washed and pelleted in PBS and resuspended in sterile saline to achieve an inoculum of 1 × 108 CFU in 50 μL for competition or 1 × 108 CFU in 100 μL for single challenge. The cochallenge inoculum was combined at a 1:1 ratio immediately prior to application on gauze. Inoculum concentration was verified by colony counting after 24 h of incubation at 37°C. C57BL/6J mice were anesthetized with 2% isoflurane, backs were shaved, and residual hair was removed with a 1-min application of Nair (Church & Dwight Co., Inc.). Mice were allowed to recover for 24 h following hair removal. Bacteria were applied to back skin for 72 h on a 2-cm2 piece of sterile gauze affixed with Tegaderm and covered with a Band-Aid. A Tewameter TM300 device (Courage & Khazaka Electronic GmbH) was used to determine changes to epithelial barrier integrity at 72 h postinfection. Two sites per lesion were measured. To enumerate bacterial CFU on skin postchallenge with AIP, the full-thickness 2-cm2 atopic lesion was excised with sterile scissors, added to 0.5 mL PBS with 1-mm zirconia/silica homogenization beads (BioSpec), and homogenized for three 1-min intervals. The suspension was serially diluted and plated on nonselective (TSA) and selective (mannitol salt agar [MSA]) media and on MSA supplemented with 5.2 μg/mL cefoxitin. Plates were incubated overnight prior to colony counting.
RNA-seq.
RNA-seq was essentially performed as described previously (54, 74). Overnight cultures of S. hominis AH5009 grown in TSB were subcultured 1:200 in fresh TSB with 100 μM apicidin diluted in DMSO (Sigma) or an equivalent volume of DMSO in biological triplicate in a 24-well plate (Corning). The plate was incubated at 37°C with shaking at 1,000 rpm in a Stuart humidified shaker for 8 h (OD600, 3). Cultures were harvested by adding 2 mL RNAprotect bacteria (2:1, vol/vol) (Qiagen) in accordance with the manufacturer’s guidelines, incubated for 5 min at room temperature, and then pelleted. Pellets were stored at −80°C until RNA purification. RNA from each sample was prepared with the Qiagen RNeasy minikit (Qiagen) in accordance with the manufacturer’s guidelines. RNA was DNase treated with the Turbo DNase kit (Thermo Scientific) in accordance with the standard manufacturer’s guidelines and sent for Illumina stranded RNA library preparation with RiboZero Plus rRNA depletion at the Microbial Genome Sequencing Center (MiGS; Pittsburg, PA). RNA-seq was performed at a depth of 25 million paired-end reads. Reads were aligned to the S. hominis C5 (AH5009) genome. RNA-seq data analysis was performed with the CLC Genomics Workbench (Qiagen version 20.0.4) using a 4-fold cutoff for differential gene expression and a false discovery rate P value of ≤0.05.
Mass spectrometric identification of S. hominis PSM-β1 in conditioned medium.
For bacterial preparation, an overnight culture of S. hominis (AH5009) grown in TSB was subcultured 1:200 in fresh TSB with 100 μM apicidin diluted in DMSO (Sigma) or an equivalent volume of DMSO in technical triplicate in a 24-well plate (Corning) in a total volume of 1 mL. Control wells of TSB plus 100 μM apicidin (Sigma), an equivalent volume of DMSO, or no addition were included in technical triplicate in the same total volume. The plate was incubated at 37°C with shaking at 1,000 rpm in a Stuart humidified shaker for 8 h (OD600, 3). Cells were pelleted out of conditioned medium at 17,000 × g for 5 min.
PSM-β1 detected with LC-MS data was obtained using a Q Exactive Plus quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific) with a heated electrospray ionization source coupled to an Acquity UPLC system (Waters), using the same UPLC method described in “LC-MS identification of S. hominis AIPs.” The mass spectrometer was operated in the positive ionization mode with the following instrument parameters: capillary temperature, 256°C; spray voltage, 3.00 kV; sheath gas, 48 arbitrary units; auxiliary gas, 12 arbitrary units; spare gas, 2 arbitrary units; and probe heater temperature, 350°C. Data were collected using four scan events: a full-scan event from m/z 500 to 5,000 with a resolving power of 35,000 and three data-dependent scan events at a resolving power of 17,500. Data-dependent scan events were triggered by ions within 5 ppm of the calculated monoisotopic m/z for PSM-β1 with five protons ([M + 5H]5+, 908.6819).
Genomic DNA preparation for whole-genome sequencing.
Genomic DNA from S. hominis AH5009 was prepared for whole-genome sequencing at the Microbial Genome Sequencing Center (MiGS; Pittsburg, PA). Four milliliters of overnight culture grown in TSB was pelleted and washed in Tris-EDTA (TE) buffer. Cells were lysed in TE buffer with lysostaphin and lysozyme for 2 h at 37°C and then homogenized in a bead beater with 0.1-mm zirconia/silica beads (BioSpec) for 1 s. Lysate was then processed with the blood and tissue kit (Qiagen) in accordance with the manufacturer’s guidelines. DNA integrity was checked on a 0.5% agarose gel before sequencing. MiGS-prepared DNA libraries were sequenced on the NextSeq 2000 Illumina platform and with Oxford Nanopore Technologies (ONT) with 300-Mbp long reads (ONT), 400-Mbp paired-end Illumina reads, genome assembly, and annotation (Small Nanopore Combo).
S. hominis AH5009 genome assembly and annotation.
Quality control and adapter trimming were performed with blc2fastq (proprietary Illumina software for conversion of bcl files to basecalls; version 2.20.0.445, default parameters) and porechop (open-source software for quality control and adapter trimming of Oxford Nanopore Technologies; version 0.2.3_seqan2.1.1, default parameters) for Illumina and Oxford Nanopore sequencing, respectively, by the MiGS facility. Unicycler (version 0.5.0, default parameters) (75) was used to assemble reads. Bandage (version 0.8.1) (76) and BUSCO (version 5.2.2) (77) were used to assess assembly completeness. Genome was annotated with the NCBI Prokaryotic Genome Annotation Pipeline (PGAP 6.0) upon submission.
Statistical analysis.
All analyses were performed using GraphPad Prism 9 (San Diego, CA) software. For multiple comparisons of in vitro data, a one-way analysis of variance (ANOVA) with Dunnett’s multiple-comparison test was chosen. Animal data were analyzed with a two-way ANOVA with Dunnett’s multiple-comparison test or with a one-way ANOVA with Dunnett’s multiple-comparison test for CFU differences. In vitro data are presented as the mean and standard deviation (SD), and in vivo data are presented as the mean and standard error of the mean (SEM); a P value of <0.05 was considered significant. Test choices are indicated in the figure legends.
Data and material availability.
All data associated with the study are present in the paper or in the supplemental material. RNA-seq data are available through the NCBI Gene Expression Omnibus (GEO) online data repository under accession number GSE199818. The S. hominis AH5009 genome sequence is available at NCBI under BioProject accession number PRJNA816516.
ACKNOWLEDGMENTS
Funding was provided by National Institute for Allergy and Infectious Diseases (NIAID) grant no. T32 AI052066 (M.M.S.), NIAID Ruth L. Kirschstein National Research Award Predoctoral Fellowship no. 1F31AI157052 (M.M.S.), NCCIH Ruth L. Kirschstein National Research Award training grant no. T32AT008938 (Z.L.B.), NIAID grant AI153185 (A.R.H., N.B.C., M.R.W., and R.L.G.), U.S. Department of Veteran Affairs grant no. BX002711 (A.R.H.), National Institute of Health Institutional Research and Academic Career Development Award grant no. 2K12GM068524-19 (M.R.W.), and National Institutes of Health grant no. zU01AI52038, R01AR07682, and R37AI052453 (R.L.G.).
Conceptualization, A.R.H. and M.M.S.; Methodology, D.A.T., N.B.C., K.Z., L.S.Z., and A.R.H.; Investigation, M.M.S., M.R.W., A.S., L.M.L., Z.L.B., and A.N.; Visualization, M.M.S., M.R.W., A.S., L.M.L., and Z.L.B.; Supervision, D.A.T., N.B.C., K.Z., L.S.Z., and R.L.G.; Writing – original draft, A.R.H. and M.M.S.; Writing – review & editing, M.M.S., M.R.W., A.S., L.M.L., Z.L.B., A.N., L.S.Z., K.Z., D.A.T., N.B.C., R.L.G., and A.R.H.
We thank the Microbial Genome Sequencing Center (MiGS; Pittsburg, PA) for their technical assistance with RNA sequencing and whole-genome sequencing.
R.L.G. is a cofounder, scientific advisor, and consultant for and has equity in MatriSys Biosciences and is a consultant for, receives income from, and has equity in Sente, Inc. The other authors declare no competing interests.
Contributor Information
Alexander R. Horswill, Email: Alexander.horswill@cuanschutz.edu.
Victor J. Torres, New York University School of Medicine
REFERENCES
- 1.Byrd AL, Belkaid Y, Segre JA. 2018. The human skin microbiome. Nat Rev Microbiol 16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 2.Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA, NISC Comparative Sequencing Program . 2016. Temporal stability of the human skin microbiome. Cell 165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker K, Heilmann C, Peters G. 2014. Coagulase-negative staphylococci. Clin Microbiol Rev 27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bay L, Barnes CJ, Fritz BG, Thorsen J, Restrup MEM, Rasmussen L, Sørensen JK, Hesselvig AB, Odgaard A, Hansen AJ, Bjarnsholt T. 2020. Universal dermal microbiome in human skin. mBio 11:e02945-19. doi: 10.1128/mBio.02945-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parlet CP, Brown MM, Horswill AR. 2019. Commensal staphylococci influence Staphylococcus aureus skin colonization and disease. Trends Microbiol 27:497–507. doi: 10.1016/j.tim.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Libertucci J, Young V. 2019. The role of the microbiota in infectious diseases. Nat Microbiol 4:35–45. doi: 10.1038/s41564-018-0278-4. [DOI] [PubMed] [Google Scholar]
- 7.Nakatsuji T, Chiang H-I, Jiang SB, Nagarajan H, Zengler K, Gallo RL. 2013. The microbiome extends to subepidermal compartments of normal skin. Nat Commun 4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, Chen YE, Li K, Farhat S, Weckel A, Krishnamurthy SR, Vujkovic-Cvijin I, Linehan JL, Bouladoux N, Merrill ED, Roy S, Cua DJ, Adams EJ, Bhandoola A, Scharschmidt TC, Aubé J, Fischbach MA, Belkaid Y. 2019. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366:eaax6624. doi: 10.1126/science.aax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scharschmidt TC, Vasquez KS, Truong HA, Gearty SV, Pauli ML, Nosbaum A, Gratz IK, Otto M, Moon JJ, Liese J, Abbas AK, Fischbach MA, Rosenblum MD. 2015. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43:1011–1021. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eyerich S, Eyerich K, Traidl-Hoffmann C, Biedermann T. 2018. Cutaneous barriers and skin immunity: differentiating a connected network. Trends Immunol 39:315–327. doi: 10.1016/j.it.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Liu Y, Du Z, Zhang L, Chen J, Shen Z, Liu Q, Qin J, Lv H, Wang H, He L, Liu J, Huang Q, Sun Y, Otto M, Li M. 2020. Skin microbiota analysis-inspired development of novel anti-infectives. Microbiome 8:85. doi: 10.1186/s40168-020-00866-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakatsuji T, Hata TR, Tong Y, Cheng JY, Shafiq F, Butcher AM, Salem SS, Brinton SL, Rudman Spergel AK, Johnson K, Jepson B, Calatroni A, David G, Ramirez-Gama M, Taylor P, Leung DYM, Gallo RL. 2021. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat Med 27:700–709. doi: 10.1038/s41591-021-01256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross AA, Müller KM, Scott Weese J, Neufeld JD. 2018. Comprehensive skin microbiome analysis reveals the uniqueness of human skin and evidence for phylosymbiosis within the class Mammalia. Proc Natl Acad Sci USA 115:E5786–E5795. doi: 10.1073/pnas.1801302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bitschar K, Wolz C, Krismer B, Peschel A, Schittek B. 2017. Keratinocytes as sensors and central players in the immune defense against Staphylococcus aureus in the skin. J Dermatol Sci 87:215–220. doi: 10.1016/j.jdermsci.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Li D, Lei H, Li Z, Li H, Wang Y, Lai Y. 2013. A novel lipopeptide from skin commensal activates TLR2/CD36-p38 MAPK signaling to increase antibacterial defense against bacterial infection. PLoS One 8:e58288. doi: 10.1371/journal.pone.0058288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li D, Wang W, Wu Y, Ma X, Zhou W, Lai Y. 2019. Lipopeptide 78 from Staphylococcus epidermidis activates β-catenin to inhibit skin inflammation. J Immunol 202:1219–1228. doi: 10.4049/jimmunol.1800813. [DOI] [PubMed] [Google Scholar]
- 17.Two AM, Nakatsuji T, Kotol PF, Arvanitidou E, Du-Thumm L, Hata TR, Gallo RL. 2016. The cutaneous microbiome and aspects of skin antimicrobial defense system resist acute treatment with topical skin cleansers. J Invest Dermatol 136:1950–1954. doi: 10.1016/j.jid.2016.06.612. [DOI] [PubMed] [Google Scholar]
- 18.Götz F, Perconti S, Popella P, Werner R, Schlag M. 2014. Epidermin and gallidermin: staphylococcal lantibiotics. Int J Med Microbiol 304:63–71. doi: 10.1016/j.ijmm.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Christensen GJM, Scholz CFP, Enghild J, Rohde H, Kilian M, Thürmer A, Brzuszkiewicz E, Lomholt HB, Brüggemann H. 2016. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genomics 17:152. doi: 10.1186/s12864-016-2489-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown MM, Horswill AR. 2020. Staphylococcus epidermidis—skin friend or foe? PLoS Pathog 16:e1009026. doi: 10.1371/journal.ppat.1009026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hon KL, Tsang YCK, Pong NH, Leung TF, Ip M. 2016. Exploring Staphylococcus epidermidis in atopic eczema: friend or foe? Clin Exp Dermatol 41:659–663. doi: 10.1111/ced.12866. [DOI] [PubMed] [Google Scholar]
- 22.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. 2014. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otto M. 2008. Staphylococcal biofilms. Curr Top Microbiol Immunol 322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paharik AE, Horswill AR. 2016. The staphylococcal biofilm: adhesins, regulation, and host response. Microbiol Spectr 4:10.1128/microbiolspec.VMBF-0022-2015. doi: 10.1128/microbiolspec.VMBF-0022-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cau L, Williams MR, Butcher AM, Nakatsuji T, Kavanaugh JS, Cheng JY, Shafiq F, Higbee K, Hata TR, Horswill AR, Gallo RL. 2021. Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. J Allergy Clin Immunol 147:955–966.e16. doi: 10.1016/j.jaci.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams MR, Cau L, Wang Y, Kaul D, Sanford JA, Zaramela LS, Khalil S, Butcher AM, Zengler K, Horswill AR, Dupont CL, Hovnanian A, Gallo RL. 2020. Interplay of staphylococcal and host proteases promotes skin barrier disruption in Netherton syndrome. Cell Rep 30:2923–2933.e7. doi: 10.1016/j.celrep.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byrd AL, Deming C, Cassidy SKB, Harrison OJ, Ng WI, Conlan S, Belkaid Y, Segre JA, Kong HH, NISC Comparative Sequencing Program . 2017. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med 9:eaal4651. doi: 10.1126/scitranslmed.aal4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kloos WE, Schleifer KH. 1975. Isolation and characterization of staphylococci from human skin. Descriptions of four new species: Staphylococcus warneri, Staphylococcus capitis, Staphylococcus hominis, and Staphylococcus simulans. Int J Syst Bacteriol 25:62–79. doi: 10.1099/00207713-25-1-62. [DOI] [Google Scholar]
- 29.Williams MR, Gallo RL. 2017. Evidence that human skin microbiome dysbiosis promotes atopic dermatitis. J Invest Dermatol 137:2460–2461. doi: 10.1016/j.jid.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kennedy EA, Connolly J, Hourihane JOB, Fallon PG, McLean WHI, Murray D, Jo JH, Segre JA, Kong HH, Irvine AD. 2017. Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol 139:166–172. doi: 10.1016/j.jaci.2016.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Sullivan JN, Rea MC, O’Connor PM, Hill C, Ross RP. 2019. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol Ecol 95:fiy241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik AV, Latif H, Kim J-N, Lockhart A, Artis K, David G, Taylor P, Streib J, Dorrestein PC, Grier A, Gill SR, Zengler K, Hata TR, Leung DYM, Gallo RL. 2017. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med 9:eaah4680. doi: 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paharik A, Parlet C, Chung N, Todd D, Rodriguez E, Van Dyke M, Cech N, Horswill A. 2017. Coagulase-negative staphylococcal strain prevents Staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe 22:746–756. doi: 10.1016/j.chom.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bannoehr J, Ben Zakour NL, Waller AS, Guardabassi L, Thoday KL, Van Den Broek AHM, Fitzgerald JR. 2007. Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains. J Bacteriol 189:8685–8692. doi: 10.1128/JB.01150-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng P, Baldry M, Gless BH, Bojer MS, Espinosa-Gongora C, Baig SJ, Andersen PS, Olsen CA, Ingmer H. 2019. Effect of co-inhabiting coagulase negative staphylococci on S. aureus agr quorum sensing, host factor binding, and biofilm formation. Front Microbiol 10:2212. doi: 10.3389/fmicb.2019.02212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown MM, Kwiecinski JM, Cruz LM, Shahbandi A, Todd DA, Cech NB, Horswill AR. 2020. Novel peptide from commensal Staphylococcus simulans blocks methicillin-resistant Staphylococcus aureus quorum sensing and protects host skin from damage. Antimicrob Agents Chemother 64:e00172-20. doi: 10.1128/AAC.00172-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fetzner S, Collin M, Canovas J, Baldry M, Ms B, Canovas J, Baldry M, Bojer MS, Andersen PS, Grzeskowiak PK, Stegger M, Damborg P, Olsen CA, Ingmer H. 2016. Cross-talk between Staphylococcus aureus and other staphylococcal species via the agr quorum sensing system. Front Microbiol 7:1733. doi: 10.3389/fmicb.2016.01733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams MR, Costa SK, Zaramela LS, Khalil S, Todd DA, Winter HL, Sanford JA, O’neill AM, Liggins MC, Nakatsuji T, Cech NB, Cheung AL, Zengler K, Horswill AR, Gallo RL. 2019. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci Transl Med 11:eaat8329. doi: 10.1126/scitranslmed.aat8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dufour P, Jarraud S, Vandenesch F, Greenland T, Novick RP, Bes M, Etienne J, Lina G. 2002. High genetic variability of the agr locus in Staphylococcus species. J Bacteriol 184:1180–1186. doi: 10.1128/jb.184.4.1180-1186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otto M, Echner H, Voelter W, Götz F. 2001. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun 69:1957–1960. doi: 10.1128/IAI.69.3.1957-1960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hersh AL, Chambers HF, Maselli JH, Gonzales R. 2008. National trends in ambulatory visits and antibiotic prescribing for skin and soft-tissue infections. Arch Intern Med 168:1585–1591. doi: 10.1001/archinte.168.14.1585. [DOI] [PubMed] [Google Scholar]
- 42.Schwan WR, Langhorne MH, Ritchie HD, Stover CK. 2003. Loss of hemolysin expression in Staphylococcus aureus agr mutants correlates with selective survival during mixed infections in murine abscesses and wounds. FEMS Immunol Med Microbiol 38:23–28. doi: 10.1016/S0928-8244(03)00098-1. [DOI] [PubMed] [Google Scholar]
- 43.Wright JS, Jin R, Novick RP. 2005. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc Natl Acad Sci USA 102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le KY, Otto M. 2015. Quorum-sensing regulation in staphylococci—an overview. Front Microbiol 6:1174. doi: 10.3389/fmicb.2015.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harraghy N, Kerdudou S, Herrmann M. 2007. Quorum-sensing systems in staphylococci as therapeutic targets. Anal Bioanal Chem 387:437–444. doi: 10.1007/s00216-006-0860-0. [DOI] [PubMed] [Google Scholar]
- 46.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 47.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. 2011. Peptide signaling in the staphylococci. Chem Rev 111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olson ME, Todd DA, Schaeffer CR, Paharik AE, Van Dyke MJ, Buttner H, Dunman PM, Rohde H, Cech NB, Fey PD, Horswill AR. 2014. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J Bacteriol 196:3482–3493. doi: 10.1128/JB.01882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otto M. 2009. Staphylococcus epidermidis—the “accidental” pathogen. Nat Rev Microbiol 7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou W, Spoto M, Hardy R, Guan C, Fleming E, Larson PJ, Brown JS, Oh J. 2020. Host-specific evolutionary and transmission dynamics shape the functional diversification of Staphylococcus epidermidis in human skin. Cell 180:454–470.e18. doi: 10.1016/j.cell.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gless BH, Bojer MS, Peng P, Baldry M, Ingmer H, Olsen CA. 2019. Identification of autoinducing thiodepsipeptides from staphylococci enabled by native chemical ligation. Nat Chem 11:463–469. doi: 10.1038/s41557-019-0256-3. [DOI] [PubMed] [Google Scholar]
- 52.Moran MC, Cahill MP, Brewer MG, Yoshida T, Knowlden S, Perez-Nazario N, Schlievert PM, Beck LA. 2019. Staphylococcal virulence factors on the skin of atopic dermatitis patients. mSphere 4:e00616-19. doi: 10.1128/mSphere.00616-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H, Archer NK, Dillen CA, Wang Y, Ashbaugh AG, Ortines RV, Kao T, Lee SK, Cai SS, Miller RJ, Marchitto MC, Zhang E, Riggins DP, Plaut RD, Stibitz S, Geha RS, Miller LS. 2017. Staphylococcus aureus epicutaneous exposure drives skin inflammation via IL-36-mediated T cell responses. Cell Host Microbe 22:653–666. doi: 10.1016/j.chom.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parlet CP, Kavanaugh JS, Crosby HA, Raja HA, El-Elimat T, Todd DA, Pearce CJ, Cech NB, Oberlies NH, Horswill AR. 2019. Apicidin attenuates MRSA virulence through quorum-sensing inhibition and enhanced host defense. Cell Rep 27:187–198.e6. doi: 10.1016/j.celrep.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Begley M, Hill C, Gahan CGM. 2006. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol 72:1729–1738. doi: 10.1128/AEM.72.3.1729-1738.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 57.Kloos WE, George CG, Olgiate J, Van Pelt L, McKinnon ML, Zimmer BL, Muller E, Weinstein MP, Mirrett S. 1998. Staphylococcus hominis subsp. novobiosepticus subsp. nov., a novel trehalose-and N-acetyl-d-gIucosamine-negative, novobiocin-and multiple-antibiotic-resistant subspecies isolated from human blood cultures. Int J Syst Bacteriol 48:799–781. doi: 10.1099/00207713-48-3-799. [DOI] [PubMed] [Google Scholar]
- 58.Götz F, Bannerman T, Schleifer KH. 2006. The genera Staphylococcus and Macrococcus, p 5–75. In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (ed), The prokaryotes. Springer, New York, NY. [Google Scholar]
- 59.King JM, Kulhankova K, Stach CS, Vu BG, Salgado-Pabón W. 2016. Phenotypes and virulence among Staphylococcus aureus USA100, USA200, USA300, USA400, and USA600 clonal lineages. mSphere 1:e00071-16. doi: 10.1128/mSphere.00071-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakatsuji T, Cheng JY, Gallo RL. 2021. Mechanisms for control of skin immune function by the microbiome. Curr Opin Immunol 72:324–330. doi: 10.1016/j.coi.2021.09.001. [DOI] [PubMed] [Google Scholar]
- 61.Lamand V, Dauwalder O, Tristan A, Casalegno JS, Meugnier H, Bes M, Dumitrescu O, Croze M, Vandenesch F, Etienne J, Lina G. 2012. Epidemiological data of staphylococcal scalded skin syndrome in France from 1997 to 2007 and microbiological characteristics of Staphylococcus aureus associated strains. Clin Microbiol Infect 18:E514–E521. doi: 10.1111/1469-0691.12053. [DOI] [PubMed] [Google Scholar]
- 62.Cheung GYC, Wang R, Khan BA, Sturdevant DE, Otto M. 2011. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun 79:1927–1935. doi: 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, Blumberg HM. 2006. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann Intern Med 144:309–317. doi: 10.7326/0003-4819-144-5-200603070-00005. [DOI] [PubMed] [Google Scholar]
- 64.Nakamura Y, Takahashi H, Takaya A, Inoue Y, Katayama Y, Kusuya Y, Shoji T, Takada S, Nakagawa S, Oguma R, Saito N, Ozawa N, Nakano T, Yamaide F, Dissanayake E, Suzuki S, Villaruz A, Varadarajan S, Matsumoto M, Kobayashi T, Kono M, Sato Y, Akiyama M, Otto M, Matsue H, Núñez G, Shimojo N. 2020. Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci Transl Med 12:eaay4068. doi: 10.1126/scitranslmed.aay4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hara T, Matsui H, Shimizu H. 2014. Suppression of microbial metabolic pathways inhibits the generation of the human body odor component diacetyl by staphylococcus spp. PLoS One 9:e111833. doi: 10.1371/journal.pone.0111833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar M, Myagmardoloonjin B, Keshari S, Negari IP, Huang CM. 2019. 5-Methyl furfural reduces the production of malodors by inhibiting sodium l-lactate fermentation of Staphylococcus epidermidis: implication for deodorants targeting the fermenting skin microbiome. Microorganisms 7:239. doi: 10.3390/microorganisms7080239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rudden M, Herman R, Rose M, Bawdon D, Cox DS, Dodson E, Holden MTG, Wilkinson AJ, James AG, Thomas GH. 2020. The molecular basis of thioalcohol production in human body odour. Sci Rep 10:12500. doi: 10.1038/s41598-020-68860-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Craney A, Dix MM, Adhikary R, Cravatt BF, Romesberg FE. 2015. An alternative terminal step of the general secretory pathway in Staphylococcus aureus. mBio 6:e01178-15. doi: 10.1128/mBio.01178-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakatsuji T, Chen TH, Butcher AM, Trzoss LL, Nam S-J, Shirakawa KT, Zhou W, Oh J, Otto M, Fenical W, Gallo RL. 2018. A commensal strain of Staphylococcus epidermidis protects against skin neoplasia. Sci Adv 4:eaao4502. doi: 10.1126/sciadv.aao4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 71.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fey PD. 2014. Staphylococcus epidermidis: methods and protocols. Humana Press, Totowa, NJ. Available at https://link.springer.com/book/10.1007/978-1-62703-736-5. [Google Scholar]
- 74.Crosby HA, Tiwari N, Kwiecinski JM, Xu Z, Dykstra A, Jenul C, Fuentes EJ, Horswill AR. 2019. The Staphylococcus aureus ArlRS two-component system regulates virulence factor expression through MgrA. Mol Microbiol 113:103–122. doi: 10.1111/mmi.14404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol 38:4647–4654. doi: 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Monk IR, Shah IM, Xu M, Tan MW, Foster TT. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boles BR, Thoende M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide- independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olson ME, Nygaard TK, Ackermann L, Watkins RL, Zurek OW, Pallister KB, Griffith S, Kiedrowski MR, Flack CE, Kavanaugh JS, Kreiswirth BN, Horswill AR, Voyich JM. 2013. Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect Immun 81:1316–1324. doi: 10.1128/IAI.01242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, Horswill AR. 2009. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods 77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol 186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of strains and plasmids used in this study. Download Table S1, DOCX file, 0.02 MB (16.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
MRSA growth in the presence of S. hominis conditioned medium. (A) Growth of MRSA agr-I P3::YFP over 24 h in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains. Growth is reported as the optical density at 600 nm. (B) Growth of MRSA agr-II P3::YFP over 24 h in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains. (C) Growth of MRSA agr-III P3::YFP over 24 h in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains. (D) Growth of MRSA agr-IV P3::YFP over 24 h in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains. Results are pooled from three independent experiments. Mean values ± SD are shown. Download FIG S1, DOCX file, 0.3 MB (273.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Synthetic AIP validation. (A) Selected-ion chromatograms (SIC) for the calculated AIP-II mass (m/z = 1,020.4137) from strain AH4553 for both the spent medium (top) and the synthetic AIP in TSB (bottom). (B) MS-MS spectra for identified AIP-II ion in both the spent medium (top) and the synthetic AIP in TSB (bottom). (C) SIC for the calculated AIP-IV mass (m/z = 957.4137) from strain AH5011 for both the spent medium (top) and the synthetic AIP in TSB (bottom). (D) MS-MS spectra for identified AIP-IV ion in both the spent medium (top) and the synthetic AIP in TSB (bottom). (E) SIC for the calculated AIP-V mass (m/z = 973.4089) from strain AH5010 for both the spent medium (top) and the synthetic AIP in TSB (bottom). (F) MS-MS spectra for identified AIP-V ion in both the spent medium (top) and the synthetic AIP in TSB (bottom). Download FIG S2, DOCX file, 0.7 MB (774.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
S. epidermidis agr-I early fluorescence is boosted by S. hominis AIP-I. Relative fluorescence (RFU) is shown compared to reporter-only control for S. epidermidis agr-I P3::sGFP incubated with increasing doses of S. hominis AIP-I over 24 h. Results were pooled from three independent experiments. Mean values ± SD are shown. Download FIG S3, DOCX file, 0.1 MB (94.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
S. hominis conditioned medium inhibits S. epidermidis agr quorum sensing. (A) S. epidermidis agr-I P3::sGFP incubated with 10% cell-free conditioned medium from S. hominis strains for 24 h. The 24-h fluorescent point is shown relative to reporter only. (B) S. epidermidis agr-I P3::sGFP growth reported as the OD600 in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains for 24 h. (C) S. epidermidis agr-II P3::sGFP 24-h fluorescent point relative to reporter only. (D) S. epidermidis agr-II P3I::sGFP growth (OD600) in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains for 24 h. (E) S. epidermidis agr-III P3::sGFP 24-h fluorescent point relative to reporter only. (F) S. epidermidis agr-III P3::sGFP growth (OD600) in the presence of 10% cell-free conditioned medium from the indicated S. hominis strains for 24 h. Significance was determined by ordinary one-way ANOVA with Dunnett’s multiple-comparison test. Results were pooled from three independent experiments. Mean values ± SD are shown. *, P < 0.05; ****, P < 0.0001. Download FIG S4, DOCX file, 0.2 MB (236.9KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Validation of S. hominis agr type distribution in healthy skin swabs. (A) Table of qPCR primers used in this study. (B) qPCR primer specificity validation for each S. hominis agr type. S. epidermidis (SE) genomic DNA (gDNA) standards were also included to determine any off-target amplification. Download FIG S5, DOCX file, 0.2 MB (209.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
S. hominis agr-I and agr-II intraspecies cross-talk with synthetic AIPs. (A) S. hominis agr-I growth (measured by OD600) with increasing concentrations of AIP-I. (B) S. hominis agr-I growth with increasing concentrations of AIP-II. (C) S. hominis agr-II growth with increasing concentrations of AIP-II. (D) S. hominis agr-II growth (OD600) with increasing concentrations of AIP-I. (E) S. hominis agr-I growth with increasing doses of AIP-III. (F) S. hominis agr-I fluorescence with increasing doses of AIP-III. (G) S. hominis agr-I growth with increasing doses of AIP-IV. (H) S. hominis agr-I fluorescence with increasing doses of AIP-IV. (I) S. hominis agr-I growth with increasing doses of AIP-V. (J) S. hominis agr-I fluorescence with increasing doses of AIP-V. (K) S. hominis agr-II growth with increasing doses of AIP-III. (L) S. hominis agr-II fluorescence with increasing doses of AIP-III. (M) S. hominis agr-II growth with increasing doses of AIP-IV. (N) S. hominis agr-II fluorescence with increasing doses of AIP-IV. (O) S. hominis agr-II growth with increasing doses of AIP-V. (P) S. hominis agr-II fluorescence with increasing doses of AIP-V. Growth curves are representative of three independent experiments. For fluorescence curves, results were pooled from three independent experiments. Mean values ± SD are shown. Download FIG S6, DOCX file, 0.6 MB (615.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
S. hominis in competition with MRSA protects skin from epicutaneous damage. (A) Transepithelial water loss 72 h postinfection for mice challenged with MRSA only, S. hominis only, or equivalent numbers of CFU of MRSA and S. hominis, or uninfected (UI; saline only). (B) Representative images of mouse back skin at 72 h postinfection with the indicated groups. Results were pooled from two independent experiments (n = 10 for all groups except UI [n = 9]). Mean ± SD is shown. Significance was determined by ordinary one-way ANOVA with the Holm-Sidak multiple-comparison test. Download FIG S7, DOCX file, 0.4 MB (426.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Apicidin inhibition of S. hominis agr-I signaling is AgrA dependent. (A) S. hominis agr-I P3::sGFP growth (OD600) and fluorescence (relative fluorescent units) in the presence of 100 μM apicidin or equivalent DMSO control over 24 h. Results were pooled from three independent experiments, and mean values ± SD are shown. (B) LC-MS selected ion chromatogram for the m/z range of 908.6774 to 908.6864 (chosen from the calculated m/z of the PSM-β1 peptide with five protons; [M + 5H]5+, 908.6819) for S. hominis AH5009 (C5). The data show PSM-β1 presence in vehicle-treated culture (DMSO) (top) and PSM-β1 absence in 100 μM apicidin-treated S. hominis AH5009 (bottom). Results are representative chromatograms from among those collected for biological replicates. (Average peak area for replicates is shown in Fig. 9 of the main text.) Download FIG S8, DOCX file, 0.4 MB (459.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
RNA-seq analysis of differentially expressed transcripts in apicidin-treated S. hominis agr-I. Download Table S2, DOCX file, 0.02 MB (16.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.