ABSTRACT
The oral microbiota is enormously diverse, with over 700 microbial species identified across individuals that play a vital role in the health of our mouth and our overall well-being. In addition, as oral diseases such as caries (cavities) and periodontitis (gum disease) are mediated through interspecies microbial interactions, this community serves as an important model system to study the complexity and dynamics of polymicrobial interactions. Here, we review historical and recent progress in our understanding of the oral microbiome, highlighting how oral microbiome research has significantly contributed to our understanding of microbial communities, with broad implications in polymicrobial diseases and across microbial community ecology. Further, we explore innovations and challenges associated with analyzing polymicrobial systems and suggest future directions of study. Finally, we provide a conceptual framework to systematically study microbial interactions within complex communities, not limited to the oral microbiota.
KEYWORDS: microbiome, microbe-microbe interactions, microbial ecology, oral microbiology
PERSPECTIVE
The oral diseases caries and periodontitis impact the majority of the adult population in the United States and are two of the most prevalent infections worldwide (1, 2). These diseases are influenced by diverse and dynamic oral microbes and their polymicrobial interactions, such as those mediated by physical attachment or metabolic cues (3–6). Thus, the study of oral diseases has spurned over a century of innovative research at the intersection of pathogenesis and microbial ecology, and the oral microbiota offers an important model to continue to increase our understanding of microbial communities.
HISTORICAL PERSPECTIVES
Foundations of oral microbiome studies.
For as long as we have known that microbes exist, we have known about the polymicrobial nature of the oral microbiota. At the discovery of microorganisms in 1683, Antonie van Leeuwenhoek observed a tartar specimen from his tooth using a primitive microscope (7), describing diverse bacterial morphologies that today would be called cocci, spirochetes, and fusobacteria (8). As microbiology research progressed in the late 1800s and early 1900s, scientists worked to discern the role of individual microbes and the collective community as the causative agents of oral diseases (9–11). Although this work was initially restricted to easy-to-culture aerobes and facultative anaerobes such as Streptococcus (11) and Actinomyces (12, 13), over time, the challenge of cultivating diverse oral bacteria was overcome by the development of anaerobic cultivation methods for obligate anaerobes and complex media to promote the growth of fastidious bacteria. The successful growth of diverse oral bacteria in the laboratory allowed for the further genotypic and phenotypic characterization of individual species and provided the foundations for polymicrobial studies.
Polymicrobial biofilm colonization, a spatiotemporal matter.
Research on microbial interactions within the oral microbiota started in 1970 when Gibbons and Nygaard mixed two pure cultures of different species together and observed clumping sedimentation within seconds, initially called interbacterial aggregation (14). While this phenomenon of physical cell-cell interactions, now termed coaggregation, was first described in the oral microbiome, it is prevalent across microbial communities, including those in the human gut, the urogenital tract, and freshwater (15). Coaggregation of oral microbes to each other and adherence to the tooth surface are key to the formation of oral biofilms (plaque) and were comprehensively characterized even before the introduction of the term “biofilm” in 1978 (16).
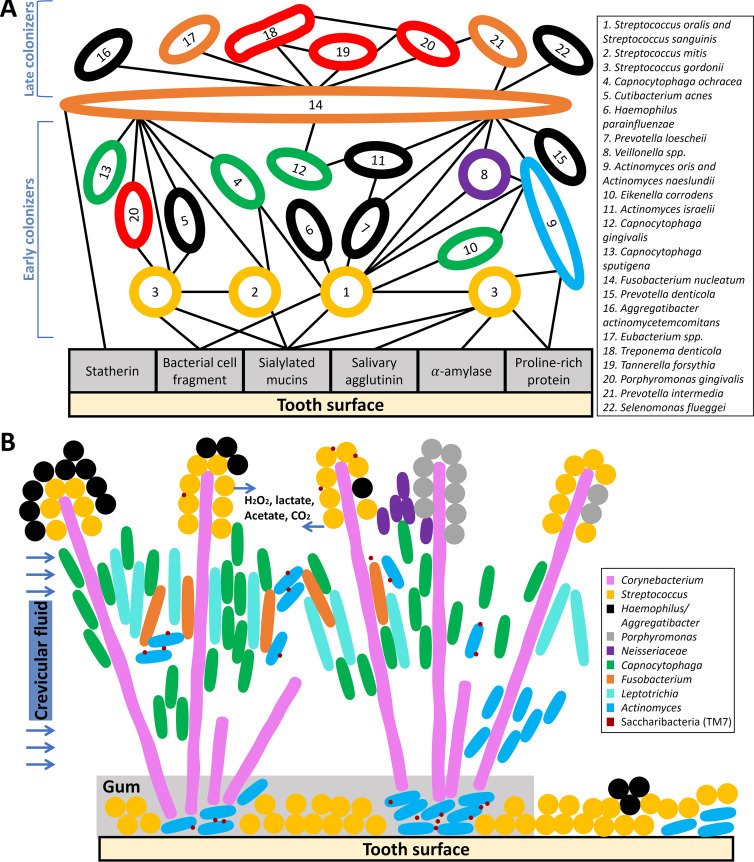
To further understand how these interactions influence how bacteria colonize the oral cavity, researchers physically removed mature human oral biofilms and monitored bacterial reappearance on the tooth surface over time. The resulting understanding of the temporal order of appearance of bacterial species combined with possible pairwise coaggregation partnerships enabled the proposal of a spatiotemporal model of oral bacterial colonization summarized by Kolenbrander in 1993 (Fig. 1A) (17–19). After 4 h, the microbes that repopulate cleaned teeth and attach to host surfaces are regarded as “early colonizers” (19). Respectively, the species that colonize at later time points ranging from hours or days to months or years are termed “late colonizers.” While coaggregation is key to tethering these species together, Kolenbrander noticed that early colonizers do not bind to late colonizers and thus proposed the existence of a bridge organism which coaggregates with both early and late colonizers (20, 21). Although rarely found in the first 12 h after professional teeth cleaning, the genus Fusobacterium is frequently isolated in both healthy and diseased dental plaques and can bind most genera of early and late colonizers. Thus, Fusobacterium is depicted as a coaggregation bridge in the oral microbiome (Fig. 1A) (19).
FIG 1.
(A) Spatiotemporal model of oral bacterial colonization and pathogenicity. A schematic of the Kolenbrander model of long-term microbial succession from the early stage of biofilm colonization on the tooth surface to the establishment of mature supragingival and subgingival biofilms and, ultimately, to the formation of diseased bacterial communities (17–19). Rods and circles indicate microbial taxa, and lines indicate physical interactions, including binding to the tooth surface or known coaggregation. This schematic integrates over 1,000 coaggregation connections found in the oral cavity, involving microbes that are primarily found both in supragingival and subgingival plaque. The colors of the microbial taxa indicate their corresponding Socransky complex from subgingival plaque, which consists of six categories, yellow, green, blue, purple, orange, and red (25). Specifically, orange and red complexes are more often associated with clinical parameters of gum disease. Species not covered in this Socransky model are colored in black. This schematic highlights that early colonizers and late colonizers are classified into different-colored complexes (57–59) and the proposed importance of F. nucleatum as a bridge species in linking early and late colonizers. Although this model has been highly influential on our understanding of biofilm formation, current work employing advanced microscopy and sequencing techniques continues to refine our understanding of oral biofilm biogeography and development. (B) Supragingival oral biofilm architecture observed using CLASI-FISH, incorporating proposed biochemical gradients and episymbiotic Saccharibacteria. In this diagram, the rods and circles indicate microbial taxa, and their locations are based on microscale imaging of supragingival plaque using CLASI-FISH (5, 46). The “hedgehogs” are structured by clusters of Corynebacterium filaments that bind to Streptococcus and Actinomyces near the base and then expand to the “corncob”-structured perimeter. This spatial patterning also divides the environment into different chemical environments, as shown. The following bacterial genera are colored with their corresponding colored complexes from mature supragingival biofilms: Streptococcus spp. (yellow), Neisseriaceae spp. (purple), Capnocytophaga spp. (green), Fusobacterium spp. (orange), and Actinomyces spp. (blue) (27). Other taxa, including Saccharibacteria, not included in the Socransky complexes, are shown in other distinct colors. Note that Porphyromonas is not included in a colored complex, as Porphyromonas here is likely aerotolerant Porphyromonas catoniae and/or Porphyromonas pasteri (46).
Together, the ability to bind to the tooth surface and the ability to coaggregate are critical for oral bacterial survival despite constant flow of saliva, regular hygiene practices, variable nutrient supply, and extreme temperature changes. Further, adhesion, coaggregation, and the environmental differences across the surfaces of the mouth result in multiple biogeographical niches in the oral cavity (17, 22). Although biofilm development is important across most ecosystems and, in fact, was first chronicled in aquatic systems in the 1930s, the oral biofilm has provided an accessible and clinically relevant biofilm system (23, 24). Research on the oral biofilm has been fundamental for our understanding of biofilm formation, microbe-microbe interactions, and the relationship between biofilms and pathogenesis (17).
Polymicrobial biofilms and species associations in vivo.
Although the physical interactions in the oral biofilm were widely studied by coaggregation, microbial interaction work specific to plaque from below the gumline (subgingival) was limited until Socransky and his colleagues performed a comprehensive and impactful study in 1998. These authors studied over 13,000 human subgingival biofilm samples from periodontally healthy and diseased patients using checkerboard DNA-DNA hybridization techniques and then grouped 40 culturable bacterial species into six “colored” complexes using cluster analysis and community ordination techniques (Fig. 1) (25). For example, the bacteria in the red complex, Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia (formerly Bacteroides forsythus), were strongly associated with each other and strongly correlated with periodontal disease. Among the colored complexes, this red complex has attracted significant interest because all three members are putative periodontal pathogens, while other complexes contain both commensals and pathogens (26). The proposal of a red complex contradicted the prevalent idea that disease resulted from the presence of a single pathogen and instead supported the alternative hypothesis that periodontal diseases are associated with communities of bacteria. In addition to identifying the association among species within each complex, Socransky et al.’s study identified the relationship among different complexes. For instance, this work showed that the species in the red complex were rarely found without the presence of orange complex bacteria. This study on subgingival species associations in vivo was pioneering and comprehensive, and a similar study was performed using plaque from above the gumline (supragingival) in 2008 (27). While these studies only identified known, culturable bacteria and were constrained by the limitations of correlative studies, they provide important context for future research to study complexes beyond the species level (i.e., strain level or genus level) and to incorporate the role of phenotypic plasticity, including virulence factor production and other responses to host factors (28).
CURRENT AND FUTURE PERSPECTIVES
Community composition and function using culture-independent sequencing.
The initial studies on species associations in vitro and in vivo provided fundamental knowledge for understanding interspecies interactions and biofilm development of oral microbiota. The Socransky et al. study showed the importance of looking beyond interactions among two or a few species. The development of next-generation sequencing techniques and bioinformatic analytic tools allows researchers to study the complexity of diverse oral communities without cultivation, although these studies are challenging due to limited bacterial genetic material in clinical samples and disagreement on the best analysis approaches. The application of 16S rRNA gene amplicon sequencing and metagenomics allows differentiation of bacteria at the species or even strain level and determination of their functional potential.
Work using these techniques has revealed enormous genetic diversity in the human oral cavity: over 700 bacterial species are found in the mouth across individuals, with approximately 200 species per person. Extensive work has resulted in the ability to culture 74% of these microbes (29–31). For instance, the candidate phylum TM7, now Saccharibacteria, was discovered 20 years ago using 16S rRNA gene sequence analyses (32), eventually leading to the recent successful cultivation of this microbe with its obligate host bacteria in lab conditions (33, 34). Thus, the initial sequencing analyses led to experimental characterizations and the finding that Saccharibacteria downregulates the pathogenicity of its host bacteria and reduces inflammatory bone loss (5).
While the metagenome shows the possible microbial function of the community, it does not reflect the community’s actual activity. A full understanding of oral microbiome dynamics requires a combined picture of microbial composition (the metagenome) and global gene expression (the metatranscriptome). Importantly, the accessibility of the human oral cavity allows for the instantaneous preservation of microbial RNA with temporal and spatial control. Studies using metatranscriptomics on oral specimens have comprehensively shaped our understanding of the oral microbiome. These studies found that although the community composition during periodontitis can vary drastically, the community’s functional activities are conserved (35). This finding suggests that rather than focusing on specific pathogens, we should consider the community as a pathogen (36). Additional support for “the community as pathogen” comes from community-wide metatranscriptomic studies that found organisms traditionally considered commensals transcribe the majority of virulence factors during periodontal diseases (37, 38). Metatranscriptomics can also provide insight into the in situ functionality of individual species (39). One striking finding is that a species can have similar relative abundance but different functional activities across different conditions. For example, Fusobacterium nucleatum does not significantly change in relative abundance between healthy and diseased samples, but metatranscriptomics shows that its metabolism is altered (40).
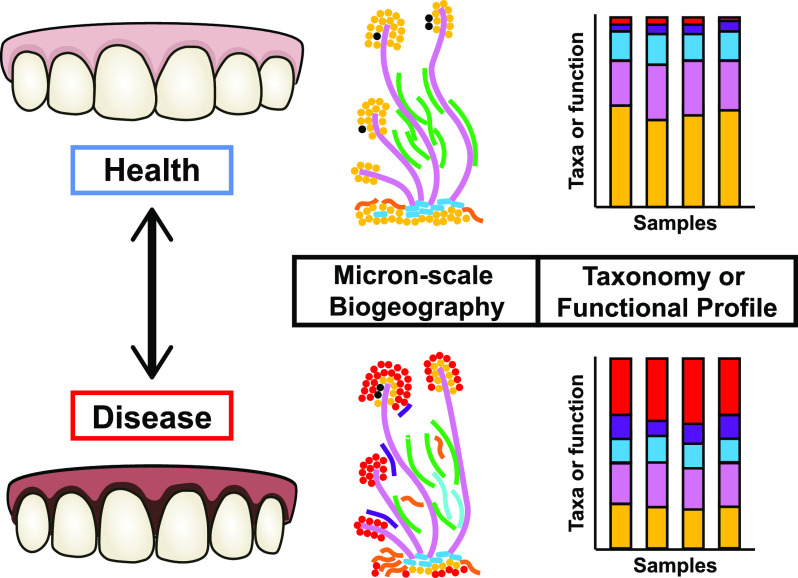
The oral cavity has become one of the best-described microbial sites in the human body due to its accessibility and clinical relevance; sequencing-driven studies of oral health and disease have broadened our understanding of the community composition and functional activities of the oral microbiota. However, these studies are only starting points, and the driving factors that lead to the progression from healthy to diseased states remain unknown (Fig. 2; Table 1). Longitudinal studies could provide clues about the initial stages of oral diseases and markers of disease progression (38, 41). Finally, there is a further need to simultaneously measure microbial and host gene expression, which could partially explain the development of disease or the shift back to health after therapeutic interventions. These future directions are important for developing therapies to prevent and treat oral infections and for broadly understanding host-associated microbial communities.
FIG 2.
Approaches to studying the dynamics of the oral microbiota during health and disease. Microscopy and sequencing-based community profiling are powerful approaches that can be leveraged for spatiotemporal studies of oral microbial ecology to further understand the relationship of the oral microbiota with health and disease. Confocal scanning microscopy of labeled strains reveals changes in micron-scale biogeography and the corresponding changes in microbial interactions. Community profiling, for instance, using metagenomics and metatranscriptomics, shows changes in the composition and functional activities of samples across space or time. These methods are important in both top-down and bottom-up approaches. For example, top-down approaches could sample the oral biofilm over time after a professional cleaning (60). For bottom-up approaches, emergent spatiotemporal dynamics can be observed using a small number of cells directly removed from oral specimens with micromanipulators or using communities constructed from pure cultures of strains (61).
TABLE 1.
Open questions and approaches in oral microbial ecology
| Questions | Bottom-up approaches | Top-down approaches |
|---|---|---|
| How to build a simplified model system for oral microbiota? What are appropriate metrics for model evaluation? | Experimental model systems can be constructed at the species level (6, 17), genus level (6, 46), or functional level. | Quantitative approaches must be used to benchmark models using the human oral community (39, 42, 56). |
| What are the similarities and differences in community dynamics between supragingival and subgingival plaque? | Models can illuminate differences in microbial interactions between the two environments. For instance, the Zurich model suggested that the subgingival plaque model could be derived from the supragingival plaque model (62). | Comparative studies using sequencing and microscopy can show intrapatient and interpatient differences between environments. |
| What are the factors that drive the progression from healthy to diseased states or from diseased to healthy states? How do oral microbes colonize and invade different oral habitats? | Model systems can test the factors that lead to pathogen abundance and the production of virulence factors. | Detailed analyses of longitudinal studies in human patients can further show how communities change over time and which healthy communities become diseased. |
| How many microenvironments exist in the oral cavity? How do microenvironments impact biodiversity? | Perturbation of laboratory models can test the importance of different environmental factors. | In situ measurement of chemical gradients and oxygen levels can indicate different niches. Also, differential abundance of microbes can indicate site specialists with distinct niches (22). |
Microbiogeography.
Despite their utility, metagenomic and metatranscriptomic approaches destroy micron-scale spatial information by homogenizing samples during nucleic acid extraction. Bacteria are micron-sized, and research on oral microbes was the first to show that micron-scale spatial patterning (microbiogeography) can impact disease progression (3, 42). Thus, to understand microbe-microbe and microbe-host interactions, it is additionally important to determine the spatial organization of bacteria with each other and with host factors at scales of microns to hundreds of microns (Fig. 1B; Fig. 2).
In 1972, morphological observation using electron scanning microscopy directly supported that dental plaques form structured biofilms (43). Imaging showed corncob structures composed of central filaments and densely packed layers of cocci attaching to filaments. The advent of fluorescence in situ hybridization (FISH) and immunofluorescence allowed researchers to distinguish the identities of microbes in complex oral communities (44). By use of combinatorial labeling and spectral imaging-FISH (CLASI-FISH) (45), the corncob structure was redefined by visualizing nine genera simultaneously in supragingival plaque. The structure was more complex than previously identified: multiple corncobs together formed “hedgehog structures” with aerotolerant taxa on the outside and anaerobic taxa on the inside toward the tooth surface (Fig. 1B) (46). This pioneering study suggested differential oxygen and nutrient usage across the plaque biofilm and provided a spatial framework to incorporate metabolic and ecological factors. Further work quantitatively assessed the proximity of microbes relative to host factors through computing microbial pairwise correlation functions using CLASI-FISH imaging of the tongue dorsum (47). These studies provide important in situ benchmarks to experimentally test hypotheses about the spatiotemporal development of the oral biofilm (22, 42).
Applications of CLASI-FISH to visualize microbial structures both in dental plaque and on the tongue show the role of micron-scale interactions in establishing distinctive oral communities. Yet much remains to be learned about how oral microbes organize themselves and respond to gradients of nutrients and oxygen in their preferred environment. Kim et al. provided an example for studying these topics by simultaneously measuring the biofilm architecture, pH microenvironment, and enamel demineralization during the formation of dental caries (6). The authors discovered a rotund-shaped biofilm architecture in intact human dental plaque and quantified the dynamics of microbial community development in lab conditions. This micron-scale patterning resulted in the protection of the oral pathogen Streptococcus mutans by surrounding oral commensals. In another example, to understand how oral microbes disperse and initiate biofilm formation, Simon-Soro et al. combined microscopy with sequencing, showing that most of the microbial biomass in saliva is composed of aggregates containing a mix of both early and late colonizers (48). These diverse aggregates, not single cells, seeded the vast majority of biofilm formation in an in vitro model of the tooth surface. This finding provides an alternative to the spatiotemporal model proposed by Kolenbrander and supports the presence of pathogenic microbes early in biofilm formation (18, 49). Together, these innovative studies highlight how research on oral microbes continues to shape our understanding of biofilm formation, microbiogeography, and disease.
There are further opportunities to use imaging approaches such as FISH-based techniques to study disease progression and community dynamics in the oral cavity (Table 1). In vitro applications of CLASI-FISH can distinguish up to 120 different species in a single image (50), and high-phylogenetic-resolution microbiome mapping by FISH (HiPR-FISH) can distinguish over 1,000 closely related bacterial strains in vitro and has been applied to study oral microbial spatial patterning across longitudinal samples (51). Finally, as metatranscriptomics showed the importance of measuring bacterial function in addition to community composition, there are increasing opportunities to measure bacterial physiology at the micron level (42, 52, 53).
CONCLUSION
Over a century of work has studied the ecology of oral microbiota, demonstrating how spatiotemporal dynamics lead to oral diseases. Despite this rich history of investigation, there remains much to learn about the emergent properties of oral biofilms and how microbe-microbe and microbe-host interactions influence disease. With diverse microenvironments influenced by dramatic nutrient and temperature changes, regular disturbances (such as hygiene practices), and host interactions, the oral cavity is a valuable model system to study biofilm development, ecological succession, and the eco-evolutionary dynamics that shape an organism’s niche. Given the enormous complexity of oral communities, we suggest that both bottom-up and top-down approaches are important for the further investigation of microbial interactions and the dynamics of oral biofilms (Fig. 2; Table 1). Bottom-up approaches using simplified experimental models provide an easy-access system to evaluate biofilm development and the outcome of clinical interventions. For example, the 10-species “Zurich model” has been used to study topics such as biofilm development, virulence factors, and microbial interactions (54, 55). The advancement of sequencing technologies and microscopy techniques offers further opportunities to design and customize in vitro models to match the oral community composition, physiology, and spatial patterning (39, 42, 56), and there are significant opportunities to use these model systems to probe diverse questions (Table 1). In contrast, top-down approaches that directly analyze human patient samples will continue to be essential for understanding the functional and spatial dynamics of oral microbial communities, as discussed in “Current and future perspectives” above.
Research on oral microbes has been critical for our overall understanding of microbial interactions, biofilm development, and spatial patterning and has driven the advancement of technologies for studying microbial communities. Undoubtedly, the oral microbiome provides an accessible and deeply studied system for further exploration of microbial interactions and the role of our microbiota in human health. The future unraveling of the complex dynamics leading to health and disease in the oral cavity will continue to result in new discoveries at the intersection of microbial ecology and pathogenesis.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01DE023193 to M.W., R01DE020100 to M.W., F32DE027281 to G.R.L., and K99DE031018 to G.R.L.
We thank our reviewers for their valuable and insightful comments and the M.W. laboratory for their feedback on the manuscript.
Contributor Information
Marvin Whiteley, Email: mwhiteley3@gatech.edu.
Gina R. Lewin, Email: glewin3@gatech.edu.
Kimberly A. Kline, Nanyang Technological University
REFERENCES
- 1.GBD 2017 Disease and Injury Incidence and Prevalence Collaborators. 2018. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC), National Center for Health Statistics (NCHS). 2018. National Health and Nutrition Examination Survey questionnaire 2017–2018. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Hyattsville, MD. [Google Scholar]
- 3.Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, Whiteley M. 2014. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci USA 111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuboniwa M, Houser JR, Hendrickson EL, Wang Q, Alghamdi SA, Sakanaka A, Miller DP, Hutcherson JA, Wang T, Beck DAC, Whiteley M, Amano A, Wang H, Marcotte EM, Hackett M, Lamont RJ. 2017. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat Microbiol 2:1493–1499. doi: 10.1038/s41564-017-0021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chipashvili O, Utter DR, Bedree JK, Ma Y, Schulte F, Mascarin G, Alayyoubi Y, Chouhan D, Hardt M, Bidlack F, Hasturk H, He X, McLean JS, Bor B. 2021. Episymbiotic Saccharibacteria suppresses gingival inflammation and bone loss in mice through host bacterial modulation. Cell Host Microbe 29:1649–1662. doi: 10.1016/j.chom.2021.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim D, Barraza JP, Arthur RA, Hara A, Lewis K, Liu Y, Scisci EL, Hajishengallis E, Whiteley M, Koo H. 2020. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc Natl Acad Sci USA 117:12375–12386. doi: 10.1073/pnas.1919099117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tal M. 1980. Periodontal disease and oral hygiene: described by Antoni van Leeuwenhoek. J Periodontol 51:668–669. doi: 10.1902/jop.1980.51.11.668. [DOI] [PubMed] [Google Scholar]
- 8.Gest H. 2004. The discovery of microorganisms by Robert Hooke and Antoni Van Leeuwenhoek, fellows of the Royal Society. Notes Rec R Soc Lond 58:187–201. doi: 10.1098/rsnr.2004.0055. [DOI] [PubMed] [Google Scholar]
- 9.Gibbons RJ. 1964. Bacteriology of dental caries. J Dent Res 43:1021–1028. doi: 10.1177/00220345640430060301. [DOI] [PubMed] [Google Scholar]
- 10.Miller WD. 1890. The micro-organisms of the human mouth: the local and general diseases which are caused by them. The S. S. White Dental Mfg. Co., Philadelphia, PA. [Google Scholar]
- 11.Clarke JK. 1924. On the bacterial factor in the aetiology of dental caries. Br J Exp Pathol 5:141–147. [Google Scholar]
- 12.Socransky SS. 1977. Microbiology of periodontal disease – present status and future considerations. J Periodontol 48:497–504. doi: 10.1902/jop.1977.48.9.497. [DOI] [PubMed] [Google Scholar]
- 13.Gibbons RJ, Houte JV. 1975. Bacterial adherence in oral microbial ecology. Annu Rev Microbiol 29:19–42. doi: 10.1146/annurev.mi.29.100175.000315. [DOI] [PubMed] [Google Scholar]
- 14.Gibbons RJ, Nygaard M. 1970. Interbacterial aggregation of plaque bacteria. Arch Oral Biol 15:1397–IN39. doi: 10.1016/0003-9969(70)90031-2. [DOI] [PubMed] [Google Scholar]
- 15.Stevens MRE, Luo TL, Vornhagen J, Jakubovics NS, Gilsdorf JR, Marrs CF, Møretrø T, Rickard AH. 2015. Coaggregation occurs between microorganisms isolated from different environments. FEMS Microbiol Ecol 91:fiv123. doi: 10.1093/femsec/fiv123. [DOI] [PubMed] [Google Scholar]
- 16.Costerton JW, Geesey GG, Cheng KJ. 1978. How bacteria stick. Sci Am 238:86–95. doi: 10.1038/scientificamerican0178-86. [DOI] [PubMed] [Google Scholar]
- 17.Kolenbrander PE, Palmer RJ, Jr., Periasamy S, Jakubovics NS. 2010. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol 8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 18.Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ. 2002. Communication among oral bacteria. Microbiol Mol Biol Rev 66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolenbrander PE, London J. 1993. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol 175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolenbrander PE, Palmer RJ, Jr., Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. 2006. Bacterial interactions and successions during plaque development. Periodontol 2000 42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 21.Kolenbrander PE. 1988. Intergeneric coaggregation among human oral bacteria and ecology of dental plaque. Annu Rev Microbiol 42:627–656. doi: 10.1146/annurev.mi.42.100188.003211. [DOI] [PubMed] [Google Scholar]
- 22.Mark Welch JL, Dewhirst FE, Borisy GG. 2019. Biogeography of the oral microbiome: the site-specialist hypothesis. Annu Rev Microbiol 73:335–358. doi: 10.1146/annurev-micro-090817-062503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henrici AT. 1933. Studies of freshwater bacteria: I. A direct microscopic technique. J Bacteriol 25:277–287. doi: 10.1128/jb.25.3.277-287.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zobell CE, Allen EC. 1935. The significance of marine bacteria in the fouling of submerged surfaces. J Bacteriol 29:239–251. doi: 10.1128/jb.29.3.239-251.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 26.Armitage GC. 2013. Learned and unlearned concepts in periodontal diagnostics: a 50‐year perspective. Periodontol 2000 62:20–36. doi: 10.1111/prd.12006. [DOI] [PubMed] [Google Scholar]
- 27.Haffajee AD, Socransky SS, Patel MR, Song X. 2008. Microbial complexes in supragingival plaque. Oral Microbiol Immunol 23:196–205. doi: 10.1111/j.1399-302X.2007.00411.x. [DOI] [PubMed] [Google Scholar]
- 28.Hajishengallis G, Lamont RJ. 2012. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol 27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cross KL, Campbell JH, Balachandran M, Campbell AG, Cooper CJ, Griffen A, Heaton M, Joshi S, Klingeman D, Leys E, Yang Z, Parks JM, Podar M. 2019. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat Biotechnol 37:1314–1321. doi: 10.1038/s41587-019-0260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. 2018. New insights into human nostril microbiome from the expanded Human Oral Microbiome Database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems 3:e00187-18. doi: 10.1128/mSystems.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson H, Rybalka A, Moazzez R, Dewhirst FE, Wade WG. 2015. In vitro culture of previously uncultured oral bacterial phylotypes. Appl Environ Microbiol 81:8307–8314. doi: 10.1128/AEM.02156-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rheims H, Rainey FA, Stackebrandt E. 1996. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol 17:159–169. doi: 10.1007/BF01574689. [DOI] [Google Scholar]
- 33.Soro V, Dutton LC, Sprague SV, Nobbs AH, Ireland AJ, Sandy JR, Jepson MA, Micaroni M, Splatt PR, Dymock D, Jenkinson HF. 2014. Axenic culture of a candidate division TM7 bacterium from the human oral cavity and biofilm interactions with other oral bacteria. Appl Environ Microbiol 80:6480–6489. doi: 10.1128/AEM.01827-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu S-Y, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, Nelson KE, Lux R, Shi W. 2015. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci USA 112:244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solbiati J, Frias-Lopez J. 2018. Metatranscriptome of the oral microbiome in health and disease. J Dent Res 97:492–500. doi: 10.1177/0022034518761644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Relman DA. 2012. The human microbiome: ecosystem resilience and health. Nutr Rev 70:S2–S9. doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duran-Pinedo AE, Chen T, Teles R, Starr JR, Wang X, Krishnan K, Frias-Lopez J. 2014. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J 8:1659–1672. doi: 10.1038/ismej.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yost S, Duran-Pinedo AE, Teles R, Krishnan K, Frias-Lopez J. 2015. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med 7:27. doi: 10.1186/s13073-015-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewin GR, Stocke KS, Lamont RJ, Whiteley M. 2022. A quantitative framework reveals traditional laboratory growth is a highly accurate model of human oral infection. Proc Natl Acad Sci USA 119:e2116637119. doi: 10.1073/pnas.2116637119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. 2014. Metatranscriptomics of the human oral microbiome during health and disease. mBio 5:e01012-14. doi: 10.1128/mBio.01012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nowicki EM, Shroff R, Singleton JA, Renaud DE, Wallace D, Drury J, Zirnheld J, Colleti B, Ellington AD, Lamont RJ, Scott DA, Whiteley M. 2018. Microbiota and metatranscriptome changes accompanying the onset of gingivitis. mBio 9:e00575-18. doi: 10.1128/mBio.00575-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azimi S, Lewin GR, Whiteley M. 2022. The biogeography of infection revisited. Nat Rev Microbiol doi: 10.1038/s41579-022-00683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones SJ. 1972. A special relationship between spherical and filamentous microorganisms in mature human dental plaque. Arch Oral Biol 17:613–616. doi: 10.1016/0003-9969(72)90081-7. [DOI] [PubMed] [Google Scholar]
- 44.Amann RI, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valm AM, Mark Welch JL, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, Borisy GG. 2011. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci USA 108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. 2016. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA 113:E791–E800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilbert SA, Mark Welch JL, Borisy GG. 2020. Spatial ecology of the human tongue dorsum microbiome. Cell Rep 30:4003–4015. doi: 10.1016/j.celrep.2020.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon-Soro A, Ren Z, Krom BP, Hoogenkamp MA, Cabello-Yeves PJ, Daniel SG, Bittinger K, Tomas I, Koo H, Mira A. 2022. Polymicrobial aggregates in human saliva build the oral biofilm. mBio 13:e00131-22. doi: 10.1128/mbio.00131-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heller D, Helmerhorst EJ, Gower AC, Siqueira WL, Paster BJ, Oppenheim FG. 2016. Microbial diversity in the early in vivo-formed dental biofilm. Appl Environ Microbiol 82:1881–1888. doi: 10.1128/AEM.03984-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valm AM, Oldenbourg R, Borisy GG. 2016. Multiplexed spectral imaging of 120 different fluorescent labels. PLoS One 11:e0158495. doi: 10.1371/journal.pone.0158495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi H, Shi Q, Grodner B, Lenz JS, Zipfel WR, Brito IL, De Vlaminck I. 2020. Highly multiplexed spatial mapping of microbial communities. Nature 588:676–681. doi: 10.1038/s41586-020-2983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jorth P, Spero MA, Livingston J, Newman DK. 2019. Quantitative visualization of gene expression in mucoid and nonmucoid Pseudomonas aeruginosa aggregates reveals localized peak expression of alginate in the hypoxic zone. mBio 10:e02622-19. doi: 10.1128/mBio.02622-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dar D, Dar N, Cai L, Newman DK. 2021. Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science 373:eabi4882. doi: 10.1126/science.abi4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ammann TW, Gmür R, Thurnheer T. 2012. Advancement of the 10-species subgingival Zurich biofilm model by examining different nutritional conditions and defining the structure of the in vitro biofilms. BMC Microbiol 12:227–213. doi: 10.1186/1471-2180-12-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bao K, Belibasakis GN, Thurnheer T, Aduse-Opoku J, Curtis MA, Bostanci N. 2014. Role of Porphyromonas gingivalis gingipains in multi-species biofilm formation. BMC Microbiol 14:258–258. doi: 10.1186/s12866-014-0258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cornforth DM, Diggle FL, Melvin JA, Bomberger JM, Whiteley M. 2020. Quantitative framework for model evaluation in microbiology research using Pseudomonas aeruginosa and cystic fibrosis infection as a test case. mBio 11:e03042-19. doi: 10.1128/mBio.03042-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carrouel F, Viennot S, Santamaria J, Veber P, Bourgeois D. 2016. Quantitative molecular detection of 19 major pathogens in the interdental biofilm of periodontally healthy young adults. Front Microbiol 7:840. doi: 10.3389/fmicb.2016.00840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Socransky SS, Haffajee AD. 2002. Dental biofilms: difficult therapeutic targets. Periodontol 2000 28:12–55. doi: 10.1034/j.1600-0757.2002.280102.x. [DOI] [PubMed] [Google Scholar]
- 59.Socransky SS, Haffajee AD. 2005. Periodontal microbial ecology. Periodontol 2000 38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 60.Palmer RJ, Jr., Wu R, Gordon S, Bloomquist CG, Liljemark WF, Kilian M, Kolenbrander PE. 2001. Retrieval of biofilms from the oral cavity. Methods Enzymol 337:393–403. doi: 10.1016/S0076-6879(01)37028-3. [DOI] [PubMed] [Google Scholar]
- 61.Foster JS, Palmer RJ, Jr., Kolenbrander PE. 2003. Human oral cavity as a model for the study of genome-genome interactions. Biol Bull 204:200–204. doi: 10.2307/1543559. [DOI] [PubMed] [Google Scholar]
- 62.Shapiro S, Giertsen E, Guggenheim B. 2002. An in vitro oral biofilm model for comparing the efficacy of antimicrobial mouthrinses. Caries Res 36:93–100. doi: 10.1159/000057866. [DOI] [PubMed] [Google Scholar]