Abstract
According to WHO warnings, the antibiotic resistance crisis is a severe health issue in the 21st century, attributed to the overuse and misuse of these medications. Consequently, the dramatic spreading rate of the drug-resistant microbial pathogens strains. The microbiological, biochemical tests and antibiotic sensitivity identified the bacteria's multi-drug resistance (MDR). About 150 different clinical samples were taken from hospitalized patients, both males, and females, ranging from 9 to 68 years. Gram-negative strains were (70.0%), while Gram-positive isolates were (30.0%). Among sixteen antibiotics, antibiotic susceptibility of imipenem was found to be the most efficient drug against most of the Gram-negative and Gram-positive isolates, followed by meropenem, depending on the culture and sensitivity results. All the experimental bacteria showed multidrug-resistant phenomena. In this study, green synthesized silver (Cur-Ag NPs) and zinc oxide (Cur-ZnO NPs) nanoparticles in the presence of curcumin extract. In addition, their physicochemical properties have been characterized using different techniques such as UV-Vis spectroscopy, transmission electron microscope (TEM), X-ray diffraction (XRD), Fourier transform infrared (FT-IR), and colloidal properties techniques. Furthermore, curcumin-capped silver nanoparticles (AgNPs) exhibited solid antimicrobial action against the experimental bacterial isolates, except Proteus vulgaris (i.e., P. vulgaris). Curcumin-capped zinc oxide nanoparticles (ZnO NPs) found antimicrobial activity against all tested strains. Finally, the minimum inhibitory concentration exhibited values from 3.9 to 15.6 μg ml−1, which is too small compared to other traditional antibiotics. In addition, the green-synthesized Cur-Ag NPs and Cur-ZnO NPs showed good biocompatibility.
Curcumin-assisted synthesized Ag and ZnO NPs showed significant antibacterial activity with lower minimum inhibitory concentration (MIC) against the multi-drug resistance bacteria and biocompatibility compared to traditionally used antibiotics.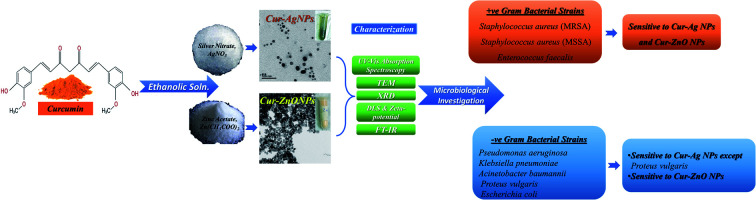
1. Introduction
The antibiotics resistance crisis is a severe health issue that has resulted from overuse and misuse of these medications; consequently, the dramatic spreading rate of the drug-resistant microbial pathogens strains such as multidrug resistance (MDR). Thus, according to WHO warnings, MDR pathogens became one of the greatest public health threats of the 21st century.1–3
Recently, the emergence of a significant increase in the presence of antibiotic-resistant bacteria has led many research groups toward using antimicrobial-based-nanomaterials.4,5 Their extraordinary physicochemical properties, such as large surface area and ease to functionalize, enhance the contact with the targeted organism. In addition, their small size facilitates their cellular membrane penetration, modulating molecular pathways and interactions for bacterial cells.6 In this regard, numerous nanomaterials based on metal oxide (e.g., titanium oxide (TiO2), silver oxide (Ag2O), silica oxide (SiO2), and copper oxide (CuO)),7 and metal nanoparticles (e.g., silver nanoparticles (AgNPs)) were used as powerful antimicrobial agents.8–10
Several studies have been devoted to using nanomaterials-based antimicrobial agents. Silver is a well-known antimicrobial agent in ancient ages, as in silver nitrate (AgNO3).11 Historically, the ancient Egyptians sterilized drinking water and wounds using metals such as silver.12 As an example of metal nanoparticles, silver nanoparticles (AgNPs) are the most potent non-toxic antimicrobial agent against many microbes, including bacteria, fungi, and viruses, even at low concentrations.13,14 Furthermore, green synthesized AgNPs via endosymbiotic marine isolated actinomycetes have been used as active anti-microbial and biofilms agents.9 Also, AgNPs with variable shapes (i.e., spheres and hexagons) exhibited a significant antibacterial activity against MDR-bacteria isolated from different water sources related to dairy farm-related sites.8 Compared to metallic nanoparticles, zinc oxide nanoparticles (ZnO NPs), as an example of metal-oxide-based nanomaterials, were used in several studies as an antimicrobial agent against pathogenic and spoilage microorganisms.15–17 In such studies, ZnO NPs have been used in conjugation with antibiotic and anti-inflammatory medicines to promote/enhance the antimicrobial activity against multi-drug pathogenic microorganisms.18,19 In addition, it could be used in sterilizing medical devices20,21 and antimicrobial surface coating.22 Finally, curcumin extract is one of the natural compounds that can be used as a capping agent and as a biocompatible reducing agent in the synthesis of nanoparticles. Curcumin is a natural extract derived from turmeric (Curcuma longa), used as one of the types of spices and as a colouring agent used for food.23 Curcumin has been widely used in traditional Chinese medicine for centuries,24,25 as it contains various therapeutic properties26 such as antioxidants,27 anti-inflammatory,28 antiseptic,29 and anticancer activities.30,31 Also, it has been approved and recognized as safe by the FDA with a dose of 12 g day−1.32–34
The current study aimed to fabricate effective antimicrobial nanoparticles based on metallic (i.e., AgNPs) and metal oxide (i.e., ZnO NPs) nanoparticles via green synthesis routes in the presence of curcumin extract. In addition, determine the optimal concentration for each curcumin capped-Ag and ZnO NPs to enhance the effective antibacterial activity against pathogenic multi-drug resistance clinical bacteria.
2. Experimental
2.1. Materials
Zinc acetate dihydrate LR (Zn(CH3COO)2·2H2O, 98.5%) is purchased from SD-Fine Chem Limited. Silver nitrate (AgNO3) was purchased from Techno Pharm-Chem 99%. Ethanol absolute (C2H5OH, EtOH Central Drug House (P) LTD. 95%), polyvinylpyrrolidone (PVP 40K, Alpha Chemika 99%), Curcuma longa powder obtained from the local market. Pure curcumin purchased from Herbal house centres, Egypt – Lot No. HHC092021). Mueller-Hinton Agar (Oxoid Limited, Cat. No. CM0337), antibiotics including amikacin, amoxycillin/clavulanic acid, tigecycline, cefepime, cefotaxime, ceftazidime, cefaclor, ciprofloxacin, gentamicin, imipenem, meropenem, levofloxacin, tetracycline, tobramycin, cefazolin, cefoxitin are purchases from Oxoid Limited.
2.2. Methodology
2.2.1. Preparation of Curcuma longa powder extract
In the present study, the extraction process of curcumin from the Curcuma longa plant was carried out by following the previously reported extraction protocol by Pothitirat and Gritsanapan35 with a bit of modification.36,37 Briefly, about 100 g of Curcuma longa powder was mixed in 1000 ml of absolute ethanol under vigorous stirring and reflux at 60C for 24 h. Then the mixture was filtered using Whatman filter paper. The net yield of curcumin extracted from Curcuma longa powder was calculated by obtaining the precipitate from the extraction process was dried at room temperature and then weighted to obtain the yield of extracted curcumin as follows;Extracted curcumin (g) = the initial used Curcuma longa – the obtained precipitate powder
In addition, the filter containing the Curcuma longa extract was dried via freeze-drying. The net yield obtained from curcumin extract was about 9.5 to 10 g (i.e., 9.5 to 10%) per 100 g of Curcuma longa.
The extracted curcumin is analyzed via two techniques to confirm the existence of curcumin in the fabricated extract. The first one is based on identifying the photophysical properties of curcumin. The UV-Vis spectra of curcumin in ethanol are scanned within the 200–900 nm wavelength range using a JASCO V730 UV-Vis spectrophotometer with an increment of 5 nm. All UV-Vis measurements are performed at 25 °C and automatically corrected for the solvent medium, the absolute ethanol. The emission (Em) spectrum of curcumin was recorded within the range from 435 to 900 nm at room temperature using the Hitachi F-7000 spectrofluorometer system. The emission spectra are measured utilizing a 450 W xenon lamp as the excitation source.
Furthermore, thin-layer chromatography (TLC) was used as another technique to identify components of the ethanolic solution for as-obtained Curcuma longa extract and pure curcumin extract powder. The separation was performed on a precoated silica gel glass plate. TLC slurry preparation for soft layer coating; silica gel (adsorber) and a small amount of calcium sulfate (gypsum, binder) is mixed with water. This mixture is spread as a thick slurry on a clean glass plate. The resultant plate is dried and activated by heating an oven for thirty minutes at 110 °C prior to analysis. The used mobile phase was composed of chloroform and methanol with a ratio of 9 : 1. Then, 20 μL of curcumin standard solution was spotted on the left side of the TLC plate, whereas; 20 μl of the as-obtained curcumin extracted from commercial Curcuma longa was spotted on the right side of the TLC plate. The TLC glass plate was placed in a closed chamber, contained the mobile phase in the bottom, and perfectly closed to be pre-saturated with the mobile phase for at least 30 minutes. Finally, the TLC plate was saturated with the vapour of the mobile phase for at least 1 h. The developed plate was dried at room temperature, after which the TLC bands were visually inspected using UV light at a wavelength of 366 nm. Under ambient conditions, the spot densities were densitometrical scanned sequentially using a high-resolution camera.
2.3.1. Fabrication of curcumin-capped silver nanoparticles (AgNPs)
Curcumin capped silver nanoparticles have been prepared via the chemical reduction method of silver ions (Ag+) via phenolic groups present in Curcuma longa extract, as reported previously38–41 with slight modifications. Typically, 25 ml of 0.2 M of ethanolic solution of AgNO3 was added to 75 ml of curcumin ethanolic extract under vigorous stirring and reflux at 60 °C. Then, 10 g of PVP 40 000 was added to the reaction vessel under vigorous stirring and reflux. The reaction temperature was kept at 60 °C for 6 h. The reaction colour changed from orange to brownish-yellow, indicating the formation of silver nanoparticles capped with curcumin.
2.3.2. Fabrication of curcumin-capped zinc oxide nanoparticles (ZnO NPs)
Curcumin-capped zinc oxide nanoparticles have been prepared via the chemical reduction method of zinc precursors (Zn2+) via phenolic groups present in Curcuma longa extract, as reported previously42–44 with minor modifications. Typically, 25 ml of 4 mM of ethanolic solution of Zn(CH3COO)2 was added to 75 ml of curcumin ethanolic extract under vigorous stirring and reflux at 70 °C for 2 h. The reaction colour changed from orange to yellowish precipitate, indicating the formation of Zn(OH)2 powder. Then, the collected precipitate was washed several times with distilled water, then dried at 100 °C, followed by calcination at 300 °C for three hours to obtain a yellowish powder of ZnO NPs.
2.3.3. Characterization
Optical properties in terms of UV-Vis absorption were obtained using a JASCO 730 double beam spectrophotometer. The absorption spectra were recorded from 200 to 900 nm, with an increment of the wavelength of about 0.2 nm. Transmission Electron Microscope (TEM) JEOL, model JEM 2100F was used to investigate the micrograph of obtained samples under an operating voltage of 160 kV. In addition, X-ray diffraction (XRD) measurements have been carried out using an Ultima IV (Rigaku, Japan) Powder Diffractometer operating with a Cu target with Kα1 = 1.54060 Å, in the 2θ range of 20–80°. X-ray scan was performed in 2θ/θ continuous mode at 2° min−1, with a step size of 0.02. The size distribution and zeta-potential for each as-prepared curcumin capped ZnO and Ag-NPs were measured using the Malvern zeta sizer Nano ZS Nano instrument with He/Ne laser (i.e., λ = 633 nm) at an angle of 173° collecting backscatter optics. Furthermore, FT-IR of green synthesized curcumin capped-ZnO and Ag NPs were obtained from 400 to 4000 cm−1 using JASCO 6700 Fourier transform infrared spectrometer (FT-IR).
2.3.4. Microbiological investigation
2.3.4.1. Sample collection
This study was performed in strict accordance with the GOTHI guidelines, ethics regulations issued by the Minister of Health & Population Cairo, Egypt: No. 238/2003, Articles 52–6121, approved by medical research ethics committee. Different clinical specimens were taken from 150 patients with complaints of different infections from both sex, and range ages from 9–68 years, admitted to the national heart institute. Cairo, Egypt, is the general organization for teaching hospitals and institutes (GOTHI). The different specimens were collected from patients according to the type of their infections and transported to the microbiology laboratory for bacterial isolation. Clinical and laboratory data of each patient were obtained and registered.
2.3.4.2. Isolation and identification
Conventional biochemical methods primarily isolated and identified Gram-negative and Gram-positive colonies according to standard microbiological techniques.45,46 According to the manufacturer's instructions, further bacterial isolates characterization was carried out by API 20kits (Biomereux, France).
2.3.4.3. Antibiotics screening test
Antimicrobial susceptibility of all pathogenic bacterial isolates was performed on Mueller-Hinton Agar (MHA) by standard disk diffusion method.47,48 Results of the antibiotic susceptibility were taken according to National Committee for Clinical Laboratory Standards (NCCLS, 1999).49 The antibiotics used and concentration of antibiotic per disc (μg per disc) are as follows: amikacin (30 μg), amoxycillin/clavulanic acid (30 μg), tigecycline (15 μg), cefepime (30 μg), cefotaxime (10 μg), ceftazidime (30 μg), cefaclor (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), imipenem (10 μg), meropenem (10 μg), levofloxacin (5 μg), tetracycline (30 μg), tobramycin (10 μg), cefazolin (30 μg), cefoxitin (30 μg) all from oxoid. The zones of bacterial isolates inhibition for individual antibiotics were measured in mm by applying an ordinary ruler. Interpretation of inhibition zone diameters was made according to Kirby–Bauer zone diameter interpretative standards as documented by Clinical Laboratory Standard Institute guidelines (2018).50–52 According to previous studies, multi-drug resistance (MDR) was defined as acquired resistance to at least one agent in three or more antimicrobial categories, according to previous studies.53
2.3.4.4. Antibacterial bioassay
The antimicrobial potential was determined by disk diffusion assay. 108 CFU ml−1 fresh microbial culture was adjusted with 0.5 McFarland standard, and 100 μl of the standardized culture of each tested organism spread on a Muller Hilton agar plate with sterile cotton swabs and were allowed to dry for some time in the laminar flow hood. Filter discs (6 mm) were impregnated with different concentrations (varying from 10 to 50 μl) of nanoparticles, then positioned on an inoculated media with tested organism.54 After incubation of plates for 24 h at 37 °C, the inhibition zone diameter surrounding the filter disc was measured. An intra-variation assay was calculated from the triplicate of each assay for the three performed independent experiments.
2.3.4.5. Minimum inhibitory concentration test
Minimum inhibitory concentration (MIC) is the lowest concentration of the drug, which will inhibit growth as measured by observed turbidity in the test tube (CLSI, 2016).55 The MIC test was determined according to a previous report by Usman et al.56 Each potential nanoparticle was determined using a micro-broth dilution technique using sterile test tubes containing sterile nutrient broth; a serial dilution from both curcumins capped-ZnO and Ag NPs were 500, 250, 125, 62.5, 31.25, 15.62, 7.81, and 3.9 μg ml−1, respectively. One hundred microliters of 108 CFU ml−1 of each tested organism were piped into each test tube and incubated at 37 °C for 24 h. broth inoculated with bacteria was used as a positive control, and nutrient broth containing nanoparticles was used as a negative control. The lowest concentration kills the organisms ultimately, where no bacterial growth is observed.
2.3.5. Biocompatibility investigation
2.3.5.1. Cell culture
In the current study, the Vero cell line derived from Kidney epithelial cells of the African green monkey used as a model of the normal cell line was obtained from ATCC (American Tissue Culture Collection). A confluent layer of the used cell line was maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (Biowest), antibiotics (2% penicillin-streptomycin (100 IU ml−1), and 0.5% fungizone (Biowest). Then the cells were sub-cultured as reported in the protocol by Schmidt and Emmons.57 Then a hemocytometer was used to count the subcultured cells that plated onto 96-well plates.58,59
2.3.5.2. MTT assay
According to Emam et al. protocol,58 Cur-Ag and Cur-ZnO NPs were sterilized under UV irradiation for three hours before application to cell culture plates. Then serial dilutions with concentrations at 5, 10, 15, 20, 25, and 30 ppm were adjusted in 2% DMEM media for curcumin-capped silver (Cur-Ag NPs) and curcumin-capped zinc oxide (Cur-ZnO NPs) nanoparticles, respectively. Here, the biocompatibility assay for all used nanomaterials was tested against Vero cells via MTT assay (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide dye (Serva Electrophoresis, Germany).60 Exponentially growing cells were collected using 0.025% Trypsin–EDTA and plated in 96-well plates at 4000–5000 cells per well. Cells were exposed to all tested materials for 48 h. At the end of exposure, MTT solution in PBS (5 mg ml−1) was added to all wells, including no cell blank, and left to incubate for 90 minutes. Then DMSO (100 ml per well) was added to dissolve the formazan crystals with shaking for 10 min, after which the absorbance was read at 590 nm against no cell blanks on a microplate reader (Biotek Model: ELX 800, USA). The relative cell viability (%) related to control wells containing cells without nanomaterials was calculated by:Cell viability percentage (%) = absorbance(sample)/absorbance(control)
Each result was the average of three wells, and 100% viability was determined from the untreated cells.
3. Results and discussion
3.1. Synthesis of curcumin capped nanostructures (Cur-Ag and Cur-ZnO NPs)
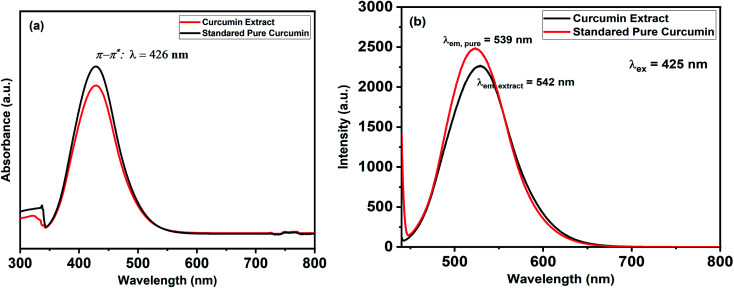
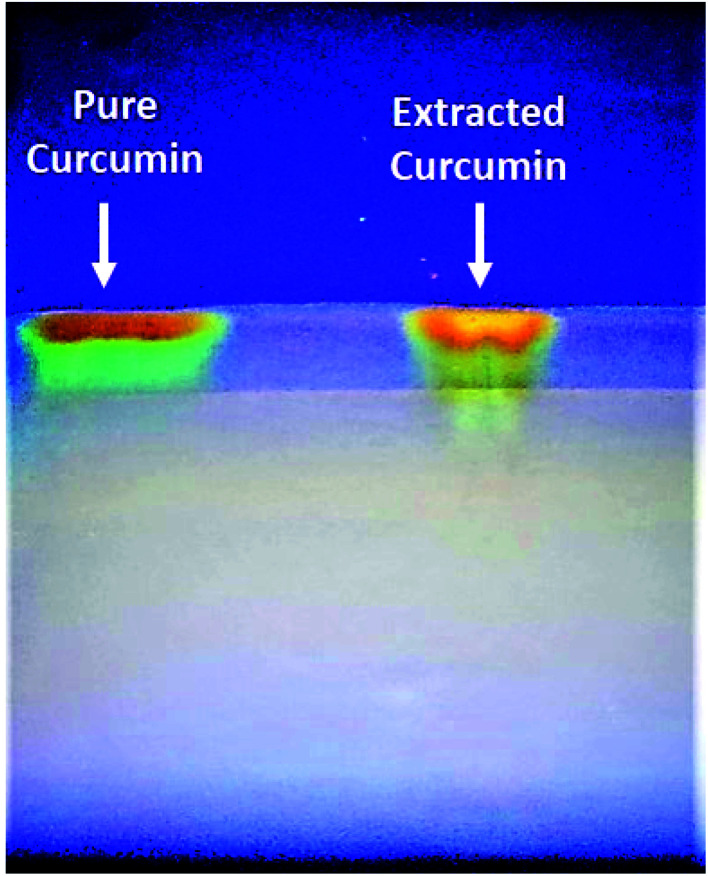
The UV-Vis spectra of curcumin extract and pure curcumin showed a broad band with a maximum absorbance peak at a wavelength of 425 nm, which could be assigned to low energy π–π* excitation of the curcumin (Fig. 1a), as previously reported.61 In addition, the emission spectra of the standard pure curcumin (red line) and curcumin extract (black line) have been recorded for comparison by excitation at the same wavelength of 425 nm in ethanolic solution, as shown in Fig. 1b. It is clear that the emission spectra are broadband at 530 nm,62 and their maximum peak shifts slightly depending on each sample. In addition, Fig. 2 shows the TLC bands of the dissolution of pure curcumin and extracted curcumin from commercial Curcuma longa powder taken after 1 h exposure to mobile phase vapour. Both curcumin samples showed a single TLC band indication of the same composition upon irradiation of TLC plate under UV light at a wavelength of 366 nm, which was in agreement with the previous report.63
Fig. 1. Shows (a) UV-Vis absorption and (b) emission spectra for the standard pure curcumin (Black-line) and curcumin extract (Red-line).
Fig. 2. TLC visual bands of pure standard curcumin (Left-side) and extracted curcumin in our lab (Right side).
Generally, plant extract-mediated nanoparticle biosynthesis may be divided into three steps; reduction, growth, and stabilization. The most crucial step is the reduction phase, in which metal ions are recovered from their salt precursors by the interaction with plant metabolites and biomolecules with reduction capabilities. Metal ions are converted from their monovalent/divalent oxidation states to zero-valent states, followed by nucleation of the reduced metal atoms. Then follows the development phase, in which the separated metal atoms combine to create metal nanoparticles, although additional biological reduction of metal ions occurs. The growth phase improves the thermodynamic stability of nanoparticles, but extensive nucleation may promote aggregation of generated nanoparticles, modifying their morphologies. The stabilization phase is the last step in the biosynthesis of nanoparticles. When nanoparticles are coated with plant metabolites, they acquire their most strongly beneficial and consistent form.64
In this study, green synthesized silver (AgNPs) and zinc oxide (ZnO NPs) have been prepared via bio-reduction in an ethanolic solution of curcumin (i.e., Curcuma longa) extract for silver nitrate (AgNO3) and zinc acetate (Zn(CH3COO)2), respectively. Green synthesis of silver nanoparticles in the presence of curcumin extract was characterized by a change in colour, from the orange colour of curcumin extract to yellowish-brown colour, indicating the formation of Cur-AgNPs.41,65 The colour change was attributed to the nucleation of Ag NPs as the interaction between Ag+ ions and the –OH− groups exists in flavonoids and Terpenoids that play a vital role in the synthesis of Ag NPs.38 Later on, with increasing the reaction time, the colour changes to brownish-yellow colour due to the maturation of Ag nanoclusters and, thereby, the formation of AgNPs.38,41,65 The same behaviour as in the formation of Cur-ZnO NPs expects the reaction colour to change from orange to light yellow due to ZnO NPs.
3.2. Characterization of curcumin capped nanostructures (Cur-Ag and Cur-ZnO NPs)
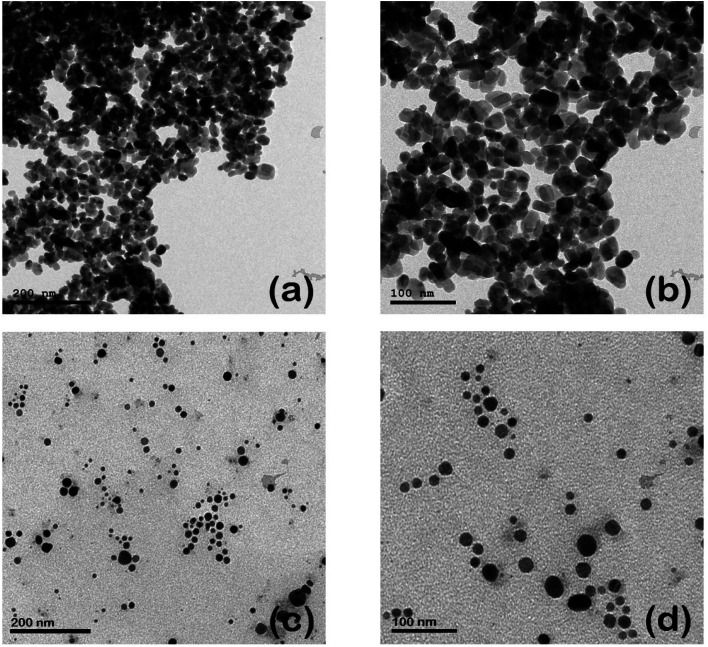
The morphological properties, including the size and shape of each as-prepared curcumin capped-silver (Cur-AgNPs) and zinc oxide (Cur-ZnO NPs) nanoparticles, are shown in Fig. 3. TEM images showed that spherical shape is dominant for Cur-AgNPs and Cur-ZnO NPs. The average particle size presented in TEM images was around 20 ± 5 nm for Cur-ZnO NPs, as shown in Fig. 3a and b. Whereas the average particle size in the case of Cur-AgNPs was about 15 ± 5 nm, as depicted in Fig. 3c and d. According to TEM images of Cur-Ag NPs and Cur-ZnO NPs nanoparticles, the coating based on curcumin cannot be noticed due to the low contrast between Ag NPs and ZnO NPs. However, the presence of curcumin-based coating could be noticed via the tendency of agglomeration for Ag NPs and ZnO NPs, as shown in Fig. 3.
Fig. 3. TEM images of Cur-ZnO NPs (a and b), and Cur-Ag NPs (c and d) at different magnification power of 100 and 200 nm, respectively.
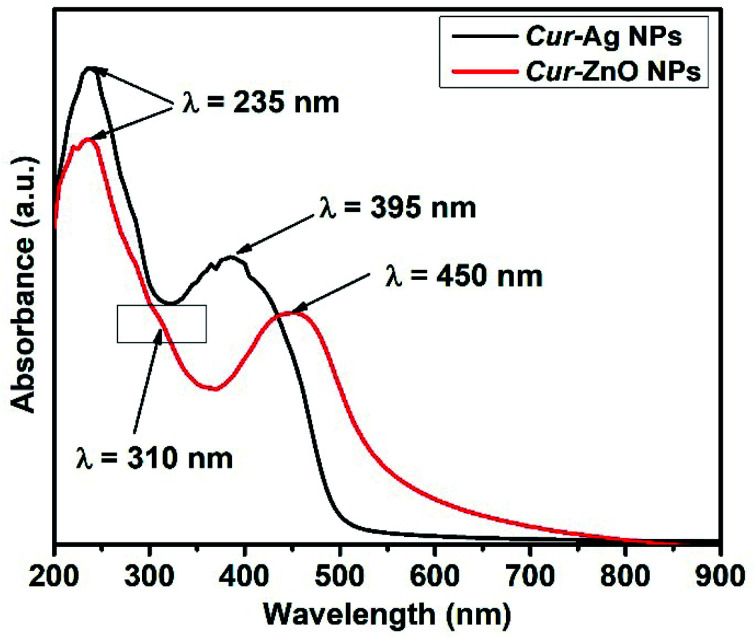
In addition, the optical properties illustrated in the monitoring of the UV-Vis absorption spectra of each of the as-prepared quasi-spherical Cur-AgNPs and Cur-ZnO NPs, as shown in Fig. 4. Cur-AgNPs exhibit a single broad surface plasmon band (SPR) at 395 nm, indicating the formation of small AgNPs (Fig. 4, Blackline). Cur-ZnO NPs showed a characteristic feature at 450 nm and shoulder at 310 nm, indicating the formation of ZnO NPs, as shown in Fig. 4, red line.
Fig. 4. Shows UV-Vis absorption spectra for the as-prepared Cur-Ag NPs (Black-line) and Cur-ZnO NPs (Red-line).
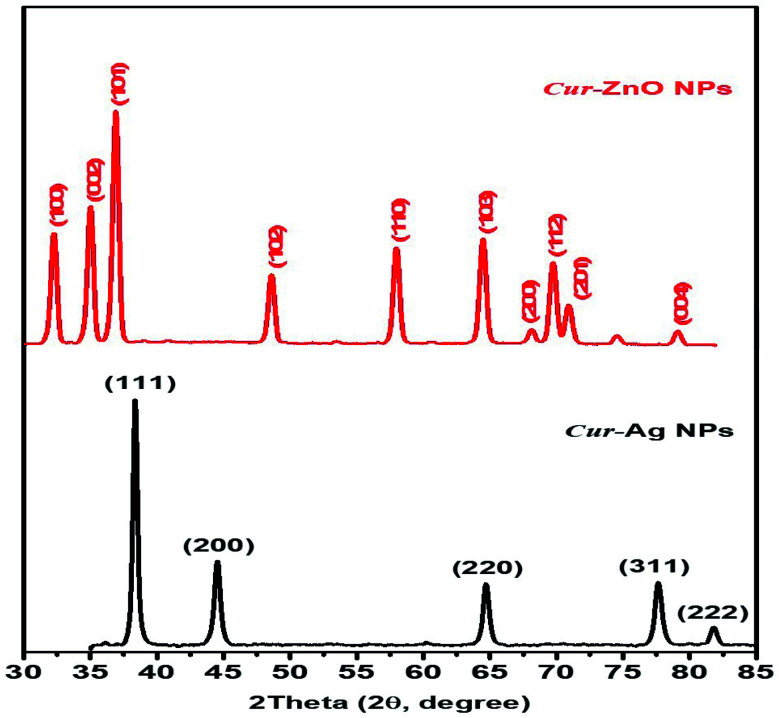
Moreover, the crystallographic structure of as-prepared biosynthesized Cur-AgNPs, and Cur-ZnO NPs have been investigated via XRD measurements, as depicted in Fig. 5. Cur-AgNPs exhibits five distinct features at 2θ = 38.27, 44.47, 64.71, 77.73 and 81.9°, respectively (see Fig. 5, Blackline), which can be assigned to the strongest line reflections from (1,1,1), (2,0,0), (2,2,0), (3,1,1), and (2,2,2) crystallographic planes of the face-centred cubic (fcc) of silver (Ag, Fm3̄m (225), see Fig. 5, Blackline). While in the case of Cur-ZnO NPs, the hexagonal structure was the predominant crystallographic structure. The typical XRD pattern for hexagonal structure shows three strongest lines at 2θ of 31.76°, 34.58°, and 36.67° due to the reflection of (100), (002), and (101) crystallographic planes. In addition, another five distinct patterns were represented at 47.89, 57.01, 63.29, 66.67, 68.25, 69.35 and 77.26° that corresponding to (102), (110), (103), (200), (112), (201) and (004) reflection plans.
Fig. 5. Shows XRD patterns of as-prepared Cur-Ag NPs (Black-line) and Cur-ZnO NPs (Red-line).
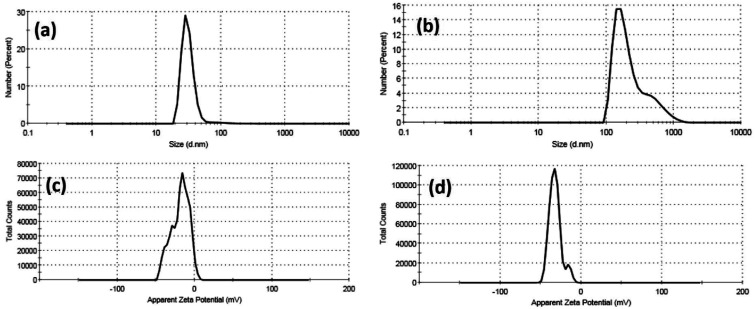
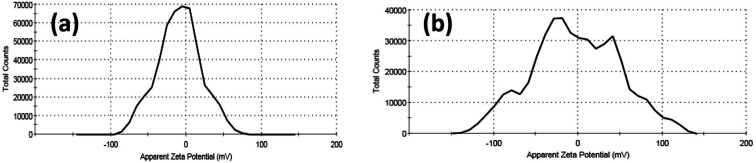
Furthermore, colloidal properties, including dynamic light scattering (DLS) and electrophoretic mobility based on zeta potential measurements, have been investigated for Cur-Ag NPs and Cur-ZnO NPs in a vehicle solution, as shown in Fig. 6 and Table 1. The hydrodynamic diameter (HD) of Cur-AgNPs was about 32.48 ± 15.98 nm with a polydispersity index (PDI) of 0.276, which is smaller than Cur-ZnO NPs. The average HD of Cur-ZnO NPs was about 261.2 ± 188.1 nm with a more extensive polydispersity index (PDI) of 0.306 (Fig. 6a and b, and Table 1). The zeta potential of Cur-AgNPs was about −18.6 mV, which is lower than the zeta-potential in Cur-ZnO NPs, which was about −32 mV, as shown in Fig. 6c and d, respectively, and Table 1. Based on the previously mentioned colloidal properties based on DLS data, the hydrodynamic particle size of as-prepared nanoparticles is enlarged, consistent with their agglomeration, as demonstrated in TEM micrographs in Fig. 3. This is because of their hydrophilicity. In addition, the intensity of the steric forces of the functional groups on the nanoparticles' surface and a steric interaction generated by the creation of a layer of water around the material.66
Fig. 6. DLS data of (a) Cur-Ag NPs and (b) Cur-ZnO NPs and zeta potential data of (c) Cur-Ag NPs and (d) Cur-ZnO NPs.
Colloidal Properties of as-prepared nano-metal oxides particles.
| Sample | Dynamic light scattering (DLS) | Zeta potential (ɳ, mV) | |
|---|---|---|---|
| Hydrodynamic diameter (HD, nm) | Polydispersity index (PDI) | ||
| Cur-Ag NPs | 32.48 ± 15.98 | 0.276 | −6.96 |
| Cur-ZnO NPs | 261.2 ± 188.1 | 0.306 | +1.39 |
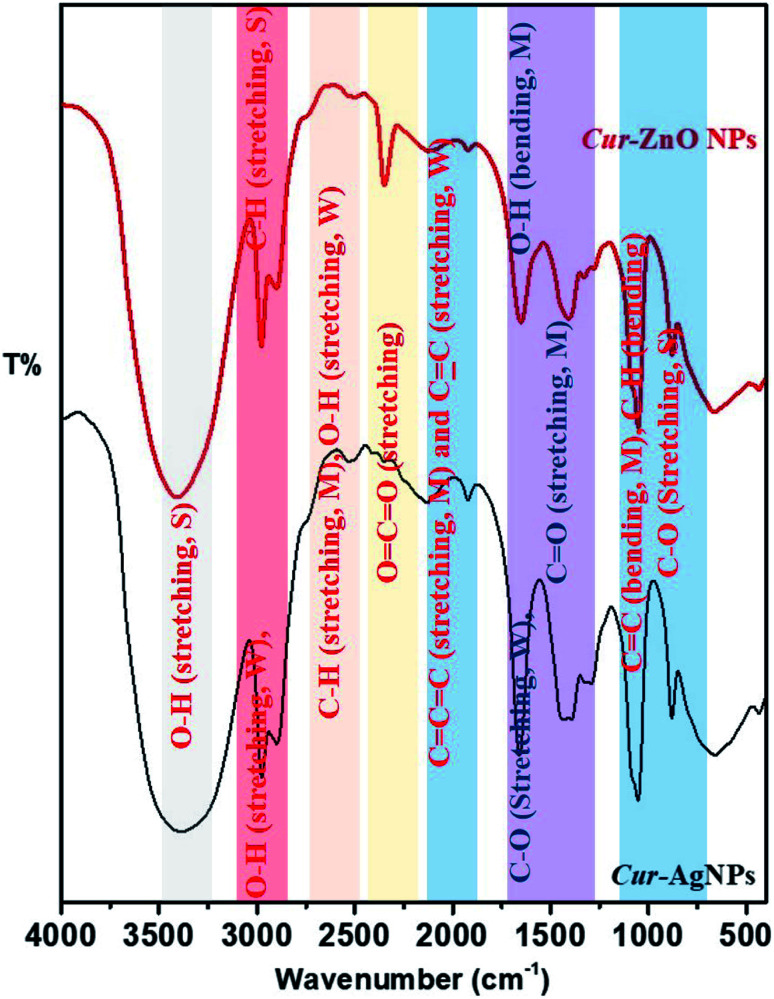
Finally, the surface properties of Cur-AgNPs and Cur-ZnO NPs have been investigated via their transmittance as function in wavenumber by Fourier-transform infrared (FT-IR), as shown in Fig. 7. Both Cur-AgNPs and Cur-ZnO NPs show the same FT-IR, which showed the same strong stretching bands at wavenumbers of 3401–3391, 2977–2975, and 1052–1053 cm−1 due to the presence of –OH intermolecular bonded of alcohol, –CH of alkyne in curcumin molecule, and –C–O of 1° alcohol, respectively (Fig. 7)—also, stretching weak band at 2898–2901 and 2133–2124 cm−1 assigned to intermolecular bonded –OH alcohol that exist in ethanolic solution, and –C C– of alkyne group present in curcumin structure of as-prepared Cur-AgNPs and Cur-ZnO NPs, respectively. The weak stretching bands 1294–1281 cm−1 were assigned to the –C–O group in bonded aromatic ester and medium stretching bands were monitored at 1659–1650 and 1921 cm−1 corresponding to conjugated –C C–, and –C C C– groups, respectively. Also, a medium bending band of bonded –C C– and C–H groups was monitored at 880 cm−1.
Fig. 7. Shows FT-IR spectra of as-prepared Cur-Ag NPs (Black-line) and Cur-ZnO NPs (Red-line).
Moreover, many differences existed between Cur-AgNPs and Cur-ZnO NPs, as follows; medium stretching bands C–H aldehyde and O–H, carboxylic acid groups in curcumin molecule, were monitored at a wavenumber of 2530 cm−1 in Cur-AgNPs, while in case of Cur-ZnO NPs was monitored at 2503 cm−1. Another difference lies in the slight position shift from 1409 cm−1 in the case of Cur-AgNPs to 1435 cm−1 in Cur-ZnO NPs, corresponding to the medium bending band of the –OH group in the ethanolic solution of curcumin extract. The same trend was detected for the medium bending band of the –OH group for phenolic species that existed in curcumin flavonoid molecules from 1330 cm−1 in Cur-AgNPs to 1394 cm−1 in the case of Cur-ZnO NPs. Finally, the remarkable increase in band intensity at 2349 cm−1 in the case of Cur-ZnO NPs compared to Cur-AgNPs, corresponds to the stretching band of the O C O group that existed in curcumin molecules.
3.3. Distribution of pathogenic bacteria isolated from clinical samples
One hundred and fifty different clinical samples (Table 2) were isolated from patients, all suffering from different infections. Table 3 shows that all bacterial isolates were characterized by phenotypic and biological characteristics.67 Based on the morphological and biochemical characterization as shown in Table 3, the distribution of isolated bacterial strains from the collected samples is tabulated in Table 4. The table showed that 45 isolates (30.0%) were Gram-positive, while Gram-negative isolates were 105 (70.0%). This result agreed with previous reports,41,42 and reported that Gram-negative isolates were more frequently distributed than Gram-positive isolates. The identified bacteria were Klebsiella pneumonia (K. pneumonia) 48 strains (32.0%), Staphylococcus aureus (S. aureus) MSSA 30 strains (20.0%), Pseudomonas aeruginosa (P. aeruginosa) 27 strains (18.0%), Escherichia coli (E. coli) 15 strains (10.0%), Proteus vulgaris (P. vulgaris) 12 strains (8.0%), Staphylococcus aureus (S. aureus) MRSA 11 strains (7.3%), Enterococcus faecalis (E. faecalis) 4 strains (2.7%) and Acinetobacter baumannii (A. baumannii) 3 strains (2.0%).
Distribution of the bacterial isolates from clinical specimens.
| Isolate source | Microorganism | |||||||
|---|---|---|---|---|---|---|---|---|
| S. aureus MSSA | S. aureus MRSA | P. aeruginosa | K. pneumonia | A. baumannii | P. Vulgaris | E. coli | E. faecalis | |
| Sputum | 0 | 0 | 0 | 14 | 0 | 0 | 4 | 0 |
| Urine | 4 | 0 | 5 | 7 | 0 | 2 | 3 | 0 |
| Wound | 21 | 7 | 21 | 25 | 3 | 10 | 7 | 4 |
| Blood | 5 | 4 | 1 | 2 | 0 | 0 | 1 | 0 |
Biochemical characteristics of bacterial isolates.
| Group | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|---|
| Isolates identification | Staphylococcus aureus | Enterococcus faecalis | Escherichia coli | Acinetobacter baumannii | Pseudomonas aeruginosa | Klebsiella pneumoniae | Proteus vulgaris | |
| Biochemical characteristics | Catalase | + | − | + | + | + | + | + |
| Coagulase | + | − | − | + | − | − | − | |
| Indole | − | − | + | − | − | − | + | |
| Methylated | + | − | + | − | − | − | + | |
| Oxidase | − | − | − | − | + | − | − | |
| Vogus Proskauer | + | + | − | − | − | + | − | |
| Citrate utilization | + | − | − | + | + | + | − | |
| H2S production | − | − | − | − | − | − | + | |
| Urease | + | − | − | − | − | + | + | |
| Motility | − | − | + | − | + | − | + | |
| Lysine decarboxylase | − | − | − | − | − | + | − | |
| Gelatin liquefaction test | + | + | − | − | + | − | + | |
| Glucose | + | + | + | + | − | + | + | |
| Lactose | + | + | + | − | − | + | − | |
| Maltose | + | + | + | − | − | + | + | |
| Mannitol | + | + | + | − | + | + | − | |
| Sucrose | + | + | − | − | − | + | + | |
| Salicin | − | − | − | − | − | + | + | |
| l-Arabinose | − | − | + | − | − | + | − | |
| d-Sorbitol | − | + | + | − | − | + | − | |
| Blood hemolysis | + | + | − | − | − | − | − | |
Frequency of bacterial agents isolated from clinical specimens.
| Organisms | No of strains | Frequency% |
|---|---|---|
| K. pneumoniae | 48 | 32 |
| S. aureus MSSA | 30 | 20 |
| P. aeruginosa | 27 | 18 |
| E. coli | 15 | 10 |
| P. vulgaris | 12 | 8 |
| S. aureus MRSA | 11 | 7.3 |
| E. faecalis | 4 | 2.7 |
| A. baumannii | 3 | 2 |
| Total | 150 | 100 |
3.4. Antibiotics inhibition effects
The antibiotic resistance patterns of all identified strains have been represented in Table 5. Methicillin-sensitive S. aureus (MSSA) strains were resistant to ceftazidime, levofloxacin, tobramycin, cefotaxime, cefaclor, and ciprofloxacin about 76.7%, 66.7%, 66.7%, 63.3%, 63.3, 56.7% respectively. However about 100% of S. aureus (MSSA) strains were sensitive to cefoxitin, furthermore tigecycline, imipenem, meropenem, cefepime, tetracycline, cefazolin and amoxycillin/clavulanic acid show sensitivity about 93.3%, 83.3%, 73.3%, 66.7%, 66.7%, 66.7%, 60.0%, respectively. Moreover, only tigecycline was effective against 100% methicillin-resistant S. aureus (MRSA), representing 100% resistance to other tested antibiotics. S. aureus is one of the most common multi-drug resistant bacterial pathogens causing different kinds of infections, especially that caused by methicillin-resistant S. aureus, which increases antimicrobial resistance in many countries.68 The mechanism of S. aureus resistance was due to genetic factors and mutations.69 About 75% of E. faecalis were sensitive to imipenem and meropenem; this was similar to a previous study reported by Asadollahi et al.70 However, E. faecalis showed 100% resistance to amikacin, cefepime, cefotaxime, ceftazidime, cefaclor, gentamicin, cefazolin, etc. cefoxitin, similarly to results found by Abdelkareem et al.71 Contrary to Gaglio et al.72 and Raafat et al.,73E. faecalis isolates showed (75%) resistance to ciprofloxacin and levofloxacin, but 50% of the same strains were resistant to amoxycillin/clavulanic acid, tigecycline, tobramycin, and tetracycline. This result is in agreement with that found by Pesavento et al.74 In addition, Pieniz et al. reported the multi-drug resistance of E. faecalis due to its ability to acquire and transfer antibiotic resistance genes.75P. aeruginosa isolates showed susceptibility to amikacin (63.0%). Furthermore, the same strain showed 100% resistance to Amoxycillin/clavulanic acid, tigecycline, cefotaxime, ceftazidime, cefaclor, and tetracycline, cefoxitin, and cefazolin, which was in agreement with previous findings that reported by Mohamed et al.76 and Bhalchandra et al.77 In many countries, the increasing distribution rate of multi-drug resistance (MDR) P. aeruginosa poses a serious therapeutic problem. In the present study, K. pneumoniae exhibited a high antibiotic resistance that agreed with previously reported results.78 In this regard; K. pneumoniae showed the highest multi-drug resistance behaviour against cefazolin by 100%, followed by cefotaxime, ciprofloxacin, gentamicin, and cefoxitin (85.4%), cefaclor (66.7%), ceftazidime, tetracycline, tobramycin (64.6%), amikacin and levofloxacin (62.5%). Moreover; K. pneumoniae exhibited (85.4%) were sensitive to imipenem followed by meropenem (77.1%) and tigecyclin (75%), which was in agreement to previously reported studies.79,80 The previous study by Babakhani et al. revealed that imipenem and meropenem are the most effective antibiotics for K. pneumonia.81A. baumannii showed 100% sensitivity to tigecycline, imipenem, and meropenem, these like previous studies.82 Tigecycline is an effective drug against MDR A. baumannii, so clinicians use it as an antibiotic treatment strategy.83 Furthermore, about 100% of A. baumannii isolates were resistant to cefotaxime, cefaclor, cefoxitin, levofloxacin, ciprofloxacin, and cefazolin. These results similar to Fayyaz et al.84 In contrast to Kassam et al., P. vulgaris showed (100%) resistance to cefaclor, cefoxitin and cefazolin. Also P. vulgaris found 100% sensitivity to imipenem and meropenem, followed by tigecyclin (75%) and amikacin (66.7%), this result was in agreement with that found by Daftary and Patel.85 The increasing levels of multi-drug resistance Proteus species isolates was reported in previous studies.86 Antimicrobial resistance in E. coli has increased over the world. E. coli were resistant to tetracycline, cefotaxime, ciprofloxacin, levofloxacin and ceftazidime about 100%, 100%, 86.7%, 86.7%, 73.3% respectively, however showed sensitivity to imipenem (93.3%), meropenem (80.0%), tigecycline (66.7%), cefoxitin (66.7%) and amikacin (60.0%) this results similar to previous studies reported by Saha et al.,87 and Yakha et al.88 Such studies stated that imipenem and amikacin are very effective drugs against E. coli isolates. In our study, the multidrug resistance ability within all identified bacteria was noticed.87,88
Antibiotic resistance pattern (%) of bacterial isolates.
| Antibiotic discs | S. aureus MSSA | S. aureus MRSA | P. aeruginosa | K. pneumonia | A. baumannii | P. vulgaris | E. coli | E. faecalis |
|---|---|---|---|---|---|---|---|---|
| Amikacin (30 μg) | 46.7 | 63.6 | 37.0 | 62.5 | 66.7 | 33.3 | 40.0 | 100 |
| Amoxycillin/Clavulanic acid (30 μg) | 40 | 100 | 100 | 50.0 | 66.7 | 50.0 | 53.3 | 50.0 |
| Tigecyclin (15 μg) | 6.7 | 0 | 100 | 25.0 | 0 | 25.0 | 33.3 | 50.0 |
| Cefepime (30 μg) | 33.3 | 100 | 51.9 | 50.0 | 33.3 | 58.3 | 53.3 | 100 |
| Cefotaxime (10 μg) | 63.3 | 100 | 100 | 85.4 | 100 | 58.3 | 100 | 100 |
| Ceftazidime (30 μg) | 76.7 | 100 | 100 | 64.6 | 66.7 | 66.7 | 73.3 | 100 |
| Cefaclor (30 μg) | 63.3 | 100 | 100 | 66.7 | 100 | 100 | 53.3 | 100 |
| Ciprofloxacin (5 μg) | 56.7 | 63.6 | 66.7 | 72.9 | 100 | 50.0 | 86.7 | 80.0 |
| Gentamicin (10 μg) | 46.7 | 100 | 51.9 | 77.0 | 66.7 | 58.3 | 53.3 | 100 |
| Imipenem (10 μg) | 16.7 | 100 | 66.7 | 14.6 | 0 | 0 | 6.7 | 25 |
| Meropenem (10 μg) | 26.7 | 100 | 55.5 | 22.9 | 0 | 0 | 20.0 | 25 |
| Levofloxacin (5 μg) | 66.7 | 100 | 70.4 | 62.5 | 100 | 41.7 | 86.7 | 75.0 |
| Tetracycline (30 μg) | 33.3 | 54.5 | 100 | 64.6 | 66.7 | 58.3 | 100 | 50.0 |
| Tobramycin (10 μg) | 66.7 | 100 | 51.9 | 64.6 | 66.7 | 50.0 | 53.3 | 50.0 |
| Cefazolin (30 μg) | 33.3 | 100 | 100 | 100 | 100 | 100 | 53.3 | 100 |
| Cefoxitin (30 μg) | 0 | 100 | 100 | 85.4 | 100 | 100 | 33.3 | 100 |
3.5. Antibacterial activity of curcumin capped zinc oxide and silver nanoparticles
The antibacterial activity of as-prepared curcumin capped ZnO, and Ag NPs has been investigated using disc diffusion assay by applying the nanoparticles with different serial dilutions, as represented in Tables 5 and 6 and Fig. 8 and 9, respectively. A gradual increase in the antibacterial effect was monitored with increased concentration (see Tables 6 and 7). Cur-AgNPs showed significant antimicrobial activity against most MDR-pathogenic bacterial strains isolated from different clinical samples except P. vulgaris.89–92 No inhibition zone was monitored upon treatment with Cur-AgNPs, similar to a previous report.93–95 In this regard, zeta-potential measurements for P. vulgaris bacterial filtrate before and after treatment with Cur-Ag NPs were performed to explore the reason for resistance against Ag NPs (Fig. 10). The bacterial filtrate of untreated P. vulgaris (−ve gram bacteria) showed a negative zeta potential of −9.04 mV (100%). In contrast, the bacterial filtrate of Cur-Ag NPs treated with P. vulgaris showed multiple zeta potential values of −19.1 (55.1%), −88.5 (11.7%) and +51.8 (33.2%) mV, respectively. Therefore, a negatively charged Cur-Ag NPs did not show any antibacterial activity against −ve gram P. vulgaris that have a negative superficial charge (Fig. 10) due to the presence of ionizable moieties leading to a repeal between the bacterial outer membrane and Cur-Ag NPs, which was in agreement with the previous reports.96
Antimicrobial activity of curcumin capped Ag NPs against pathogenic bacterial isolatesa.
| Microorganism | Mean of zone inhibition in mm (mean ± SD) | |||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | ||
| +ve Gram bacterial strains | Staphylococcus aureus (MSSA) | 11.7 ± 1.53 | 13.7 ± 1.53 | 14.7 ± 0.58 | 16.3 ± 1.15 | 18.7 ± 3.2 |
| Staphylococcus aureus (MRSA) | 11.0 ± 1.0 | 12.7 ± 1.53 | 14.7 ± 0.58 | 16.0 ± 1.0 | 17.7 ± 1.53 | |
| Enterococcus faecalis | 8.7 ± 2.5 | 10.0 ± 2.6 | 12.7 ± 2.5 | 14.7 ± 1.15 | 15.7 ± 1.15 | |
| −ve Gram bacterial strains | Pseudomonas aeruginosa | 11.7 ± 0.58 | 13.7 ± 1.53 | 15.7 ± 1.53 | 20.3 ± 0.58 | 22.3 ± 1.15 |
| Klebsiella pneumoniae | 10.3 ± 0.58 | 12.3 ± 0.58 | 15.0 ± 1.0 | 17.7 ± 2.08 | 19.7 ± 0.58 | |
| Acinetobacter baumannii | 9.7 ± 0.58 | 11.3 ± 0.58 | 14.3 ± 0.58 | 16.7 ± 1.53 | 18.7 ± 3.21 | |
| Proteus vulgaris | — | — | — | — | — | |
| Escherichia coli | 10.3 ± 0.58 | 12.3 ± 1.53 | 13.7 ± 1.53 | 14.7 ± 0.58 | 16.3 ± 0.58 | |
A: 10 μl ∼5 ppm; B: 20 μl ∼10 ppm; C: 30 μl ∼15 ppm; D: 40 μl ∼20 ppm; E: 50 μl ∼25 ppm of curcumin capped-Ag NPs. (−): no inhibition.
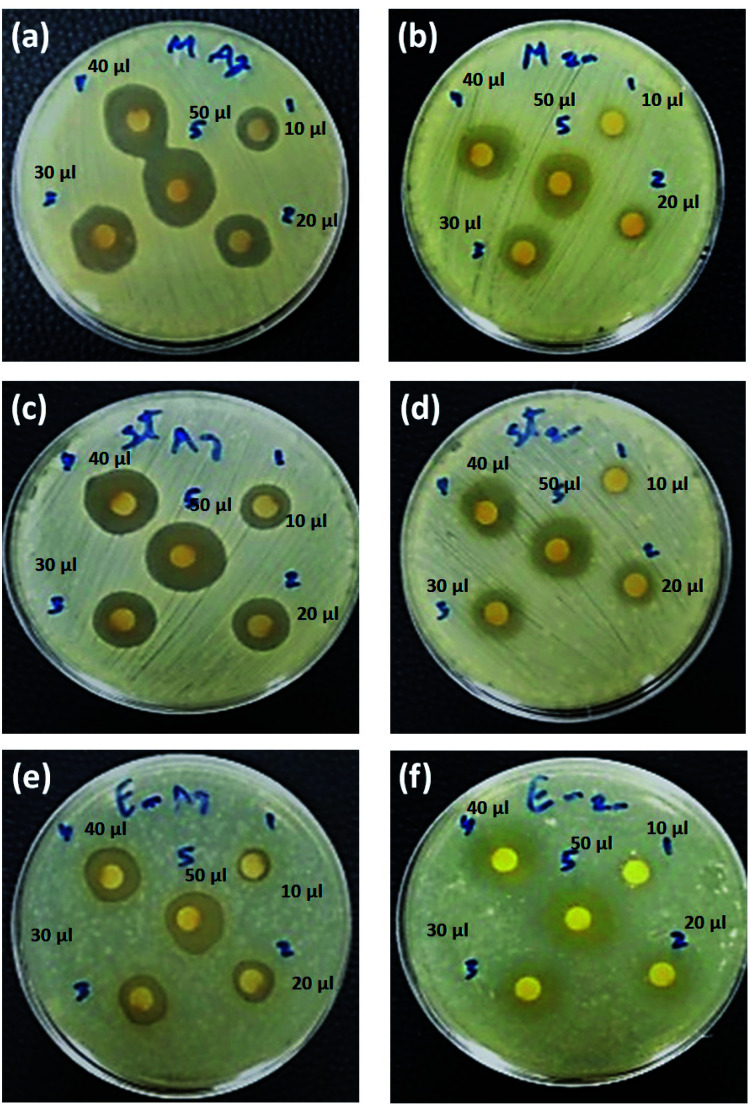
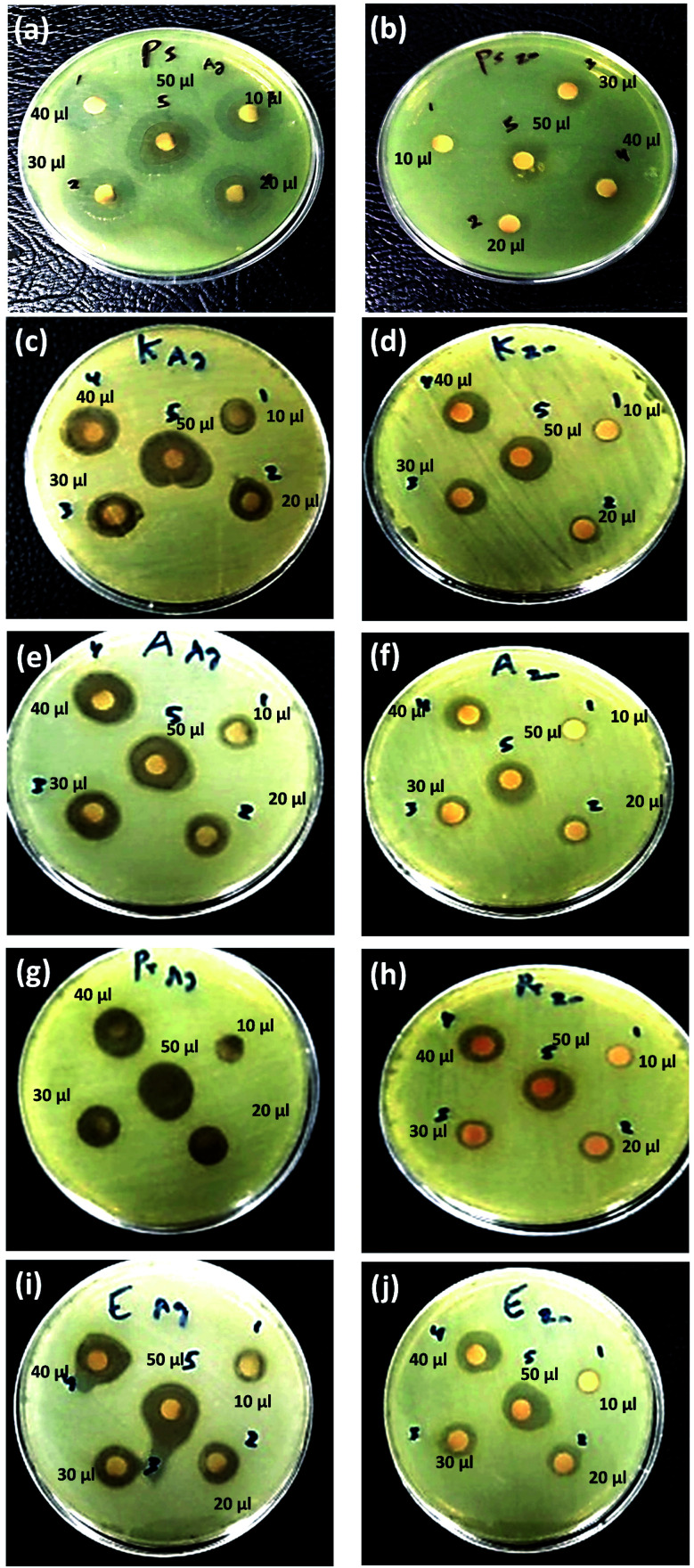
Fig. 8. Antibacterial activity of Cur-Ag NPs (a, c and e) and Cur-ZnO NPs (b, d and f) against +ve gram bacteria including S. aureus (MRSA) (a and b), S. aureus (MSSA) (c and d) and E. faecalis (e and f). Note: 10 μl ∼5 ppm; 20 μl ∼10 ppm; 30 μl ∼15 ppm; 40 μl ∼20 ppm; and 50 μl ∼25 ppm.
Fig. 9. Antibacterial activity of Cur-Ag NPs (a, c e, g and h) and Cur-ZnO NPs (b, d, f, h and j) against −ve gram bacteria including P. aeruginosa (a and b), K. pneumoniae (c and d), A. baumannii (e and f), P. vulgaris (g and h) and E. coli (i and j). Note: 10 μl ∼5 ppm; 20 μl ∼10 ppm; 30 μl ∼15 ppm; 40 μl ∼20 ppm; and 50 μl ∼25 ppm.
Antimicrobial activity of curcumin capped ZnO NPs against pathogenic bacterial isolatesa.
| Microorganism | Mean of zone inhibition in mm (mean ± SD) | |||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | ||
| +ve Gram bacterial strains | Staphylococcus aureus (MSSA) | 7.7 ± 1.53 | 10.3 ± 0.58 | 12.0 ± 1.0 | 14.0 ± 1.0 | 16.0 ± 1.0 |
| Staphylococcus aureus (MRSA) | 7.0 ± 1.0 | 9.7 ± 0.58 | 11.7 ± 0.58 | 13.3 ± 1.15 | 15.3 ± 0.58 | |
| Enterococcus faecalis | 11.0 ± 1.0 | 13.3 ± 1.15 | 15.3 ± 0.58 | 18.0 ± 2.0 | 20.3 ± 2.08 | |
| −ve Gram bacterial strains | Pseudomonas aeruginosa | 7.3 ± 0.58 | 9.0 ± 1.0 | 10.7 ± 0.58 | 12.3 ± 1.15 | 14.7 ± 0.58 |
| Klebsiella pneumoniae | 7.7 ± 0.58 | 9.0 ± 1.0 | 11.0 ± 1.0 | 13.0 ± 1.0 | 14.7 ± 1.15 | |
| Acinetobacter baumannii | 7.0 ± 1.0 | 9.0 ± 1.0 | 11.0 ± 1.0 | 13.3 ± 1.15 | 15.0 ± 1.0 | |
| Proteus vulgaris | 7.0 ± 1.0 | 9.3 ± 1.15 | 12.0 ± 1.0 | 14.0 ± 1.0 | 16.0 ± 1.0 | |
| Escherichia coli | 8.0 ± 1.0 | 10.3 ± 0.58 | 12.0 ± 1.0 | 14.0 ± 1.0 | 16.0 ± 1.0 | |
A: 10 μl ∼5 ppm; B: 20 μl ∼10 ppm; C: 30 μl ∼15 ppm; D: 40 μl ∼20 ppm; E: 50 μl ∼25 ppm of curcumin capped-ZnO NPs. (−): no inhibition.
Fig. 10. The zeta potential data for the bacterial filtrate of (a) untreated and (b) Cur-Ag NPs treated Proteus vulgaris.
In such a case, the highest antibacterial activity of Cur-AgNPs was monitored against P. aeruginosa with all applied dosing concentrations compared to other MDR-pathogenic organisms (see Table 6).90–92,97 Moreover, the current study's inhibition zone of used Cur-AgNPs was higher than AgNPs embedded in gum/Gelatin by Khan et al.98 Canvassing et al. stated that AgNPs have highly antibacterial action against multiple Gram-positive (i.e., +ve gram) and Gram-negative (i.e., −ve gram) bacteria.99
In addition, Cur-AgNPs have an antimicrobial activity of AgNPs showed an inhibitory effect against all +ve gram bacterial strains (see Fig. 8a, c, and e); however, E. faecalis showed a minimum inhibition zone even at high dosing concentration (i.e., 50 μl) 15.7 ± 1.15 mm (Fig. 7e). While methicillin-sensitive S. aureus (MSSA) showed an inhibition zone of about ∼18.7 ± 3.2 mm at 50 μl (Fig. 8c), and methicillin-resistant S. aureus (MRSA) showed an inhibition zone of about ∼17.7 ± 1.53 mm at 50 μl (Fig. 8a); these results were agreed with previous reports.100,101 The antibacterial efficiency of AgNPs may be related to the features of bacterial species. Inhibitory potential of AgNPs due to physicochemical properties, including size and surface, allow AgNPs to pass through cell walls or membranes and directly reacts with intracellular components causing inactivation of bacteria.102,103 Thus, AgNPs can be used as effective broad-spectrum antibacterial agents for Gram-positive and Gram-negative MDR bacteria, including the genera Enterococcus, Staphylococcus, Acinetobacter, Escherichia, and Pseudomonas, as shown in Fig. 9a, c, e, g, and i, respectively.
The as-prepared Cur-ZnO NPs showed antibacterial activity against the whole isolated bacterial strains from the same clinical samples, as shown in Table 7. In addition, the highest antibacterial activity of Cur-ZnO NPs was monitored against E. faecalis with all applied dosing concentrations compared to other MDR-pathogenic organisms (see Table 7). In such case, the antibacterial activity of Cur-ZnO NPs against +ve gram pathogenic bacterial strains is represented as follows; E. faecalis > S. aureus (MSSA) > S. aureus (MRSA) (see Table 7, and Fig. 8b, d, and f) that in agreement with reported studies.104–106 Also, for the −ve gram bacterial strains, the antibacterial activity of as-prepared Cur-ZnO NPs descends as follows; E. coli > P. vulgaris > A. baumannii > P. aeruginosa > K. pneumonia, which is similar to previous studies,107,108 (see Table 7 and Fig. 9b, d, f, h, and j). Generally, the antibacterial activity of Cur-ZnO NPs against both +ve and −ve gram bacterial strains depends on the microorganism's susceptibility. In addition, the difference in susceptibility of +ve and −ve gram bacteria was attributed to the differences in their cell wall structure.109,110 As well as AgNPs, the antibacterial activity increased with an increase in the ZnO NP's applied concentrations.105 Thus, ZnO NPs have antibiotic properties and could be used in biomedical applications for combating multi-drug resistance bacteria.111 Furthermore, the antimicrobial activity of nanoparticles is perfect for treating infections and eliminating pathogens.112
Furthermore, the antibacterial activity of plain curcumin was evaluated to find any synergistic activity of as-prepared green synthesized silver (Ag NPs) and zinc oxide (ZnO NPs) nanoparticles, as shown in Table 8. Plain curcumin showed no antibacterial effect against both Gram-positive and Gram-negative pathogenic bacteria. In contrast, upon the capping of Ag NPs and ZnO NPs with curcumin, significant antibacterial activity was monitored against all tested pathogenic bacteria, as shown in Tables 6 and 7, respectively.
Antimicrobial activity of plain curcumin against pathogenic bacterial isolatesa.
| Microorganism | Mean of zone inhibition in mm (mean ± SD) | |||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | ||
| +ve Gram bacterial strains | Staphylococcus aureus (MSSA) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Staphylococcus aureus (MRSA) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| Enterococcus faecalis | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| −ve Gram bacterial strains | Pseudomonas aeruginosa | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Klebsiella pneumoniae | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| Acinetobacter baumannii | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| Proteus vulgaris | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| Escherichia coli | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
A: 10 μl ∼5 ppm; B: 20 μl ∼10 ppm; C: 30 μl ∼15 ppm; D: 40 μl ∼20 ppm; E: 50 μl ∼25 ppm of plain curcumin. (−): no inhibition.
Several studies presented the possible antibacterial mechanism for both Cur-Ag NPs and Cur-ZnO NPs.113–116 Curcumin is poorly soluble in water because of its hydrophobic property, which could strongly bind to lipid layers (i.e., peptidoglycans) present in the bacterial cellular membrane. This binding could facilitate and provide the diffusion of both Ag NPs and ZnO NPs into the cellular membrane for more antibacterial activity.113 In addition, curcumin destabilises bacterial replication and inhibits the DNA damage repair processes through the interaction with the FtsZ protein.115,117,118 Alves et al. suggested that AgNPs could enhance the antibacterial efficacy of curcumin in the form of Cur-Ag NPs nanocomposites.114,115 Therefore, the major antibacterial mechanism of Cur-Ag NPs and Cur-ZnO NPs could be described as follows; (i) curcumin act as a reducing and capping agent for each Ag NPs and ZnO NPs, (ii) Cur-AgNPs and Cur-ZnO nanocomposites bind to the bacterium and release Ag+ and Zn2+ ions. Finally, the Ag+ and Zn2+ ions are released, leading to the generation of reactive oxygen species, which cause membrane damage, and bacterial lipases and induce leakage of intracellular contents followed by bacterial death.113,114
3.6. Determination of minimum inhibitory concentration (MIC)
As shown in Table 9; the minimum inhibitory concentration (MIC) of Cur-AgNPs was determined to be 7.8 μg ml−1 for E. faecalis and 3.9 μg ml−1 for S. aureus (MSSA), S. aureus (MRSA), P. aeruginosa, K. Pneumoniae, A. baumannii, P. vulgaris, and E. coli. These results were nearly similar to those reported by Mortazavi-Derazkola et al.119 and Lkhagvajav et al.120 and lower than the study reported by Jaswil et al.121 The MIC of Cur-ZnO NPs was 15.6 μg ml−1 for P. aeruginosa and K. pneumonia, 7.8 μg ml−1 for S. aureus (MSSA), and S. aureus (MRSA), A. baumannii and P. vulgaris. In addition, the MIC values were about 3.9 μg ml−1 for E. faecalis and E. coli. These MIC values were lower than those found in previous studies.122–124
Minimum inhibitory concentration (MIC) evaluation of used nanoparticles.
| Microorganism | Nanoparticles | |
|---|---|---|
| Curcumin capped silver nanoparticles (Cur-AgNPs) in ppm | Curcumin capped zinc oxide nanoparticles (Cur-ZnONPs) in ppm | |
| Staphylococcus aureus (MSSA) | 3.9 | 7.8 |
| Staphylococcus aureus (MRSA) | 3.9 | 7.8 |
| Enterococcus faecalis | 7.8 | 3.9 |
| Pseudomonas aeruginosa | 3.9 | 15.6 |
| Klebsiella pneumoniae | 3.9 | 15.6 |
| Acinetobacter baumannii | 3.9 | 7.8 |
| Proteus vulgaris | — | 7.8 |
| Escherichia coli | 3.9 | 3.9 |
In addition, MIC assays have been performed for each of the plain curcumin and AgNPs and ZnO NPs as controls to emphasize the synergistic effect of curcumin on the antimicrobial activity of AgNPs and ZnO NPs nanoparticles if it exists or exists is insufficient to enhance their antimicrobial activity, as shown in Table 10. A slight increase in the MIC value for both Ag NPs and ZnO NPs alone without capping with curcumin compared to Cur-AgNPs and Cur-ZnO NPs, as shown in Table 9. In addition, plain curcumin has a MIC value at a high concentration of 375–750 ppm. The used curcumin concentration in Cur-AgNPs and Cur-ZnO NPs not exceed 150 ppm. Therefore, curcumin has a slight synergetic effect on the antimicrobial efficacy of silver and zinc oxide nanoparticles, compared to Ag and ZnO NPs alone, as reported in Table 9. Also, our finding agreed with other studies that reported that the antimicrobial activity of metal-curcumin complex such as silver-curcumin was lower than curcumin itself.125
Minimum inhibitory concentration (MIC) evaluation of bare individual nanoparticles and plain curcumin.
| Microorganism | Nanoparticles | ||
|---|---|---|---|
| Bare silver nanoparticles (AgNPs) in ppm | Bare zinc oxide nanoparticles (ZnONPs) in ppm | Plain curcumin extract in ppm | |
| Staphylococcus aureus (MSSA) | 5.3 ± 0.47 | 7.7 ± 0.59 | 375 ± 0.00 |
| Staphylococcus aureus (MRSA) | 5.1 ± 0.24 | 9.5 ± 0.33 | 375 ± 0.00 |
| Enterococcus faecalis | 7.7 ± 0.94 | 4.2 ± 0.45 | 375 ± 0.00 |
| Pseudomonas aeruginosa | 5.6 ± 0.94 | 14.5 ± 0.55 | 375 ± 0.00 |
| Klebsiella pneumoniae | 5.0 ± 0.00 | 16.1 ± 0.29 | 375 ± 0.00 |
| Acinetobacter baumannii | 4.1 ± 0.63 | 7.5 ± 0.55 | 750 ± 0.00 |
| Proteus vulgaris | 0.0 ± 0.0 | 8.5 ± 0.55 | 375 ± 0.00 |
| Escherichia coli | 6.1 ± 0.82 | 8.0 ± 0.50 | 750 ± 0.00 |
3.7. Biocompatibility assay
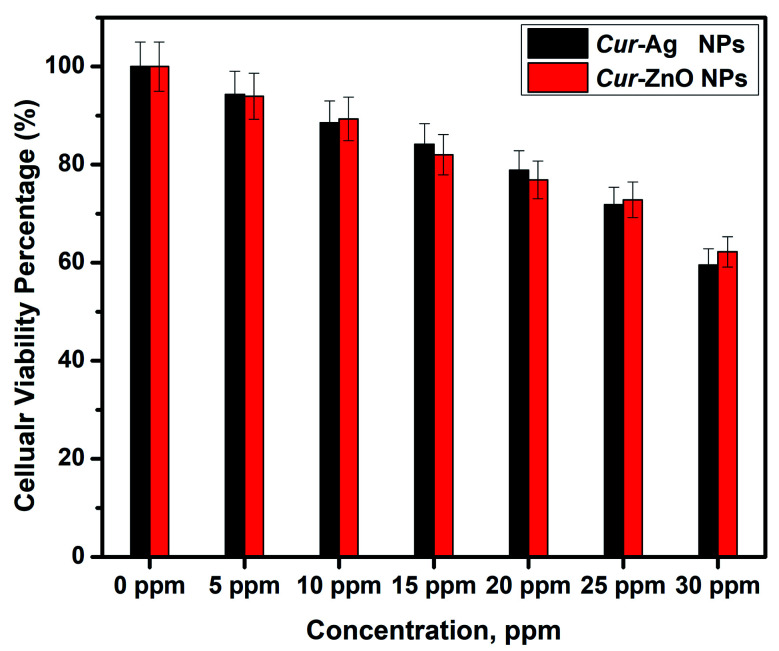
The biocompatibility assay was performed by exposing Vero cells as an in vitro test model of a normal cell line to a safety issue for both Cur-Ag NPs and Cur-ZnO NPs, respectively. In such a study, various concentrations of 10, 15, 20, 25, and 30 ppm from Cur-Ag NPs and Cur-ZnO NPs, respectively, are subjected to MTT screening colorimetric assay at 48 h time interval, as shown in Fig. 11. Our results showed that Cur-Ag NPs at the highest concentration of 30 ppm could kill about 40.5% of cells, whereas when applied at lower concentrations of 5 and 10 ppm killed only 5.7 and 11.5%, respectively, of cells after 48 h of cell exposure (Fig. 10). Treatment of cells with the highest concentration of 30 ppm Cur-ZnO NPs kills about 38.5% of cells, whereas treatment of cells with lower concentrations of 5 and 10 ppm kills about 6.5 and 10.7% of cells after 48 h of cell exposure (Fig. 11).
Fig. 11. Biocompatibility assay of Cur-Ag NPs and Cur-ZnO NPs at different levels of exposure doses.
4. Conclusions
In this study, Ag NPs and ZnO NPs were successfully green synthesized in the presence of Curcuma longa ethanolic extract forming Cur-AgNPs and Cur-ZnO NPs, respectively. The average particle sizes were about 15 ± 5 (Cur-AgNPs), and 20 ± 5 nm (Cur-ZnO NPs), with spherical like-shape. Cur-AgNPs and Cur-ZnO NPs showed excellent antibacterial efficacy against MDR-pathogenic bacterial isolates from the clinical sample. Curcumin-capped AgNPs exhibited solid antimicrobial action against the experimental bacterial isolates, except P. vulgaris. Whereas curcumin-capped ZnO NPs found antimicrobial activity against all tested strains. Finally, the minimum inhibitory concentration exhibited values from 3.9 to 15.6 μg ml−1, which is too small compared to other traditional antibiotics. Thus, the present green synthesized nanomaterials could act as effective antimicrobial agents and prove an alternative for developing new antimicrobial agents to combat the problem of antibiotic resistance. In addition, the green-synthesized Cur-Ag NPs and Cur-ZnO NPs showed good biocompatibility versus normal Vero cell lines.
Our findings indicate that Cur-Ag NPs and Cur-ZnO NPs showed a remarkable MIC and antimicrobial activity against both Gram-positive and Gram-negative pathogenic bacterial isolates compared to traditionally used antibiotics. In addition, the presence of curcumin in situ green synthesis of nanoparticles such as silver and zinc oxide nanoparticles facilitate to increase in the bio-aviablibty of curcumin than curcumin itself. In such a case, the cellular uptake efficiency of Cur-capped ZnO or Cur-Ag NPs is dramatically increased due to the increased solubility of curcumin after loading on the nanostructure, which increases the anti-inflammatory applications of nanoparticles.126 Also, their efficient antimicrobial activity allowed us to use wound healing applications, as reported by Gupta and co-workers.127 They reported that Cur-capped AgNPs upon loaded in bacterial cellulose-based (BC) hydrogels increase efficiency and minimize the potential failure of previously used precisely structured biomaterials. In addition, the high moisture content and the good level of transparency further advocate the potential application of Cur-AgNPs loaded in BC hydrogels in the management of chronic wounds with high microbial bioburden.127 Finally, and not the end; Stati et al. developed a new formulation of eye drops based on Cur-Ag NPs to be used in the in vivo treatment of human pterygium, which is known as an invasive and quite severe human disease, which is now difficult to treat, let alone eliminate.128 Furthermore, the biocompatibility assay results guide us to recommend the use of the as-green synthesized nanoparticles in disinfectants or to be used as an active ingredient in antimicrobial coating paints, which will be present in further studies and experiments.
Author contributions
Noura El-Kattan; conceptualization, perform the microbiological studies against +ve and −ve gram bacteria. Ahmed Nabile Emam (corresponding author); performing characterization experiments including UV-Vis optical absorption spectroscopy, TEM, XRD, FTIR, and dynamic light scattering (DLS) & zeta potential, data analysis, and formulating and revision of the manuscript. Ahmed Sadek Mansour; synthesis of nanomaterials. Mostafa Ahmed Ibrahim; synthesis of nanomaterials. Ashraf Bakry Abd El-Razik; conceptualization of the manuscript topic and supports the third and fourth authors with chemicals used in the fabrication of nanomaterials. Kamilia A. M. Allam and Nadia Youssef Riad; help the co-authors with the basic steps for getting the ethical approval and support the first author in collecting the clinical samples from patients, and Samir A. Ibrahim; assists the first author in the microbiological investigation studies.
Consent to publications
All authors accepted the publication of the manuscript in the present form.
Availability of data and materials
Not available.
Funding
There are no funds for this study (not applicable).
Ethical approval/consent to participate
This study was performed in strict accordance with the GOTHI guidelines, ethics regulations issued by the Minister of Health & Population, Cairo, Egypt: No. 238/2003, Articles 52–6121, approved by the medical research ethics committee. In addition, the approval for performing this study has been registered under No. IME 00057 on 25/8/2021 and is valid for one year.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors are grateful to the Faculty of Postgraduate Studies for Nanotechnology, Cairo University, for getting financial discounts for performing an X-Ray diffraction crystallography laboratory. In addition, special acknowledgement to NanoFab Technology for its support with the chemical used in the fabrications of nanomaterials used in this study.
Notes and references
- Jansen K. U. Anderson A. S. Hum. Vaccines Immunother. 2018;14:2142–2149. doi: 10.1080/21645515.2018.1476814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfensberger A. Kuster S. P. Marchesi M. Zbinden R. Hombach M. Antimicrob. Resist. Infect. Control. 2019;8:1–9. doi: 10.1186/s13756-018-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadgostar P. Infect. Drug Resist. 2019;12:3903. doi: 10.2147/IDR.S234610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista P. V. McCusker M. P. Carvalho A. Ferreira D. A. Mohan N. M. Martins M. Fernandes A. R. Front. Microbiol. 2018;9:1441. doi: 10.3389/fmicb.2018.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W. Zhang L. Nat. Rev. Microbiol. 2021;19:5–6. doi: 10.1038/s41579-020-00469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N.-Y. Ko W.-C. Hsueh P.-R. Front. Pharmacol. 2019;10:1153. doi: 10.3389/fphar.2019.01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachaiappan R. Rajendran S. Show P. L. Manavalan K. Naushad M. Chemosphere. 2021;272:128607. doi: 10.1016/j.chemosphere.2020.128607. [DOI] [PubMed] [Google Scholar]
- Ali H. R. Emam A. N. Koraney N. F. Hefny E. G. Ali S. F. J. Nanoparticle Res. 2020;22:1–15. doi: 10.1007/s11051-019-4718-8. [DOI] [Google Scholar]
- Hamed A. A. Kabary H. Khedr M. Emam A. N. RSC Adv. 2020;10:10361–10367. doi: 10.1039/C9RA11021F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanje A. S. Sharma S. J. Pode R. B. J. Chem. Pharm. Res. 2010;2:478–483. [Google Scholar]
- Salman H. D. J. Glob. Pharma Technol. 2017;9:238–248. [Google Scholar]
- Marin E. Boschetto F. Pezzotti G. J. Biomed. Mater. Res., Part A. 2020;108:1617–1633. doi: 10.1002/jbm.a.36930. [DOI] [PubMed] [Google Scholar]
- Singh A. Gautam P. K. Verma A. Singh V. Shivapriya P. M. Shivalkar S. Sahoo A. K. Samanta S. K. Biotechnol. Rep. 2020;25:e00427. doi: 10.1016/j.btre.2020.e00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous Z. Nemmar A. Int. J. Mol. Sci. 2020;21:2375. doi: 10.3390/ijms21072375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusof H. M. Mohamad R. Zaidan U. H. J. Anim. Sci. Biotechnol. 2019;10:1–22. doi: 10.1186/s40104-018-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I. Viswanathan K. Kasi G. Thanakkasaranee S. Sadeghi K. Seo J. Food Rev. Int. 2020:1–29. [Google Scholar]
- Gudkov S. V. Burmistrov D. E. Serov D. A. Rebezov M. B. Semenova A. A. Lisitsyn A. B. Front. Physiol. 2021;9:641481. doi: 10.3389/fphy.2021.641481. doi: 10.3389/fphy.2021.641481. [DOI] [Google Scholar]
- Van Giau V. An S. S. A. Hulme J. Drug Des. Dev. Ther. 2019;13:327. doi: 10.2147/DDDT.S190577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-López E. Gomes D. Esteruelas G. Bonilla L. Lopez-Machado A. L. Galindo R. Cano A. Espina M. Ettcheto M. Camins A. Nanomaterials. 2020;10:292. doi: 10.3390/nano10020292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Carmona M. Gun’Ko Y. Vallet-Regí M. Nanomaterials. 2018;8:268. doi: 10.3390/nano8040268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milionis A. Tripathy A. Donati M. Sharma C. S. Pan F. Maniura-Weber K. Ren Q. Poulikakos D. Ind. Eng. Chem. Res. 2020;59:14323–14333. doi: 10.1021/acs.iecr.0c01998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Saeed A. M. Abd El-Fattah M. Azzam A. M. Dyes Pigm. 2015;121:282–289. doi: 10.1016/j.dyepig.2015.05.037. [DOI] [Google Scholar]
- Goel A. Kunnumakkara A. B. Aggarwal B. B. Biochem. Pharmacol. 2008;75:787–809. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Xie M., Song Y., Wang W., Zhao H., Tian Y., Wang Y., Bai S., Zhao Y. and Chen X., Evidence-Based Complementary and Alternative Medicine, 2016, 2016 [Google Scholar]
- Hewlings S. J. Kalman D. S. Foods. 2017;6:92. doi: 10.3390/foods6100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karandish M. Mozaffari-Khosravi H. Mohammadi S. M. Azhdari M. Cheraghian B. Trials. 2020;21:1–11. doi: 10.1186/s13063-020-04923-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M. J. Pharm. Pharmacol. 1997;49:105–107. doi: 10.1111/j.2042-7158.1997.tb06761.x. [DOI] [PubMed] [Google Scholar]
- Brouet I. Ohshima H. Biochem. Biophys. Res. Commun. 1995;206:533–540. doi: 10.1006/bbrc.1995.1076. [DOI] [PubMed] [Google Scholar]
- Cikrikci S. Mozioglu E. Yilmaz H. Record Nat. Prod. 2008;2:19. [Google Scholar]
- Maheshwari R. K. Singh A. K. Gaddipati J. Srimal R. C. Life Sci. 2006;78:2081–2087. doi: 10.1016/j.lfs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Anand P. Kunnumakkara A. B. Newman R. A. Aggarwal B. B. Mol. Pharm. 2007;4:807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- Gupta S. C. Patchva S. Aggarwal B. B. AAPS J. 2013;15:195–218. doi: 10.1208/s12248-012-9432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan A. Senthil N. Min T. Front. Pharmacol. 2020;11:487. doi: 10.3389/fphar.2020.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi-Rad J. El Rayess Y. Abi Rizk A. Sadaka C. Zgheib R. Zam W. Sestito S. Rapposelli S. Neffe-Skocińska K. Zielińska D. Front. Pharmacol. 2020:11. doi: 10.3389/fphar.2020.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothitirat W. Gritsanapan W. J. Pharmaceut. Sci. 2005;32:23–30. [Google Scholar]
- Hashim F. J. Shawkat M. S. Al-Rikabi A. A. N. Malaysian J. Fund. Appl. Sci. 2013;9:105–109. [Google Scholar]
- Surojanametakul V. Satmalee P. Saengprakai J. Siliwan D. Wattanasiritham L. Agric. Nat. Resour. 2010;44:123–130. [Google Scholar]
- Alsammarraie F. K. Wang W. Zhou P. Mustapha A. Lin M. Colloids Surf. B Biointerfaces. 2018;171:398–405. doi: 10.1016/j.colsurfb.2018.07.059. [DOI] [PubMed] [Google Scholar]
- Viana A. V. Viana D. d. S. F. Figueirêdo G. S. d. Brito J. E. d. Viana V. G. F. Res., Soc. Dev. 2021;10:e11310615512. doi: 10.33448/rsd-v10i6.15512. [DOI] [Google Scholar]
- Muniyappan N. Pandeeswaran M. Amalraj A. J. Environ. Chem. Ecotoxicol. 2021;3:117–124. doi: 10.1016/j.enceco.2021.01.002. [DOI] [Google Scholar]
- Patra D. El Kurdi R. Green Chem. Lett. Rev. 2021;14:474–487. doi: 10.1080/17518253.2021.1941306. [DOI] [Google Scholar]
- Khalil M. I. Al-Qunaibit M. M. Al-Zahem A. M. Labis J. P. Arab. J. Chem. 2014;7:1178–1184. doi: 10.1016/j.arabjc.2013.10.025. [DOI] [Google Scholar]
- Jayandran M. Haneefa M. Int. J. Pharm. Sci. Res. 2016;7:4117–4124. [Google Scholar]
- Bekele B. Degefa A. Tesgera F. Jule L. T. Shanmugam R. Priyanka Dwarampudi L. Nagaprasad N. Ramasamy K. J. Nanomater. 2021:2021. [Google Scholar]
- Forbes B. A., Sahm D. F. and Weissfeld A. S., Study Guide for Bailey and Scott's Diagnostic Microbiology-E-Book, Elsevier Health Sciences, 2016 [Google Scholar]
- Forbes B., Sahm D. and Weissfeld A., Bailey and Scotts' Diagnostic Microbiology, Mosby, Missouri, 10th edn, 1998, pp. 134–149 [Google Scholar]
- Mueller J. H. Hinton J. Proc. Soc. Exp. Biol. Med. 1941;48:330–333. doi: 10.3181/00379727-48-13311. [DOI] [Google Scholar]
- Raja N. S. Singh N. N. J. Microbiol. Immunol. Infect. 2007;40:45–49. [PubMed] [Google Scholar]
- Standards N. C. f. C. L. and Barry A. L., Methods for determining bactericidal activity of antimicrobial agents: approved guideline, National Committee for Clinical Laboratory Standards Wayne, PA, 1999 [Google Scholar]
- O'Halloran C. Walsh N. O'Grady M. Barry L. Hooton C. Corcoran G. Lucey B. Br. J. Biomed. Sci. 2018;75:24–29. doi: 10.1080/09674845.2017.1392736. [DOI] [PubMed] [Google Scholar]
- Shoaib M. Satti L. Hussain A. Khursheed N. Sarwar S. Shah A. H. Cureus. 2021:13. doi: 10.7759/cureus.16777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries R. M. Ambler J. Mitchell S. L. Castanheira M. Dingle T. Hindler J. A. Koeth L. Sei K. J. Clin. Microbiol. 2018;56:e01934. doi: 10.1128/JCM.01934-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A.-P. Srinivasan A. Carey R. Carmeli Y. Falagas M. Giske C. Harbarth S. Hindler J. Kahlmeter G. Olsson-Liljequist B. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- El-Mahmood A. Doughari J. Afr. J. Pharmacy Pharmacol. 2009;3:185–190. [Google Scholar]
- Tawfick M. M. El-Borhamy M. I. Am. J. Microb. Res. 2017;5:37–43. [Google Scholar]
- Usman J. G. Sodipo O. Sandabe U. Int. J. Phytomed. 2014;6:268. [Google Scholar]
- Schmidt N. J., General principles of laboratory diagnostic methods for viral, rickettsial and chlamydial infections, 1995, pp. 1–35 [Google Scholar]
- Emam A. Loutfy S. A. Mostafa A. A. Awad H. Mohamed M. B. RSC Adv. 2017;7:23502–23514. doi: 10.1039/C7RA01423F. [DOI] [Google Scholar]
- Loutfy S. A. El-Din H. M. A. Elberry M. H. Allam N. G. Hasanin M. Abdellah A. M. Adv. Nat. Sci. Nanosci. Nanotechnol. 2016;7:035008. doi: 10.1088/2043-6262/7/3/035008. [DOI] [Google Scholar]
- Bruinink A. and Luginbuehl R., in Tissue Engineering III: Cell-Surface Interactions for Tissue Culture, Springer, 2011, pp. 117–152 [Google Scholar]
- Ruhane T. Islam M. T. Rahaman M. S. Bhuiyan M. Islam J. M. Newaz M. Khan K. Khan M. A. Optik. 2017;149:174–183. doi: 10.1016/j.ijleo.2017.09.024. [DOI] [Google Scholar]
- Stati G. Rossi F. Trakoolwilaiwan T. Tung L. D. Mourdikoudis S. Thanh N. T. K. Di Pietro R. Molecules. 2022;27:282. doi: 10.3390/molecules27010282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setyaningsih D. Murti Y. B. Fudholi A. Hinrichs W. L. Mudjahid R. Martono S. Hertiani T. Indonesian J. Pharmaceut. Sci. 2017;14:147–157. [Google Scholar]
- Barzinjy A. A. Azeez H. H. SN Appl. Sci. 2020;2:1–14. [Google Scholar]
- Selvan D. A. Mahendiran D. Kumar R. S. Rahiman A. K. J. Photochem. Photobiol. B Biol. 2018;180:243–252. doi: 10.1016/j.jphotobiol.2018.02.014. [DOI] [PubMed] [Google Scholar]
- Marimón-Bolívar W. González E. E. Dyna. 2018;85:19–26. doi: 10.15446/dyna.v85n205.68245. [DOI] [Google Scholar]
- Bam B. A. M., Bacteriological Analytical Manual, 2001, pp. 53–67 [Google Scholar]
- Chambers H. F. DeLeo F. R. Nat. Rev. Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu K. Katayama Y. Matsuo M. Sasaki T. Morimoto Y. Sekiguchi A. Baba T. J. Infect. Chemother. 2014;20:593–601. doi: 10.1016/j.jiac.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Asadollahi P. Razavi S. Asadollahi K. Pourshafie M. Talebi M. J. Infect. Chemother. 2018;26:92–99. doi: 10.1016/j.nmni.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelkareem M. Z. Sayed M. Hassuna N. A. Mahmoud M. S. Abdelwahab S. F. J. Chemother. 2017;29:74–82. doi: 10.1080/1120009X.2016.1182358. [DOI] [PubMed] [Google Scholar]
- Gaglio R. Couto N. Marques C. Lopes M. d. F. S. Moschetti G. Pomba C. Settanni L. Int. J. Food Microbiol. 2016;236:107–114. doi: 10.1016/j.ijfoodmicro.2016.07.020. [DOI] [PubMed] [Google Scholar]
- Raafat S. A. J. Med. Microbiol. 2016;25:47–55. [Google Scholar]
- Pesavento G. Calonico C. Ducci B. Magnanini A. Nostro A. L. Food Microbiol. 2014;41:1–7. doi: 10.1016/j.fm.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Pieniz S. de Moura T. M. Cassenego A. P. V. Andreazza R. Frazzon A. P. G. de Oliveira Camargo F. A. Brandelli A. Food Control. 2015;51:49–54. doi: 10.1016/j.foodcont.2014.11.012. [DOI] [Google Scholar]
- Mohamed A. Abdelhamid F. Zagazig J. Pharmaceut. Sci. 2020;28:10–17. [Google Scholar]
- Bhalchandra M. Naik S. Verma P. K. Int. J. Curr. Microbiol. Appl. Sci. 2018;7:1668–1679. doi: 10.20546/ijcmas.2018.706.198. [DOI] [Google Scholar]
- Tonkic M. Goic Barisic I. Punda-Polic V. Int. Microbiol. 2005;8:119–124. [PubMed] [Google Scholar]
- Naqid I. A. Hussein N. R. Balatay A. A. Saeed K. A. Ahmed H. A. J. Kermanshah Univ. Med. Sci. 2020:24. [Google Scholar]
- Gomatheswari S. N. Jeyamurugan T. Int. J. Curr. Microbiol. Appl. Sci. 2017;6:1405–1413. doi: 10.20546/ijcmas.2017.610.166. [DOI] [Google Scholar]
- Babakhani S. Shokri S. Baharvand M. Rep. Health Care. 2015;1:55–59. [Google Scholar]
- Perez F. Hujer A. M. Hujer K. M. Decker B. K. Rather P. N. Bonomo R. A. Antimicrob. Agents Chemother. 2007;51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergogne-Berezin E. Towner K. Clin. Microbiol. Rev. 1996;9:148–165. doi: 10.1128/CMR.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayyaz M. Khan I. U. Hussain A. Mirza I. A. Ali S. Akbar N. J. Coll. Phys. Surg. Pakistan. 2015;25:346–349. [PubMed] [Google Scholar]
- Daftary N. Patel D. Saudi J. Pathol. Microbiol. 2020;5:512–515. doi: 10.36348/sjpm.2020.v05i12.009. [DOI] [Google Scholar]
- Newman M. J. Frimpong E. Donkor E. S. Opintan J. A. Asamoah-Adu A. Infect. Drug Resist. 2011;4:215. doi: 10.2147/IDR.S21769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A. Nandi S. Dhar P. MGM J. Med. Sci. 2017;4:10–18. doi: 10.5005/jp-journals-10036-1130. [DOI] [Google Scholar]
- Yakha J. K. Sharma A. R. Dahal N. Lekhak B. Banjara M. R. Nepal J. Sci. Technol. 2014;15:91–96. doi: 10.3126/njst.v15i2.12121. [DOI] [Google Scholar]
- Rai M. K. Deshmukh S. Ingle A. Gade A. J. Appl. Microbiol. 2012;112:841–852. doi: 10.1111/j.1365-2672.2012.05253.x. [DOI] [PubMed] [Google Scholar]
- Nanda A. Saravanan M. Nanomed.: Nanotechnol. Biol. Med. 2009;5:452–456. doi: 10.1016/j.nano.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Lara H. H. Ayala-Núnez N. V. Turrent L. d. C. I. Padilla C. R. World J. Microbiol. Biotechnol. 2010;26:615–621. doi: 10.1007/s11274-009-0211-3. [DOI] [Google Scholar]
- Bonde S. Rathod D. Ingle A. Ade R. Gade A. Rai M. Nanosci. Methods. 2012;1:25–36. doi: 10.1080/17458080.2010.529172. [DOI] [Google Scholar]
- Abbaszadegan A. Ghahramani Y. Gholami A. Hemmateenejad B. Dorostkar S. Nabavizadeh M. Sharghi H. J. Nanomater. 2015;2015:1–8. doi: 10.1155/2015/720654. [DOI] [PubMed] [Google Scholar]
- Salas-Orozco M. Niño-Martínez N. Martínez-Castañón G.-A. Méndez F. T. Jasso M. E. C. Ruiz F. J. Nanomater. 2019;2019:1–11. [Google Scholar]
- Mohandass C. Vijayaraj A. Rajasabapathy R. Satheeshbabu S. Rao S. Shiva C. De-Mello I. Indian J. Pharmaceut. Sci. 2013;75:606. [PMC free article] [PubMed] [Google Scholar]
- Tormena R. P. I. Rosa E. V. Mota B. d. F. O. Chaker J. A. Fagg C. W. Freire D. O. Martins P. M. da Silva I. C. R. Sousa M. H. RSC Adv. 2020;10:20676–20681. doi: 10.1039/D0RA03240A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adebayo-Tayo B. Salaam A. Ajibade A. Heliyon. 2019;5:e02502. doi: 10.1016/j.heliyon.2019.e02502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N. Kumar D. Kumar P. Colloid Interface Sci. Commun. 2020;35:100242. doi: 10.1016/j.colcom.2020.100242. [DOI] [Google Scholar]
- Cavassin E. D. de Figueiredo L. F. P. Otoch J. P. Seckler M. M. de Oliveira R. A. Franco F. F. Marangoni V. S. Zucolotto V. Levin A. S. S. Costa S. F. J. Nanobiotechnol. 2015;13:1–16. doi: 10.1186/s12951-015-0120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namasivayam S. Ganesh S. Avimanyu B. Int. J. Med. Res. 2011;1:131–136. [Google Scholar]
- Ansari M. Khan H. Khan A. Sultan A. Azam A. Shahid M. Shujatullah F. Int. J. Appl. Biol. Pharm. Technol. 2011;2:34–42. [Google Scholar]
- Durán N. Durán M. De Jesus M. B. Seabra A. B. Fávaro W. J. Nakazato G. Nanomed.: Nanotechnol. Biol. Med. 2016;12:789–799. doi: 10.1016/j.nano.2015.11.016. [DOI] [PubMed] [Google Scholar]
- Percival S. L. Bowler P. G. Dolman J. Int. Wound J. 2007;4:186–191. doi: 10.1111/j.1742-481X.2007.00296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashajyothi C. Manjunath N. R. Kelmani C. Int. J. ChemTech Res. 2014;6:3131–3136. [Google Scholar]
- Wahab R. Kim Y.-S. Mishra A. Yun S.-I. Shin H.-S. Nanoscale Res. Lett. 2010;5:1675–1681. doi: 10.1007/s11671-010-9694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadiyala U. Turali-Emre E. S. Bahng J. H. Kotov N. A. VanEpps J. S. Nanoscale. 2018;10:4927–4939. doi: 10.1039/C7NR08499D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozyen H. Ibrahim E. Khairy E. El-Dek S. Vet. World. 2019;12:1225. doi: 10.14202/vetworld.2019.1225-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MH Al-Kordy H. Sabry S. A. Mabrouk M. E. M. Egypt. J. Aquat. Biol. Fish. 2020;24:43–56. doi: 10.21608/ejabf.2020.94732. [DOI] [Google Scholar]
- Bajaj H. Acosta Gutierrez S. Bodrenko I. Malloci G. Scorciapino M. A. Winterhalter M. Ceccarelli M. ACS Nano. 2017;11:5465–5473. doi: 10.1021/acsnano.6b08613. [DOI] [PubMed] [Google Scholar]
- Shinde V. V. Dalavi D. S. Mali S. S. Hong C. K. Kim J. H. Patil P. S. Appl. Surf. Sci. 2014;307:495–502. doi: 10.1016/j.apsusc.2014.04.064. [DOI] [Google Scholar]
- Jin S.-E. Jin H.-E. nanomaterials. 2021;11:263. doi: 10.3390/nano11020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavin Y. N. Asnis J. Häfeli U. O. Bach H. J. Nanobiotechnol. 2017;15:1–20. doi: 10.1186/s12951-017-0308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumya K. Snigdha S. Sugathan S. Mathew J. Radhakrishnan E. 3 Biotech. 2017;7:1–10. doi: 10.1007/s13205-017-0861-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z. Wu Y. Wang H. Han H. Mater. Sci. Eng. C. 2019;99:255–263. doi: 10.1016/j.msec.2018.12.053. [DOI] [PubMed] [Google Scholar]
- Alves T. F. Chaud M. V. Grotto D. Jozala A. F. Pandit R. Rai M. Dos Santos C. A. Aaps Pharmscitech. 2018;19:225–231. doi: 10.1208/s12249-017-0832-z. [DOI] [PubMed] [Google Scholar]
- Perera W. Dissanayake R. K. Ranatunga U. Hettiarachchi N. Perera K. Unagolla J. M. De Silva R. Pahalagedara L. RSC Adv. 2020;10:30785–30795. doi: 10.1039/D0RA05755J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teow S.-Y. Liew K. Ali S. A. Khoo A. S.-B. Peh S.-C. J. Trop. Med. 2016:2016. doi: 10.1155/2016/2853045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morão L. G. Polaquini C. R. Kopacz M. Torrezan G. S. Ayusso G. M. Dilarri G. Cavalca L. B. Zielińska A. Scheffers D. J. Regasini L. O. MicrobiologyOpen. 2019;8:e00683. doi: 10.1002/mbo3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi-Derazkola S. Ebrahimzadeh M. A. Amiri O. Goli H. R. Rafiei A. Kardan M. Salavati-Niasari M. J. Alloys Compd. 2020;820:153186. doi: 10.1016/j.jallcom.2019.153186. [DOI] [Google Scholar]
- Lkhagvajav N. Yasa I. Celik E. Koizhaiganova M. Sari O. Dig. J. Nanomater. Biostructures. 2011;6:149–154. [Google Scholar]
- Jaiswal S. Mishra P. Med. Microbiol. Immunol. 2018;207:39–53. doi: 10.1007/s00430-017-0525-y. [DOI] [PubMed] [Google Scholar]
- Hassani S. M. Nakhaei M. M. Forghanifard M. M. Nanomed. J. 2015;2:121–128. [Google Scholar]
- Shinde S. S. Sci. Med. Central. 2015;3:1033. [Google Scholar]
- Aflatoonian M. Khatami M. Sharifi I. Pourseyedi S. Yaghobi H. Naderifar M. Tehran Univ. Med. J. 2017;75:562–569. [Google Scholar]
- Syed H. K. Iqbal M. A. Haque R. A. Peh K.-K. J. Coord. Chem. 2015;68:1088–1100. doi: 10.1080/00958972.2014.1003051. [DOI] [Google Scholar]
- Somu P. Paul S. New J. Chem. 2019;43:11934–11948. doi: 10.1039/C9NJ02501D. [DOI] [Google Scholar]
- Gupta A. Briffa S. M. Swingler S. Gibson H. Kannappan V. Adamus G. Kowalczuk M. Martin C. Radecka I. Biomacromolecules. 2020;21:1802–1811. doi: 10.1021/acs.biomac.9b01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stati G. Rossi F. Trakoolwilaiwan T. Tung L. D. Mourdikoudis S. Thanh N. T. K. Di Pietro R. Molecules. 2022;27:282. doi: 10.3390/molecules27010282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not available.