ABSTRACT
Honey bees are globally important pollinators threatened by many different pathogens, including viruses. We investigated the virome of honey bees collected at the end of the beekeeping season (August/September) in Czechia, a Central European country. Samples were examined in biological replicates to assess the homogeneity, stability, and composition of the virome inside a single hive. By choice of healthy workers from colonies, where Varroa destructor was under control, we could identify ubiquitous bee viruses. Deformed wing virus (DWV) was highly prevalent, even though the bees were healthy, without any noticeable disease signs. The overall virome composition (consisting of honey bee-, plant-, and bacterium-infecting viruses) was driven primarily by the hive and its location. However, honey bee-specific viruses showed an uneven distribution within the same hive. In addition, our results point to an unusual cooccurrence between two rhabdoviruses and reveal the presence of five distinct lineages of Lake Sinai viruses (LSVs) clustering with other LSV strains described globally. Comparison of our results with the virome of Australian honey bees, the last truly Varroa- and DWV-free population, showed a strong difference with respect to DWV and a set of diverse members of the Picornavirales, of which the latter were absent in our samples. We hypothesize that the occurrence of DWV introduced by Varroa strongly affects the virome structure despite the mite being under control.
IMPORTANCE The Western honey bee, Apis mellifera, is a vital part of our ecosystem as well as cultural heritage. Annual colony losses endanger beekeeping. In this study, we examined healthy bees from the heart of Central Europe, where honey bee colonies have been commonly affected by varroosis over 5 decades. Our virome analysis showed the presence of ubiquitous viruses in colonies where the mite Varroa destructor was under control and no honey bee disease signs were observed. Compared to previous studies, an important part of our study was the analysis of multiple replicates from individual hives. Our overall results indicate that the virome structure (including bee-infecting viruses, plant-infecting viruses, and bacteriophages) is stable within hives; however, the bee-infecting viruses varied largely within interhive replicates, suggesting variation of honey bee viruses within individual bees. Of interest was the striking difference between the viromes of our 39 pools and 9 pools of honey bee viromes previously analyzed in Australia. It could be suggested that Varroa not only affects DWV spread in bee colonies but also affects diverse members of the Picornavirales, which were strongly decreased in Czech bees compared to the Varroa- and DWV-naive Australian bees.
KEYWORDS: Apis mellifera, Picornavirales, rhabdovirus, Lake Sinai virus, metagenomics, viruses
INTRODUCTION
The European honey bee, Apis mellifera Linnaeus 1758, is used for the production of honey, propolis, beeswax, venom, pollen, and royal jelly (1). However, the most crucial beneficial feature of honey bees lies in pollination in both agricultural (2, 3) and natural (4) habitats. Annual colony losses jeopardize these benefits provided by honey bees (5). In temperate zones of Europe, the main colony losses occur over winter and are thus referred to as wintering losses (6). Another problem, not yet fully understood, is colony losses in the United States, also known as colony collapse disorder (CCD) (7). This phenomenon is probably due to a combination of several factors, mainly Varroa destructor (8), other viral pathogens, and their interaction (7, 9). Conservation of honey bees is difficult, especially in countries where the density of managed colonies is very high, and this is precisely the case in Czechia, in the heart of Central Europe (10).
The global spread of V. destructor has had a severe effect on the transmission and virulence of certain honey bee viruses such as deformed wing virus (DWV); DWV variant B (DWV-B), also described as Varroa destructor virus 1 (11, 12); and viruses belonging to the acute-Kashmir-Israeli complex (13–15). However, without high mite infestations, DWV infections are often benign or asymptomatic. Important from this point of view is the presence of diverse viruses in Australia, where honey bees are free of both V. destructor and DWV, although some viruses from the Picornavirales order can be found (16). Interestingly, the interaction between viruses and the mite can affect virus strain distribution, as described previously for DWV (17–20). The DWV-A/B strain ratio is affected by the level of mite infestation in a colony (21). Another parasite, Nosema ceranae, has been shown to aggravate black queen cell virus (BQCV) infection (22). However, the synergistic effect of N. ceranae in combination with different viruses such as DWV was negated (23). Thus, for other known or newly identified viruses, similar or unexpected interactions may exist. Various nonviral pathogens may play an important role in the prevalence and severity of diseases.
Until lately, honey bee virus research focused mainly on 23 described viral species, as reviewed in 2015 (24). In the last few years, more viruses that demonstrably or presumably infect honey bees were discovered due to the increased use of next-generation sequencing (NGS) technologies (16, 25, 26). Moreover, it was recently shown that the honey bee gut virome contains many bacteriophages (27, 28). Previous knowledge was limited to phages from pathogens such as Paenibacillus larvae (27). In contrast to the best-characterized bee-infecting viruses, which belong to the Picornavirales or other positive-sense single-stranded RNA (+ssRNA) virus groups (e.g., Iflaviridae and Dicistroviridae), some novel viruses belonging to viral families like the Rhabdoviridae or Orthomyxoviridae have recently been reported in honey bees (29, 30) and the parasite V. destructor (30). Most infections with these novel viruses are not yet known to manifest symptomatically but could impact colony health through fitness costs, even though subtle, for the host and/or through interactions with the host and other pathogens/parasites associated with honey bees. Furthermore, the spread of viral infections from honey bees to wild pollinators is also of great concern (31, 32).
In this study, we explored the diversity and composition of the virome in honey bees from healthy colonies from beekeepers breeding various honey bee genetic lines in Czechia. To see the robustness of the virome analyses, we analyzed three biological repeats from each hive. To our knowledge, this is the first such analysis performed on honey bees. We focused on the virome composition (common versus new viruses, plant viruses, and bacteriophages). In addition, we compared our results with those for nine Australian viromes from bees with no exposure to V. destructor or DWV.
RESULTS
Composition and similarity of virome samples.
NGS of 39 samples (3 replicates of 9 pooled bees from 13 colonies) yielded a total of 398,231,288 reads, with an average of 10 million reads (range, 1,920,148 to 30,170,502; median, 11,650,042) per sample containing 9 bees. The reads were classified as follows: (i) 46.66% eukaryotic, (ii) 30.98% viral, (iii) 13.14% bacterial, and (iv) 9.21% not identified as homologous to any reference sequence.
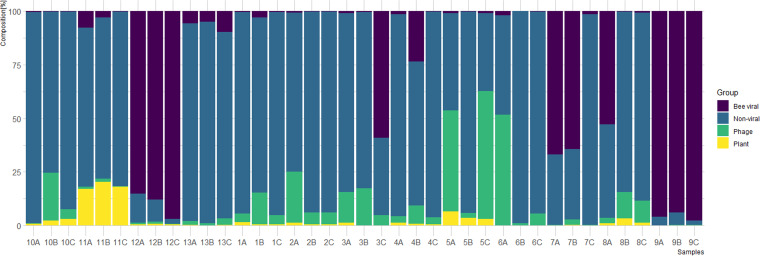
First, we analyzed the composition of sequencing reads in each of the 39 samples by determining the proportion of reads originating from nonviral sequences (e.g., honey bee genome and bacterial microbiome), known bee viruses (33), bacteriophages, and plant viruses (Fig. 1). Although some replicates seemed to be rather consistent, notable heterogeneity was detected among several other samples (e.g., samples 3, 5, 6, 7, and 8). Next, we analyzed the taxonomic composition of the bee virome with respect to the (i) abundance and (ii) diversity of viral families (see Fig. S1 in the supplemental material).
FIG 1.
Composition of sequencing reads in all 39 analyzed samples. Each hive was analyzed in 3 independent replicates consisting of nine individual bees. The percentages of reads of different taxonomic affiliations (plant viruses, bacteriophages, bee viruses, and nonviral) are denoted in different colors.
Additional figures and data (map, plant heatmap, viral family distribution in the virome, comparison of samples from Australia and the rest of the world, BRV phylogeny, and additional information on k-means clustering). Download FIG S1, PDF file, 0.7 MB (670.3KB, pdf) .
Copyright © 2022 Kadlečková et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
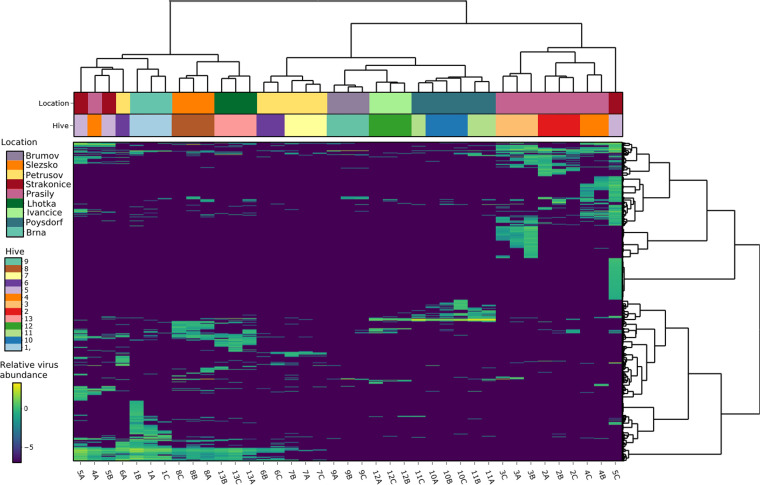
To visualize the virome similarity of the 39 samples, a heatmap was constructed from the relative abundances of all viral sequences (Fig. 2; Table S1). For almost all samples, replicates originating from the same hive clustered together and thus exhibited similar total viromes. Samples obtained from different sites (hives and apiaries) at the same location also exhibited related viromes. The Adonis test confirmed the highly significant association of virome composition with hive and location (P < 0.0001 by an Adonis-Bray test; R2, 0.62964 and 0.45389). k-means clustering had for k 13 (representing hives) an adjusted mutual information score (taking on values from 1 for identical to around 0 for random) of 0.14, suggesting the existence of a pattern in clustering (Fig. S1).
FIG 2.
Heatmap constructed from all classified viral sequences in all 39 samples. The viral sequences are taxonomically assigned to the family or species level. Relative abundances (viruses per 1 million sequencing reads) are shown on a log10 scale. Samples (columns) and contigs (rows) are clustered by Ward’s minimum variance method; both columns and rows are seriated by optimal leaf ordering.
Several sheets of raw and transformed data used to create heatmaps, also including metadata (replicate, subtype of honey bee, and location, etc.) for each sample. Download Table S1, XLSX file, 1.6 MB (1.6MB, xlsx) .
Copyright © 2022 Kadlečková et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bee viruses.
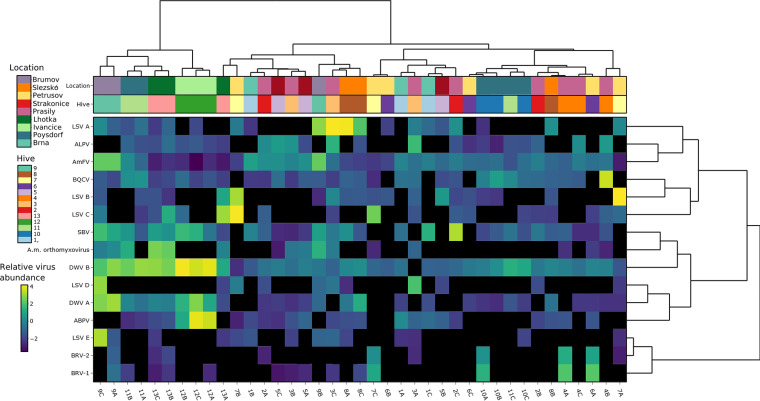
Furthermore, we focused our analysis on eukaryotic viruses that were demonstrated or predicted to infect honey bees (33). Altogether, the analyzed samples revealed the presence of one DNA and nine RNA viruses. Besides the well-known viruses belonging to the Dicistroviridae and Iflaviridae, we found viruses belonging to the families Rhabdoviridae and Orthomyxoviridae as well as several variants of Lake Sinai virus (LSV) and the DNA virus Apis mellifera filamentous virus (AmFV) (Fig. 3).
FIG 3.
Diversity of viruses infecting honey bees in healthy bee colonies from Czechia. Relative abundances (viruses per 1 million sequencing reads, calculated from reference genome coverage [see Materials and Methods]) are shown on a log10 scale. Samples (columns) and viruses (rows) are clustered by Ward’s minimum variance method algorithm and seriated by optimal leaf ordering. ABPV, acute bee paralysis virus; ALPV, aphid lethal paralysis virus; AmFV, Apis mellifera filamentous virus; BRV, bee rhabdovirus; BQCV, black queen cell virus; DWV, deformed wing virus; SBV, Sacbrood virus; LSV, Lake Sinai virus.
Overall distribution of bee viruses.
A heatmap was constructed based on the relative abundances of all detected bee-infecting viruses (Fig. 3). The most commonly present viruses were Deformed wing virus variant A (DWV-A) and variant B (DWV-B), Black queen cell virus (BQCV), Aphid lethal paralysis virus (ALPV), AmFV, and Sacbrood virus (SBV). In sharp contrast to the heatmap constructed from all viral sequences (Fig. 2), clustering between bee viruses of most replicate samples was no longer discernible (Fig. 3). We presume that the lack of geographic clustering can be attributed to the absence of bacteriophages and plant viruses (see below) in this analysis. For bee-infecting viruses, the adjusted mutual information score for k 13 (hive) was low (−0.006), further confirming that clustering between samples is absent.
High differences in abundance among the replicate samples were observed for all known bee-infecting viruses, even though each sample consisted of nine pooled bees. This implies that the pooling of nine bees per hive is not sufficient to compensate for the variability in the occurrence of viruses in individual bees. Importantly, in some samples, a single virus accounted for over 50% of the total sequencing reads (58.8% LSV-A reads in pool 3C and 59.4% acute bee paralysis virus [ABPV] reads in pool 12C) while being present in negligible quantities (<1% reads) in each of the two remaining replicate samples. This indicates the (sporadic) presence of individual bees with very high viral loads compared to those in other bees within the same honey bee colony.
Virome comparison with the Varroa- and DWV-naive honey bees (Australia).
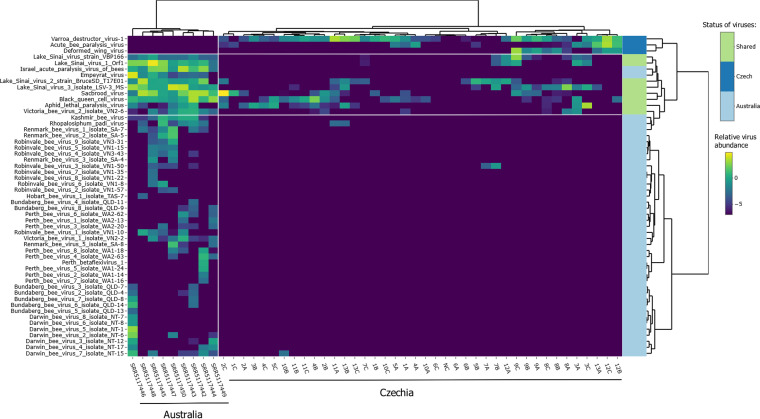
Since we used healthy asymptomatic bees where varroosis was under control, we compared our data with those reported previously by Roberts et al. (16). The heatmap in Fig. 4 shows that the virome of Varroa- and DWV-naive Australian honey bees is different from that of the Czech samples. All the Australian viromes clustered together and were separated from the Czech viromes. The Australian viromes included several abundant and diverse viruses belonging to the Picornavirales (e.g., Perth bee virus, Darwin bee virus, or Robinvale bee virus). In contrast, we did not detect any of the diverse Picornavirales (Fig. 4). Conversely, DWV-A/B and ABPV found in Czech honey bees were absent in the nine Australian viromes. Finally, several viruses (BQCV, variants of LSV, SBV, and ALPV) were present in both Australian and Czech viromes.
FIG 4.
Heatmap of bee-infecting viruses in a comparison between the data from our study and Australian bees (SRA accession numbers SRR5117442 to SRR5117450). Relative abundances are shown on a log10 scale. Samples (columns) and viruses (rows) are clustered by Ward’s minimum variance method algorithm and seriated by optimal leaf ordering. Row colors show if the virus is present in Australia, Czechia, or both countries. Czech, viruses found only in Czechia; Australia, viruses found only in Australia; shared, viruses present in both regions. White lines separate the heatmap into several parts, Australian/Czech samples and viruses present/absent in the given regions.
Prompted by these results, we decided to investigate if this difference was also discernible in other studies of non-Australian viromes. We used public NGS data from Belgium (34); Israel (30); South Africa, The Netherlands, and Tonga (29); and the United States, Central America, Europe, Kenya, India, and New Zealand (26). This allowed us to compare the bee populations where V. destructor is present (most of the world) with Varroa-naive populations (16). The results showed that the difference observed between the Czech and Australian bee viromes can be generalized to other parts of the world: the above-mentioned diverse Picornavirales were absent from the honey bee viromes in all other geographic regions except Australia (the trace amount can be attributed to read misalignment), whereas DWV-A and DWV-B were globally abundant. Australia clustered aside from other interleaved samples (for the principal-coordinate analysis [PCoA] and heatmap, see Fig. S1; for raw data, see Table S3).
Raw data from comparison of Australian and other viromes, including accession numbers used for viral genomes. Download Table S3, XLSX file, 0.05 MB (52.8KB, xlsx) .
Copyright © 2022 Kadlečková et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Emerging viruses in samples from Czechia.
The heatmap of bee virus abundances (Fig. 3) revealed a conspicuous pattern of cooccurrence of two recently discovered rhabdoviruses, bee rhabdovirus 1 (BRV-1) and BRV-2, which possibly infect both the honey bee and the mite V. destructor (29). Among our samples, BRV-1 and BRV-2 were always present together (pools 4A, 6A, 7C, 9A, and 10A), whereas both identified rhabdoviruses were absent from the remaining replicates (Fig. 3). The positivity of one out of three replicates probably implicates a low prevalence of BRV-positive bees within hives. In addition, BRV-1 always showed a higher abundance than BRV-2 in individual samples (Fig. 3). These viruses, albeit related, are phylogenetically distinct and display very limited sequence similarity (Fig. S1).
Furthermore, we reanalyzed NGS data from BRV-1-positive samples reported in three previous studies (26, 29, 35) by differentially mapping the sequencing reads to BRV-1 and BRV-2 reference genomes. Despite differences in multiple sample characteristics (i.e., bee pooling, nucleic acid isolation, and library preparation), we detected BRV-2 in all BRV-1-positive samples (Table 1). As in the Czech samples, BRV-2 was always present at a lower abundance. We consider this to be an indicator of an unusual relationship between the two rhabdoviruses, which has been left unnoticed previously (see Discussion).
TABLE 1.
Cooccurrence of bee rhabdoviruses 1 and 2 in NGS samples here and in three other studies where BRV-1 was identifieda
| Sample ID | No. of BRV-1 reads |
No. of BRV-2 reads |
BRV-1/BRV-2 ratio |
Country | SRA accession no. |
No. of bees/sample |
NA for library construction |
|---|---|---|---|---|---|---|---|
| SRR3927497 | 9,509 | 765 | 12.4 | Israel | SRR3927497 | 30 | Total RNA |
| DWV | 41,335 | 1,024 | 40.4 | USA | SRR6033679 | 10 | Virus-enriched (encapsulated) DNA + RNA |
| NE_AWD_1442 | 34,115 | 737 | 46.3 | The Netherlands | SRR5109823 | 5 | Total RNA (rRNA depleted) |
| SA_RI_49 | 83,757 | 6,791 | 12.3 | South Africa | SRR5109831 | 5 | Total RNA (rRNA depleted) |
| T_V9 | 104,233 | 181 | 575.9 | Tonga | SRR5109822 | 5 (thoraces only) | Total RNA (rRNA depleted) |
| T_V10 | 38,738 | 12,120 | 3.2 | Tonga | SRR5109821 | 5 (thoraces only) | Total RNA (rRNA depleted) |
| T_T12 | 449,293 | 4,501 | 99.8 | Tonga | SRR5109828 | 5 (thoraces only) | Total RNA (rRNA depleted) |
| T_T23 | 331,169 | 113 | 2,930.7 | Tonga | SRR5109834 | 5 (thoraces only) | Total RNA (rRNA depleted) |
| 4A | 44,492 | 6,741 | 6.6 | Czechia | SRS11094606 | 9 | Virus-enriched (encapsulated) DNA + RNA |
| 6A | 83,517 | 4,475 | 18.7 | Czechia | SRS11094614 | 9 | Virus-enriched (encapsulated) DNA + RNA |
| 7C | 7,029 | 1,072 | 6.6 | Czechia | SRS11094620 | 9 | Virus-enriched (encapsulated) DNA + RNA |
| 9A | 127 | 40 | 3.2 | Czechia | SRS11094624 | 9 | Virus-enriched (encapsulated) DNA + RNA |
| 10A | 16,770 | 1,493 | 11.2 | Czechia | SRS11094627 | 9 | Virus-enriched (encapsulated) DNA + RNA |
Sequencing reads were mapped to a hybrid reference sequence consisting of combined BRV-1 and BRV-2 genomes (BRV-1, GenBank accession number MH267692; BRV-2, GenBank accession number KY354234) to prevent the interference of multiple mapped reads. The BRV-1/BRV-2 ratio was calculated from read counts.
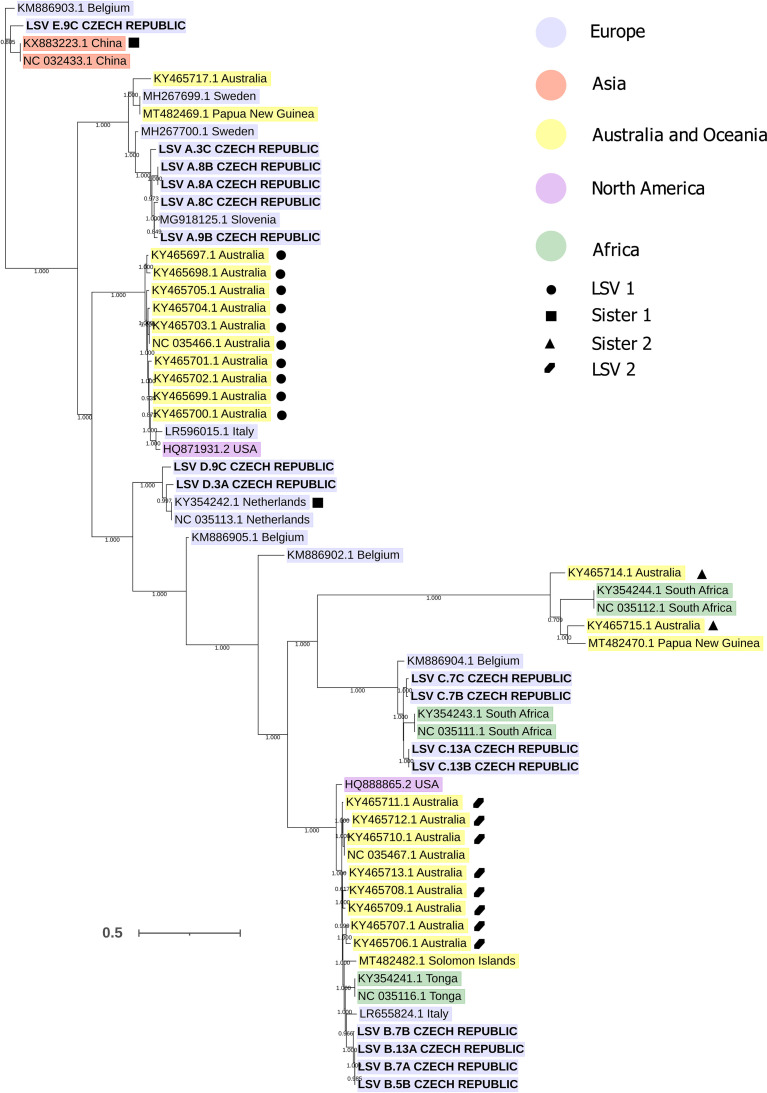
Surprisingly, our analysis also revealed a large diversity of LSVs. The LSV genomes identified in the Czech samples were distributed in five positions in the global LSV phylogenetic tree (Fig. 5). Five variants were thus designated de novo as LSV-A to LSV-E for the purpose of this study. The interlineage nucleotide identity among Czech genomic sequences ranged from 72% to 79%, and the intralineage identity ranged from 86% to 99%. As shown in Fig. 5, among the closest relatives of LSV variants A, B, C, and E were sequences originating from different continents, suggesting that these variants belonged to LSV lineages with an intercontinental or even a global distribution. The RNA-dependent RNA polymerase (RdRp) phylogeny places LSV-D among a dozen European and one Iranian LSV genotypes; however, the resolution of the RdRp fragment phylogeny is insufficient, as indicated by the level of bootstrap support (Fig. S2).
FIG 5.
Phylogenetic tree of full LSV genomic sequences. LSV sequences are highlighted in color according to the continent of origin. Symbols mark isolates assigned to LSV lineages by Cornman (72). Sequences obtained in this study are in boldface capital letters. Bootstrap values are shown.
Phylogeny of RdRp regions of LSVs. Download FIG S2, PDF file, 0.3 MB (286.2KB, pdf) .
Copyright © 2022 Kadlečková et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In general, the distribution of LSV variants in Czechia was variable in both between- and within-hive comparisons. Interestingly, two or more variants of LSV were detected in individual hives (LSV-B and LSV-C in hives 7 and 12 and LSV-A, LSV-D, and LSV-E in hive 3).
Bacteriophages and clustering.
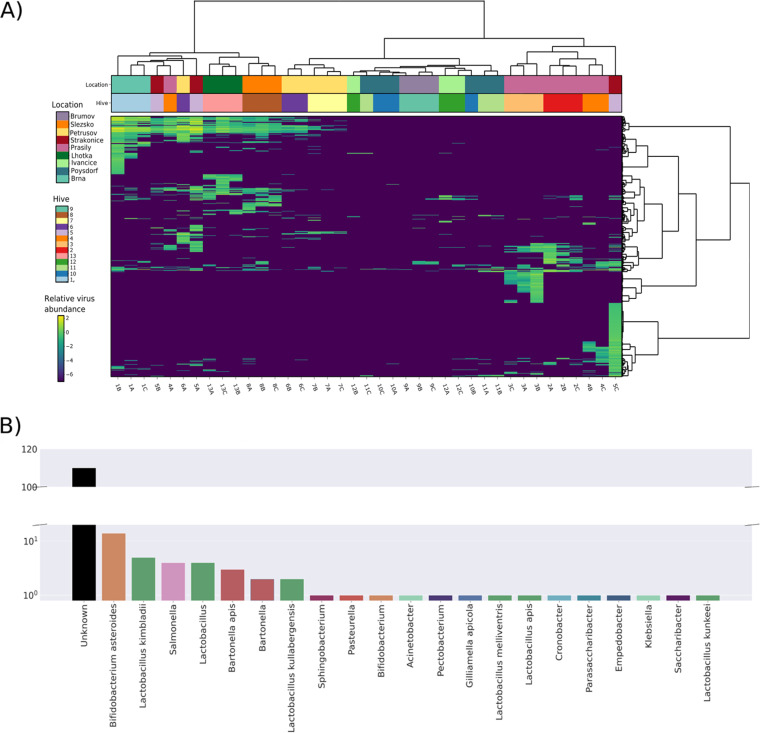
The results showed the high diversity and abundance of bacteriophages (Fig. S1). Therefore, we analyzed the bee phageome in detail. A heatmap was created by mapping reads to the set of phages identified by VirSorter2 (Table S1). Clustering of the phageome by the geographic origins of the samples was weaker than that in the heatmap of all viral sequences (Fig. 2), but still, replicate samples from seven hives clustered together. In five other hives, two out of three replicate samples clustered together; the replicate samples from hive 13 did not cluster (Fig. 6A). The Adonis test showed a highly significant correlation between the phageome and hive/location (P < 0.0001 by an Adonis-Bray test; R2, 0.47355/0.34024).
FIG 6.
Host calling and clustering of prokaryotic viruses. (A) Heatmap of all contigs classified as bacteriophages. Relative abundances (viruses per 1 million sequencing reads) are shown on a log10 scale. Samples (columns) and contigs (rows) are clustered by Ward’s minimum variance method; both columns and rows are seriated by optimal leaf ordering. (B) Count of predicted hosts for 158 bacteriophage contigs identified by VirSorter2. Contigs with no prediction are in the “Unknown” category.
Since the plant viruses originating from pollen were abundant (Fig. S1), we further explored their clustering. The plant viruses clustered almost perfectly (Fig. S1) (P < 0.0001 by an Adonis-Bray test; R2, 0.70959 for hive and 0.53102 for location). Interestingly, k-means clustering had higher adjusted mutual information scores for both bacteriophages and plant viruses than for all viral sequences (0.22 and 0.23, respectively, against 0.14) (Fig. S1).
To classify the phage genomes, which were predicted to be more than 50% complete (by CheckV), we used vConTACT2, which clustered the sequences with phages in the RefSeq database by their encoded protein profile. The resulting network had 398 individual viral clusters (roughly equivalent to genus-level assignment). Visualization of the resulting sequence similarity network (Fig. S3) shows the distribution of putative phage contigs through the network. Out of 158 individual phage contigs, 71 were unambiguously clustered. These formed 22 clusters, 15 of which were composed entirely of putative phage contigs from this study; 22 viral clusters (representing 26 putative bacteriophage contigs) were clustered with at least one reference sequence. Thanks to the reference sequences, the clusters could be tentatively classified as belonging to the family Myoviridae but also as belonging to the Siphoviridae, Podoviridae, and Microviridae. A single cluster contained strains from more than one viral family.
Similarity network for bacteriophages. Download FIG S3, PDF file, 0.2 MB (174.7KB, pdf) .
Copyright © 2022 Kadlečková et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Host calling for each of the 158 detected phage contigs was performed through matches with CRISPR spacers identified using MinCED (36) and an additional analysis with CrisprOpenDB (37). Over 200 spacers matched the detected viral contigs, yet due to duplicate assignments (one contig matching spacers from multiple strains of one bacterial host species), only 22 (14%) of the phage contigs could be assigned to a host. The most common phage hosts were Lactobacillus species, Bifidobacterium, Bartonella, and Salmonella (Fig. 6B).
DISCUSSION
This study is the first to analyze the honey bee virome in Czechia, a country located in the heart of Central Europe. In addition, to our knowledge, this is the first work where the honey bee virome was examined in biological replicate samples within hives/colonies. Virome variation in sample replicates raises the question of to what extent single bees can affect the virome structure of an entire colony. However, the open question that remains is how NGS-based virome analysis of pools of bees can be affected by a single bee with a distinct virome. It is relevant to what we have attempted: to examine the homogeneity of virus infection within hives. A major factor that could affect the virome is that honey bee colonies were from a Central European country with one of the highest colony densities worldwide (10). Thus, despite the fact that V. destructor occurrence was low in all investigated colonies, and no signs of varroosis were observed, a virus(es) introduced by the mite (17, 38) was expected to be found. Furthermore, we compared our viromes with previously described viromes of Australian honey bees, which have never been exposed to varroosis (DWV and Varroa mite) (16, 39). When it came to bee-infecting viruses, our sets of viruses were diametrically different from those in Australia. Notably, one substantial difference was the lack of diverse members of the Picornavirales in our data set, which could be explained by Varroa-DWV interaction pressure indicated previously (17–22). Further investigation will be necessary because of methodological differences in this study compared to the study of Roberts et al. (16). For future comparability with other studies, it is also of importance to note that our samples were collected at the end of the beekeeping season in August/September because seasonal variations in virus occurrence have been previously described (40), whereas the Australian samples were collected between a longer period (August 2013 to April 2015, but the majority were collected in August).
Traditional and new bee viruses.
The following viruses were detected in Czech samples: BQCV, AmFV, DWV-A and -B, SBV, ALPV, ABPV, BRV-1 and -2, Apis mellifera orthomyxovirus, and variants of LSV. The prevailing honey bee virus, in both abundance and prevalence, was DWV-B. Importantly, this result is in accordance with its recent global spread (20). Notably, the prevalence of DWV-B observed in asymptomatic (healthy) hives/colonies is in agreement with the results of a study by Norton et al., who showed DWV-B persistence in colonies with low V. destructor levels and those treated with miticides (21). Finally, the prevalence of DWV-A was low in our virome samples, consistent with the fact that varroosis was under control in our colonies (21).
We determined the viromes in 39 pools, each consisting of nine individual honey bees. Importantly, we show that none of the structures of our Czech virome resembled those of the Australian viromes (16). Moreover, unique viruses were identified in each data set. These results may correspond to the fact that the virome structure is affected by the presence/absence of V. destructor since Australian honey bees are Varroa-free (16, 39), while the Czech viromes originated from regions where V. destructor has been widely distributed since the late 1970s (41, 42). Although we worked with “healthy” honey bee colonies, where V. destructor was under control, the effect of the mite is still to be considered. In Czech samples, the diverse set of viruses belonging to the Picornavirales detected in Australian honey bees was absent. Another important observation is the complete absence of DWV in the Australian viromes (16) and the contrary wide DWV presence in our samples. This shift from diverse Picornavirales members to a primarily DWV-dominated virome could signify that the presence of V. destructor and its interaction with DWV change the virome of honey bees. We suggest that the absence of diverse Picornavirales in other samples from honey bee populations around the world, even though varroosis was under control in some, adds support to this assumption.
The two rhabdoviruses (BRV-1 and -2) exhibited a conspicuous pattern of distribution among our samples: (i) both viruses were always present simultaneously; (ii) in each sample, BRV-1 was more abundant than BRV-2; and (iii) BRV-1 and -2 were always present in only one out of three replicate nine-bee samples per hive. Reanalysis of previously reported BRV-positive NGS samples (26, 29, 35) independently validated both the first and second phenomena (see above). So far unnoticed, this relationship may indicate a type of (inter)dependence between these two viruses. It is possible that an interaction takes place, either a nondirect one (e.g., through the immune system) or a direct one (e.g., a defect in protein synthesis) (43). This phenomenon can also be caused by a common transmission route or another unknown mechanism. Either way, the precise nature of the relationship between BRV-1 and BRV-2 should be confirmed by a single-bee analysis.
Since the first description of LSVs by Runckel et al. (44), various variants of this group of new viruses have been discovered in different regions around the world, including Australia (39, 45–48). Such diversity, in combination with a global distribution, could indicate a long coexistence of LSV with honey bees (39). According to current knowledge, LSV infections occur asymptomatically, with no described adverse effect, which might have hindered their discovery in the pre-omics era. It was suggested that, based on the specific antibody detection LSV was described as bee virus X or Y (39, 49, 50). Among 39 Czech samples, we detected five distinct variants (designated LSV-A to -E) that were distributed in 17/39 samples (read count of ≥1 per million reads). Several cases of the simultaneous presence of several variants in one colony/hive were observed. A single-bee analysis is needed to determine the precise prevalence of LSV variants and the frequency of their cooccurrence as the simultaneous presence of several LSV variants in a single bee has been reported previously (47). The phylogenetic analysis of LSV sequences suggests that the LSV variants identified in the Czech samples belong to lineages that are globally distributed. Given the variable distribution and extraordinary global diversity of LSV, additional LSV variants are likely to be found in future studies. However, the available LSV sequences, both partial and complete, suffer from a heavy geographic sampling bias, implying that the true diversity might be even (much) larger than currently observed.
Virome structure stability.
When all viral sequences present in our samples were analyzed, most samples clustered in accordance with their site of origin. Clustering was also observed for samples originating from different hives from the same apiary and for hives from different apiaries located in the same municipality. It appears that this clustering is driven mainly by plant viruses and to a lesser extent by bacteriophages (see Fig. S1 in the supplemental material). In this study, bacteriophage sequences were one of the most diverse and abundant virome components in our samples. However, bacteriophages still represent a novel topic in honey bee research (27, 28) and are often excluded from analyses. In the bacteriophage heatmap, replicate samples still clustered based on hive and location, like the whole virome (plant viruses plus bacteriophages and other viral sequences). This is in accordance with previously reported findings that the phageome should be relatively stable in the location for the given year (28). The plant viruses in the total bee virome display the strongest geographic dependence (Fig. S1). Unfortunately, to our knowledge, no studies examining the stability and geographic differences of the pollen virome have been carried out yet.
The robust overall clustering pattern was lost when only viruses known to infect honey bees were analyzed. The uneven distribution of bee-infecting viruses among replicate nine-bee samples from the same hive raises serious questions for the NGS study design. NGS studies are typically carried out on pools of a small number (~1 to 5 [e.g., see reference 34]) or large numbers (>30 [e.g., see reference 35]) of individual bees.
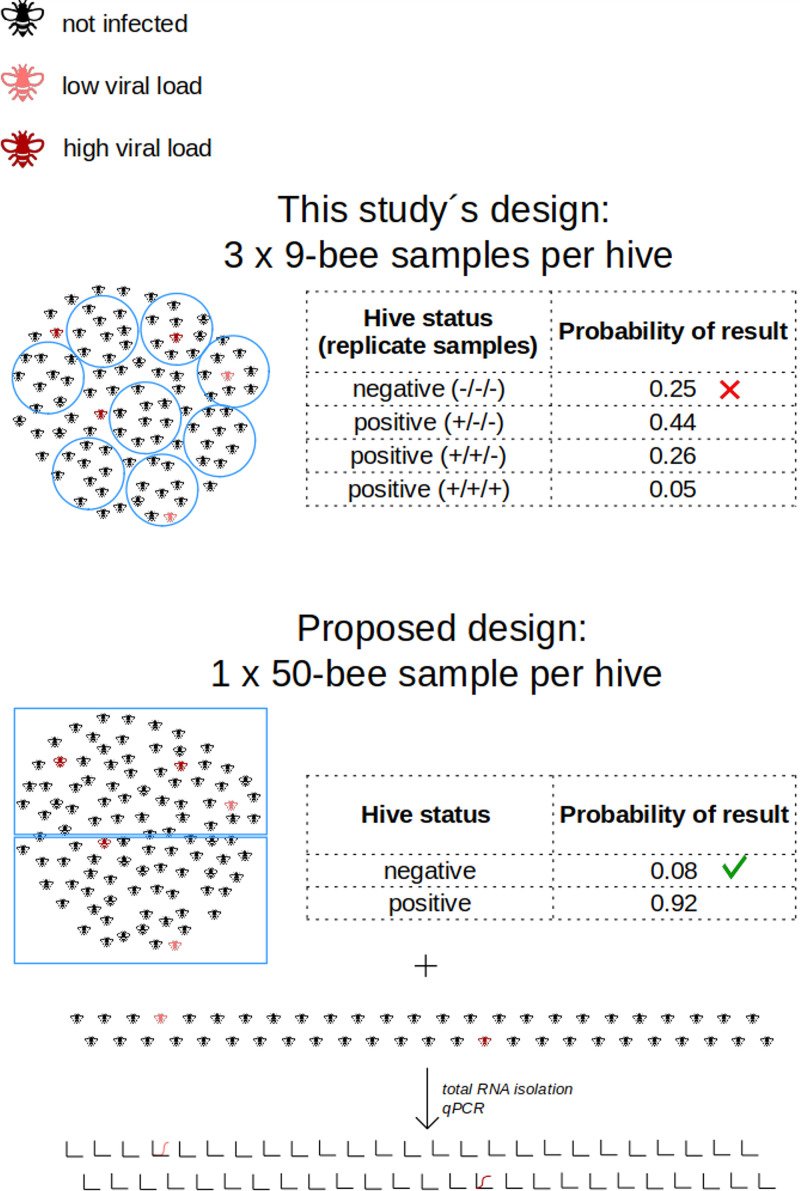
In our case, the pooling of nine bees per sample was not enough to balance the uneven representation of bee viruses among individuals. The recurring observations that only one nine-bee pool out of three yielded positivity for some viruses (e.g., BRV-1/2 [see “Traditional and new bee viruses,” above) indicate that the actual prevalence of infected worker bees per hive is low. In addition, we suspect that individual bees with relatively high viral loads are present in hives, and when they are randomly included in a pool for NGS virome analysis, this virus may dominate the virome of this pool. To accurately determine the abundance and prevalence of bee viruses in a hive, NGS of a large number of libraries, each prepared from an individual bee, appears to be the methodologically correct solution, which unfortunately is labor-intensive, expensive, and, thus, often beyond feasibility. Another possibility is to pool a large number of bees into a single master NGS library per hive (Fig. 7) and carry out NGS at a sequencing depth that is the same as or higher than the one in this study. The actual number of bees per master NGS library pool, our results suggest, should optimally be 50 individuals or more. Such an approach is expected to (i) detect the diversity of the viruses present, including those of low prevalence, and (ii) determine the actual genotypes of the viruses present in an affordable fashion (even though it can be challenging to disentangle closely related genomes). In this case, however, the viral prevalence in hives would remain to be assessed by analyzing similar quantities of individual bees, preferably by a rapid method like quantitative PCR (qPCR) (Fig. 7).
FIG 7.
Effect of the NGS experimental design on hive status determination (positive/negative) for low-prevalence bee viruses (5%; 1/20 bees infected). + or − shows positivity or negativity, respectively, for that replicate sample (9-bee sample).
Bacteriophages.
The phages identified in the Czech viromes belonged to several families, with Myoviridae, Podoviridae, Microviridae, and Siphoviridae being the most common. Out of the total of 158 (at least 50% complete) phage genomes, 26 could be directly clustered with the reference sequences from the database (approximately genus-level assignment), while the majority of contigs could not be assigned to a known genus or family (132), suggesting that multiple novel phage genera are present in honey bees. Host calling through CRISPR spacers predicted bacterial hosts for around 15% of bacteriophages. The predicted hosts (mainly Lactobacillus species, Bifidobacterium, Bartonella, and Salmonella) are those residing in the honey bee gut (51, 52). Even though this assignment level is higher than, e.g., that in the human gut, it did not reach the levels described previously for honey bees (27), with the majority of contigs being assigned to their hosts. This difference may be due to our approach for assigning a host to individual phage sequences instead of a viral cluster or to our stringent setting when comparing CRISPR spacers with phage sequences.
Conclusion.
In conclusion, we identified an important aspect of the total bee virome: the bee-infecting viruses vary widely among individual bees, while the complete virome, which is composed predominantly of bacteriophages and plant viruses, is largely stable and geographically dependent. As the samples originate from the heart of Central Europe, our virome could be considered representative for the region. We revealed the absence of diverse Picornavirales in Czech and other global non-Australian honey bees, probably resulting from the cooccurrence/interaction of V. destructor and DWV. We report the wide presence of LSVs among Czech bees and their unexpected diversity, consisting of five globally represented variants. We provide the first description of a tentative close relationship between two related honey bee-infecting rhabdoviruses. Finally, we provide a preliminary characterization of bacteriophages present in the Czech honey bee virome samples.
MATERIALS AND METHODS
Sample origin.
The worker bees were collected from 8 August to 29 September 2018 with owner permission from 13 representative bee colonies of nine beekeepers, some of whom provided more subspecies/subtype information (see Fig. S1 and Table S1 in the supplemental material). The honey bee workers were shaken off the brood combs into plastic bags, which were immediately placed into a box with dry ice for transport. The samples were then stored at −80°C until analyses. An overview of sampling sites (13 hives, 11 apiaries, and 9 locations) is shown in Fig. S1. All apiaries enrolled in this study were healthy; i.e., they did not exhibit symptoms of common pathogen infections or V. destructor infestation. All hives (except hive 9) were previously treated against V. destructor by either organic acids or amitraz. Three subspecies/subtypes of honey bee as provided by the beekeeper were included: Buckfast honey bee (A. m. buckfast), a hybrid of dark honey bee (A. m. mellifera Linnaeus 1758), and Carniolan honey bee (A. m. carnica Pollman 1879). Some bees originated from different apiaries from the same location or from different hives of the same apiary (metadata are available in Table S1).
Nucleic acid extraction and sequencing library preparation.
Bees were processed according to the NetoVir protocol (28, 53). Three randomly chosen bees from each hive were homogenized in a tube with 2.8-mm ceramic beads (zirconium oxide) (Bertin Technologies, Montigny-le-Bretonneux, France) and 1 mL of sterile phosphate-buffered saline (PBS) using a Minilys homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France) at 3,000 rpm for 5 min. After centrifugation at 17,000 × g for 3 min, the supernatant was filtered through a 0.8-μm filter (polyethersulfone [PES]) (Sartorius, Göttingen, Germany) and centrifuged again at 17,000 × g for 1 min. The supernatant from three homogenates, each consisting of three individuals, gave a pool of nine bees after mixing in one tube. Overall, three distinct replicates of the pooled nine individuals from a hive, denoted replicates A, B, and C, were used in further analyses.
For nuclease treatment, 260 μL of each pooled sample was used and treated with 4 μL of Benzonase nuclease and 2 μL of micrococcal nuclease (New England BioLabs, Ipswich, MA, USA). The total nucleic acids were extracted using the QIAamp viral RNA minikit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, without using carrier RNA. Extracted nucleic acids were reverse transcribed and amplified with the WTA2 kit using 17 amplification cycles (Sigma-Aldrich, St. Louis, MO, USA). The concentration of samples was measured by the Qubit dsDNA (double-stranded DNA) HS (high-sensitivity) assay kit (Thermo Fisher Scientific, Waltham, MA, USA) on the Qubit 2.0 fluorometer. Three nanograms of isolated DNA was processed with the Nextera XT library preparation kit (Illumina, San Diego, CA, USA). The quality of the DNA libraries and size distribution were evaluated using a high-sensitivity DNA assay on a Bioanalyzer 2100 instrument (Agilent Technologies, CA, USA), and the concentration was measured on the Qubit 2.0 fluorometer. The libraries were sent on dry ice to the KU Leuven Nucleomics Core (VIB), Leuven, Belgium, for analysis. Sequencing was performed on the HiSeq2000 platform (Illumina, CA, USA) for 2× 150-bp paired-end cycles.
Bioinformatic analysis. (i) Sequencing data processing and assembly.
Sequencing quality was assessed using FastQC (Babraham Bioinformatics, Cambridge, UK) both before and after trimming. Adapters and low-quality bases were removed using Trimmomatic (54) with settings of 4, 20; leading of 19; tailing of 15; and minlen of 50. The assembly of trimmed reads was done with SPAdes (55) with the metagenomic option and using the following k-mers: 21, 33, 55, and 77. Contigs larger than 500 nucleotides (nt) and with identities of >95% and coverage of at least 80% of the length of the shortest scaffold were merged with ClusterGenomes (https://bitbucket.org/MAVERICLab/docker-clustergenomes). DIAMOND (56) with the settings -sensitive and -c 1 was used to compare sequences against the nonredundant protein database (NCBI) downloaded on 30 September 2018 and subsequently annotated via Kronatools (57). Individual reads were mapped against the nonredundant contigs with BWA-MEM (58), and BamM was used to determine coverage (tpmean) (https://github.com/Ecogenomics/BamM).
(ii) Targeted analysis of bee-infecting viruses.
To obtain precise mapping information about the viruses that infect honey bees, we performed an additional analysis targeted on individual bee viruses. Reference genomic sequences of all currently recognized viral species known to infect honey bees (33) were retrieved from the GenBank database. Sequencing reads were mapped to these reference sequences under conditions of a maximum of 20% mismatches and a maximum of 20% gaps using the Geneious 6.0.3 Read Mapper (59). Consensus nucleotide sequences (majority rule) were called for viruses with complete or nearly complete coverage of reference sequences; the terminal and low-coverage regions of consensus sequences were visually inspected and manually curated. Virus abundance values (virus reads per 1 million sequencing reads) were determined from the sequencing read coverage of the actual viral sequences present in the samples from Czechia.
Comparison with other studies.
For comparisons, we addressed data from previous honey bee virome studies (16, 26, 29, 30, 34). FastQ files were retrieved with the prefetch and fasterq-dump tools available in the SRA toolkit (NCBI). Reads were mapped against viruses known to infect bees (33) with BWA-MEM (58), and coverage was extracted with BamM using the tpmean method. Only RNA viruses (DNA viruses were not investigated in the Australian samples) with sum tpmean values over all samples of ≥20 were included for downstream analysis. Furthermore, we specifically screened our data for novel viruses from the order Picornavirales.
Phylogenetic analysis.
For phylogenetic analysis, complete and partial (>500-bp; RdRp) LSV sequences (as of 6 January 2021) were retrieved from the NCBI database and combined with the LSV sequences obtained in this study. Sequence alignment was done with MAFFT with –localpair; –maxiterate 1000 (60). Alignments were trimmed with trimAL -automated1 (61), and the best model was determined by ModelTest-NG (62). Phylogenetic trees were created with PhyML (63) with the model best suited for alignment.
Bacteriophage identification.
Bacteriophages were identified from all nonredundant contigs (>500 bp) with VirSorter2 (64), ignoring the groups Nucleocytoplasmic large DNA viruses and Lavidaviridae. Presumed phage contigs were checked with CheckV (65) to determine their “completeness.” Putative phage sequences that were at least 50% complete were then classified with VConTACT2 (66) with the BLASTP mode using the Prokaryotic Viral RefSeq 88 database MCL for protein clustering and ClusterONE for genome clustering. Host calling was performed using CRISPR spacers retrieved from a set of 304 genomes of bacterial species described to reside in the honey bee gut (NCBI and JGI IMG/M) (Table S2). Bacterial genomic sequences were processed with MinCED (36), and the predicted spacers were pulled and analyzed by BLAST against phage contigs with the stringent settings -ungapped and -perc_identity 100. A complementary host-calling approach was performed by utilizing a CrisprOpenDB (37) search against all complete bacterial genomes to identify bacterial hosts outside the common spectrum (only level 1 predictions). All predicted hosts from CrisprOpenDB were bee-infecting bacteria less frequently mentioned in the literature and therefore were not included in our database (MinCED). The results of the two predictions were merged.
Two sheets showing bacteria that were used for host prediction, one for NCBI and the second for IMG, containing accession numbers and IMG genome identifiers. Download Table S2, XLSX file, 0.01 MB (14.3KB, xlsx) .
Copyright © 2022 Kadlečková et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistical analysis and visualization.
Statistical analysis was done in R with the Adonis test (permutations, 10,000) implemented in the vegan package (67). Heatmaps were created using the heatmaply package (68), and trees were visualized with iTOL (69). The VConTACT2 network was visualized in Python with the graph-tool library (70). k-means clustering was done in Python with sklearn (71). For each data set, data were first scaled by a standard scaler, and clusters were predicted with k-means (13 clusters). Labels of these predicted clusters were compared with labels of real clusters (1 to 13). Gained scores are available in Fig. S1.
Data availability.
The sequencing reads were deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA781422. The assembled viral genomic sequences were deposited in GenBank (accession numbers OL803813 to OL803870).
ACKNOWLEDGMENTS
We thank M. Simonovsky for valuable help with sample collection. We thank beekeepers for allowing us to collect bee samples. We also thank N. Vaclavikova for technical support.
This study was supported by CELSA [Metagenomic Analysis of Honey Bee (Apis mellifera) in the Czech Republic, 2017 to 2019], grant number QK1910018 of the Ministry of Agriculture of the Czech Republic (http://www.eagri.cz), and the MICOBION project funded from EU H2020 (number 810224).
Contributor Information
Jelle Matthijnssens, Email: jelle.matthijnssens@kuleuven.be.
Seth Bordenstein, Vanderbilt University.
REFERENCES
- 1.Schmidt JO. 1997. Bee products, p 15–26. In Mizrahi A, Lensky Y (ed), Bee products: properties, applications, and apitherapy. Springer, Boston, MA. [Google Scholar]
- 2.Klein A-M, Vaissière BE, Cane JH, Steffan-Dewenter I, Cunningham SA, Kremen C, Tscharntke T. 2007. Importance of pollinators in changing landscapes for world crops. Proc Biol Sci 274:303–313. doi: 10.1098/rspb.2006.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calderone NW. 2012. Insect pollinated crops, insect pollinators and US agriculture: trend analysis of aggregate data for the period 1992-2009. PLoS One 7:e37235. doi: 10.1371/journal.pone.0037235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hung K-LJ, Kingston JM, Albrecht M, Holway DA, Kohn JR. 2018. The worldwide importance of honey bees as pollinators in natural habitats. Proc Biol Sci 285:20172140. doi: 10.1098/rspb.2017.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann P, Carreck NL. 2010. Honey bee colony losses. J Apic Res 49:1–6. doi: 10.3896/IBRA.1.49.1.01. [DOI] [Google Scholar]
- 6.Gray A, Adjlane N, Arab A, Ballis A, Brusbardis V, Charrière J-D, Chlebo R, Coffey MF, Cornelissen B, da Costa CA, Dahle B, Danihlík J, Dražić MM, Evans G, Fedoriak M, Forsythe I, Gajda A, de Graaf DC, Gregorc A, Ilieva I, Johannesen J, Kauko L, Kristiansen P, Martikkala M, Martín-Hernández R, Medina-Flores CA, Mutinelli F, Patalano S, Raudmets A, Martin GS, Soroker V, Stevanovic J, Uzunov A, Vejsnaes F, Williams A, Zammit-Mangion M, Brodschneider R. 2020. Honey bee colony winter loss rates for 35 countries participating in the COLOSS survey for winter 2018-2019, and the effects of a new queen on the risk of colony winter loss. J Apic Res 59:744–751. doi: 10.1080/00218839.2020.1797272. [DOI] [Google Scholar]
- 7.vanEngelsdorp D, Evans JD, Saegerman C, Mullin C, Haubruge E, Nguyen BK, Frazier M, Frazier J, Cox-Foster D, Chen Y, Underwood R, Tarpy DR, Pettis JS. 2009. Colony collapse disorder: a descriptive study. PLoS One 4:e6481. doi: 10.1371/journal.pone.0006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson DL, Trueman J. 2000. Varroa jacobsoni (Acari: Varroidae) is more than one species. Exp Appl Acarol 24:165–189. doi: 10.1023/A:1006456720416. [DOI] [PubMed] [Google Scholar]
- 9.Cornman RS, Tarpy DR, Chen Y, Jeffreys L, Lopez D, Pettis JS, vanEngelsdorp D, Evans JD. 2012. Pathogen webs in collapsing honey bee colonies. PLoS One 7:e43562. doi: 10.1371/journal.pone.0043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De la Rúa P, Jaffé R, Dall’Olio R, Muñoz I, Serrano J. 2009. Biodiversity, conservation and current threats to European honeybees. Apidologie 40:263–284. doi: 10.1051/apido/2009027. [DOI] [Google Scholar]
- 11.Möckel N, Gisder S, Genersch E. 2011. Horizontal transmission of deformed wing virus: pathological consequences in adult bees (Apis mellifera) depend on the transmission route. J Gen Virol 92:370–377. doi: 10.1099/vir.0.025940-0. [DOI] [PubMed] [Google Scholar]
- 12.Wilfert L, Long G, Leggett HC, Schmid-Hempel P, Butlin R, Martin SJM, Boots M. 2016. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 351:594–597. doi: 10.1126/science.aac9976. [DOI] [PubMed] [Google Scholar]
- 13.Brødsgaard CJ, Ritter W, Hansen H, Brødsgaard HF. 2000. Interactions among Varroa jacobsoni mites, acute paralysis virus, and Paenibacillus larvae larvae and their influence on mortality of larval honeybees in vitro. Apidologie 31:543–554. doi: 10.1051/apido:2000145. [DOI] [Google Scholar]
- 14.Martin SJ. 2001. The role of Varroa and viral pathogens in the collapse of honeybee colonies: a modelling approach. J Appl Ecol 38:1082–1093. doi: 10.1046/j.1365-2664.2001.00662.x. [DOI] [Google Scholar]
- 15.Francis RM, Nielsen SL, Kryger P. 2013. Varroa-virus interaction in collapsing honey bee colonies. PLoS One 8:e57540. doi: 10.1371/journal.pone.0057540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts JMK, Anderson DL, Durr PA. 2018. Metagenomic analysis of Varroa-free Australian honey bees (Apis mellifera) shows a diverse Picornavirales virome. J Gen Virol 99:818–826. doi: 10.1099/jgv.0.001073. [DOI] [PubMed] [Google Scholar]
- 17.Martin SJ, Highfield AC, Brettell L, Villalobos EM, Budge GE, Powell M, Nikaido S, Schroeder DC. 2012. Global honey bee viral landscape altered by a parasitic mite. Science 336:1304–1306. doi: 10.1126/science.1220941. [DOI] [PubMed] [Google Scholar]
- 18.Ryabov EV, Wood GR, Fannon JM, Moore JD, Bull JC, Chandler D, Mead A, Burroughs N, Evans DJ. 2014. A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog 10:e1004230. doi: 10.1371/journal.ppat.1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Posada-Florez F, Childers AK, Heerman MC, Egekwu NI, Cook SC, Chen Y, Evans JD, Ryabov EV. 2019. Deformed wing virus type A, a major honey bee pathogen, is vectored by the mite Varroa destructor in a non-propagative manner. Sci Rep 9:12445. doi: 10.1038/s41598-019-47447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norton AM, Remnant EJ, Buchmann G, Beekman M. 2020. Accumulation and competition amongst deformed wing virus genotypes in naïve Australian honeybees provides insight into the increasing global prevalence of genotype B. Front Microbiol 11:620. doi: 10.3389/fmicb.2020.00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norton AM, Remnant EJ, Tom J, Buchmann G, Blacquiere T, Beekman M. 2021. Adaptation to vector-based transmission in a honeybee virus. J Anim Ecol 90:2254–2267. doi: 10.1111/1365-2656.13493. [DOI] [PubMed] [Google Scholar]
- 22.Doublet V, Labarussias M, de Miranda JR, Moritz RFA, Paxton RJ. 2015. Bees under stress: sublethal doses of a neonicotinoid pesticide and pathogens interact to elevate honey bee mortality across the life cycle. Environ Microbiol 17:969–983. doi: 10.1111/1462-2920.12426. [DOI] [PubMed] [Google Scholar]
- 23.Martin SJ, Hardy J, Villalobos E, Martín‐Hernández R, Nikaido S, Higes M. 2013. Do the honeybee pathogens Nosema ceranae and deformed wing virus act synergistically? Environ Microbiol Rep 5:506–510. doi: 10.1111/1758-2229.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMenamin AJ, Genersch E. 2015. Honey bee colony losses and associated viruses. Curr Opin Insect Sci 8:121–129. doi: 10.1016/j.cois.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 25.McMenamin AJ, Flenniken ML. 2018. Recently identified bee viruses and their impact on bee pollinators. Curr Opin Insect Sci 26:120–129. doi: 10.1016/j.cois.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Galbraith DA, Fuller ZL, Ray AM, Brockmann A, Frazier M, Gikungu MW, Martinez JFI, Kapheim KM, Kerby JT, Kocher SD, Losyev O, Muli E, Patch HM, Rosa C, Sakamoto JM, Stanley S, Vaudo AD, Grozinger CM. 2018. Investigating the viral ecology of global bee communities with high-throughput metagenomics. Sci Rep 8:8879. doi: 10.1038/s41598-018-27164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonilla-Rosso G, Steiner T, Wichmann F, Bexkens E, Engel P. 2020. Honey bees harbor a diverse gut virome engaging in nested strain-level interactions with the microbiota. Proc Natl Acad Sci USA 117:7355–7362. doi: 10.1073/pnas.2000228117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deboutte W, Beller L, Yinda CK, Maes P, de Graaf DC, Matthijnssens J. 2020. Honey-bee-associated prokaryotic viral communities reveal wide viral diversity and a profound metabolic coding potential. Proc Natl Acad Sci USA 117:10511–10519. doi: 10.1073/pnas.1921859117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remnant EJ, Shi M, Buchmann G, Blacquière T, Holmes EC, Beekman M, Ashe A. 2017. A diverse range of novel RNA viruses in geographically distinct honey bee populations. J Virol 91:e00158-17. doi: 10.1128/JVI.00158-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin S, Sela N, Erez T, Nestel D, Pettis J, Neumann P, Chejanovsky N. 2019. New viruses from the ectoparasite mite Varroa destructor infesting Apis mellifera and Apis cerana. Viruses 11:94. doi: 10.3390/v11020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin SJ, Brettell LE. 2019. Deformed wing virus in honeybees and other insects. Annu Rev Virol 6:49–69. doi: 10.1146/annurev-virology-092818-015700. [DOI] [PubMed] [Google Scholar]
- 32.Alger SA, Burnham PA, Boncristiani HF, Brody AK. 2019. RNA virus spillover from managed honeybees (Apis mellifera) to wild bumblebees (Bombus spp.). PLoS One 14:e0217822. doi: 10.1371/journal.pone.0217822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beaurepaire A, Piot N, Doublet V, Antunez K, Campbell E, Chantawannakul P, Chejanovsky N, Gajda A, Heerman M, Panziera D, Smagghe G, Yañez O, de Miranda JR, Dalmon A. 2020. Diversity and global distribution of viruses of the Western honey bee, Apis mellifera. Insects 11:239. doi: 10.3390/insects11040239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deboutte W, Beller L, Yinda CK, Shi C, Smets L, Vanmechelen B, Conceição-Neto N, Dallmeier K, Maes P, de Graaf DC, Matthijnssens J. 2020. Hymenoptera associated eukaryotic virome lacks host specificity. bioRxiv 10.1101/2020.09.15.298042. [DOI]
- 35.Levin S, Sela N, Chejanovsky N. 2016. Two novel viruses associated with the Apis mellifera pathogenic mite Varroa destructor. Sci Rep 6:37710. doi: 10.1038/srep37710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P. 2007. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pourcel C, Touchon M, Villeriot N, Vernadet J-P, Couvin D, Toffano-Nioche C, Vergnaud G. 2020. CRISPRCasdb a successor of CRISPRdb containing CRISPR arrays and cas genes from complete genome sequences, and tools to download and query lists of repeats and spacers. Nucleic Acids Res 48:D535–D544. doi: 10.1093/nar/gkz915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowen-Walker PL, Martin SJ, Gunn A. 1999. The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasitic mite Varroa jacobsoni Oud. J Invertebr Pathol 73:101–106. doi: 10.1006/jipa.1998.4807. [DOI] [PubMed] [Google Scholar]
- 39.Roberts JMK, Anderson DL, Durr PA. 2017. Absence of deformed wing virus and Varroa destructor in Australia provides unique perspectives on honeybee viral landscapes and colony losses. Sci Rep 7:6925. doi: 10.1038/s41598-017-07290-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gauthier L, Tentcheva D, Tournaire M, Dainat B, Cousserans F, Colin ME, Bergoin M. 2007. Viral load estimation in asymptomatic honey bee colonies using the quantitative RT-PCR technique. Apidologie 38:426–435. doi: 10.1051/apido:2007026. [DOI] [Google Scholar]
- 41.Thompson HM, Brown MA, Ball RF, Bew MH. 2002. First report of Varroa destructor resistance to pyrethroids in the UK. Apidologie 33:357–366. doi: 10.1051/apido:2002027. [DOI] [Google Scholar]
- 42.Roth MA, Wilson JM, Tignor KR, Gross AD. 2020. Biology and management of Varroa destructor (Mesostigmata: Varroidae) in Apis mellifera (Hymenoptera: Apidae) colonies. J Integr Pest Manag 11:1. doi: 10.1093/jipm/pmz036. [DOI] [Google Scholar]
- 43.DaPalma T, Doonan BP, Trager NM, Kasman LM. 2010. A systematic approach to virus-virus interactions. Virus Res 149:1–9. doi: 10.1016/j.virusres.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Runckel C, Flenniken ML, Engel JC, Ruby JG, Ganem D, Andino R, DeRisi JL. 2011. Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS One 6:e20656. doi: 10.1371/journal.pone.0020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravoet J, Maharramov J, Meeus I, De Smet L, Wenseleers T, Smagghe G, de Graaf DC. 2013. Comprehensive bee pathogen screening in Belgium reveals Crithidia mellificae as a new contributory factor to winter mortality. PLoS One 8:e72443. doi: 10.1371/journal.pone.0072443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Granberg F, Vicente-Rubiano M, Rubio-Guerri C, Karlsson OE, Kukielka D, Belák S, Sánchez-Vizcaíno JM. 2013. Metagenomic detection of viral pathogens in Spanish honeybees: co-infection by aphid lethal paralysis, Israel acute paralysis and Lake Sinai viruses. PLoS One 8:e57459. doi: 10.1371/journal.pone.0057459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravoet J, De Smet L, Wenseleers T, de Graaf DC. 2015. Genome sequence heterogeneity of Lake Sinai virus found in honey bees and Orf1/RdRP-based polymorphisms in a single host. Virus Res 201:67–72. doi: 10.1016/j.virusres.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 48.Daughenbaugh KF, Martin M, Brutscher LM, Cavigli I, Garcia E, Lavin M, Flenniken ML. 2015. Honey bee infecting Lake Sinai viruses. Viruses 7:3285–3309. doi: 10.3390/v7062772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey L, Ball B, Carpenter J, Woods R. 1980. Small virus-like particles in honey bees associated with chronic paralysis virus and with a previously undescribed disease. J Gen Virol 46:149–155. doi: 10.1099/0022-1317-46-1-149. [DOI] [Google Scholar]
- 50.de Miranda JR, Bailey L, Ball BV, Blanchard P, Budge GE, Chejanovsky N, Chen Y-P, Gauthier L, Genersch E, de Graaf DC, Ribière M, Ryabov E, De Smet L, van der Steen JJM. 2013. Standard methods for virus research in Apis mellifera. J Apic Res 52:1–56. doi: 10.3896/IBRA.1.52.4.22. [DOI] [Google Scholar]
- 51.Ellegaard KM, Engel P. 2019. Genomic diversity landscape of the honey bee gut microbiota. Nat Commun 10:446. doi: 10.1038/s41467-019-08303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anjum SI, Aldakheel F, Shah AH, Khan S, Ullah A, Hussain R, Khan H, Ansari MJ, Mahmoud AH, Mohammed OB. 2021. Honey bee gut an unexpected niche of human pathogen. J King Saud Univ Sci 33:101247. doi: 10.1016/j.jksus.2020.101247. [DOI] [Google Scholar]
- 53.Conceição-Neto N, Zeller M, Lefrère H, De Bruyn P, Beller L, Deboutte W, Yinda CK, Lavigne R, Maes P, Ranst MV, Heylen E, Matthijnssens J. 2015. Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis. Sci Rep 5:16532. doi: 10.1038/srep16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 57.Ondov BD, Bergman NH, Phillippy AM. 2011. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katoh K. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol 37:291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 64.Guo J, Bolduc B, Zayed AA, Varsani A, Dominguez-Huerta G, Delmont TO, Pratama AA, Gazitúa MC, Vik D, Sullivan MB, Roux S. 2021. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome 9:37. doi: 10.1186/s40168-020-00990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nayfach S, Camargo AP, Schulz F, Eloe-Fadrosh E, Roux S, Kyrpides NC. 2021. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol 39:578–585. doi: 10.1038/s41587-020-00774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bin Jang H, Bolduc B, Zablocki O, Kuhn JH, Roux S, Adriaenssens EM, Brister JR, Kropinski AM, Krupovic M, Lavigne R, Turner D, Sullivan MB. 2019. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat Biotechnol 37:632–639. doi: 10.1038/s41587-019-0100-8. [DOI] [PubMed] [Google Scholar]
- 67.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2019. vegan: community ecology package (2.5-6). https://CRAN.R-project.org/package=vegan. Accessed 20 July 2020.
- 68.Galili T, O’Callaghan A, Sidi J, Sievert C. 2018. heatmaply: an R package for creating interactive cluster heatmaps for online publishing. Bioinformatics 34:1600–1602. doi: 10.1093/bioinformatics/btx657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Letunic I, Bork P. 2019. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peixoto TP. 2017. The graph-tool python library. figshare.
- 71.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830. [Google Scholar]
- 72.Cornman RS. 2019. Relative abundance and molecular evolution of Lake Sinai virus (Sinaivirus) clades. PeerJ 7:e6305. doi: 10.7717/peerj.6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional figures and data (map, plant heatmap, viral family distribution in the virome, comparison of samples from Australia and the rest of the world, BRV phylogeny, and additional information on k-means clustering). Download FIG S1, PDF file, 0.7 MB (670.3KB, pdf) .
Copyright © 2022 Kadlečková et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Several sheets of raw and transformed data used to create heatmaps, also including metadata (replicate, subtype of honey bee, and location, etc.) for each sample. Download Table S1, XLSX file, 1.6 MB (1.6MB, xlsx) .
Copyright © 2022 Kadlečková et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Raw data from comparison of Australian and other viromes, including accession numbers used for viral genomes. Download Table S3, XLSX file, 0.05 MB (52.8KB, xlsx) .
Copyright © 2022 Kadlečková et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogeny of RdRp regions of LSVs. Download FIG S2, PDF file, 0.3 MB (286.2KB, pdf) .
Copyright © 2022 Kadlečková et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Similarity network for bacteriophages. Download FIG S3, PDF file, 0.2 MB (174.7KB, pdf) .
Copyright © 2022 Kadlečková et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Two sheets showing bacteria that were used for host prediction, one for NCBI and the second for IMG, containing accession numbers and IMG genome identifiers. Download Table S2, XLSX file, 0.01 MB (14.3KB, xlsx) .
Copyright © 2022 Kadlečková et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The sequencing reads were deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA781422. The assembled viral genomic sequences were deposited in GenBank (accession numbers OL803813 to OL803870).