ABSTRACT
The transcription factor AdpA is a key regulator controlling both secondary metabolism and morphological differentiation in Streptomyces. Due to its critical functions, its expression undergoes multilevel regulations at transcriptional, posttranscriptional, and translational levels, yet no posttranslational regulation has been reported. Sulfane sulfur, such as hydro polysulfide (HSnH, n ≥ 2) and organic polysulfide (RSnH, n ≥ 2), is common inside microorganisms, but its physiological functions are largely unclear. Here, we discovered that sulfane sulfur posttranslationally modifies AdpA in Streptomyces coelicolor via specifically reacting with Cys62 of AdpA to form a persulfide (Cys62-SSH). This modification decreases the affinity of AdpA to its self-promoter PadpA, allowing increased expression of adpA, further promoting the expression of its target genes actII-4 and wblA. ActII-4 activates actinorhodin biosynthesis, and WblA regulates morphological development. Bioinformatics analyses indicated that AdpA-Cys62 is highly conserved in Streptomyces, suggesting the prevalence of such modification in this genus. Thus, our study unveils a new type of regulation on the AdpA activity and sheds a light on how sulfane sulfur stimulates the production of antibiotics in Streptomyces.
KEYWORDS: sulfane sulfur, AdpA, polyketides, actinorhodin, Streptomyces
INTRODUCTION
Streptomyces spp. are Gram-positive bacteria with a filamentous form which colonize a wide range of terrestrial and aquatic niches. The most famous characteristic of Streptomyces is the ability to produce a myriad of secondary metabolites, including antibiotics, antifungals, antivirals, anthelmintic agents, antitumoral drugs, antihypertensives, herbicides, and valuable pigments (1–3). Much effort has been spent on searching, identifying, and modifying the gene clusters responsible for biosynthesis of these secondary metabolites (4). In contrast, much less energy has been invested in illustrating the transcriptional/translational regulation of these gene clusters. One reason is that Streptomyces have a complex life cycle that includes sporulation, a vegetative or substrate state, and aerial mycelial growth. The biosynthesis of secondary metabolites is closely linked to the stages of the life cycle (5, 6), which makes relative studies challenging.
AdpA is a transcriptional regulator universally present in Streptomyces (7). It is located in the second layer of the A-factor-dependent transcriptional network in Streptomyces griseus; the first layer is the A-factor receptor, which activates AdpA expression at the presence of A-factor (γ-butyrolactone, a quorum sensing hormone). Therefore, AdpA expression is indirectly controlled by the quorum sensing signal. Aside from A-factor, there are at least four other players in AdpA expression regulation—the master developmental regulator BldD regulating at the transcriptional level (8, 9), the cis-antisense RNA regulating at the posttranscriptional level (10), the rare tRNA (tRNAUUALeu)-encoding gene bldA, and the posttranscriptional tRNA modifications regulating at the translational level (11, 12). It was also reported that AdpA can be transcriptionally self-inhibited (13). One reason why regulation of AdpA expression is so complicated is that AdpA is a key regulator of both secondary metabolism and morphological differentiation (14). Considering the critical functions it conducts, whether there are other players regulating at different levels on AdpA expression or activity is unclear but worthy of further investigation.
Sulfane sulfur-containing compounds, such as persulfide (HSSH and RSSH) and polysulfide (HSSnH, Sn, RSSnH, RSSnR, n ≥ 2), are commonly present in both eukaryotic and prokaryotic cells (15). In the past 2 decades, intensive studies of sulfane sulfur have been performed with mammalian cells because it was found that sulfane sulfur is involved in the regulation of diverse physiological and pathological processes, including apoptosis, carcinogenesis, and redox maintenance (16–19). On the other hand, studies of microorganism sulfane sulfur are traditionally focused on its metabolism and its role in the global sulfur cycle (20, 21). In recent years, the physiological functions of sulfane sulfur in microorganisms also got attentions. For instance, Peng et al. (22) found that sulfane sulfur regulates the expression of virulence factors in Staphylococcus aureus, and Liu et al. (23) reported that sulfane sulfur is involved in photosynthesis regulation in Synechococcus. Although they have been noticed, the functions of sulfane sulfur in microorganisms are largely obscure.
In a previous study, we discovered that sulfane sulfur functions as a signal to activate actinorhodin (ACT) production in S. coelicolor M145, a model strain of Streptomyces. In addition, the spore formation process is accelerated by endogenously accumulated sulfane sulfur (24). These phenomena suggest that sulfane sulfur affects both secondary metabolism and the cell cycle in S. coelicolor M145. Based on these findings, we studied the underlying mechanism of how sulfane sulfur performs such functions. We found that AdpA is the key medium of sulfane sulfur signaling. AdpA senses the level of intracellular sulfane sulfur and adjusts ACT production and spore formation. Even the expression of AdpA itself is affected by sulfane sulfur; i.e., sulfane sulfur is a new regulator of AdpA. Thus, this study unveils one way via which sulfane sulfur signals in Streptomyces.
RESULTS
AdpA is a key regulator of ACT production and morphological development in S. coelicolor.
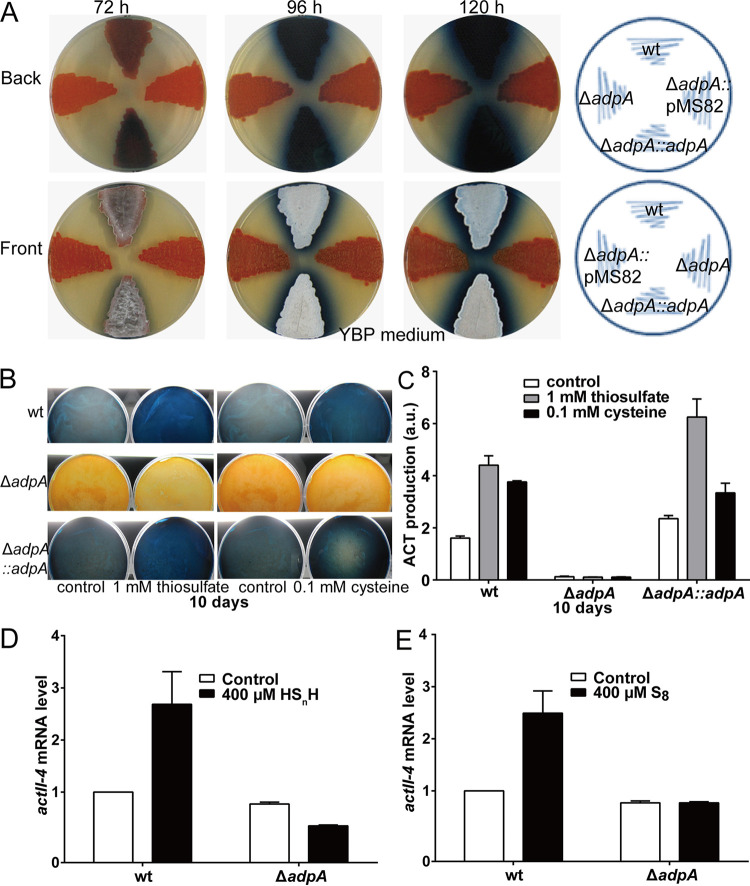
Previous studies demonstrated that AdpA is involved in the regulation of ACT production and morphological development in S. coelicolor (25, 26). Here, we constructed an adpA-disrupted S. coelicolor M145 strain (ΔadpA). It exhibited a phenotype of no ACT but high undecylprodigiosin (RED) production when cultured on yeast-beef-peptone (YBP) agar medium (Fig. 1A). Complementary expression of the adpA gene using a plasmid, pMS82-adpA (ΔadpA::adpA), restored ACT production, while the control, ΔadpA harboring an empty plasmid (ΔadpA::pMS82), showed no change. In addition, we noticed that both ΔadpA and the control showed a bald and nonspore form on YBP medium (Fig. 1A), while ΔadpA::adpA restored the spore formation. These results verified that AdpA controls ACT production and morphological development in S. coelicolor strain M145.
FIG 1.
AdpA is required for ACT production and morphological development in S. coelicolor M145. (A) Phenotypes of the WT, ΔadpA, ΔadpA::adpA, and ΔadpA::pMS82 strains grown on YBP medium at 30°C. Images were taken at the indicated times. (B) First, 1 mM thiosulfate or 0.1 mM cysteine was added to YBP agar plates before inoculation. The plates were incubated at 30°C for 10 days, and images were captured from the reverse side of the plates. (C) Quantitative determination of ACT produced by the wt, ΔadpA, and ΔadpA::adpA strains on YBP containing thiosulfate or cysteine. The plates were incubated at 30°C for 10 days. Data are from three independent repeats. (D and E) WT and ΔadpA strains were grown on YBP liquid medium. At 36 h, 400 μM HSnH or S8 was added, and after 1 h of induction, RNA samples were isolated. Real-time PCR data are from three independent repeats and shown as the average ± standard deviation (SD).
Sulfane sulfur performing ACT activation requires the presence of AdpA.
Since the ΔadpA strain displayed opposite phenotypes as that of the sulfane sulfur-treated strain (24), we suspected that AdpA had interwound functions with sulfane sulfur. We performed sulfane sulfur induction experiments using the S. coelicolor M145 (wild type [WT]), ΔadpA, and ΔadpA::adpA strains. The strains were spread on YBP medium containing 1 mM thiosulfate or 0.1 mM cysteine, which can be converted to sulfane sulfur in vivo (27), and cultured at 30°C for 10 days. For the WT, the production of ACT was significantly increased by thiosulfate/cysteine treatment (Fig. 1B and C). For ΔadpA, no production of ACT was observed with or without thiosulfate/cysteine treatment. For ΔadpA::adpA, the induction effects were similar to those in the WT (Fig. 1B and C). These results indicated that AdpA is required for sulfane sulfur to execute the ACT production-activating function.
ActII-4 is the ACT production “pathway-specific” activator (26, 28). We analyzed transcription of actII-4 using the real-time quantitative reverse transcription-PCR (RT-qPCR) method. The WT and ΔadpA strains were treated with two sulfane sulfur-containing chemicals, hydrogen polysulfides (HSnH, n ≥ 2) and sublimed sulfur (S8). For the WT, the transcription level of actII-4 was much higher in the treated strain than that in the untreated one (Fig. 1D and E), whereas for ΔadpA, the transcription level of actII-4 had no obvious change after sulfane sulfur treatment (Fig. 1D and E). These results indicated that sulfane sulfur can increase ActII-4 expression, which subsequently activates ACT production, but this process requires the presence of AdpA.
Sulfane sulfur affects the interaction between AdpA and its cognate promoters.
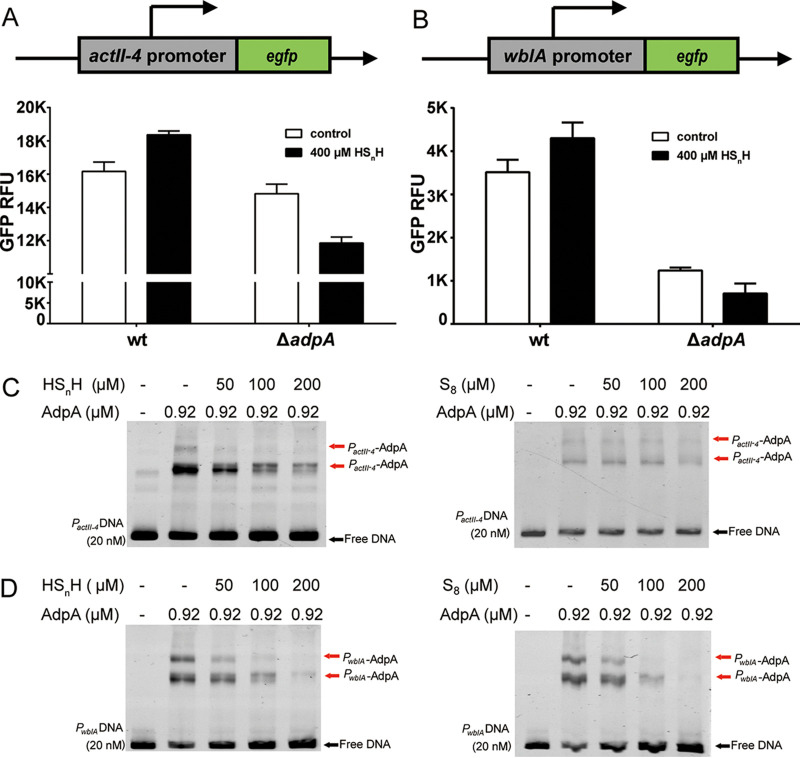
AdpA controls the transcription of actII-4 and wblA (whiB-like gene A, which controls morphological development in S. coelicolor) via binding to their promoters (29). Using these two promoters and an enhanced green fluorescence protein-encoding gene (egfp), we constructed two reporter systems (Fig. 2A and B). These reporter systems were introduced into the WT and ΔadpA strains. HSnH (400 μM) was used to treat the strains containing the reporter systems. After 30 min of treatment, the mycelium was collected by centrifugation, and the fluorescence was read by a fluorophotometer. For the WT strain, HSnH treatment enhanced the strength of both the actII-4 promoter (PactII-4) and wblA promoter (PwblA), evidenced by the increased EGFP expression, whereas, in the ΔadpA strain, EGFP expression was not increased but decreased after HSnH treatment, indicating that HSnH treatment lost the enhancing effect on these promoters. These results suggested that sulfane sulfur may affect the interaction between AdpA and its cognate promoters.
FIG 2.
Sulfane sulfur is involved in the process of AdpA regulating target genes. (A) HSnH was used to treat WT and ΔadpA strains harboring pMS82-actII-4p-egfp. (B) HSnH was used to treat WT and ΔadpA strains harboring pMS82-wblAp-egfp. Data are from three independent repeats and shown as the average ± SD. (C and D) EMSA analysis of the AdpA affinity to PactII-4 promoter DNA (C) and the PwblA promoter DNA (D). All lanes contained 20 nM probe DNA, lanes 2 to 5 contained protein with the indicated concentration, and lanes 3 to 5 contained the HSnH (left) or S8 (right). The black arrow indicates the free DNA probe, and red arrows indicate the PactII-4-AdpA or PwblA-AdpA complex.
We then performed electrophoretic mobility shift assays (EMSA) to investigate the interaction. The AdpA protein was expressed in Escherichia coli BL21(DE3) and purified. The DNA probes of the wblA and actII-4 promoters were obtained by PCR. When AdpA was mixed with the PactII-4 or PwblA DNA probe, it bound to them (Fig. 2C and D). When HSnH (200 μM) or S8 (200 μM) was also added, the fraction of the AdpA-probe complexes decreased (Fig. 2C and D). These results indicated that sulfane sulfur decreased the affinity of AdpA to PactII-4 and PwblA.
To test whether the influence of sulfane sulfur can be reversed by a reductant, we added dithiothreitol (DTT) into the mixture of sulfane sulfur (1 mM), AdpA, and the PwblA probe (the DTT dosage was 2-fold of HSnH/S8). After DTT treatment, AdpA restored the high affinity with the PwblA probe (Fig. 3), which had been attenuated by sulfane sulfur. These phenomena demonstrated that the affinity attenuation of AdpA to its cognate DNA caused by sulfane sulfur was reversible.
FIG 3.
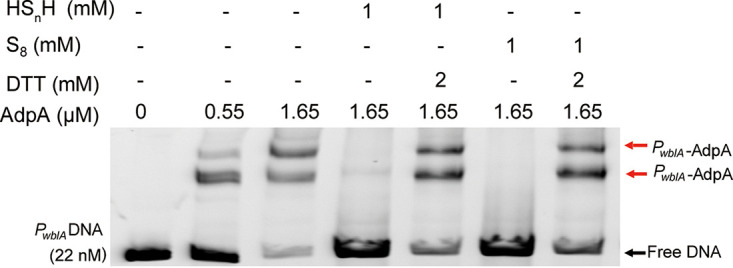
EMSA analysis of AdpA binding to PwblA promoter DNA. All lanes contained 22 nM PwblA DNA, lanes 2 to 7 contained AdpA, lanes 4 and 5 contained HSnH, lanes 6 and 7 contained S8, lanes 5 and 7 contained DTT.
Sulfane sulfur also affects the transcription of AdpA itself.
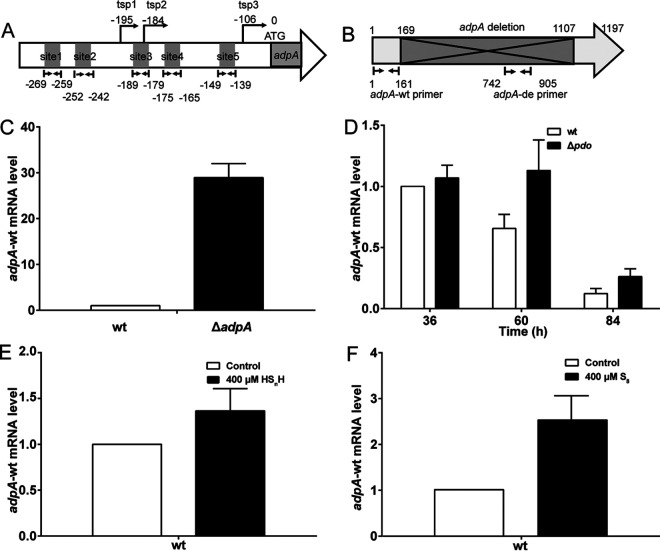
The adpA gene is transcriptionally self-controlled (30). There are five AdpA binding sites in the PadpA promoter (Fig. 4A). We designed a pair of primers (adpA-wt) from the undeleted part of the adpA gene (Fig. 4B) and used these primers to analyze the transcription change of adpA in the WT and ΔadpA. The transcription level of the undeleted part was ~30-fold higher in ΔadpA than that in the WT, indicating that in the absence of AdpA, the strength of PadpA was higher, i.e., AdpA acted as a repressor for its own transcription (Fig. 4C).
FIG 4.
Sulfane sulfur affects the transcription of adpA itself. (A) Schematic diagram of the AdpA binding sites in the adpA promoter region. (B) Schematic diagram of the AdpA coding sequence. The fragment covering 169 bp to 1,107 bp was deleted in ΔadpA. The adpA-wt and adpA-de primers were used to test the undeleted and deleted sequences, respectively. (C) RT-qPCR analysis of the adpA-wt mRNA level in the WT and ΔadpA. (D) RT-qPCR analysis of adpA-wt mRNA level in the WT and Δpdo. Data are from three independent repeats and shown as the average ± SD. (E and F) RT-qPCR analysis of adpA-wt in the WT and ΔadpA after induction by HSnH (400 μM) (E) and S8 (400 μM) (F). Data are from three independent repeats and shown as the average ± SD.
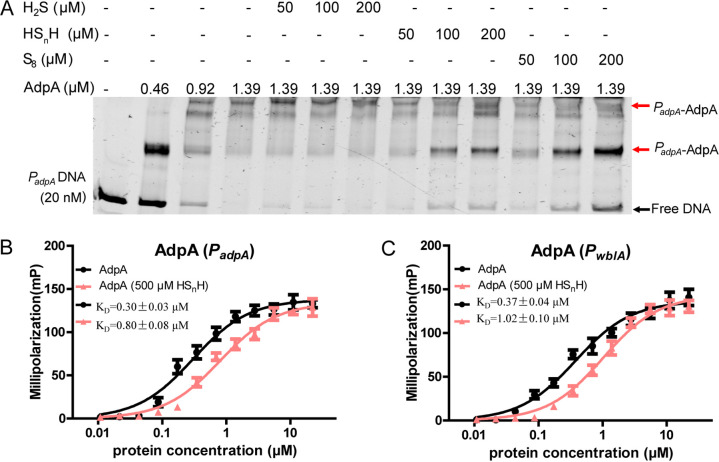
To test whether sulfane sulfur can affect this self-repression, we compared the transcription levels of adpA in the WT stain and the Δpdo strain. In the latter, intracellular sulfane sulfur is accumulated due to a lack of the persulfide oxidation gene (pdo) (24). Results showed that the adpA transcription levels were higher in Δpdo than in the WT (Fig. 4D). We then used exogenous sulfane sulfur to treat the WT strain and found that both HSnH (400 μM) and S8 (400 μM) can increase adpA transcription (Fig. 4E and F). EMSA showed that sulfane sulfur (100 to 200 μM) also reduced the affinity of AdpA to PadpA probe, as the unbound probe increased after the addition of HSnH and S8 (Fig. 5A). Fluorescence polarization (FP) analysis was performed, and the results showed that HSnH (500 μM) obviously increased the KD value (the equilibrium dissociation constant) of AdpA to the PadpA probe, as well as to the PwblA probe (Fig. 5B and C), indicating that the affinities of AdpA to these promoters were attenuated by HSnH.
FIG 5.
EMSA and FP analysis of AdpA binding to DNA probes. (A) EMSA analysis of AdpA binding to the PadpA probe. All lanes contained 20 nM probe, lanes 2 to 13 contained AdpA, lanes 5 to 7 contained H2S (50, 100, and 200 μM, respectively), lanes 8 to 10 contained HSnH (50, 100, and 200 μM, respectively), and lanes 11 to 13 contained S8 (50, 100, and 200 μM, respectively). (B and C) FP analysis of AdpA binding to the PadpA probe (B) and PwblA probe (C). First, 1 nM FAM-labeled PadpA or PwblA was incubated with increasing amounts of AdpA or HSnH (500 μM)-treated AdpA. The KD values were calculated based on FP data using GraphPad Prism 5 software. Data are from three independent experiments and shown as the average ± SD.
Simulating the dynamics of the AdpA-controlled promoter system with a simplified model.
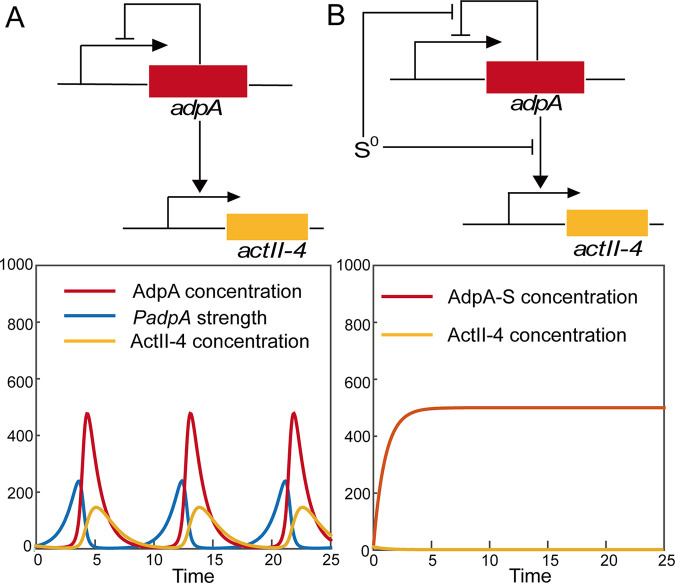
For the regulation of AdpA on PadpA strength, the logic is easy to understand; PadpA and AdpA compose a classic closed negative-feedback loop. AdpA is a repressor of PadpA. When AdpA is abundant, it binds with PadpA to turn it off/down. The off/down state lasts until the AdpA concentration becomes low due to degradation, and then PadpA turns on/up again. Therefore, without other interference, the strength of PadpA fluctuates, leading to a wave-like expression pattern of AdpA. Since AdpA is an activator of Pact-4, the expression of Act-4 also fluctuates following the concentration wave of AdpA. The principle of these dynamics can be simulated with a simplified mathematical model (Fig. 6A).
FIG 6.
Modeling principles of how AdpA regulates adpA and actII-4 expression. (A) In the absence of sulfane sulfur, PadpA is self-repressed by AdpA to form a negative-feedback loop, and hence, both PadpA strength and AdpA amount show a wave-like pattern. (B) Sulfane sulfur temporarily breaks the negative-feedback loop, which leads to a higher and longer expression of AdpA but not ActII-4 at the initial stage. After sulfane sulfur is consumed, the high AdpA level will lead to high expression of ActII-4. Equations and related parameters used for modeling are provided in Text S1.
Mathematical modelling. Download Text S1, PDF file, 0.3 MB (274.3KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
For the regulation of AdpA on PactII and PwblA, there is a paradoxical phenomenon; reporter system and RT-qPCR experiments indicated that AdpA enhanced the strength of these two promoters in the presence of sulfane sulfur, but EMSA and FP experiments indicated that sulfane sulfur decreased the AdpA affinities to them. There are several possible reasons for this paradox:
-
1.
There is another player, possibly a transcription factor (TF), involved in this system. This unknown TF takes the place of AdpA in the presence of sulfane sulfur and then further increases PactII/PwblA strength.
-
2.
Sulfane sulfur leads to increased AdpA production, and when the concentration of AdpA is higher than that of sulfane sulfur, free AdpA is more abundant than the sulfane sulfur-modified AdpA. In this case, free AdpA binds to PactII/PwblA and enhances their strength.
-
3.
The affinities of sulfane sulfur-modified AdpA to PadpA, PactII, and PwblA are different. These differences lead to variations in the transcriptional levels of these genes.
Previously, we observed that the concentration of intracellular sulfane sulfur of S. coelicolor changed along with expression levels of its metabolic genes (24). Therefore, it is highly possible that the ratio of sulfane sulfur to AdpA (S0/AdpA) is dynamic (a scenario in item 2). To understand how S0/AdpA influences the strength of PactII-4 and PadpA, we developed another mathematical model; at the initial stage, S0/AdpA is high, so the sulfane sulfur-reacted AdpA (AdpA-S) is the dominant form (more abundant than apo AdpA), which leads to enhanced expression of adpA but not actII-4 (Fig. 6B). Before S0 is completely consumed, AdpA is continuously produced, leading to a higher level of AdpA than that in the wave-like expression pattern. However, along with consumption of S0, S0/AdpA gradually reduces, and finally apo AdpA becomes the dominant form; then the AdpA system returns to its closed negative-feedback loop as shown in Fig. 6A. Based on this simulation, we proposed that sulfane sulfur can temporarily break the self-inhibition in AdpA expression, allowing AdpA to accumulate to a higher level for a longer period (compared with the no-sulfane sulfur scenario), which finally leads to more ActII-4 expression.
The cysteine residue Cys62 is critical for AdpA sensing sulfane sulfur.
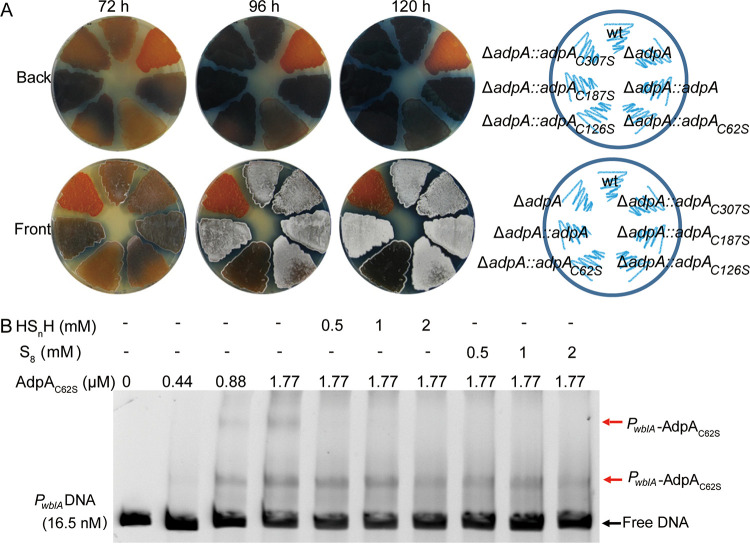
Sulfane sulfur can react with cysteine residues of certain proteins to change their configurations (31, 32). AdpA contains four cysteine residues, Cys62, Cys126, Cys187, and Cys307. To find out which cysteine residue involves in the AdpA-sulfane sulfur interaction, we made a cysteine-to-serine mutation on each cysteine residue of AdpA. The mutated adpA genes were introduced into the ΔadpA strain. When growing in YBP agar medium, the ΔadpA::adpAC126S, ΔadpA::adpAC187S, and ΔadpA::adpAC307S strains did not show obvious difference from the WT and ΔadpA::adpA strains. However, the ΔadpA::adpAC62S strain was distinct from the others. It lost the ability to generate spores, and its ACT production was also apparently lower (Fig. 7A). These phenotype changes indicated that Cys62 was critical for AdpA performing its regulatory function.
FIG 7.
Cys62 residue is critical for AdpA sensing sulfane sulfur. (A) Phenotypes of WT, ΔadpA, and complementary strains (ΔadpA::adpAC62S, ΔadpA::adpAC126S, ΔadpA::adpAC187S, and ΔadpA::adpAC307S) grown on YBP medium. Images were captured from both sides of the plates. (B) EMSA analysis of AdpAC62S binding to PwblA DNA. All lanes contained 16.5 nM probe DNA, lanes 2 to 10 contained AdpA, lanes 5 to 7 contained HSnH, and lanes 8 to 10 contained S8. The black arrow indicates the free DNA probe, and red arrows indicate the PwblA-AdpAC62S complex.
EMSA was then performed to examine whether the C62S mutation affects the binding of AdpA to its cognate promoter. AdpAC62S still bound to the PwblA DNA fragment, and two main PwblA-AdpAC62S complexes with different molecular weights (MW) were observed. The complex with lower MW was no longer influenced by sulfane sulfur even when sulfane sulfur was added at high concentrations (>1,000-fold higher than that of AdpAC62S) (Fig. 7B), whereas the complex with higher MW disappeared when high concentrations of sulfane sulfur were added. Since the lower-MW complex was the most abundant one formed by AdpAC62S and the PwblA DNA fragment, we proposed that the sulfane sulfur sensing ability was at least partially impaired by the C62S mutation.
To check how sulfane sulfur reacts with AdpA, purified AdpA was treated with HSnH (200 μM) or DTT (200 μM). The treated-AdpA was labeled with iodoacetamide (IAM) and then subjected to trypsin digestion, followed by LTQ-Orbitrap tandem mass spectrometry analysis. For the HSnH-treated AdpA, two peptides (1 and 2, Fig. 8) were identified. In peptide 1 (1,299.67 Da), the Cys62 residue was directly blocked by IAM to form Cys62-AM (acetamide) (Fig. 8 and Fig. S1). In peptide 2 (1,331.64 Da), a mass increase of 32 (+32 MW) was identified. A secondary mass spectrometry(MS2) spectrum indicated that the +32 MW happened on the thiol group of Cys62 to form peptide-S-AM (Fig. 8 and Fig. S2). The MS1 signal intensity ratio of peptide 1/peptide 2 was 17%. As the control, only a peptide with Cys62-AM (1,299.67 Da, peptide 3) was identified from DTT-reacted AdpA, corresponding to a direct blockage of IAM on the Cys62 residue (Fig. 8 and Fig. S3). These results indicated that sulfane sulfur can modify Cys62-SH to form Cys62-SSH.
FIG 8.
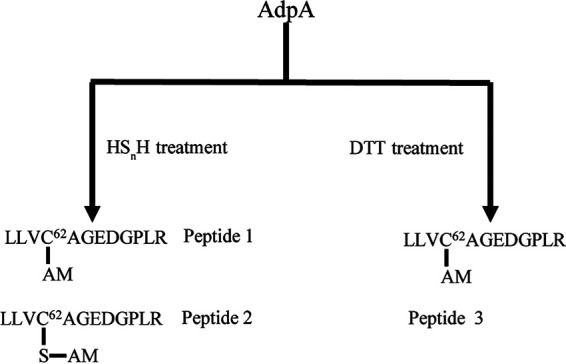
LC-MS/MS analysis of HSnH-treated and DTT-treated AdpA. MS2 data of the peptides are provided in Fig. S1 to S3.
MS2 data of peptide 1 (Cys62-AM) (from HSnH-treated AdpA). Download FIG S1, PDF file, 0.4 MB (431.2KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MS2 data of peptide 2 (Cys62-SH) (from HSnH-treated AdpA). Download FIG S2, PDF file, 0.4 MB (402.4KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MS2 data of peptide 1 (Cys62-AM) (from DTT-treated AdpA). Download FIG S3, PDF file, 0.4 MB (400.1KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The thiol group of Cys62 is accessible to solution due to its location on the AdpA 3D structure.
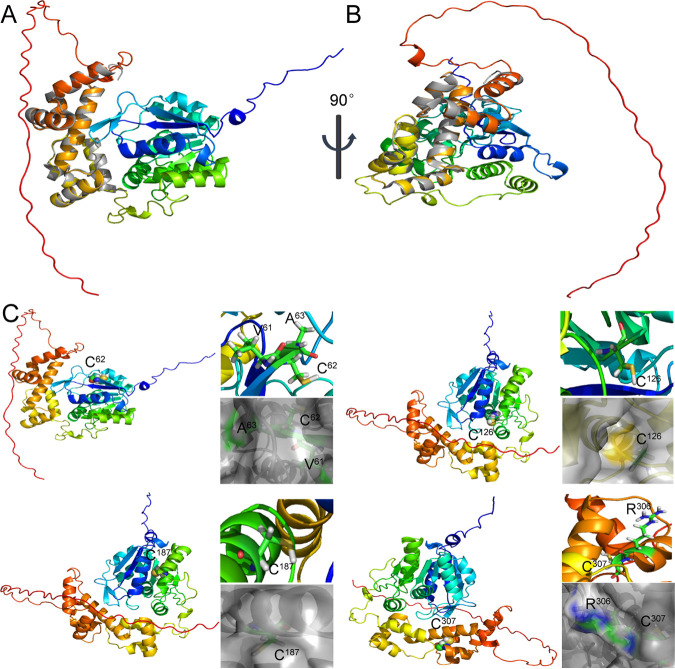
The 3D structure of AdpA was modeled by using AlphaFold 2 (https://www.hpc.caltech.edu/documentation/software-and-modules/alphafold-2). The crystal structure of a truncated AdpAsg containing only the DNA binding domain, which is from Streptomyces griseus, is available in the PDB database (PDB: 3w6v). We aligned AdpAsg with the modeled AdpA, and the alignment parameter RMSD was 0.365, indicating a high confidence of the predicted structure of AdpA (Fig. 9A and B).
FIG 9.
AlphaFold 2-predicted 3D structure of AdpA. (A and B) Alignment of the predicted AdpA structure (multicolor) with the AdpAsg crystal structure (gray). (C) Locations of the cysteine residues in AdpA. Yellow spheres represent the sulfur atoms.
We then analyzed the locations of the four cysteine residues in AdpA (Fig. 9C). Cys62, Cys126, and Cys187 are located in the ThiJ/PfpI/DJ-1-like dimerization domain, and Cys307 is located in the AraC/XylS-type DNA binding domain (DBD). Cys187 and Cys307 fold into the interior of AdpA, and hence they are protected from sulfane sulfur attack. In contrast, Cys62 is located near the protein surface, and its thiol group is exposed to solution, which may explain why Cys62 can be modified by sulfane sulfur. It is noteworthy that Cys126 is located on the interior surface of a tunnel through the dimerization domain. Therefore, sulfane sulfur compounds may not enter this tunnel to react with its thiol group. In addition, the distance between any two cysteine residues is too far to form a disulfide (S-S) or tri-sulfide (S-S-S) bond.
The cysteine residues are conserved in Streptomyces AdpAs.
We analyzed AdpA and its homologues in the Streptomyces genus. In the PATRIC database, 2,752 of 3,033 sequenced Streptomyces strains contain AdpA, accounting for a 90.73% prevalence. We selected some representative AdpA sequences to construct a phylogenetic tree. The results revealed that AdpA homologues were not on one evolutionary branch (Fig. S4). However, when we performed multiple sequence comparisons with them, we found that their four cysteine residues were highly conserved, including Cys62 (Fig. S5). These results suggested that using cysteine residues to sense sulfane sulfur may be a common mechanism for AdpA functioning in Streptomyces.
Bioinformatics analysis of AdpA distribution. Phylogenetic analysis of AdpA and its homologues in Streptomyces and Actinomycetes. AdpA sequence accession numbers in NCBI: Streptomyces coelicolor (SCO2792, CAB87229.1), Streptomyces xiamenensis 318 (SXIM_38050, WP_030737153.1), Streptomyces roseosporus NRRL 11379 (CP979_13280, KAA6217800.1), Streptomyces lincolnensis NRRL 2936 (SLINC_3216, ANS65440.1), Streptomyces cyanogenus S136 (S1361_15285, QTD98720.1), Streptomyces ansochromogenes 7100 (AdpA-L, ABY86620.1), Streptomyces diastatochromogenes 1628 (AdpAdi, AFX97763.1), Streptomyces chattanoogensis L10 (AdpAch, ACY78399.1), Streptomyces fradiae CGMCC 4.7387 (BG846_05140, OSY49238.1), Streptomyces hygroscopicus subsp. jinggangensis 5008 (SHJG_4295, AEY89567.1), Streptomyces lividans 1326 (SLI_3139, EOY47852.1), Streptomyces ghanaensis ATCC 14672 (AdpAgh, EFE69329.1), Streptomyces griseus (SGR_4742, BAG21571.1), Streptomyces clavuligerus ATCC 27064 (SCLAV_1957, EFG07030.1), Streptomyces sp. FR-008 (SFR_4905, ALM41520.1), Streptomyces venezuelae ATCC 10712 (SVEN_2580, CCA55866.1), Streptomyces avermitilis MA-4680 (SAV_5261, BAC72973.1), Streptomyces bingchenggensis BCW-1 (SBI_06911, ADI10031.1), Streptomyces albus J1074 (XNR_4181, AGI90514.1), Saccharopolyspora erythraea NRRL2338 (SACE_4523, CAM03792.1), Actinoplanes teichomyceticus NRRL-B16726 (AdpA19AT, AKR67211.1), Micromonospora echinospora DSM 43036 (FHU28_002617, WP_184684050.1), Micromonospora aurantiaca ATCC 27029 (MICAU_1999, WP_013285179.1), and Amycolatopsis mediterranei U32 (AdpAamed, AMED_7695, ADJ49403.1). The phylogenetic tree was constructed using MEGA 5.0. Download FIG S4, PDF file, 0.3 MB (346.8KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence alignment of AdpA with its homologues in Streptomyces and Actinomycetes. AdpA amino acid sequences of Streptomyces xiamenensis 318 (AdpAsxi, SXIM_38050, GenBank accession no. WP_030737153.1), Streptomyces roseosporus NRRL 11379 (AdpAsrose, CP979_13280, KAA6217800.1), Streptomyces lincolnensis NRRL 2936 (AdpAslinc, SLINC_3216, ANS65440.1), Streptomyces hygroscopicus subsp. jinggangensis 5008 (AdpAshjg, SHJG_4295, AEY89567.1), Streptomyces griseus (AdpAsg, SGR_4742, BAG21571.1), Saccharopolyspora erythraea NRRL2338 (AdpAsace, SACE_4523, CAM03792.1), and Amycolatopsis mediterranei U32 (AdpAamed, AMED_7695, ADJ49403.1) were used. Download FIG S5, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
The global transcription factor AdpA plays an important role in regulation of secondary metabolism and morphological development in the Streptomyces genus (33–37). Its own expression is controlled by multiple factors. In this study, we discovered that sulfane sulfur affects AdpA activity via the posttranslational modification. After reacting with sulfane sulfur, the affinity of AdpA to its cognate promoters, PadpA, PactII-4, and PwblA, is attenuated. We constructed a simplified model to help understand the effect of sulfane sulfur on the AdpA-controlled promoters. As shown in our simulation (Fig. 6), PadpA is under the control of a negative feedback loop of self-repression. Without the presence of sulfane sulfur and/or other disturbing factors, activation of AdpA on actII-4 and wblA expression cannot last long due to the negative feedback. Sulfane sulfur modifies AdpA to temporarily break the self-repression, and hence, AdpA can accumulate to a higher level for a longer time until sulfane sulfur is consumed. The accumulated AdpA finally activates expression of actII-4 and wblA. Thus, the effect of sulfane sulfur on the AdpA regulon may represent a fine-tuned regulation for the production of antibiotics and morphological development.
Furthermore, we found that Cys62 is critical for AdpA sensing sulfane sulfur. Our in vitro experiments showed that sulfane sulfur treatment can lead to a sulfhydration modification in Cys62 (Cys62-SSH), and this modification was not observed in the other three cysteine residues of AdpA. A limitation of this work is that such modification has not been examined in vivo due to the lack of a trustable method. The AlphaFold 2 predicted structure shows that the thiol of Cys62 is accessible to solution, while the other thiols are not, which may explain why Cys62 is easily sulfhydrated by sulfane sulfur. However, we also noticed that AdpA and AdpAC62S formed different complexes with the PwblA DNA fragment even in the absence of sulfane sulfur, suggesting that the binding pattern was affected by C62S mutation. Since Cys62 is located in the dimerization domain, it may affect the dimerization and then alter the AdpA binding pattern. Therefore, C62S mutation may result in multiple influences, including both DNA binding and sulfane sulfur sensing.
It is noteworthy that the AdpA complemented strain (ΔadpA::adpA) shows a peculiar pattern of ACT production—the center of the plate lacks the characteristic blue color—in the plates containing 0.1 M cysteine (Fig. 1B). A similar pattern was described in a recent report (38), and the authors linked the pattern formation to AdpA. Therefore, the peculiar pattern observed in our experiment may be caused by both AdpA expression alteration and cysteine addition. In the same report the authors discovered that expression of adpA and other genes controlled by it displayed a spatiotemporally separated wave-like pattern when S. coelicolor was cultured in solid medium and found that this pattern was driven by a combination of physiological gradients and regulatory network architecture (38). The finding is consistent with our simulation. From a genetic architecture viewpoint, the negative feedback loop inevitably leads to wave-like expression of adpA. However, the frequency (or wavelength) of the “wave” can be altered by environmental factors such as sulfane sulfur or siderophore. Understanding how the pattern forms and its determinants surely are important for interpreting the complicated differentiation process of Streptomyces and hence worth further study.
Streptomyces mainly exist in terrestrial soils, but they also have been detected in extreme environments such as deep seas, the north and south poles, hydrothermal fluids, hot springs, etc. In some environments (such as sea/lake bed) sulfane sulfur levels can be high, up to ~400 μM. Therefore, the possibility that Streptomyces live in sulfane sulfur-rich conditions cannot be excluded. In addition, sulfane sulfur has been recognized as a common intracellular chemical nowadays, and its concentration varies from 10 μM to ~500 μM (39). Hence, both self-produced and environmental sulfane sulfur may affect secondary metabolism and morphological development of Streptomyces.
In recent years, a few transcription factors that can be modified by sulfane sulfur have been identified from different microorganisms (40, 41). These transcription factors can be categorized into two groups. Group I consists of specific regulators for genes related to sulfur metabolism, including BigR (42), CstR (43), FisR (44), CsoR (24), and SqrR (45, 46). They control the expression of sulfane sulfur oxidation enzyme PDO and sulfane sulfur transferase RhoD (BigR in this case). Since H2S oxidation enzyme SQR is often located in the same operon with PDO and RhoD, they also control SQR expression (CstR, FisR, and SqrR in this case) (47). Group I regulators can sense the intracellular level of sulfane sulfur via their cysteine residues; when the sulfane sulfur level is high, cysteine residues are sulfhydrated to form an RSnH (n ≥ 2) or RSnR (n ≥ 3) bond, which leads to a configuration change of the regulator and subsequently high expression of PDO and other genes, and then the sulfane sulfur level is decreased through being oxidized to sulfite (20). Therefore, group I regulators mainly function as managers to maintain the homeostasis of intracellular sulfane sulfur.
Group II includes global or multifunctional transcription factors currently including MgrA (22), MexR (48), and OxyR (40). MgrA is a global virulence regulator of Staphylococcus aureus. It senses the intracellular level of sulfane sulfur to regulate the expression of virulence factors (22). MexR controls the multiple-antibiotic resistance process in Pseudomonas aeruginosa, and it senses intracellular sulfane sulfur to regulate the expression of the mexAB-oprM multidrug efflux operon (48). OxyR is a global antioxidation regulator in many bacteria. Recently, it was found that OxyR also senses sulfane sulfur and controls the expression of sulfane sulfur-reducing enzymes (40). Like group I, group II regulators also sense sulfane sulfur via their cysteine residues.
AdpA is deemed a group II regulator since it senses sulfane sulfur and accordingly adjusts the ACT production and spore formation in S. coelicolor. Bioinformatics analyses indicated that AdpA and its Cys residues are highly conserved in Streptomyces spp. Further investigation of this protein and its homologues should provide insights into how sulfane sulfur regulates the production of secondary metabolites and morphological developments in this genus. The widespread existence of AdpA implies that sulfane sulfur may play a wide range of regulatory functions in Streptomyces, providing unlimited possibilities for sulfane sulfur working as a signal molecule to stimulate increased production of important secondary metabolites, such as antibiotics, antitumor drugs, immunosuppressants, and antibiotics.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains and plasmids used or constructed in this work are summarized in Table S1.
Strains and plasmids used in this study. Download Table S1, PDF file, 0.3 MB (285.9KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Streptomyces strains cultivated at 30°C on mannitol soya flour (MS) solid medium (49) or yeast-beef-peptone (YBP) solid or liquid medium (50) were used for different experiments, including spore suspension preparation, intergeneric conjugation, growth assay, RNA isolation, and phenotypic observation. All E. coli strains were cultured at 37°C on solid or liquid Luria-Bertani (LB) medium. The E. coli DH5α and E. coli BL21(DE3) strains were used as hosts for plasmid construction and protein expression, respectively. E. coli ET12567 (pUZ8002) was used as a medium for transferring nonmethylated DNA to Streptomyces. When required, ampicillin (100 μg/mL), apramycin (50 μg/mL), chloramphenicol (25 μg/mL), kanamycin (50 μg/mL), hygromycin (50 μg/mL), or nalidixic acid (25 μg/mL) was added into the medium.
Preparation of sulfane sulfur species and other sulfur-containing compounds.
Sodium hydrosulfide (NaHS, H2S donor), cysteine, sulfur power, and thiosulfate were purchased from Sigma-Aldrich. S8 solution was prepared by dissolving excess sulfur powder in acetone to saturation. The concentration of saturated acetone sulfur is determined as 17 mM as reported previously (51). The stock solution of HSnH was prepared by mixing sulfur powder, NaOH, and NaHS (40 mM each chemical) in degassed distilled water at 30°C until the powder was completely dissolved as previously described (48, 52). The concentrations of HSnH were determined with the cyanolysis method (53) and calibrated by using thiosulfate as the standard. Specifically, pipetting 550 μL 1% boric acid into a 1.5-mL Eppendorf (EP) tube and removing dissolved oxygen by putting the EP tube in boiling water for 1 min and then adding 250 μL sample and 200 μL 1 M potassium cyanide. After boiling in a water bath (100°C) for 1 min, the EP tube was taken out and cooled down to room temperature, and 100 μL ferric nitrate color solution was added to form Fe(SCN)3. The A460nm absorbance value was detected. Thiosulfate was used to make a standard curve.
Construction of S. coelicolor ΔadpA.
All primers used in this experiment are listed in Table S2. The strain ΔadpA was constructed using a homologous recombination method (54). Briefly, a 939-bp region was deleted from the open reading frame (ORF) of adpA, leaving the upstream 168 bp (relative to the start codon) and the downstream 90-bp (relative to the stop codon) coding sequence of adpA. The knockout region was replaced by the apramycin resistance gene. The conjugation transfer was accomplished using the methylation-sensitive strain E. coli ET12567/pUZ8002 (containing the mutant plasmid pJTU-adpA) and S. coelicolor M145 following a previously reported protocol (55). The deletion mutant was verified by resistance screening and colony PCR with the primers VeradpA-F/R.
Primers used in this study. Download Table S2, PDF file, 0.2 MB (163.7KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of ΔadpA::adpA, ΔadpA::pMS82, ΔadpA::adpAC62S, ΔadpA::adpAC126S, ΔadpA::adpAC187S, and ΔadpA::adpAC307S.
A DNA fragment carrying the adpA ORF (1,197 bp) and its promoter (500 bp) was obtained using PCR amplification and was connected to the ΦBT1 integrative vector pMS82 (56) to generate pMS82-adpA plasmid (Table S1). This plasmid was then integrated into the attP site of the ΔadpA genome by intergeneric conjugation. To construct the negative-control strains, empty pMS82 vector was also transformed into ΔadpA; these derivative strains were selected and confirmed by PCR and DNA sequencing.
To construct other AdpA complementary strains, we used a point mutation strategy (57) to construct plasmids pMS82-adpAC62S, pMS82-adpAC126S, pMS82-adpAC187S, and pMS82-adpAC307S. The same method was used to obtain complementary strains ΔadpA::adpAC62S, ΔadpA::adpAC126S, ΔadpA::adpAC187S, and ΔadpA::adpAC307S. The primers used in this process are shown in Table S2.
AdpA protein overexpression, purification, and mutation.
To construct the AdpA expression strain, the coding sequence of adpA was amplified from WT genomic DNA with the primers ExadpA-F/R. The PCR product was purified and ligated into the pET15b vector with a C-terminal His tag to create plasmid pET-AdpA by using the ClonExpress II one-step cloning kit (TaKaRa). The plasmid was transformed into E. coli BL21(DE3) cells, which were grown in LB medium at 37°C to an optical density at 600 nm (OD600) of 0.6, and then a total of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added and an additional overnight cultivation was continued at 16°C. Cultures were collected by centrifugation and disrupted though a pressure cell homogenizer (SPCH-18) in sonication buffer (50 mM NaH2PO4, 250 mM NaCl, 20 mM imidazole, pH 8.0); 1 mM DTT was added before breaking the cells. Purification of the AdpA His-tagged proteins was performed with a Ni-NTA-Sefinose column (Sangon) as described previously (24). The protein purification process was conducted in an anaerobic glove box, which was filled with mixed gas (N2, 85%; H2, 10%; CO2, 5%). The purity of the protein was assessed by SDS-PAGE gel, and its concentration was determined using the bicinchoninic acid (BCA) protein assay reagent (Thermo Fisher Scientific). The same method was used for purification of AdpA mutants.
Electrophoretic mobility shift assay (EMSA).
The DNA probes containing AdpA binding sequences were amplified from genomic DNA. Different sulfane sulfur compounds were reacted with purified AdpA (and its mutants) in the binding buffer (20 mM Tris-HCI, 2 mM EDTA, 20 mM KCI, 0.5 mM dithiothreitol [DTT], pH 8.0) at room temperature for 20 min. Then DNA probe was added, and the binding reaction was performed at 30°C for 20 min. The binding complexes were separated on an 8% nondenaturing polyacrylamide gel at 120 V for 2 h in ice (58). The gel was dyed with SYBR green I (Sangon) for 20 min (44). All images were captured with a FluorChemQ system (Alpha Innotech).
RNA preparation, RT-PCR, and RT-qPCR.
To extract RNA, spores (2 × 107) of WT and ΔadpA strains were inoculated into the liquid YBP medium and incubated at 30°C with shaking (220 rpm) for 36 h to the mid-exponential phase. HSnH (400 μM) or S8 (400 μM) was added. After another 30-min cultivation, these mycelia were harvested and ground into powder with liquid nitrogen. Similarly, the cultures of WT, ΔadpA, and Δpdo were collected at the indicated times. All RNAs were isolated with a SteadyPure universal RNA extraction kit (Accurate Biology) following the manufacturer’s instructions, and their quality and concentration were determined using a NanoDrop ND-1000 device (Thermo Fisher). RT-PCR was carried out using a reverse transcriptase kit (Invitrogen) and SYBR premix Ex Taq (TaKaRa) following the manufacturers’ recommendations. The Roche LightCycler 480 thermal cycler was used (59). The expression of hrdB mRNA was used as the internal standard to normalize the relative quantities of cDNA. The relative expression abundance of the target gene was analyzed using a relative quantification method (2ΔCT, test gene-hrdB). Three independent replicates were performed.
Phenotypic analysis and ACT production assay.
S. coelicolor strains were cultured on solid YBP medium at 30°C for phenotypic analysis. ACT production was determined following a previously reported method (24, 60, 61). Briefly, Streptomyces strains were incubated on YBP medium for 7 or 10 days, and mycelia were harvested from the plate. KOH (1 M final concentration) was added to treat the mycelia for 4 h. Then the mixtures were centrifuged. The ACT concentration in the supernatant was determined by a spectrophotometer. Three independent biological experiments were replicated.
Construction and testing of EGFP reporter systems.
To construct the reporter plasmids, promoter fragments (−400 to −1 upstream of actII-4 and −460 to −1 upstream of wblA) were amplified using primers pMS82-actII-4p-egfp S1-F/R and pMS82-wblAp-egfp S1-F/R (Table S2). Then these promoter fragments and a DNA fragment encompassing the egfp gene were cloned into the pMS82 vector to generate pMS82-actII-4p-egfp and pMS82-wblAp-egfp. Next, we introduced these reporter plasmids into the WT and ΔadpA.
Strains containing reporter plasmids were precultured in liquid YBP medium for 36 h at 30°C. Subsequently, equal amounts mycelia of each strain were transferred to the fluted bottle, and inducer (400 μM HSnH or 400 μM S8) was added. After 60 min of induction, the bacteria were collected by centrifugation, and mycelia were resuspended in 200 μL phosphate-buffered saline (PBS) buffer (OD450, 2). EGFP fluorescence was measured using the microplate reader Synergy H1. The excitation wavelength and emission wavelength were set to 485 nm and 515 nm, respectively. The EGFP fluorescence intensity was normalized against cell density (fluorescence/OD450 of mycelia).
LC-MS/MS analysis of AdpA.
The analysis was performed following a previous report (24). Freshly purified protein AdpA (<100 μg) was treated with 10-fold amounts of HSnH (200 μM) or DTT (200 μM). After reacting at room temperature for 40 min. The reacted protein was treated with denaturing buffer (0.5 M Tris-HCl, 2.75 mM EDTA, 6 M guanidine-HCl, pH 8.0) containing 1 M iodoacetamide (IAM). The treatment was carried out in the dark for 1 h, and then the sample was digested with trypsin (1:25, wt/wt) at 37°C for 20 h. The digestion products were filtered by C18 Zip-Tip (Millipore) and vacuum-dried. The obtained peptides were resuspended in 10 μL double-distilled water (ddH2O).
The Prominence nano-LC system (Shimadzu) equipped with a custom-made silica column (75 μm by 15 cm) packed with 3 μm ReproSil-Pur 120 C18-AQ was used. Positive electrospray ionization was performed, and the ions were scanned with an LTQ-Orbitrap Velos Pro CID mass spectrometer (Thermo Scientific); the data were analyzed using a data-dependent acquisition mode with Xcalibur 2.2.0 software (Thermo Scientific). Full-scan MS spectra (from 400 to 1,800 m/z) were detected and assessed with the Orbitrap at a resolution of 60,000 at 400 m/z.
AdpA structure modeling.
The AlphaFold 2 algorithm (62) was used to predict the tertiary structure of AdpA. This method used the custom multiple sequence alignment (MSA) option and was accessed via the Colab server on GitHub (https://github.com/sokrypton/ColabFold). The structural model of AdpA was analyzed and visualized with PyMOL.
Fluorescence polarization (FP) analysis.
FP analysis experiments were performed following a reported protocol (63). DNA probes were amplified by PCR and labeled by 5′6-FAM (carboxyfluorescein) (Sangon). Purified AdpA (treated with 1 mM HSnH for 10 min or not) was diluted to different concentrations (0.01 μM to ~22.5 μM). The reaction buffer contained 10 mM Tris–HCl and 75 mM NaCl, pH 7.5. After mixing diluted AdpA and labeled DNA in the reaction buffer, the solution was incubated at 37°C for 15 min in the dark. The fluorescence was detected with a BioTek Synergy HT instrument. The KD value was calculated using GraphPad Prism 5 software.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (91951202) and the National Key R&D Program of China (2018YFA0901200).
We declare no conflict of interest regarding the publication of this article.
Contributor Information
Luying Xun, Email: luying_xun@vetmed.wsu.edu.
Huaiwei Liu, Email: liuhuaiwei@sdu.edu.cn.
Christiane Dahl, University of Bonn, Germany.
Caroline S. Harwood, University of Washington
REFERENCES
- 1.Sanchez J, Yague P, Manteca A. 2012. New insights in Streptomyces fermentations. Ferment Technol 1:e105. doi: 10.4172/2167-7972.1000e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olmos E, Mehmood N, Husein LH, Goergen JL, Fick M, Delaunay S. 2013. Effects of bioreactor hydrodynamics on the physiology of Streptomyces. Bioprocess Biosyst Eng 36:259–272. doi: 10.1007/s00449-012-0794-1. [DOI] [PubMed] [Google Scholar]
- 3.Hasani A, Kariminik A, Issazadeh K. 2014. Streptomycetes: characteristics and their antimicrobial activities. Int J Adv Biol Biomed Res 2:63–75. [Google Scholar]
- 4.Lee N, Hwang S, Kim J, Cho S, Palsson B, Cho BK. 2020. Mini review: genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput Struct Biotechnol J 18:1548–1556. doi: 10.1016/j.csbj.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honma S, Ito S, Yajima S, Sasaki Y. 2021. Nitric oxide signaling for actinorhodin production in Streptomyces coelicolor A3(2) via the DevS/R two-component system. Appl Environ Microbiol 87:e00480-21. doi: 10.1128/AEM.00480-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee N, Hwang S, Kim W, Lee Y, Kim JH, Cho S, Kim HU, Yoon YJ, Oh MK, Palsson BO, Cho BK. 2021. Systems and synthetic biology to elucidate secondary metabolite biosynthetic gene clusters encoded in Streptomyces genomes. Nat Prod Rep 38:1330–1361. doi: 10.1039/d0np00071j. [DOI] [PubMed] [Google Scholar]
- 7.Ohnishi Y, Yamazaki H, Kato JY, Tomono A, Horinouchi S. 2005. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci Biotechnol Biochem 69:431–439. doi: 10.1271/bbb.69.431. [DOI] [PubMed] [Google Scholar]
- 8.den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol 78:361–379. doi: 10.1111/j.1365-2958.2010.07338.x. [DOI] [PubMed] [Google Scholar]
- 9.Yan H, Lu XR, Sun D, Zhuang S, Chen Q, Chen Z, Li JL, Wen Y. 2020. BldD, a master developmental repressor, activates antibiotic production in two Streptomyces species. Mol Microbiol 113:123–142. doi: 10.1111/mmi.14405. [DOI] [PubMed] [Google Scholar]
- 10.Setinova D, Smidova K, Pohl P, Music I, Bobek J. 2017. RNase III-binding-mRNAs revealed novel complementary transcripts in Streptomyces. Front Microbiol 8:2693. doi: 10.3389/fmicb.2017.02693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takano E, Tao M, Long F, Bibb MJ, Wang L, Li W, Buttner MJ, Bibb MJ, Deng ZX, Chater KF. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol Microbiol 50:475–486. doi: 10.1046/j.1365-2958.2003.03728.x. [DOI] [PubMed] [Google Scholar]
- 12.Koshla O, Yushchuk O, Stash I, Dacyuk Y, Myronovskyi M, Jager G, Sussmuth RD, Luzhetskyy A, Bystrom A, Kirsebom LA, Ostash B. 2019. Gene miaA for post-transcriptional modification of tRNAXXA is important for morphological and metabolic differentiation in Streptomyces. Mol Microbiol 112:249–265. doi: 10.1111/mmi.14266. [DOI] [PubMed] [Google Scholar]
- 13.Kato J, Ohnish Y, Horinouchi S. 2005. Autorepression of AdpA of the AraC/XylS family, a key transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. J Mol Biol 350:12–26. doi: 10.1016/j.jmb.2005.04.058. [DOI] [PubMed] [Google Scholar]
- 14.Rabyk M, Yushchuk O, Rokytskyy I, Anisimova M, Ostash B. 2018. Genomic insights into evolution of AdpA family master regulators of morphological differentiation and secondary metabolism in Streptomyces. J Mol Evol 86:204–215. doi: 10.1007/s00239-018-9834-z. [DOI] [PubMed] [Google Scholar]
- 15.Lau N, Pluth MD. 2019. Reactive sulfur species (RSS): persulfides, polysulfides, potential, and problems. Curr Opin Chem Biol 49:1–8. doi: 10.1016/j.cbpa.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Filipovic MR, Zivanovic J, Alvarez B, Banerjee R. 2018. Chemical biology of H2S signaling through persulfidation. Chem Rev 118:377–461. doi: 10.1021/acs.chemrev.7b00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukuto JM, Ignarro LJ, Nagy P, Wink DA, Kevil CG, Feelisch M, Cortese-Krott MM, Bianco CL, Kumagai Y, Hobbs AJ, Lin J, Ida T, Akaike T. 2018. Biological hydropersulfides and related polysulfides: a new concept and perspective in redox biology. FEBS Lett 592:2140–2152. doi: 10.1002/1873-3468.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toohey JI, Cooper AJL. 2014. Thiosulfoxide (sulfane) sulfur: new chemistry and new regulatory roles in biology. Molecules 19:12789–12813. doi: 10.3390/molecules190812789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang SSS, Chen YHH, Chen N, Wang LJJ, Chen DXX, Weng HLL, Dooley S, Ding HGG. 2017. Hydrogen sulfide promotes autophagy of hepatocellular carcinoma cells through the PI3K/Akt/mTOR signaling pathway. Cell Death Dis 8:e2688. doi: 10.1038/cddis.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin YF, Gao R, Cui FF, Lü CJ, Liu HL, Liu HW, Xia YZ, Xun LY. 2020. The heterotrophic bacterium Cupriavidus pinatubonensis JMP134 oxidizes sulfide to sulfate with thiosulfate as a key intermediate. Appl Environ Microbiol 86:e01835-20. doi: 10.1128/AEM.01835-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wunder LC, Aromokeye DA, Yin XR, Richter-Heitmann T, Willis-Poratti G, Schnakenberg A, Otersen C, Dohrmann I, Romer M, Bohrmann G, Kasten S, Friedrich MW. 2021. Iron and sulfate reduction structure microbial communities in (sub-)Antarctic sediments. ISME J 15:3587–3604. doi: 10.1038/s41396-021-01014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng H, Zhang YX, Palmer LD, Kehl-Fie TE, Skaar EP, Trinidad JC, Giedroc DP. 2017. Hydrogen sulfide and reactive sulfur species impact proteome S-sulfhydration and global virulence regulation in Staphylococcus aureus. ACS Infect Dis 3:744–755. doi: 10.1021/acsinfecdis.7b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu DX, Zhang JJ, Lü CJ, Xia YZ, Liu HW, Jiao NZ, Xun LY, Liu JH. 2020. Synechococcus sp. strain PCC7002 uses sulfide:quinone oxidoreductase to detoxify exogenous sulfide and to convert endogenous sulfide to cellular sulfane sulfur. mBio 11:e03420-19. doi: 10.1128/mBio.03420-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu T, Cao Q, Pang XH, Xia YZ, Xun LY, Liu HW. 2020. Sulfane sulfur-activated actinorhodin production and sporulation is maintained by a natural gene circuit in Streptomyces coelicolor. Microb Biotechnol 13:1917–1932. doi: 10.1111/1751-7915.13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park SS, Yang YH, Song E, Kim EJ, Kim WS, Sohng JK, Lee HC, Liou KK, Kim BG. 2009. Mass spectrometric screening of transcriptional regulators involved in antibiotic biosynthesis in Streptomyces coelicolor A3(2). J Ind Microbiol Biotechnol 36:1073–1083. doi: 10.1007/s10295-009-0591-2. [DOI] [PubMed] [Google Scholar]
- 26.Liu G, Chater KF, Chandra G, Niu GQ, Tan HR. 2013. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura Y, Koike S, Shibuya N, Lefer D, Ogasawara Y, Kimura H. 2017. 3-Mercaptopyruvate sulfurtransferase produces potential redox regulators cysteine- and glutathione-persulfide (Cys-SSH and GSSH) together with signaling molecules H2S2, H2S3 and H2S. Sci Rep 7:10459. doi: 10.1038/s41598-017-11004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arias P, Fernandez-Moreno MA, Malpartida F. 1999. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol 181:6958–6968. doi: 10.1128/JB.181.22.6958-6968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee HN, Kim JS, Kim P, Lee HS, Kim ES. 2013. Repression of antibiotic downregulator WblA by AdpA in Streptomyces coelicolor. Appl Environ Microbiol 79:4159–4163. doi: 10.1128/AEM.00546-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolanski M, Donczew R, Kois-Ostrowska A, Masiewicz P, Jakimowicz D, Zakrzewska-Czerwinska J. 2011. The level of AdpA directly affects expression of developmental genes in Streptomyces coelicolor. J Bacteriol 193:6358–6365. doi: 10.1128/JB.05734-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mustafa AK, Gadalla MM, Sen N, Kim S, Mu WT, Gazi SK, Barrow RK, Yang GD, Wang R, Snyder SH. 2009. H2S signals through protein S-sulfhydration. Sci Signal 2:ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul BD, Snyder SH. 2015. Protein sulfhydration. Methods Enzymol 555:79–90. doi: 10.1016/bs.mie.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 33.Bu XL, Weng JY, He BB, Xu MJ, Xu J. 2019. A novel AdpA homologue negatively regulates morphological differentiation in Streptomyces xiamenensis 318. Appl Environ Microbiol 85:e03107-18. doi: 10.1128/AEM.03107-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang YJ, Wang YY, Hou BB, Wang RD, Ye J, Zhu XY, Wu HZ, Zhang HZ. 2019. AdpAlin, a pleiotropic transcriptional regulator, is involved in the cascade regulation of lincomycin biosynthesis in Streptomyces lincolnensis. Front Microbiol 10:2428. doi: 10.3389/fmicb.2019.02428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yushchuk O, Ostash I, Vlasiuk I, Gren T, Luzhetskyy A, Kalinowski J, Fedorenko V, Ostash B. 2018. Heterologous AdpA transcription factors enhance landomycin production in Streptomyces cyanogenus S136 under a broad range of growth conditions. Appl Microbiol Biotechnol 102:8419–8428. doi: 10.1007/s00253-018-9249-1. [DOI] [PubMed] [Google Scholar]
- 36.Xu JJ, Zhang JH, Zhuo JM, Li Y, Tian YQ, Tan HR. 2017. Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. J Biol Chem 292:19708–19720. doi: 10.1074/jbc.M117.809145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao XM, Luo S, Zhou RC, Wang F, Yu P, Sun N, Chen XX, Tang Y, Li YQ. 2015. Transcriptional regulation of the daptomycin gene cluster in Streptomyces roseosporus by an autoregulator, AtrA. J Biol Chem 290:7992–8001. doi: 10.1074/jbc.M114.608273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zacharia VM, Ra Y, Sue C, Alcala E, Reaso JN, Ruzin SE, Traxler MF. 2021. Genetic network architecture and environmental cues drive spatial organization of phenotypic division of labor in Streptomyces coelicolor. mBio 12:e00794-21. doi: 10.1128/mBio.00794-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ran MX, Wang TQ, Shao M, Chen ZG, Liu HW, Xia YZ, Xun LY. 2019. Sensitive method for reliable quantification of sulfane sulfur in biological samples. Anal Chem 91:11981–11986. doi: 10.1021/acs.analchem.9b02875. [DOI] [PubMed] [Google Scholar]
- 40.Hou NK, Yan ZZ, Fan KL, Li HJ, Zhao R, Xia YZ, Xun LY, Liu HW. 2019. OxyR senses sulfane sulfur and activates the genes for its removal in Escherichia coli. Redox Biol 26:101293. doi: 10.1016/j.redox.2019.101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan KL, Chen ZG, Liu HW. 2020. Evidence that the ProPerDP method is inadequate for protein persulfidation detection due to lack of specificity. Sci Adv 6:eabb6477. doi: 10.1126/sciadv.abb6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Lira NPV, Pauletti BA, Marques AC, Perez CA, Caserta R, de Souza AA, Vercesi AE, Leme AFP, Benedetti CE. 2018. BigR is a sulfide sensor that regulates a sulfur transferase/dioxygenase required for aerobic respiration of plant bacteria under sulfide stress. Sci Rep 8:3508. doi: 10.1038/s41598-018-21974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luebke JL, Shen JC, Bruce KE, Kehl-Fie TE, Peng H, Skaar EP, Giedroc DP. 2014. The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol Microbiol 94:1343–1360. doi: 10.1111/mmi.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li HJ, Li J, Lü CJ, Xia YZ, Xin YF, Liu HL, Xun LY, Liu HW. 2017. FisR activates σ54-dependent transcription of sulfide-oxidizing genes in Cupriavidus pinatubonensis JMP134. Mol Microbiol 105:373–384. doi: 10.1111/mmi.13725. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu T, Shen JC, Fang MX, Zhang YX, Hori K, Trinidad JC, Bauer CE, Giedroc DP, Masuda S. 2017. Sulfide-responsive transcriptional repressor SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proc Natl Acad Sci USA 114:2355–2360. doi: 10.1073/pnas.1614133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Capdevila DA, Walsh BJC, Zhang YF, Dietrich C, Gonzalez-Gutierrez G, Giedroc DP. 2021. Structural basis for persulfide-sensing specificity in a transcriptional regulator. Nat Chem Biol 17:65–70. doi: 10.1038/s41589-020-00671-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giedroc DP. 2017. A new player in bacterial sulfide-inducible transcriptional regulation. Mol Microbiol 105:347–352. doi: 10.1111/mmi.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xuan GH, Lü CJ, Xu HW, Chen ZG, Li K, Liu HL, Liu HW, Xia YZ, Xun LY. 2020. Sulfane sulfur is an intrinsic signal activating MexR-regulated antibiotic resistance in Pseudomonas aeruginosa. Mol Microbiol 114:1038–1048. doi: 10.1111/mmi.14593. [DOI] [PubMed] [Google Scholar]
- 49.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics, 2nd ed. John Innes Foundation, Norwich, UK. [Google Scholar]
- 50.Zhu YP, Lu T, Zhang J, Zhang PP, Tao MF, Pang XH. 2020. A novel XRE family regulator that controls antibiotic production and development in Streptomyces coelicolor. Appl Microbiol Biotechnol 104:10075–10089. doi: 10.1007/s00253-020-10950-z. [DOI] [PubMed] [Google Scholar]
- 51.Visser JM, Robertson LA, Van Verseveld HW, Kuenen JG. 1997. Sulfur production by obligately chemolithoautotrophic Thiobacillus species. Appl Environ Microbiol 63:2300–2305. doi: 10.1128/aem.63.6.2300-2305.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xin YF, Liu HL, Cui FF, Liu HW, Xun LY. 2016. Recombinant Escherichia coli with sulfide:quinone oxidoreductase and persulfide dioxygenase rapidly oxidises sulfide to sulfite and thiosulfate via a new pathway. Environ Microbiol 18:5123–5136. doi: 10.1111/1462-2920.13511. [DOI] [PubMed] [Google Scholar]
- 53.Flavin M. 1962. Microbial transsulfuration: the mechanism of an enzymatic disulfide elimination reaction. J Biol Chem 237:768–777. doi: 10.1016/S0021-9258(18)60371-0. [DOI] [PubMed] [Google Scholar]
- 54.Lu T, Zhu YP, Zhang PP, Sheng DH, Cao GX, Pang XH. 2018. SCO5351 is a pleiotropic factor that impacts secondary metabolism and morphological development in Streptomyces coelicolor. FEMS Microbiol Lett 365:fny150. doi: 10.1093/femsle/fny150. [DOI] [PubMed] [Google Scholar]
- 55.Zhu YP, Xu WH, Zhang J, Zhang PP, Zhao ZL, Sheng DH, Ma W, Zhang YZ, Bai LQ, Pang XH. 2020. A hierarchical network of four regulatory genes controlling production of the polyene antibiotic candicidin in Streptomyces sp. strain FR-008. Appl Environ Microbiol 86:e00055-20. doi: 10.1128/AEM.00055-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gregory MA, Till R, Smith MCM. 2003. Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J Bacteriol 185:5320–5323. doi: 10.1128/JB.185.17.5320-5323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia YZ, Chu WQ, Qi QS, Xun LY. 2015. New insights into the QuikChangeTM process guide the use of Phusion DNA polymerase for site-directed mutagenesis. Nucleic Acids Res 43:e12. doi: 10.1093/nar/gku1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu YP, Zhang PP, Zhang J, Xu WH, Wang XY, Wu LL, Sheng DH, Ma W, Cao GX, Chen XA, Lu YH, Zhang YZ, Pang XH. 2019. The developmental regulator MtrA binds GlnR boxes and represses nitrogen metabolism genes in Streptomyces coelicolor. Mol Microbiol 112:29–46. doi: 10.1111/mmi.14252. [DOI] [PubMed] [Google Scholar]
- 59.Zhu YP, Zhang PP, Lu T, Wang XY, Li AY, Lu YH, Tao MF, Pang XH. 2021. Impact of MtrA on phosphate metabolism genes and the response to altered phosphate conditions in Streptomyces. Environ Microbiol 23:6907–6923. doi: 10.1111/1462-2920.15719. [DOI] [PubMed] [Google Scholar]
- 60.Liu M, Zhang PP, Zhu YP, Lu T, Wang YM, Cao GX, Shi M, Chen XL, Tao MF, Pang XH. 2019. Novel two-component system MacRS is a pleiotropic regulator that controls multiple morphogenic membrane protein genes in Streptomyces coelicolor. Appl Environ Microbiol 85:e02178-18. doi: 10.1128/AEM.02178-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu YP, Zhang PP, Zhang J, Wang J, Lu YH, Pang XH. 2020. Impact on multiple antibiotic pathways reveals MtrA as a master regulator of antibiotic production in Streptomyces spp. and potentially in other actinobacteria. Appl Environ Microbiol 86:e01201-20. doi: 10.1128/AEM.01201-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang FY, Li BQ, Dong HJ, Chen M, Yao S, Li JW, Zhang HH, Liu XG, Wang HW, Song NN, Zhang KD, Du N, Xu SJ, Gu LC. 2020. YdiV regulates Escherichia coli ferric uptake by manipulating the DNA-binding ability of Fur in a SlyD-dependent manner. Nucleic Acids Res 48:9571–9588. doi: 10.1093/nar/gkaa696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mathematical modelling. Download Text S1, PDF file, 0.3 MB (274.3KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MS2 data of peptide 1 (Cys62-AM) (from HSnH-treated AdpA). Download FIG S1, PDF file, 0.4 MB (431.2KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MS2 data of peptide 2 (Cys62-SH) (from HSnH-treated AdpA). Download FIG S2, PDF file, 0.4 MB (402.4KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MS2 data of peptide 1 (Cys62-AM) (from DTT-treated AdpA). Download FIG S3, PDF file, 0.4 MB (400.1KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bioinformatics analysis of AdpA distribution. Phylogenetic analysis of AdpA and its homologues in Streptomyces and Actinomycetes. AdpA sequence accession numbers in NCBI: Streptomyces coelicolor (SCO2792, CAB87229.1), Streptomyces xiamenensis 318 (SXIM_38050, WP_030737153.1), Streptomyces roseosporus NRRL 11379 (CP979_13280, KAA6217800.1), Streptomyces lincolnensis NRRL 2936 (SLINC_3216, ANS65440.1), Streptomyces cyanogenus S136 (S1361_15285, QTD98720.1), Streptomyces ansochromogenes 7100 (AdpA-L, ABY86620.1), Streptomyces diastatochromogenes 1628 (AdpAdi, AFX97763.1), Streptomyces chattanoogensis L10 (AdpAch, ACY78399.1), Streptomyces fradiae CGMCC 4.7387 (BG846_05140, OSY49238.1), Streptomyces hygroscopicus subsp. jinggangensis 5008 (SHJG_4295, AEY89567.1), Streptomyces lividans 1326 (SLI_3139, EOY47852.1), Streptomyces ghanaensis ATCC 14672 (AdpAgh, EFE69329.1), Streptomyces griseus (SGR_4742, BAG21571.1), Streptomyces clavuligerus ATCC 27064 (SCLAV_1957, EFG07030.1), Streptomyces sp. FR-008 (SFR_4905, ALM41520.1), Streptomyces venezuelae ATCC 10712 (SVEN_2580, CCA55866.1), Streptomyces avermitilis MA-4680 (SAV_5261, BAC72973.1), Streptomyces bingchenggensis BCW-1 (SBI_06911, ADI10031.1), Streptomyces albus J1074 (XNR_4181, AGI90514.1), Saccharopolyspora erythraea NRRL2338 (SACE_4523, CAM03792.1), Actinoplanes teichomyceticus NRRL-B16726 (AdpA19AT, AKR67211.1), Micromonospora echinospora DSM 43036 (FHU28_002617, WP_184684050.1), Micromonospora aurantiaca ATCC 27029 (MICAU_1999, WP_013285179.1), and Amycolatopsis mediterranei U32 (AdpAamed, AMED_7695, ADJ49403.1). The phylogenetic tree was constructed using MEGA 5.0. Download FIG S4, PDF file, 0.3 MB (346.8KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence alignment of AdpA with its homologues in Streptomyces and Actinomycetes. AdpA amino acid sequences of Streptomyces xiamenensis 318 (AdpAsxi, SXIM_38050, GenBank accession no. WP_030737153.1), Streptomyces roseosporus NRRL 11379 (AdpAsrose, CP979_13280, KAA6217800.1), Streptomyces lincolnensis NRRL 2936 (AdpAslinc, SLINC_3216, ANS65440.1), Streptomyces hygroscopicus subsp. jinggangensis 5008 (AdpAshjg, SHJG_4295, AEY89567.1), Streptomyces griseus (AdpAsg, SGR_4742, BAG21571.1), Saccharopolyspora erythraea NRRL2338 (AdpAsace, SACE_4523, CAM03792.1), and Amycolatopsis mediterranei U32 (AdpAamed, AMED_7695, ADJ49403.1) were used. Download FIG S5, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and plasmids used in this study. Download Table S1, PDF file, 0.3 MB (285.9KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S2, PDF file, 0.2 MB (163.7KB, pdf) .
Copyright © 2022 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.