Abstract
Antibiotic resistance plasmids were exogenously isolated in biparental matings with piggery manure bacteria as plasmid donors in Escherichia coli CV601 and Pseudomonas putida UWC1 recipients. Surprisingly, IncQ-like plasmids were detected by dot blot hybridization with an IncQ oriV probe in several P. putida UWC1 transconjugants. The capture of IncQ-like plasmids in biparental matings indicates not only their high prevalence in manure slurries but also the presence of efficiently mobilizing plasmids. In order to elucidate unusual hybridization data (weak or no hybridization with IncQ repB or IncQ oriT probes) four IncQ-like plasmids (pIE1107, pIE1115, pIE1120, and pIE1130), each representing a different EcoRV restriction pattern, were selected for a more thorough plasmid characterization after transfer into E. coli K-12 strain DH5α by transformation. The characterization of the IncQ-like plasmids revealed an astonishingly high diversity with regard to phenotypic and genotypic properties. Four different multiple antibiotic resistance patterns were found to be conferred by the IncQ-like plasmids. The plasmids could be mobilized by the RP4 derivative pTH10 into Acinetobacter sp., Ralstonia eutropha, Agrobacterium tumefaciens, and P. putida, but they showed diverse patterns of stability under nonselective growth conditions in different host backgrounds. Incompatibility testing and PCR analysis clearly revealed at least two different types of IncQ-like plasmids. PCR amplification of total DNA extracted directly from different manure samples and other environments indicated the prevalence of both types of IncQ plasmids in manure, sewage, and farm soil. These findings suggest that IncQ plasmids play an important role in disseminating antibiotic resistance genes.
Plasmid-mediated gene exchange between bacteria plays an important role in bacterial adaptation and flexibility. Plasmids have greatly contributed to the rapid spread of antibiotic resistance genes in bacterial populations due to antibiotic selective pressures resulting from the intensive use of antibiotics in human therapy and agriculture (24, 48, 53, 54). However, our knowledge of the prevalence and diversity of plasmids in bacteria from different environments is very limited. Moreover, the screening of bacteria isolated by standard cultivation techniques for the presence of plasmids only allows for analysis of a small proportion of bacteria accessible to standard cultivation techniques (39). Systematic studies on the prevalence and diversity of plasmids in different environmental niches have not yet been performed. The application of new approaches, such as the exogenous plasmid isolation techniques (2, 19) or the PCR-based detection of mobile genetic elements in total community DNA (6, 13, 37), now broadens our view of the plasmid pool present in bacteria from different environmental habitats. Furthermore, PCR and DNA hybridization were shown to be extremely valuable tools for plasmid characterization and classification (5, 13, 27, 37, 38, 42, 49).
IncQ plasmids are small mobilizable plasmids which have an extremely broad host range, including gram-negative and even gram-positive bacteria (1, 9, 11). Plasmid mobilization frequency varies with the mobilizing plasmid employed, and mobilization is particularly efficient when IncP, IncIα, IncM, and IncX are used as mobilizers (14). The molecular biology of the IncQ plasmid RSF1010 and the similar, if not identical, plasmids R300B and R1162 has been studied intensively (9, 16, 33). RSF1010-like plasmids have been detected in numerous gram-negative genera of commensal and pathogenic bacteria (9). However, limited information is available on the prevalence and the genetic diversity of IncQ plasmids, particularly in bacteria of different environments. Recently, primers for PCR amplification of IncQ-specific sequences were shown to be well suited for the detection of IncQ-plasmids in environmental bacteria and DNA extracted directly from different environments (12, 13). Oligonucleotide primers to amplify different regions of the plasmid backbone were designed on the basis of the complete sequence of RSF1010 (33). Seven IncQ plasmids analyzed in the study of Götz et al. (13) yielded PCR products of the expected sizes with primer pairs specific for oriV, repB, and oriT of RSF1010, respectively. PCR amplification of IncQ oriV and repB sequences from total community DNA extracted from manure and different soils showed that IncQ-like plasmids might be relatively abundant in manure slurries (13). However, a further characterization of those plasmids depends upon their isolation.
Here we describe the capture of antibiotic resistance plasmids exogenously isolated in biparental matings with piggery manure bacteria as plasmid donors in Escherichia coli CV601 and Pseudomonas putida UWC1. IncQ-like plasmids in P. putida UWC1 transconjugants were identified by dot blot hybridization with an IncQ oriV probe prepared by PCR from RSF1010. However, some of these plasmids showed no or only weak hybridizations with IncQ repB or IncQ oriT probes. In order to elucidate these unusual hybridization data, four out of these IncQ-like plasmids, each representing a unique EcoRV restriction pattern different from that of the IncQ “type plasmid” RSF1010, were selected for a more thorough plasmid characterization. One of these plasmids, pIE1107, has been completely sequenced, and its unusual unidirectional incompatibility towards RSF1010 has been elucidated recently (43). Briefly summarized, pIE1107 contains two copies of the origin of vegetative replication (oriV) of IncQ plasmids. These two copies of IncQ-oriV DNA are slightly different from each other. One of them (oriVα) is identical with the oriV of RSF1010 but is dispensable for replication of pIE1107. This follows from the regular replication of a deletion derivative of pIE1107, pIE1108, which contains only oriVβ. Moreover, pIE1108 and RSF1010 stably coexist in a bacterial host cell, indicating that the two plasmids have distinct, compatible replicons. Therefore, pIE1107 was considered an IncQβ plasmid in order to distinguish it from the RSF1010-type IncQα replicons (43). From the nucleotide sequence of pIE1107 additional primer sets were derived and tested for the amplification of IncQβ-specific plasmid DNA. The complete nucleotide sequence of another plasmid reported here, pIE1130, was recently deposited in the database under EMBL, GenBank, and DBJ accession number AJ271879 (E. Tietze and K. Smalla, unpublished data). PCR analysis, DNA-hybridization, incompatibility testing and host range studies were used to compare the new IncQ-like plasmids pIE1130, pIE1120 and pIE1115 with the IncQα plasmid RSF1010 and the IncQβ plasmid pIE1107. Moreover, PCR amplification of total DNA extracted directly from different manure samples and other environments was performed to provide data on the prevalence of IncQ-like plasmids in these samples.
MATERIALS AND METHODS
Strains and plasmids.
P. putida UWC1 (Rifr) and E. coli K-12 CV601 (Rifr Thr− Leu− Thi−) were used as recipients in exogenous plasmid isolations. P. putida UWC1 was obtained from J. Fry, Cardiff, Wales. E. coli CV601 was provided by H. Tschäpe, Wernigerode, Germany. Both recipients are restriction-negative strains. Plasmids isolated in P. putida UWC1 were transferred into E. coli K-12 strain DH5α by transformation (32). E. coli DH5α served as the general host for incompatibility testing, preparation of plasmid DNA, and determining antibiotic resistance patterns conferred by the plasmids. Rifampin (RIF)-resistant P. putida UWC1, Agrobacterium tumefaciens DSM30150, Ralstonia eutropha JMP 228, and Acinetobacter sp. strain BD413 (23) (obtained from J. D. van Elsas, Wageningen, The Netherlands) were used as recipients for plasmid host range determinations. E. coli J53(pTH10) (15) (obtained from H. Tschäpe) was used as a helper strain in triparental filter matings. Plasmids pMMB600 and RSF1010 were obtained from M. Bagdasarian. While the IncQ prototype RSF1010 is a natural plasmid, pMMB600 is an RSF1010 derivative lacking the sul and str genes but containing a bla gene conferring an ampicillin resistance (10). Plasmid pIE1108 is an in vitro-generated deletion derivative of the natural IncQ-like plasmid pIE1107, which lacks the IncQα-specific incompatibility determinant (oriVα) of the parental plasmid (43).
Media and growth conditions.
Putative P. putida UWC1 and E. coli CV601 transconjugants were isolated from M9 medium (32) supplemented with calcium pantothenate (1 μg/ml), nicotinamide (1 μg/ml), and thiamine (1 μg/ml) or leucine (20 μg/ml), threonine (20 μg/ml), and thiamine (2 μg/ml), respectively, and the following antibiotics: rifampicin (RIF) (100 μg/ml) and either streptomycin (SM) (50 μg/ml), kanamycin (KM) (100 μg/ml), gentamicin (GM) (20 μg/ml), tetracycline (TC) (50 μg/ml) or streptothricin (ST) (100 μg/ml). In addition, cycloheximide (200 μg/ml) was added to the selective media to suppress fungal growth. Mueller-Hinton agar (Merck, Darmstadt, Germany) was used to determine plasmid-conferred antibiotic resistances. Yeast extract medium (peptone, 15 g; yeast extract, 10 g; NaCl, 6 g; agar, 13 g; H2O, 1 liter [pH 7.5]) was used for biparental matings. E. coli strains containing the respective plasmids and recipient strains were grown in Luria-Bertani (LB) (32) broth overnight at 28°C with agitation (200 rpm). Plate count agar (PCA) (Merck) supplemented with SM (20 μg/ml) or ST (100 μg/ml) was used in host range studies.
Exogenous plasmid isolation in biparental matings.
Exogenous plasmid isolations were essentially done as described previously (2, 21). The bacterial fraction of manure was obtained by stomacher treatment (Seward Medical) of manure resuspended in sterile saline. Debris were removed by a low-speed centrifugation (2 min at 500 × g and 20°C). The bacteria were harvested from the supernatant (20 min at 5,000 × g and 20°C). The pellet was resuspended in LB broth and used as plasmid donor, while RIF-resistant P. putida UWC1 or E. coli CV601 (grown overnight in 5 ml of nutrient broth) served as the recipient in biparental matings. One milliliter each of donor cells and recipient cells was mixed and plated onto a yeast extract agar plate. Controls were made by incubating the bacterial fraction from manure slurry and the recipient separately. After overnight growth, the cell lawn was resuspended in 5 ml of sterile saline, and transconjugants were obtained after plating serial 10-fold dilutions on selective media. After 48 h incubation at 28°C CFU were counted and putative transconjugants picked for further characterization.
Verification of recipients.
Transconjugants obtained after biparental mating were confirmed by repetitive extragenic palindromic (REP) fingerprints obtained using the primers and PCR conditions described by Rademaker et al. (30). Genomic DNA extracted from freshly grown colonies according to Wilson (52) was used as a template.
Antibiotic resistance patterns.
Plasmid-encoded antibiotic resistances were determined by disk diffusion assay according to the manufacturer's specification (BioMérieux, Mary l'Etoile, France). Appropriate dilutions of E. coli CV601 or P. putida UWC1 transconjugants or E. coli DH5α cells containing the plasmids pIE1107, pIE1115, pIE1120, and pIE1130 were plated onto Mueller-Hinton agar, and disks containing the antibiotics ampicillin (10 μg), chloramphenicol (CM) (30 μg), GM (10 μg), KM (30 μg), SM (10 μg), sulfonamide (SU) (25 μg), TC (30 μg), and ST (100 μg) were applied with the BioMérieux distributor 8C. Inhibition zones were measured after 24 h of incubation at 28°C and compared with respective zones observed for E. coli CV601, P. putida UWC1, or E. coli DH5α.
Dot blot and Southern blot analysis.
Plasmid DNA extracted from transconjugants was analyzed by dot blot hybridizations for the presence of broad-host-range plasmids using PCR generated probes (13). Dot and Southern blotting was performed by standard protocols (32). Hybridizations of Southern- or dot-blotted plasmid DNA or PCR products with digoxigenin-labeled probes were performed according to the manufacturer's instructions (Boehringer, Mannheim, Germany).
Transformation into E. coli DH5α.
Prior to further characterization, the exogenously isolated plasmids in P. putida were introduced into E. coli DH5α using the standard CaCl2 transformation protocol (32).
Plasmid isolations.
Plasmid DNA was extracted according to the method of Holmes and Quigley (22) or according to the method of Birnboim and Doly (3) modified as follows. A loop of material from a freshly grown colony was resuspended in 2 ml of MgSO4 and centrifuged at 3,000 × g for 5 min. Pellets were resuspended in 200 μl of lysozyme solution (5 mg in 25 mM Tris, 50 mM glucose, 10 mM EDTA) and incubated for 30 min at 37°C. After addition of 400 μl of 0.2 M NaOH–1% sodium dodecyl sulfate, the lysates were incubated on ice for 5 min. Subsequently, 300 μl of potassium acetate (pH 5.2; 5 M) was added and carefully mixed, and the mixture was kept on ice for another 15 min before centrifugation at 15,000 × g for 20 min. The lysate was then extracted with 900 μl of phenol (pH 8)-chloroform followed by a chloroform extraction. DNA was precipitated with 0.7 volume of isopropanol and kept at room temperature for 1 h. The pellet obtained by centrifugation at 25,000 × g for 30 min was washed with 70% ethanol.
Characterization of IncQ-like plasmids by PCR. (i) Replicon-related sequences.
Using the strategy described by Götz et al. (13), two additional primer sets designed on the basis of the published sequences of RSF1010 (33) and pIE1107 (43) were used in addition to the recently published IncQ primer sets (Table 1). Primer sequences, the annealing temperatures, and sizes of the PCR products are summarized in Table 1. PCR was performed with either heat-denatured cells (1 μl of a 1:10 diluted overnight broth) or plasmid DNA as template. PCR mixtures contained Taq polymerase (Stoffel fragment; Perkin-Elmer), Stoffel buffer, a 0.2 mM concentration of each deoxynucleoside triphosphate, 3.75 mM MgCl2, and 0.2 μmol of each primer. After a 5-min denaturation step at 94°C, 35 cycles consisting of 1 min at 94°C, 1 min of primer annealing, and 1 min at 72°C were performed, followed by a final 10-min extension step at 72°C. PCR products (10 μl) were analyzed on 1% agarose gels with Tris-borate-EDTA buffer, Southern blotted (32), and hybridized using digoxigenin-labeled probes. Probes were generated by PCR using a deoxynucleoside triphosphate mixture containing 0.13 mM dTTP and 0.07 mM digoxigenin-labeled dUTP (Boehringer).
TABLE 1.
Primer systems used to amplify different parts of the IncQ replicon and of the SM resistance genes strA and strB
| Primer | Primer sequence (5′-3′)a | Annealing temp (°C) | Product size (bp) | Primer position | Accession no. (reference) |
|---|---|---|---|---|---|
| PI | CTC CCG TAC TAA CTG TCA CG | 57 | 436 | 2328 | M2882935 |
| RSF1010 oriV | ATC GAC CGA GAC AGG CCC TGC | 2743 | |||
| PII | TTC GCG CTC GTT GTT CTT CGA GC | 57 | 191 | 2988 | M28829 |
| RSF1010 oriT | GCC GTT AGG CCA GTT TCT CG | 3159 | |||
| PIII | TCG TGG TCG CGT TCA AGG TAC G | 62 | 1,160 | 2816 | M28829 |
| RSF1010 repB | CTG TAA GTC GAT GAT CTG GGC GTT | 3952 | |||
| PIV | GAC ATG AGG GCA GCC GCA GGG GAA C | 60 | 1,476 | Z7478745 | |
| pIE1107 mobA | CGC CAT GCG CCG CCT AGT CAG TG | ||||
| PV | CGG TGA TGA CCT CAG CAA GTG CGA C | 65 | 1,156/1,003 | ||
| RSF1010/pIE1107 | GGC CGT CGA ACC ACT CCG GG | Z74787 | |||
| mobA/repA | |||||
| strA | GCC CCG AGG TCA TCA ACT | 54 | 469 | M28829 | |
| TTC TCT TCG GCG TTA GCA | |||||
| strB | CTG TTT TTC CTG CTC ATT | 54 | 613 | M28829 | |
| CAA AGG TCG TCT CTG TCA | |||||
| strA-strB | AAC GCC GAA GAG AAC TGG | 54 | 186 | M28829 | |
| AGG TGT CCG CAA TGA GAA |
In each pair of sequences, the first and second sequences listed are forward and reverse, respectively.
(ii) Detection of SM resistance genes strA and strB.
Based on the sequence of RSF1010 (33), two sets of primers were selected to amplify parts of either the SM resistance genes strA or strB and a third set with the forward primer annealing in the strA and the reverse primer in the strB gene (Table 1).
Incompatibility testing.
Two methods of incompatibility testing were used as described by Tietze (43). The ability of an incoming plasmid to establish in a host which already contains another, resident plasmid was measured in reciprocal transformation experiments by comparing the efficiency of transformation with that of a plasmid-free recipient (set at 100%). In case of transformants which contained two plasmids several colonies were tested for segregation of plasmid markers under nonselective conditions.
Host range determination.
The host range of the plasmids under investigation (Table 2) was determined in triparental matings with E. coli DH5α(pTH10) as helper, and the RIF-resistant recipients A. tumefaciens DSM30150 (α-proteobacterium), R. eutropha JMP 228 (β-proteobacterium), Acinetobacter sp. strain BD413 (γ-proteobacterium), and P. putida UWC1 (γ-proteobacterium). All strains used in matings were grown in nutrient broth supplemented with the appropriate antibiotics to the late log phase (∼20 h) and harvested by centrifugation at 14,000 × g for 5 min. The cell pellets were resuspended in fresh nutrient broth. One hundred microliters of each donor and helper was mixed with 500 μl of recipient, and the mixture was centrifuged at 14,000 × g for 5 min. The pellet resuspended in 100 μl of LB broth was applied to a Millipore filter (pore size, 0.22 μm, type GV) placed on yeast extract agar. After overnight incubation the filter was resuspended in 10 ml of sterile saline by vigorous vortexing, and 10-fold serial dilutions were plated on PCA supplemented with RIF (50 μg/ml) to determine the number of recipients. Transconjugants were selected on PCA supplemented with RIF (50 μg/ml) and SM (50 μg/ml) for the matings with RSF1010-, pIE1120-, pIE1115-, and pIE1130-containing donors and on PCA with RIF (50 μg/ml) and ST (50 μg/ml) for matings with donors containing pIE1107. CFU numbers were determined after 2 days' incubation at 28°C. Transconjugants were verified by their REP-PCR fingerprints.
TABLE 2.
EcoRV restriction fragments, size, and antibiotic resistances conferred by IncQ-like plasmids
To determine the stability of the plasmids in A. tumefaciens DSM30150, R. eutropha JMP228, Acinetobacter sp. strain BD413, P. putida UWC1, and E. coli DH5α under nonselective growth conditions, two colonies of each host grown on selective plates were randomly picked and used to inoculate 10 ml of nutrient broth. After ∼20 h of incubation at 28°C, serial 10-fold dilutions were plated onto PCA and 10 μl of the culture was added to 10 ml of fresh nutrient broth. Plating onto PCA and inoculation of a fresh nutrient broth was performed for another round. Twenty-four colonies were picked from dilutions with approximately 50 to 100 colonies and streaked on PCA (SM50 RIF50 or ST50 RIF50, respectively). After 2 days' incubation the percentage of colonies grown on the selective medium was determined. To check for the presence of pTH10 and the IncQ-like plasmids, Southern-blotted plasmid DNA was hybridized with PCR-generated probes for IncPα trfA2 and IncQ oriV according to the method of Götz et al. (13).
Community DNA extraction.
Total DNA directly extracted from different environments was used. DNA from manure and rhizosphere samples was extracted and purified essentially as described by Smalla et al. (35). Briefly, the DNA was extracted from manure slurries and soil and rhizosphere samples by lysozyme, bead beating, and alkaline sodium dodecyl sulfate treatments followed by phenol, phenol-chloroform, and chloroform extraction. The crude DNA was purified by two salt precipitation steps (cesium chloride and potassium acetate precipitation) and Wizard DNA cleanup (Promega, Madison, Wis.) (50). Further DNA samples extracted directly from different environments were provided by the participants of an EU-funded MECBAD (Mobile Genetic Elements' Contribution to Bacterial Adaptability and Diversity) workshop (for further information see http://mecbad.bba.de). PCR amplifiability was checked using 16S ribosomal DNA primers (F984GC, R1378) and denaturing gradient gel electrophoresis analysis as described by Heuer et al. (17, 18).
RESULTS
Exogenous isolation of antibiotic resistance plasmids from piggery manure.
Mobile genetic elements conferring antibiotic resistances were exogenously isolated from manure bacteria with P. putida UWC1 and E. coli CV601 as recipients. Putative transconjugants were obtained on selective media supplemented with SM, TC, KM, GM, or ST. CM-resistant transconjugants were obtained only with E. coli CV601 because P. putida UWC1 is intrinsically resistant to CM. Frequencies of antibiotic resistance gene transfers were 1.7 × 10−9 to 9.3 × 10−8 for nonenriched manure. Transfer frequencies were much higher (1.1 × 10−5 to 3.4 × 10−7) with manure samples that had been enriched with LB broth prior to the mating overnight at 28°C. Background growth required that putative transconjugants be confirmed by REP-PCR fingerprinting by comparing the fingerprints with those of the respective recipients. Antibiotic resistance patterns of the transconjugants and the respective recipient determined by the disk diffusion method showed that most of the transconjugants acquired multiple antibiotic resistances. Transferable elements captured in E. coli CV601 conferred resistance to CM (98.5%), TC (84%), SM (73%), SU (70%), KM (41%), and ampicillin (70%) in a total of 70 transconjugants analyzed. Resistance to SM, TC, and KM was detected for 88, 48, and 36%, respectively in a collection of 130 P. putida UWC1 transconjugants. Plasmid DNAs extracted from transconjugants were screened by dot blot hybridizations using PCR-generated probes (13) to assess whether these belonged to plasmids of the IncQ, IncN, IncP, and IncW groups. Only two plasmids isolated in E. coli CV601 hybridized with the IncN probe; one plasmid isolated in P. putida UWC1 gave a strong hybridization with the IncPα probe. However, a number of plasmids captured in P. putida UWC1 after a biparental mating with bacteria from fresh manure and isolated on selective media supplemented with either KM, SM, TC, or ST showed DNA homology with the IncQ oriV probes. None of the plasmids isolated in E. coli CV601 exhibited homology to the IncQ oriV probe. Hybridization with the IncQ oriT probe indicated the absence of oriT-specific sequences in several plasmids that gave strong hybridization signals with the IncQ oriV probe and less-intensive hybridization with the IncQ repB probe. Hybridization results indicated that the plasmids exogenously isolated in P. putida UWC1 were related to IncQ plasmids but showed an unusual variability in IncQ backbone DNA. Therefore, after transfer of these plasmids to E. coli DH5α, they were further characterized with respect to their molecular size, plasmid-encoded antibiotic resistances, restriction digestion profiles, incompatibility, and host range.
Characterization of IncQ-like plasmids.
Plasmid DNA digested with EcoRV revealed four different restriction patterns (Table 2). Four plasmids, each representing a different EcoRV restriction fragment length polymorphism type, were selected for further studies and named pIE1107, pIE1120, pIE1115, and pIE1130. The detailed molecular and genetic analysis of pIE1107 was published elsewhere (43). The complete nucleotide sequence of pIE1130 and part of the nucleotide sequence of plasmid pIE1120 are available from the databases under the accession numbers AJ271879 and AF070999, respectively.
(i) Characterization by different PCR primer sets.
Five different primer systems were used to characterize the collection of exogenously isolated IncQ-like plasmids. In addition to the three recently published primer sets (PI to PIII), which did not or only partly amplify pIE1107, pIE1115, and pIE1130, two new primer sets based on the sequences of pIE1107 and RSF1010 were developed (Table 1). Primer set PIV is specific for pIE1107 because primer annealing sites are absent in RSF1010. In contrast, amplification products with both RSF1010 and pIE1107 plasmids were obtained using primer set V. However, the sizes of the PCR products differed by 153 bp. Thus, primer set V with primer annealing sites in the repA and mobA region is suitable for a differentiation between the RSF1010-like (IncQα) and pIE1107-like (IncQβ) plasmids. Plasmid analysis with primer sets I to V (Table 3) revealed that PCR products of identical sizes were obtained with primer systems I, II, III, and V when pIE1120 or RSF1010 served as templates. Thus pIE1120 can be assigned to the IncQα group. In contrast, plasmid pIE1115 gave PCR products identical in size to those of pIE1107, suggesting that pIE1115 might be an IncQβ plasmid. For pIE1130, a PCR product was obtained with primer set I only. This is in agreement with the DNA sequence of this plasmid (AJ271879), which reveals differences from the typical IncQα and IncQβ plasmids at those sites in the plasmid backbone targeted by the primers of sets II, III, IV, and V.
TABLE 3.
PCR characterization of IncQ-like plasmids amplified with primer systems I to V
| Plasmid | Size (bp) of plasmid amplified with
|
||||
|---|---|---|---|---|---|
| PI | PII | PIII | PIV | PV | |
| RSF1010 | 430 | 190 | 1,100 | 1,150 | |
| pIE1120 | 430 | 190 | 1,100 | 1,150 | |
| pIE1107 | 1,500 | 1,000 | |||
| pIE1115 | 1,500 | 1,000 | |||
| pIE1130 | 430 | ||||
(ii) Antibiotic resistance patterns and determinants.
Plasmid-encoded resistance patterns in E. coli DH5α as determined by disk diffusion assay are listed in Table 2. Three of the exogenously isolated plasmids conferred resistance to SM. PCR amplification with three different primer sets homologous to strA and strB of RSF1010 and subsequent Southern blot hybridization showed that pIE1120, pIE1115, and pIE1130 contain the strA and strB genes arranged as in RSF1010 (Fig. 1). Plasmids pIE1115 and pIE1130 encoded SU resistance in addition to SM resistance, as does the IncQ-prototype plasmid RSF1010. The DNA sequence of pIE1130 (AJ271879) reveals the presence of a sulII gene, but in a location different from that seen with RSF1010. Moreover, the CM and KM resistance determinants of pIE1130 are typical catIII and aph(3′)-Id genes (34), respectively. Plasmid pIE1120, in addition to the SM resistance genes, carries a novel efflux-type TC resistance gene, tetY, as determined by DNA sequencing (AF070999). The sat3 gene for ST resistance and the aph(3′)-Id gene for KM resistance on plasmid pIE1107 have been described elsewhere (43, 44).
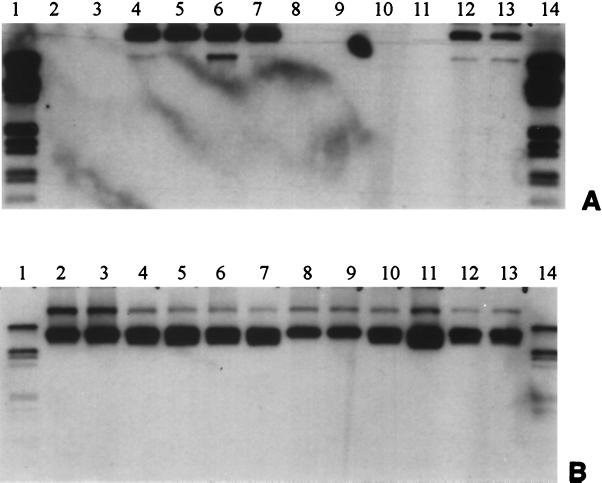
FIG. 1.
Hybridization of Southern-blotted plasmid DNA extracted from Acinetobacter sp. strain BD413, A. tumefaciens, and R. eutropha transconjugants with RSF1010 before (lanes 2 to 7) and after (lanes 8 to 13) stability testing with IncPα trfA2 probe (A) and with IncQ oriV probe (B) (12). Lanes 1 and 14, digoxigenin-labeled ladder; lanes 2, 3, 8, and 9, Acinetobacter sp.; lanes 4, 5, 10, and 11, A. tumefaciens; lanes 6, 7, 12, and 13, R. eutropha.
Antibiotic resistance analysis revealed four different types of antibiotic resistance patterns conferred by IncQ-like plasmids which were retrieved from a single piggery manure slurry sample. This is the first report that TC or CM resistance genes or an aph(3′)-I-type KM resistance gene are carried on IncQ-like plasmids.
(iii) Incompatibility testing.
Plasmids pIE1115, pIE1120, and pIE1130 were tested for incompatibility with pMMB600 and pIE1108, which served as type plasmids for the IncQα and IncQβ subgroup, respectively. Plasmid pIE1108 is an in vitro-generated deletion derivative of the natural IncQ-like plasmid pIE1107, which lacks the IncQα-specific incompatibility determinant (oriVα) of the parental plasmid (43). The results are summarized in Table 4.
TABLE 4.
Summary of incompatibility testinga
| Incoming plasmid | Resident plasmid | Relative transformation frequency | % of transformants with plasmid marker of:
|
% of colonies with plasmid marker ofb:
|
|||
|---|---|---|---|---|---|---|---|
| Incoming only | Both | Incoming only | Resident only | Both | |||
| pIE1115 | pMMB600 | 83 | 100 | 0 | NTc | NT | NT |
| pMMB600 | pIE1115 | <0.001 | NT | NT | NT | NT | NT |
| pIE1115 | pIE1108 | 14 | 8 | 92 | 61 | 34 | 5 |
| pIE1108 | pIE1115 | 3 | 11 | 89 | 48 | 40 | 12 |
| pIE1120 | pMMB600 | 24 | 1 | 99 | 38 | 34 | 28 |
| pMMB600 | pIE1120 | 17 | 0 | 100 | 47 | 30 | 23 |
| pIE1120 | pIE1108 | 111 | 0 | 100 | 0 | 0 | 100 |
| pIE1108 | pIE1120 | 120 | 0 | 100 | 0 | 0 | 100 |
| pIE1130 | pMMB600 | 129 | 98 | 2 | 100 | 0 | 0 |
| pMMB600 | pIE1130 | 2 | 0 | 100 | 0 | 100 | 0 |
| pIE1130 | pIE1108 | <0.001 | NT | NT | NT | NT | NT |
| pIE1108 | pIE1130 | 19 | 100 | 0 | NT | NT | NT |
The ability of the incoming plasmid to establish in a bacterial strain containing another resident plasmid was measured in reciprocal transformation experiments by comparing the efficiency of transformation with that of a plasmid-free recipient (set at 100). About 200 fresh transformant colonies were tested for the markers of both plasmids by subculture onto agar plates containing the respective antibiotics separately or combined in one plate. In case of growth on agar selective for both plasmids, such extransformant clones were tested for segregation of plasmid markers during nonselective growth. Per transformation experiment two macrocolonies grown from independent extransformants were inoculated separately into nonselective broth and grown for about 30 generations with three successive subcultures. Appropriate dilutions were then plated onto nonselective agar. About 200 colonies from each culture were tested again for the respective plasmid markers.
Segregation during nonselective growth.
NT, not tested.
When plasmid pIE1115 was tested as the incoming plasmid, a strong displacement of the resident plasmid pMMB600 was observed, while recipients containing pIE1115 could not be transformed by pMMB600. This is a remnant of the unidirectional incompatibility exerted by the IncQβ plasmid pIE1107 towards the IncQα plasmid RSF1010 (43). E. coli cells containing pIE1108 could be transformed by pIE1115 and vice versa, but the transformation frequency was only low. The majority of fresh transformants contained both plasmids. A symmetrical segregation during nonselective growth indicated incompatibility of pIE1115 with plasmid pIE1108.
E. coli DH5α containing the IncQα type plasmid pMMB600 could be transformed at low frequency by pIE1120 and vice versa. During nonselective growth of transformants which contained both plasmids, a symmetrical segregation indicated the incompatibility of pIE1120 and pMMB600. E. coli cells containing pIE1120 and a coresident pIE1108 stably maintained both plasmids under nonselective growth conditions. Thus, pIE1108 and pIE1120 are compatible.
E. coli DH5α cells containing pMMB600 could be transformed at high frequency by pIE1130, and a rapid displacement of the resident plasmid was observed. In contrast, recipients containing pIE1130 were transformed poorly by pMMB600 and the incoming plasmid was unable to stably establish in the transformants. Testing pIE1130 with pIE1108 revealed that the two plasmids were unable to coexist in a host. pIE1108, as an incoming plasmid, rapidly displaced pIE1130, whereas pIE1130 never could transform a host with a resident pIE1108. Obviously, the IncQα as well as the IncQβ type plasmid is incompatible with pIE1130. However, the unidirectional incompatibility reactions are different from the mutual incompatibility patterns observed when testing typical IncQα or IncQβ plasmids, respectively.
(iv) Determination of the host range.
The IncQ-like plasmids pIE1107, pIE1115, pIE1120, and pIE1130 were considered to be capable of replication among a range of γ-proteobacteria due to their exogenous isolation in P. putida and stable maintenance in E. coli DH5α under selective growth conditions. To obtain additional information on their host range and, in particular, on plasmid stability under nonselective conditions, four RIF-resistant soil bacteria, Acinetobacter sp. strain BD413, R. eutropha JMP228, A. tumefaciens DSM30150, and P. putida UWC1, were used as recipients in triparental filter matings. Mobilization of the IncQ-like plasmids by the IncP plasmid pTH10 was observed for all plasmids supporting their broad host range. Highest numbers of transconjugants were observed for all plasmids when A. tumefaciens DSM30150, R. eutropha JMP228, or P. putida UWC1 was used as the recipient. Transfer frequencies of 10−2 and 10−3 were observed for most of the plasmids with R. eutropha JMP228, A. tumefaciens DSM30150, and P. putida UWC1 as recipients. The mobilization of the plasmids into Acinetobacter sp. strain BD413 with transconjugant CFU numbers of 5 × 103 to 3 × 104 and transfer frequencies ranging from 5 × 10−7 to 4 × 10−6 was less efficient. To test the stable maintenance of the plasmids in different host backgrounds, two colonies each of A. tumefaciens DSM30150, R. eutropha JMP228, E. coli DH5α, and P. putida UWC1 were used to study the stability of the plasmid under nonselective growth conditions. After ∼30 generations of nonselective growth in LB broth, serial dilutions were plated onto PCA without antibiotics added. From each of the replicates 24 colonies were picked and restreaked on selective media. Based on the assumption that the resistance genes are stably expressed and maintained on the plasmid replicon, we correlated no growth of colonies on the selective medium with a loss of the plasmid. The two parallel experiments per plasmid-host combination consistently showed identical or similar percentages for colonies growing on selective media (Table 5). While all plasmids stably replicated in E. coli DH5α under nonselective growth conditions for ∼30 generations, all P. putida colonies originally carrying pIE1107 or pIE1115 failed to grow under selective conditions owing to loss of the plasmid. In contrast, plasmids RSF1010, pIE1120, and pIE1130 showed a high stability in P. putida UWC1. More than 90% of A. tumefaciens colonies carrying pIE1120, pIE1130, or RSF1010 were growing on the selective media, indicating a rather high stability, while for A. tumefaciens pIE1107 and pIE1115 growth was only observed for 23 and 79%, respectively, of the picked colonies. While plasmids pIE1107, pIE1115, and pIE1120 were not maintained in R. eutropha, pIE1130 and RSF1010 were. Most striking was the high stability of plasmid pIE1130 shown in all hosts studied. Overall, different patterns of plasmid stability in the different host backgrounds tested were found. Plasmid DNA extracted from two colonies selected per plasmid-host combination before and after stability testing were analyzed by Southern blot hybridization with IncPα trfA2 and IncQ oriV probes for the presence of pTH10 and the mobilized plasmids (Fig. 1). Before stability testing the mobilizing plasmid pTH10 was detected by hybridization with the IncP trfA2 probe only in R. eutropha and A. tumefaciens but not in Acinetobacter sp. strain BD413. After nonselective growth, the IncPα plasmid pTH10 was detectable only in R. eutropha recipients. The IncQ oriV sequence was detectable in all transconjugants before and after stability testing.
TABLE 5.
Stability of IncQ-like plasmids under nonselective growth conditions in different host backgrounds
| Plasmid | % Colonies growing on selective medium after replicating from a nonselective medium
|
||||
|---|---|---|---|---|---|
| E. coli DH5α | P. putida UWC1 | Acinetobacter sp. strain BD413 | R. eutropha JMP228 | A. tumefaciens DSM30150 | |
| RSF1010 | 100 | 87 | 63 | 100 | 100 |
| pIE1120 | 100 | 100 | 4 | 2 | 98 |
| pIE1107 | 100 | 0 | 100 | 2 | 23 |
| pIE1115 | 100 | 0 | 81 | 2 | 79 |
| pIE1130 | 100 | 100 | 92 | 94 | 90 |
Detection of IncQα and IncQβ replicon-related sequences in total community DNA.
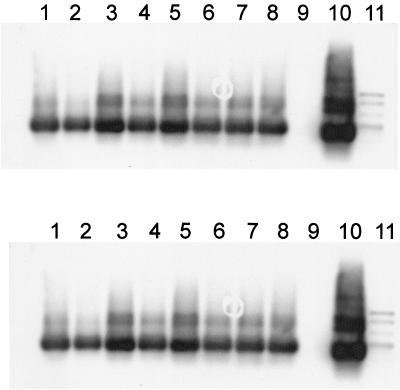
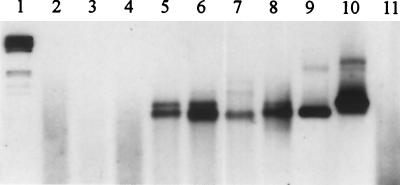
PCR amplification of total community DNA extracted from piggery manure samples of which the bacterial fractions had served as donors for exogenous plasmid isolations showed very intense signals for IncQ oriV and less intense PCR products for IncQ repB (Fig. 2). A different set of total community DNA extracted directly from various environments, such as soil, wastewater, and chicken, pig, and cattle manure, was analyzed using primer V for the presence of IncQ-specific sequences. Primer set V was used to allow the simultaneous detection of IncQα- and IncQβ-like plasmids in total DNA. Bands of the expected sizes could be detected after Southern blot hybridization in DNA from cattle and pig manure, wastewater, and farm soil (Fig. 3). Interestingly, hybridizing bands corresponding in size with pIE1107 (IncQβ) were stronger, indicating that the IncQβ sequence might be more abundant than the IncQα-like one.
FIG. 2.
Southern blot hybridizations of PCR products obtained after amplification of directly extracted community DNA from three piggery manure samples (samples 3 to 5) with IncQ oriV (A) and IncQ repB (B) primers (12). (A) Lanes 1 and 2, manure 3; lanes 3 and 4, manure 4; lanes 5 to 8, manure 5; lane 9, negative control; lane 10, RSF1010; lane 11, digoxigenin-labeled ladder. (B) Lane 1, digoxigenin-labeled ladder; lanes 2 and 3, manure 3; lanes 4 and 5, manure 4; lanes 6 to 9, manure 5; lane 10, negative control; lane 11, RSF1010.
FIG. 3.
Southern blot hybridization of PCR products amplified from community DNA directly extracted from different environmental samples with primer set V, which allows simultaneous detection of IncQα and IncQβ plasmids. Lanes: 1, digoxigenin-labeled ladder; 2, soil A; 3, soil CD2; 4, chicken feces; 5, cattle manure; 6, piggery manure; 7, wastewater; 8, farm soil; 9, pIE1107; 10, RSF1010; 11, negative control.
DISCUSSION
The exogenous plasmid isolation approach facilitates the capture of indigenous plasmids without the need to cultivate the original host. Plasmid capture relies upon self-transmission or mobilization of indigenous plasmids and expression of a selectable marker (e.g., antibiotic or heavy metal resistance) from the bacterial fraction retrieved from environmental samples in a well-defined recipient strain. Exogenous plasmid isolation has been primarily used to isolate plasmids conferring mercury resistance (7, 8, 19, 25) or carrying biodegradative genes (47). Lilley and Bailey (26) showed the acquisition of indigenous self-transferable mercury resistance plasmids by a genetically marked recipient colonizing the sugar beet phytosphere and thus demonstrated biparental exogenous isolation under in situ conditions. Here we report the isolation of transferable antibiotic resistance plasmids in biparental matings using the bacteria from piggery manure slurries as donors and E. coli CV601 or P. putida UWC1 as recipients. The majority of mobile genetic elements captured conferred multiple antibiotic resistances to their host. The rather frequent isolation of antibiotic resistance plasmids exhibiting homology to IncQ plasmids in biparental matings indicates not only a high prevalence of IncQ-like plasmids but also a high abundance of mobilizing plasmids in manure since IncQ plasmids can be only transferred when tra functions are provided by mobilizing plasmids in trans. This confirms observations of Götz and Smalla (12) on IncQ plasmid mobilization by indigenous bacteria in manure-treated soils under field conditions and corresponds with recent reports of gene mobilizing capacity in various environments (12, 20, 36, 46, 51). All plasmids exhibiting homology with IncQ plasmids were isolated in P. putida UWC1 but not in E. coli, and this might indicate that the mobilizing plasmids originate from Pseudomonas or related populations. Gene mobilizing capacity could be shown also for all manure samples analyzed here (unpublished results). In triparental matings performed as described by Hill et al. (19) the IncQ plasmid pIE639 (45) was mobilized by mobilizing plasmids of manure-derived bacteria into P. putida UWC1 but not into E. coli CV601. However, none of the transconjugants carried the mobilizing plasmid. Even more surprising was the high diversity of IncQ-like plasmids in terms of their phenotypic and genotypic characteristics. Four different plasmids, pIE1107, pIE1115, pIE1120, and pIE1130, were isolated from a single pig manure sample. Each of the plasmids was slightly different in size and had a unique set of antibiotic resistance genes. The genetic and molecular characterization of pIE1107 (43) revealed a new type of IncQ-like plasmid (IncQβ) which differs from the prototype IncQ plasmid RSF1010 not only in the antibiotic resistance genes but also in the replication and incompatibility functions. Incompatibility testing and comparative PCR analysis of pIE1115 and pIE1107 disclosed that these two plasmids have the same kind of replication system. Therefore, it seems appropriate to subdivide the family of IncQ-like replicons into RSF1010-type (IncQα) and a pIE1107-type (IncQβ) plasmids. As concluded on the basis of PCR analyses and incompatibility testing, pIE1120 turned out to be a typical IncQα plasmid which, however, differs from RSF1010 in the antibiotic resistance genes (Table 2). Generally, the results of PCR analyses with primer sets PI to PV (Table 3) were in accordance with incompatibility data (Table 4). PCR typing, therefore, might serve as a rapid method for classification of IncQ-like plasmids. Most interesting, plasmid pIE1130, which was neither a typical IncQα nor a typical IncQβ plasmid according to incompatibility testing, also could not be assigned to either group by PCR. The analysis of the recently finished nucleotide sequence of pIE1130 (AJ271879 [Tietze and Smalla, unpublished data]) suggests that this plasmid might represent a third type of IncQ-like plasmids. It will be interesting to derive PCR primers from the DNA sequence of pIE1130 in order to extend the typing set for IncQ-like plasmids beyond primer pairs PI to PV. This plasmid showed the highest plasmid stability in all host backgrounds under nonselective growth conditions.
Plasmids pIE1107, pIE1115, pIE1120, and pIE1130 carried by E. coli DH5α could be mobilized by the RP4 derivative pTH10 into Acinetobacter sp. and P. putida UWC1 (both γ-proteobacteria), R. eutropha (β-proteobacterium), and A. tumefaciens (α-proteobacterium). All plasmids under investigation were of broad host range. Recent studies conducted mainly with RSF1010 showed that it is the special mode of replication rather than an individual broad host range function which allows RSF1010 as well as probably other IncQ plasmids to replicate in nearly all gram-negative bacteria (9). Most studies describing the host range of novel plasmids analyzed whether or not a plasmid under investigation is transferable to different recipients. The question of whether or not the plasmid is stably maintained under nonselective growth conditions is only rarely addressed. Despite the fact that plasmids pIE1107, pIE1115, pIE1120, and pIE1130 were mobilized to Acinetobacter sp., P. putida, R. eutropha and A. tumefaciens, they clearly showed different patterns of stability under nonselective growth conditions in different host backgrounds. Despite the fact that all IncQ-like plasmids were originally isolated in P. putida UWC1, the two IncQβ plasmids pIE1107 and pIE1115 were not stably maintained in this host under nonselective growth conditions. While none of the IncQ-like plasmids was captured in E. coli CV601, all IncQ-like plasmids were perfectly stable in E. coli DH5α under nonselective conditions. The most stable plasmid in all five host backgrounds under nonselective growth conditions was pIE1130. This plasmid is neither an IncQα nor a typical IncQβ plasmid but might represent a third type of IncQ-like plasmids. Interestingly, the mobilizing plasmid pTH10 was detectable only in A. tumefaciens and R. eutropha transconjugants but not in Acinetobacter sp. After plasmid stability testing (growth under nonselective conditions) for all host-plasmid combinations the IncPα trfA2 sequence was only detected in plasmid DNA extracted from R. eutropha. This observation indicates that R. eutropha is a potential reservoir of IncP plasmids.
Although all IncQ-like plasmids pIE1107, pIE1115, pIE1120, and pIE1130 were isolated from one fresh manure sample and two additional pIE639-like IncQ plasmids (45) were recovered from another manure sample (data not presented), IncQ PCR-based detection of IncQ oriV and IncQ repB indicated a comparable abundance of the IncQ-specific sequences in manure samples 3 to 5. However, the isolation of IncQ plasmids in biparental exogenous plasmid isolations clearly depends not only on the presence of IncQ plasmids but also on the presence of efficiently mobilizing plasmids and the metabolic state of their hosts. While recently published IncQ primer sets (13) would have failed to detect plasmid pIE1107, pIE1115, or pIE1130 (except for primer set I), primer V, which was designed based on the sequences of RSF1010 and pIE1107, allowed us to simultaneously amplify both IncQα and IncQβ plasmids from total community DNA. Southern blot hybridizations indicated the prevalence of both plasmid types in pig and cattle manure DNA and in farm soil.
In view of the broad host range of IncQ plasmids, their load of antibiotic resistance genes is of particular interest. All IncQ-like plasmids analyzed in this study conferred different combinations of antibiotic resistances. Although pIE1107 and pIE1115 both have an IncQβ-group backbone, they carried different antibiotic resistance genes. Plasmid pIE1115 had a similar organization of antibiotic resistance genes (strA-strB and sulII) as found in the IncQα prototype RSF1010 while plasmid pIE1107 does not carry strA and strB and lacks part of the sulII gene (43). The organization of the ST and KM resistance gene block carried on pIE1107 is identical with that found on the IncQα plasmid pIE639 (EMBL database accession no. Z48231 [43–45]). Three of the plasmids analyzed contained linked SM resistance genes strA and strB, which appear to be widely distributed (40). The strA-strB genes are often located on small nonconjugative plasmids belonging to the IncQ group and are usually linked with the SU resistance gene sulII (40). In plant-associated bacteria the identical strA-strB genes were mainly detected on large conjugative plasmids located on the transposon Tn5393 (4, 41). The 81-bp right-inverted repeat of Tn5393, which is located downstream of strB, is the only relic of Tn5393 on RSF1010 (40). A group of SM- and SU-resistant Erwinia amylovora strains isolated from an apple orchard in California was described which harbor a small plasmid which hybridized to a strA probe (28). Sequencing data indicated that indeed pEa8.7 is closely related, if not identical, to RSF1010 (29). Recchia and Hall (31) reported that the GM resistance of plasmid pIE723 (45) was acquired by insertion of an aadB gene cassette at a secondary site in the IncQ plasmid RSF1010.
The characterization of IncQ-like plasmids which were exogenously isolated from a single piggery manure slurry sample revealed an astonishingly high diversity with regard to phenotypic and genotypic properties. The replicons have certainly evolved from a common ancestral plasmid. Tietze (43) proposed that cointegrate formation of two different IncQ-like plasmids might have been important for diversification and evolution of IncQ-like plasmids. It is supposed that IncQ plasmids play an important role in disseminating antibiotic resistance genes, given their high abundance in manure slurries and other environments, their broad host range, and the different combinations of resistance genes carried by these plasmids.
ACKNOWLEDGMENTS
This work was supported by EU-BIOTECH grants BIO2-CT92-0491 and BIO4-CT98-0053 (Reservoir) and the European Union-funded Concerted Action MECBAD (BIO4-CT98-0099).
We are grateful to Michael Bagdasarian, Michigan State University, East Lansing, for reading and discussion of the manuscript.
REFERENCES
- 1.Bagdasarian M, Lurz R, Rückert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 2.Bale M J, Day M J, Fry J C. Novel method for studying plasmid transfer in undisturbed river epilithon. Appl Environ Microbiol. 1988;54:2756–2758. doi: 10.1128/aem.54.11.2756-2758.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiou C S, Jones A L. Nucleotide sequence analysis of a transposon Tn5393 carrying streptomycin resistance genes in Erwinia amylovora and other gram-negative bacteria. J Bacteriol. 1993;175:723–740. doi: 10.1128/jb.175.3.732-740.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couturier M, Bex F, Berquist P L, Maas W K. Identification and classification of bacterial plasmids. Microbiol Rev. 1988;52:375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahlberg C, Hermansson M. Abundance of Tn3, Tn21, and Tn501 transposase (trpA) in bacterial community DNA from marine environments. Appl Environ Microbiol. 1995;61:3051–3056. doi: 10.1128/aem.61.8.3051-3056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahlberg C, Linberg C, Torsvik V L, Hermansson M. Conjugative plasmids isolated from bacteria in marine environments show various degrees of homology to each other and are not closely related to well-characterized plasmids. Appl Environ Microbiol. 1997;63:4692–4697. doi: 10.1128/aem.63.12.4692-4697.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drønen A K, Torsvik V, Goksøyr J, Top E M. Effect of mercury addition on plasmid incidence and gene mobilizing capacity in bulk soil. FEMS Microbiol Ecol. 1999;27:381–394. [Google Scholar]
- 9.Frey J, Bagdasarian M. The molecular biology of IncQ plasmids. In: Thomas C M, editor. Promiscuous plasmids of gram-negative bacteria. London, United Kingdom: Academic Press; 1989. pp. 79–94. [Google Scholar]
- 10.Fürste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 11.Gormley E P, Davies J. Transfer of plasmid RSF1010 by conjugation from Escherichia coli to Streptomyces lividans and Mycobacterium smegmatis. J Bacteriol. 1991;173:6705–6708. doi: 10.1128/jb.173.21.6705-6708.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Götz A, Smalla K. Manure enhances plasmid mobilization and survival of Pseudomonas putida introduced into field soil. Appl Environ Microbiol. 1997;63:1980–1986. doi: 10.1128/aem.63.5.1980-1986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Götz A, Pukall R, Tietze E, Prager R, Tschäpe H, van Elsas J D, Smalla K. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl Environ Microbiol. 1996;62:2621–2628. doi: 10.1128/aem.62.7.2621-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guiney D G. Broad host range conjugative and mobilizable plasmids in gram-negative bacteria. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 75–103. [Google Scholar]
- 15.Harayama S, Tsuda M, Iino T. High frequency mobilization of the chromosome of Escherichia coli by a mutant of plasmid RP4 temperature-sensitive for maintenance. Mol Gen Genet. 1980;180:47–56. doi: 10.1007/BF00267351. [DOI] [PubMed] [Google Scholar]
- 16.Haring V, Scherzinger E. Replication proteins of the IncQ plasmid RSF1010. In: Thomas C M, editor. Promiscuous plasmids of gram-negative bacteria. London, United Kingdom: Academic Press; 1989. pp. 95–124. [Google Scholar]
- 17.Heuer H, Krsek M, Baker P, Smalla K, Wellington E M H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heuer H, Hartung K, Wieland G, Kramer I, Smalla K. Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl Environ Microbiol. 1999;65:1045–1049. doi: 10.1128/aem.65.3.1045-1049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill K E, Weightman A J, Fry J C. Isolation and screening of plasmids from the epilithon which mobilize recombinant plasmid pD10. Appl Environ Microbiol. 1992;58:1292–1300. doi: 10.1128/aem.58.4.1292-1300.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill K E, Fry J C, Weightman A J. Gene transfer in the aquatic environment: persistence and mobilization of the catabolic recombinant plasmid pD10 in the epilithon. Microbiology. 1994;140:1555–1563. [Google Scholar]
- 21.Hill K E, Marchesi J R, Fry J C. Conjugation and mobilization in the epilithon. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 5.2.2/1–5.2.2/28. [Google Scholar]
- 22.Holmes D S, Quigley M. Isolation of plasmid DNA by mini quick method. Annu Rev Biochem. 1983;114:193–197. [Google Scholar]
- 23.Juni E. Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J Bacteriol. 1972;112:917–931. doi: 10.1128/jb.112.2.917-931.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy S B. Environmental dissemination of microbes and their plasmids. Swiss Biotechnol. 1987;5:32–37. [Google Scholar]
- 25.Lilley A K, Bailey M J, Day M J, Fry J C. Diversity of mercury resistance plasmids obtained by exogenous isolation from the bacteria of sugar beet in three successive years. FEMS Microbiol Ecol. 1996;20:211–227. [Google Scholar]
- 26.Lilley A K, Bailey M J. The acquisition of indigenous plasmids by a genetically marked pseudomonad population colonizing the sugar beet phytosphere is related to local environmental conditions. Appl Environ Microbiol. 1997;63:1577–1583. doi: 10.1128/aem.63.4.1577-1583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Llanes C, Gabant P, Couturier M, Michel-Briand Y. Cloning and characterization of Inc A/C plasmid RA1 replicon. J Bacteriol. 1994;176:3403–3407. doi: 10.1128/jb.176.11.3403-3407.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McManus P S, Jones A L. Genetic fingerprinting of Erwinia amylovora strains isolated from tree fruit crops and Rubus spp. Phytopathology. 1995;85:1547–1553. [Google Scholar]
- 29.Palmer E L, Teviotdale B L, Jones A L. A relative of the broad-host-range plasmid RSF1010 detected in Erwinia amylovora. Appl Environ Microbiol. 1997;63:4604–4607. doi: 10.1128/aem.63.11.4604-4607.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rademaker J L W, Louws F J, de Bruijn F J. Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 7.1.3/1–7.1.3/33. [Google Scholar]
- 31.Recchia G D, Hall R M. Plasmid evolution by aquisition of mobile gene cassettes: plasmid pIE723 contains the aadB gene cassette precisely inserted at a secondary site in the IncQ plasmid RSF1010. Mol Microbiol. 1995;15:179–187. doi: 10.1111/j.1365-2958.1995.tb02232.x. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 34.Shaw K J, Rather P N, Hare R S, Miller G H. Molecular genetics of aminoglycoside resistance genes and familial relationships of aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57:138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smalla K, Cresswell N, Mendonca-Hagler L C, Wolters A, van Elsas J D. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J Appl Bacteriol. 1993;74:78–85. [Google Scholar]
- 36.Smit E, Wolters A, van Elsas J D. Self-transmissible mercury resistance plasmids with gene-mobilizing capacity in soil bacterial populations: influence of wheat roots and mercury addition. Appl Environ Microbiol. 1998;64:1210–1219. doi: 10.1128/aem.64.4.1210-1219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobecky P A, Mincer T J, Chang M C, Helinski D E. Plasmids isolated from marine sediment microbial communities contain replication and incompatibility regions unrelated to those of known plasmid groups. Appl Environ Microbiol. 1997;63:888–895. doi: 10.1128/aem.63.3.888-895.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobecky P A, Mincer T J, Chang M C, Toukdarian A, Helinski D E. Isolation of broad-host-range replicons from marine sediment bacteria. Appl Environ Microbiol. 1998;64:2822–2830. doi: 10.1128/aem.64.8.2822-2830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staley J T, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 40.Sundin G W, Bender C L. Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals and plants. Mol Ecol. 1996;5:133–143. doi: 10.1111/j.1365-294x.1996.tb00299.x. [DOI] [PubMed] [Google Scholar]
- 41.Sundin G W, Monks D E, Bender C L. Distribution of the streptomycin-resistance transposon Tn5393 among phylloplane and soil bacteria from managed agricultural habitats. Can J Microbiol. 1995;41:792–799. doi: 10.1139/m95-109. [DOI] [PubMed] [Google Scholar]
- 42.Thomas C M, Thorsted P. PCR probes for promiscuous plasmids. Microbiology. 1994;140:1. doi: 10.1099/13500872-140-1-1. [DOI] [PubMed] [Google Scholar]
- 43.Tietze E. Nucleotide sequence and genetic characterization of the novel IncQ-like plasmid pIE1107. Plasmid. 1998;39:165–181. doi: 10.1006/plas.1998.1343. [DOI] [PubMed] [Google Scholar]
- 44.Tietze E, Brevet J. Nucleotide sequence of bacterial streptothricin resistance gene sat3. Biochim Biophys Acta. 1995;1263:176–178. doi: 10.1016/0167-4781(95)00103-n. [DOI] [PubMed] [Google Scholar]
- 45.Tietze E, Tschäpe H, Voigt W. Characterization of new resistance plasmids belonging to incompatibility group Q. J Basic Microbiol. 1989;29:129–136. doi: 10.1002/jobm.3620291013. [DOI] [PubMed] [Google Scholar]
- 46.Top E, De Smet I, Verstraete W, Dijkmans R, Mergeay M. Exogenous isolation of mobilizing plasmids from polluted soils and sludges. Appl Environ Microbiol. 1994;60:831–839. doi: 10.1128/aem.60.3.831-839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Top E M, Holben W E, Forney L J. Characterization of diverse 2,4-dichlorophenoxyacetic acid degradative plasmids isolated from soil by complementation. Appl Environ Microbiol. 1995;61:1691–1698. doi: 10.1128/aem.61.5.1691-1698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tschäpe H. The spread of plasmids as a function of bacterial adaptability. FEMS Microbiol Ecol. 1994;15:23–32. [Google Scholar]
- 49.Turner S L, Rigottier-Gois L, Poer R S, Amarger N, Young J P W. Diversity of repC plasmid-replication sequences in Rhizobium leguminosarum. Microbiology. 1996;142:1705–1713. doi: 10.1099/13500872-142-7-1705. [DOI] [PubMed] [Google Scholar]
- 50.van Elsas J D, Smalla K. Extraction of microbial community DNA from soils. 1995. p. 1.3.3. /1–1.3.3/11. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 51.van Elsas J D, McSpadden Gardener B B, Wolters A C, Smit E. Isolation, characterization, and transfer of cryptic gene-mobilizing plasmids in the wheat rhizosphere. Appl Environ Microbiol. 1998;64:880–889. doi: 10.1128/aem.64.3.880-889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, et al., editors. Current protocols in molecular biology. J. New York, N.Y: Wiley and Sons; 1987. pp. 2.10–2.12. [DOI] [PubMed] [Google Scholar]
- 53.Witte W. Impact of antibiotic use in animal feeding on bacterial pathogens in humans. In: Levy S B, editor. Antibiotic resistance: origins, evolution, selection and spread. Ciba Foundation Symposium 207. Chichester, United Kingdom: Wiley; 1997. pp. 61–75. [DOI] [PubMed] [Google Scholar]
- 54.Witte W. Medical consequences of antibiotic use in agriculture. Science. 1998;279:996–997. doi: 10.1126/science.279.5353.996. [DOI] [PubMed] [Google Scholar]