Abstract
Polyploidy, a cell status defined as more than two sets of genomic DNA, is a conserved strategy across species that can increase cell size and biosynthetic production, but the functional aspects of polyploidy are nuanced and vary across cell types. Throughout Drosophila developmental stages (embryo, larva, pupa and adult), polyploid cells are present in numerous organs and help orchestrate development while contributing to normal growth, well-being and homeostasis of the organism. Conversely, increasing evidence has shown that polyploid cells are prevalent in Drosophila tumors and play important roles in tumor growth and invasiveness. Here, we summarize the genes and pathways involved in polyploidy during normal and tumorigenic development, the mechanisms underlying polyploidization, and the functional aspects of polyploidy in development, homeostasis and tumorigenesis in the Drosophila model.
Keywords: polyploidy, cancer, development, drosophila, endoreplication, endocycle
Introduction
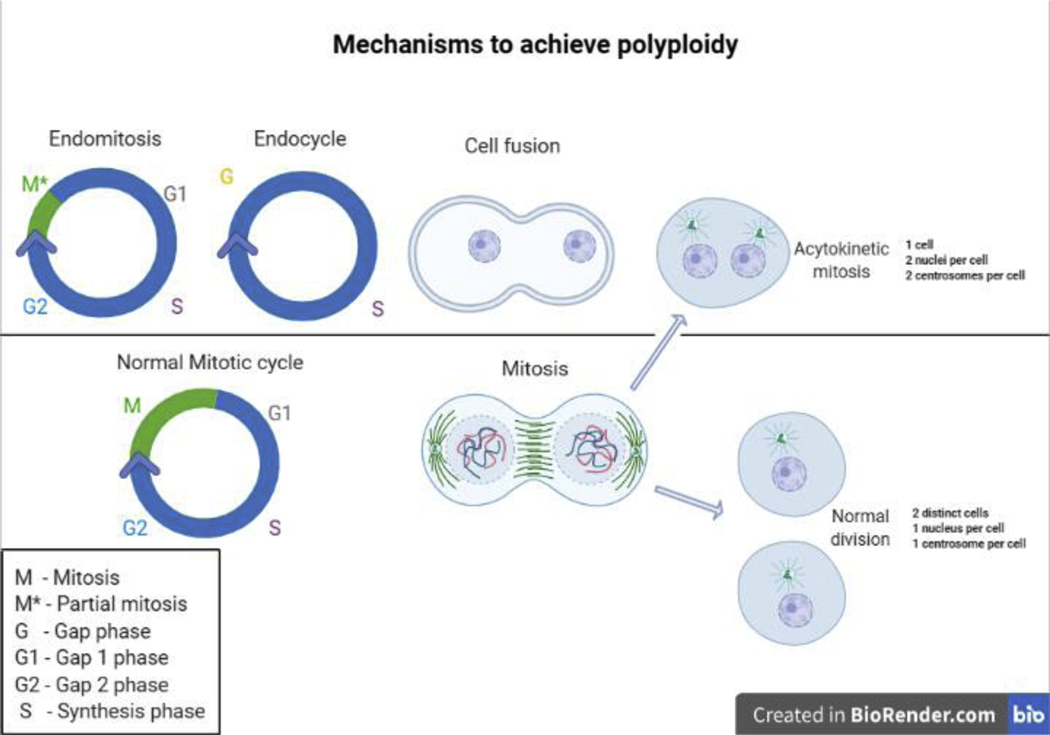
Polyploidy is relevant to specific tissues and their development and is a strategy used by cells to increase cell size and transcriptional output in eukaryotes [1–4]. Cells increase their ploidy level by a variety of processes, including acytokinetic mitosis, cell fusion, endomitosis and endocycle (Fig. 1). In cell fusion, cells merge to form a single multinucleated cell; for example, myoblasts fuse and form polynucleated muscle fibers [2, 5–7]. Acytokinetic mitosis is associated with failure of cytokinesis: cells undergo all cell cycle stages (G1-S-G2-M) but cytokinesis fails to occur, which results in a multinucleated cell [2, 5, 7]. Endomitosis involves a partial mitosis in the absence of both karyokinesis and cytokinesis, as seen in megakaryocytes [2, 5]. A fourth mechanism for increasing ploidy is called the endocycle, where mitosis is entirely evaded and genomic DNA is doubled or nearly doubled between an intervening gap (G) phase [2, 5–7]. Endoreplication refers to replication of the genome via actyokinetic mitosis, endomitosis or the endocycle. In Drosophila, the endocycle can produce polytene chromosomes, which are a fusion of sister chromatids that have not undergone segregation. These chromosomes have densely packed heterochromatin interspaced with loosely packed euchromatin, forming distinct banding patterns visible with electron microscopy [8, 9]. Because the endocycle can increase DNA content without increasing the chromosome set number (N), the ploidy of polytene cells is measured in multiples of the total DNA content in a gamete (C). In this review, we will discuss studies that have used Drosophila melanogaster as a model system to uncover the intricacies that promote polyploidization, primarily focusing on the endocycle and its role in normal and tumorigenic tissues.
Figure 1.
Endomitosis encompasses G1, S, G2 and partial M phase without karyokinesis and cytokinesis. Endocycle comprises the G and S phase. In cell fusion, cells merge, resulting in a multinucleated cell. The mitotic cycle includes G1, S, G2 and M phase and ends with two single nucleated cells.
Polyploidy affects development by impacting organ size and function. In mammals, cytokinetic failure frequently occurs soon after fertilization. The resulting polyploid cells exist as multinucleated blastomeres in the developing embryo [10–13]. Polyploidization is also inherent to the developing placenta, as villous trophoblasts fuse to form multinucleated syncytiotrophoblasts and as trophoblast stem cells differentiate by endoreplication into trophoblast giant cells, which are vital for placenta growth, implantation of the conceptus into the uterus, lactogenesis, regulation of maternal blood flow, and synthesis of cytokines and hormones like progesterone [14–16]. Polyploidization is also seen in megakaryopoiesis, where megakaryoblasts perform endomitosis to mature into megakaryocytes. Polyploidy in megakaryocytes is supportive of large cell size and high transcriptional output appropriate for platelet biogenesis [17–19]. In liver parenchyma, stochastic cytokinetic failure of cells occurs during postnatal growth, patterning hepatic tissue with mosaic ploidy levels [20, 21]. In various human solid tumors, histopathologists have recognized Polyploid Giant Cancer Cells (PGCCs), which can be identified by a comparatively large nucleus size and cell size three times or greater than regular cancer cells [22, 23]. Additionally, the high prevalence of polyploidy in tumors has been inferred from clinical sequencing data of several thousands of cancer patients [24]. The occurrence of genomic doubling is closely associated with worse prognosis, making the presence of polyploidy a predictor for patient survival and tumor evolution [25]. Genomic doublings and PGCCs appear early in tumorigenesis, implying a role for tumor initiation. Polyploidy in human tumors have also been associated with drug resistance and metastasis [22, 26–28].
Drosophila is a well-established model to study polyploidy as polyploid cells exist across distinct organs such as the ovary, gut, brain, and salivary glands [29–33]. Besides this advantage, the Drosophila model also includes more than a century of research, and a great number of genetic tools and techniques are therein described [34]. These advantages set Drosophila apart as a leading model to explore genes and mechanisms involved in the regulation of polyploidization particularly via endoreplication. In line with this argument, this review focuses on polyploidization primarily in reference to endocycling Drosophila cells. The first section starts off by describing the various instances of polyploidy during Drosophila development. The middle sections discuss distinct genes and pathways that influence the endocycle in different tissues. The review concludes with a discussion of polyploidy on wound repair and tumorigenesis.
1. Polyploidy in Drosophila development
Polyploid cells are present across a wide variety of organs and tissues in Drosophila and are found throughout all four stages of the insect’s holometabolous life cycle, comprising embryo, larva, pupa, and adult. These polyploid cells have assorted features and functions.
In the embryo, DNA is endoreplicated through endocycles between 8 and 15 hours after fertilization at specific times and domains of the salivary gland, Malphigian tubules, and gut [29, 35]. Polyteny is a property of these cells and refers to chromosomes formed as a fusion of undivided chromatids [36]. In the salivary gland, the polytene cells include the secretory cells, which undergo asynchronous endoreplications at embryogenesis to become giant cells [33].
In Drosophila larvae, large size and ample DNA content of the salivary gland cells is appropriate for the synthesis and secretion of digestive enzymes and adhesive substance for attachment to the substrate. Polytene cells of the Malpighian tubules remove toxic chemicals and nitrogenous waste from the hemolymph [37], making these structures analogous in function to the human kidney [38, 39]. Drosophila larvae also contain polytene cells in regions of the fat bodies, epidermis, trachea, brain, and prothoracic glands [29, 40–43]. The larval prothoracic glands regulate growth and maturation by secreting the steroid molting hormone ecdysone [44]. Ecdysone synthesis is endocycle-dependent and essential for larval development; inhibition of the endocycle in prothoracic gland cells reduces ecdysone synthesis and results in larval developmental arrest [45].
Many larval cells and tissues that are no longer useful are removed by programmed cell death at metamorphosis [46]. The hormone ecdysone contributes to this process by inducing histolysis of cells in the ileum, midgut, ventral nerve cord, anterior and abdominal muscle, optic lobe, and salivary gland [47–54]. During pupation, other modifications that concern or give rise to polyploid cells take place as well. For example, though senescence usually follows endoreplication, rectal papillar cells re-enter mitosis after having undergone endoreplication during the larval stage, making these cells susceptible to chromosome abnormality [48]. Additionally, cells of the sensory organ lineage such as the socket and shaft cells that form the hair bristles will undergo endocycles, with the size of the hair shaft dependent on the number of endoreplications [55].
In the adult, polyploid cells are included in the ovary, spermatheca, Malpighian tubules, gut and brain. The cell cycle program of the ovaries and the timing of endoreplication are well characterized. The somatic follicle cells surrounding germline cells form egg chambers that show distinct cell cycle programs during oogenesis, including the mitotic cycle (stages 1–6), the endocycle (stages 7–10a), and chorion gene amplification (stages 10b-13). During the endocycle phase, main body follicle cells undergo three rounds of endoreplication, an essential step that promotes polyploidy and cell growth [56–60]. Nurse cells also undergo multiple rounds of endoreplication. At stage 6, the nurse cell polytene chromosomes disassemble via a mitotic-like process into 32 pairs of chromatids that disperse throughout the nucleus. Oogenesis proceeds with subsequent endoreplications, increasing nuclear size and leading to a final cell ploidy of around 1500C [61]. Nurse cells support oocyte growth by providing mRNA, proteins, and organelles through specialized ring channels [62-64]. Included within the female reproductive tract is the spermatheca, which is lined with polyploid secretory cells. These secretory cells contribute to large-scale bioproduction of secretory discharge, which helps with attraction of sperm to the sperm receptacle and regulation of ovulation [65–67].
During adult gut development, intestinal stem cells and enteroblasts require Notch signaling to decrease cell proliferation and to initiate differentiation into mature polyploid enterocytes [68, 69]. In the brain, neurons and glia are diploid at eclosion, except for the subperineurial glia, which undergo endocycles and endomitosis in the larval and pupal stages to form the blood-brain barrier. As the adult ages, several of the diploid cells acquire polyploidy as well, which serves as a protective effect against DNA damage-induced cell death [32].
2. Genes and pathways involved in polyploidy regulation
2.1. The cell cycle machinery
The cell cycle alternates among four sequential phases (G1-S-G2-M). Phase progression and transitions are correlated with the concentration of ancillary cyclins, which are regulatory proteins that form active complexes with cyclin-dependent kinases (CDKs). Cyclin concentrations oscillate while CDK concentrations remain relatively constant, providing temporal occurrence of cyclin-CDK complexes. These complexes activate or inhibit substrates by phosphorylation, catalyzing the cascade of events that lead to phase progression or transitions [70–72]. Cyclins are promoted for degradation by enzymes such as Fizzy-related (Fzr), which is auxiliary to the E3 ubiquitin ligase Anaphase Promoting Complex/Cyclosome (APC/C) [73, 74]. Activity of Cyclin-CDK complexes can further be regulated by cyclin-dependent kinase inhibitors like Dacapo (Dap). With specific expression of cell cycle regulators, the cell can bypass the M phase and enter the endocycle (M/E switch). Differential expression of regulatory factors allows for unique variation on endocycle progression.
CycE
CycE is the primary cyclin driving the endocycle. In contrast, mitotic cyclins are expressed at near undetectable levels throughout the endocycle, and while cyclin D is expressed during endocycles, it is primarily involved in cell growth, thus only affecting the endocycle indirectly [75]. Therefore, the endocycle implements CycE fluctuations in a relatively static backdrop of Cdk2 without much convolution by additional cyclins or CDKs [75].
Low levels of CycE-Cdk2 complex are correlated with DNA replication licensing during G phase because of a repressive role of CycE-Cdk2 on the pre-replication complex (pre-RC). Specifically, high levels of CycE-Cdk2 suppress pre-RC formation in part through Fzr inhibition, leading to Geminin (Gem) accumulation, as demonstrated in the salivary gland [74, 76–78]. Accordingly, constitutive expression of cycE affects endocycles by inhibiting DNA licensing and causing a reduction of DNA content of salivary gland cell nuclei. The role of cycE in mitotic cycles differs, as mitotic cycles were largely unaffected by constitutive cycE expression [76, 77, 79]. CycE-Cdk2 is also required for DNA origin firing and progression through S phase [76, 80–82]. Ectopic expression can induce pre-mature S phase initiation. Furthermore, CycE regulates S phase duration, as its diminished expression heralds the completion of DNA replication. This role has been demonstrated in germline Nurse cells carrying a hypomorphic allele of cycE that shows persistent expression at late S phase. These mutant nurse cells have a lengthened S phase and ectopic replication of satellite DNA, these regions being normally under-represented in the wild-type [82].
Given that periodic activity of CycE-Cdk2 alternates the endocycle through DNA replicative and dormant phases, inputs and processes are maintained to regulate the activation and inactivation of this complex. CycE is embedded in an autoregulatory loop, with high cycE expression being a threshold for initiating its degradation [82]. Apontic and E2f1 have been shown to initiate G phase transcription of cycE, while CycE destruction is mediated by the Skp/Cullin/F-box (SCF) ubiquitin ligase complexed with Archipelago, the F-box specificity component [79, 83–85]. In the ovary and salivary gland, in addition to CycE oscillation, CycE-Cdk2 dynamics are reinforced by oscillatory expression of Dap, the CycE-Cdk2 inhibitor [86, 87].
CycA and CycB
CycA and CycB promote mitosis and progress the canonical cell cycle and are expressed in proliferating cells throughout development [88]. CycA forms complexes with several CDKs, and the CycA-Cdk1 complex likely drives mitosis through phosphorylation of the Myb-MuvB complex, which subsequently activates the transcription of M phase genes [89]. In the Schneider D2 (S2) cell line, knockdown of CycA, but not CycB, was sufficient to induce endoreplication [89]. Additionally, CycA knockdown in follicle cells induced M phase evasion and precocious entry into the endocycle [90].
Despite its role in promoting mitosis, CycA has been detected at low levels of oscillation in endocycling cells. This low level of expression is not entirely insignificant, as loss of CycA localized to cells of the bristle lineage and salivary gland decreases final ploidy of the fully differentiated cells. As a possible explanation, subnuclear re-localization of Orc2, a member of the Pre-RC complex, is dependent upon the minimal accumulation of CycA at mid S phase. The perturbance on timing of early and late S phase, where euchromatin and heterochromatin are replicated, respectively, may lead to a reduction of final cell ploidy [55].
Fzr
Fzr, the equivalent of the Cdh1 in mammals, is a positive regulator of APC/C, which targets and ubiquitinates M-phase components and cyclins such as CycA and CycB for degradation [91]. In the adult pylorus, Fzr mediates endocycle entry of cells neighboring empty spaces left by cell loss after injury, and endocycle entry leads to protection against tissue architectural and permeability disruptions [92]. In follicle cells, Notch signaling induces Fzr, which promotes the degradation of CycA and CycB, promoting cell transition from the mitotic to endoreplication cycle [59]. Similarly, in the embryonic salivary glands and midgut, Fzr is required to promote the degradation of CycA and CycB, ending the proliferative phase and beginning the endoreplication phase [73].
Dap
Dap protein, a cyclin kinase inhibitor with a similar binding-domain and function to p21 and p27 (mammalian homologs), specifically suppresses the CycE-Cdk2 complex [93]. In the embryonic epidermis, eye disc, and nervous system, Dap serves an essential role as an S phase inhibitor, and transient high expression of Dap coincides with permanent cell cycle arrest at G0 [93, 94].
Oscillatory Dap expression is required for proper endocycle progression in the ovary [86], and its oscillation is also needed for normal progression of the endocycle in the salivary gland [87]. Dap facilitates replication licensing by inhibiting CycE-Cdk2 activity at G phase [86]. Dap can also suppress CycE-Cdk2 activity during late S phase, which contributes to early S-phase truncation, meaning DNA replication is often incomplete during a normal endocycle, particularly in heterochromatic regions [86, 87]. In both the ovary and salivary gland, oscillatory Dap expression is due to S phase-dependent ubiquitylation of Dap by the E3 ubiquitin ligase CRL4Cdt2. Dap has a PIP degron region that binds to DNA-bound PCNA, the sliding clamp that is active during DNA replication, and preferential Dap ubiquitination occurs when Dap is bound to DNA-bound PCNA [87, 95].
Cdc6
Cdc6 serves as a primary DNA replication licensing factor. Cdc6 together with Cdt1/Double-parked (Dup) initiates DNA replication via recruitment of the replicative helicase MCM2–7 and other initiation factors to the origin recognition complex (ORC), forming the pre-RC [96]. In this context, polyploid cells, which require multiple rounds of DNA replication, are reliant upon Cdc6 for maturation. Indeed, loss of function studies confirmed that Cdc6 is pertinent to the onset of DNA replication in both diploid and polyploid cells; quantitative analysis by flow cytometry on fat body and follicle cells with loss-of-Cdc6 shows significantly decreased levels of ploidy [97]. Additionally, in the adult central nervous system, where glia and mature neurons exhibit high levels of ploidy as a cushion for accumulative DNA stress, knockdown of Cdc6 inhibits or reduces polyploidy [32].
For proper DNA replication during either the mitotic cycle or endocycle, licensing is only initiated once. While evidence using yeast cells suggests that Cdk complexes phosphorylate Cdc6 and inhibit re-licensing [98], in Drosophila, a more significant factor to re-licensing inhibition is negative regulation by Geminin, which can bind and sequester Dup [99, 100].
2.2. Transcription factors
Transcription factors (TF) are essential regulators of gene expression, making them essential to cancer and organ development. Some TFs display oncogenic activity and are targets of drugs in cancer therapies [101, 102]. Many functional aspects of Drosophila TFs are conserved in humans. For example, overexpression of Myc is able to enhance endoreplication in p53 mutated melanoma cells that were M-phase arrested by the damaging agent paclitaxel [103]. And in Drosophila follicle cells, overexpression of Myc promotes extra rounds of endoreplication and increases nuclear sizes [85, 104, 105]. This section reviews relevant studies that involve Drosophila TFs involved in polyploidization of cells.
E2f1
E2f1, an evolutionarily conserved TF from the E2f family, is required for S phase entry in both mitotic and polytene tissues [106]. Specifically, E2f1 forms a transcriptional heterodimer with Dp to regulate genes involved in the G1 to S phase transition such as CycE, PCNA and RnrS [82, 107–109]. E2f1 protein oscillates during endocycling salivary gland cells, which contrasts with the ubiquitous expression of e2f1 mRNA. Stabilized E2f1 blocks DNA replication by deregulating genes such as CycE, Cdk1 and mitotic cyclins [79]. E2f1 levels are high at G phase, causing CycE expression and entry into S phase. In the salivary gland, S phase entry is delayed by the second isoform of E2f1, E2f1b, which upregulates Dap [110]. Upon entry into S phase, CRL4Cdt2 ubiquitin ligase mediates E2f1 degredation, which is coupled to DNA synthesis by interaction of E2f1 PIP box with PCNA [111]. PCNA expression is dependent upon E2f1b, establishing a negative feedback loop [110].
Apt
Apontic (Apt), a basic leucine zipper (bZIP) TF, is involved in the development of Drosophila organs such as the head, heart, tracheae, nervous system and imaginal discs [112–118]. apt and another TF-encoding-gene e2f1 activate expression of each other in a positive feedback loop in wing discs and salivary glands. Additionally, the genomic binding motifs of both Apt and E2f1 are clustered in the first intron of cycE, and Apt and E2f1 upregulate CycE expression levels in endocycling cells of the salivary gland, advancing S-phase [83]. Apt also promotes transcription of Rbf1, the Drosophila counterpart to the mammalian Retinoblastoma (Rb), which promotes chromatin compaction [119–121]. Rbf1 suppresses cycE, but only becomes active after de-phosphorylation upon S-phase initiation. Given these interactions with E2f1 and Rbf1, Apt is essential for mediating both the rapid rise and fall of CycE expression [83]. Consistent with this information, apt null mutation causes salivary gland cells to show less DNA content and decreased chromosome condensation. FSBP, the human homolog of Apt, regulates E2f1, CycE (CCNE1, CCNE2), and Rb1 in mammalian cells (human 293T and mouse 3T3 cells) to ensure S phase entry and chromatin condensation, suggesting the evolutionarily conserved roles of Apt [83, 118].
Snail
Snail is a zinc-finger DNA binding TF involved in establishing the mesoderm-neuroectoderm boundary during embryogenesis [122–126]. Snail is also involved in development of the prothoracic gland cells of the ring gland, as its loss or overexpression causes endoreplication cessation cell-autonomously with consequential whole organismal larval arrest [127]. In prothoracic gland cells, Snail is regulated by Target of Rapamycin (TOR) signaling, as disrupted TOR signaling reduces Snail expression but only during and prior to the critical weight check point, which coincides with assessment on nutritional status at the early third instar and determination of whether the larva commits to pupation. Some evidence suggests that Snail expression oscillates in the endocycle of the prothoracic gland cells, with high and low expression coinciding with G and S phases, respectively [127]. The mechanism of Snail regulation on the endocycle may involve transcriptional regulation of dup [127]. Altogether, at least in prothoracic gland cells, modulated Snail levels are important to endocycle progression.
Myb
Myb, the equivalent of MYBL2 in humans, belongs to the Myb-MuvB complex, which encompasses the dE2f2-dDp DNA-binding proteins together with the tumor suppressors Rbf1 and Rbf2. This complex is activated by Cdk1-CycA kinase, and generally suppresses endoreplication by promoting mitosis through transcriptional activation of M-phase genes, which includes Aurora B (AurB), a component of the chromosomal passenger complex [128, 129]. Knockdown of Myb promotes endoreplication and increases ploidy in Drosophila S2 cells [89]. In the ovarian follicle cells, knockdown of Myb also increases cell ploidy and some cells acquire two nuclei, suggesting failure of cytokinesis [89]. In females expressing a null allele for Mip120, a component of the multi-protein MuvB core, ovarian nurse cells fail to undergo the mitosis-like disassembly of their polytene chromosomes at stage 5, and oogenesis is arrested between stages 7 and 8 [61].
Lov
Lov is encoded by the gene Jim Lovell (Lov) and contains the conserved BTB/POZ (Bric-a-Brac/Tramtrack/Broad/ Pox virus and Zinc finger) domain. Lov is involved in chromatin folding and ring canal formation [130]. During larval development, Lov is required to promote growth and endoreplication of trachea epithelial cells [131]. Absence of lov leads to an increased number of epithelial cells, which are consequently smaller and show loss of endopolyploidization [131, 132].
Myc
Myc can promote cell proliferation, cell competition and endoreplication and is conserved in vertebrates as a family of three related Myc proto-oncogenes [104, 105, 133, 134]. Transcription of Myc activated genes was enhanced by the ubiquitin ligase dHUWE1 (CG8184) [135]. In salivary glands, knockdown of either Myc or dHUWE1 via RNAi leads to decreased nuclear size and reduced endoreplication [136]. In follicle cells, although ectopic expression of Myc does not induce precocious entry into the endocycle, Myc overexpression during the endocycle phase causes additional rounds of endoreplication and larger nuclei [85]. Myc positively regulates expression of CTP synthase, an enzyme involved in pyrimidine biosynthesis, which provides partial explanation of these phenomena [137].
2.3. Additional factors involved in polyploidization
CTPsyn
Endocycling cells are heavily reliant upon S phase replication and rapid growth, but for a round of DNA synthesis to occur, the appropriate nucleotides are required. CTP synthase (CTPsyn) is an enzyme that catalyzes UTP conversion into CTP, and it is an essential regulator of purine to pyrimidine balance in the nucleotide pool. Interestingly, CTPsyn arranges itself into a filamentous structure called the cytoophidium, a dynamic structure that reinforces proper CTPsyn activity. Immunostaining of CTPsyn in various Drosophila endocycling cells reliably shows the presence of large cytoophidia. Maintenance and stability of the CTPsyn filament is likely dependent on ubiquitination by the proto-oncogene Casitas B-lineage lymphoma (Cbl), an E3 ligase [137]. Cbl null mutants have smaller salivary gland and salivary gland cells have reduced nuclear sizes. In ovarian follicle cells, while Cbl heterozygous mutants do not show significant change in DNA replication, with CTPsyn knockdown, DNA replication and nuclear size is significantly reduced as compared to CTPsyn knockdown alone. Therefore, Cbl and CTPsyn interact to regulate the endocycle, possibly by maintaining balance of the nucleotide pool [137].
The chromosomal passenger complex
The Chromosomal passenger complex is a conserved hetero-tetrameric complex that regulates cell division and chromosome segregation, locating to the central spindle and centromere, respectively. The four distinct components of this complex are AurB, Incenp, Borealin-related (Borr) and the Drosophila Survivin ortholog Deterin (Det). Localization of each component is dependent upon the others, and alteration of the components can result in polyploid cell formation [138]. For example, the kinase AurB is required to promote phosphorylation of Histone H3, and when AurB is depleted by RNAi in the S2 cell line, polyploidy is promoted. Chromosomal alterations such as extensive chromatin bridging at anaphase, partial chromosome condensation and abnormal chromosome segregation resulted from lagging chromatids, were also observed [139]. In ovarian follicle cells, knockdown of aurB, borr, or det resulted in large cells with polyploid nuclei. In aurb knockdown, some cells acquired two nuclei each with increased DNA content, indicating cytokinesis failure followed by endoreplication [89]. Regarding Mammalia, studies showed that AurB is dispensable to megakaryocyte endomitosis; however, AurB localizes to the centromere and contributes to the normal endomitotic process, making its deficiency unlikely to explain the mechanism of megakaryocyte endomitosis [140].
Dind
diamond (dind), encoding a non-conserved protein, is related to mitotic division and male meiosis [141]. In larvae with hypomorphic or null alleles of dind, abnormalities in chromosome morphology and number were observed in cells of the brain tissue, such as mitotic spindle disorganization, centriole fragmentation, defective chromosome segregation, chromosome rearrangements and breaks. An abnormal percentage of polyploid and hyperploid cells were also observed. Similar defects are present in dind mutant spermatogonia and spermatocytes. How dind mutants manifest this pleiotropic phenotype remains unanswered [141].
Epigenetic regulators
Epigenetic regulators such as histone methyltransferases control gene expression by modifying the condensation state of chromatin. Histone modifications are required for control of DNA replication. In salivary gland polytene chromosomes, latest replicating regions are often characterized by pericentric heterochromatin with methylation enriched on histone H3 Lysine K9 (H3K9) residues [142]. These regions are often left unreplicated during endoreplication, which results in decreased ploidy in this region. When the histone residue H3K9 residue is mutated through histone gene replacement strategy (H3K9R), DNA copy number increases in under-replicated regions of the salivary gland, indicating that under-replication in the latest replicating regions is dependent on H3K9 methylation [142].
Additionally, under-replicated regions of salivary gland polyploid cells are controlled by Suppressor of Under-Replication (SuUR), a chromatin binding protein that associates with H1 to establish proper underreplication. Interestingly, during the S phase of the endocycle, H1 is located in the late replicating regions and gets redistributed as endoreplication proceeds, which increases DNA copy number in the late replicating regions [8].
The Lysine demethylase 5 (KDM5) is a histone demethylase involved in chromatin-mediated regulation of transcription, and promotes endoreplication in prothoracic gland cells. KDM5 promotes endoreplication by activating the expression of the receptor tyrosine kinase Torso, which results in the activation of MAPK signaling pathway in prothoracic gland cells [143]. Hat-trick (htk), a chromodomain protein, is involved in chromatin remodeling and mostly expressed in the oocyte nucleus. When htk is null mutated in ovarioles, egg chambers display extranumerary nurse cell nuclei with decreased ploidy, as evidenced by decreased nuclear size [144].
LINC Complex
The Linker of Nucleoskeleton and Cytoskeleton (LINC) connects the cytoskeleton to the nuclear lamina and allows the transmission of mechanical inputs from the cytoplasm to nuclear membrane components. Null mutation of LINC components klarsicht (klar) and klaroid (koi) increases DNA content in myonuclei, disrupts cell cycle progression, and promotes recurring endoreplication [145]. Knockdown of β-PS-integrin distorts adhesion between tendons and muscle fibers, which also increases DNA content in myonuclei, suggesting that increased DNA content in LINC mutant myonuclei is caused by changes in nuclear mechanotransduction [145]. Barrier-to-autointegration factor (BAF) bridges the nuclear lamina to chromatin, and possibly acts downstream to LINC complex-induced nuclear mechanotransduction, leading to chromatin remodeling and changes in cell cycle gene expression [145].
2.4. Signaling pathways involved in the endocycle
Cells use established signaling pathways to communicate information, such that specific signals can produce predictable and reproducible results within a target tissue. Signaling pathways are vital in the orchestration of tissue development and cell differentiation, including those select cells that enter the endocycle. In Drosophila, a wide variety of cell types programmed for polyploidy are affected by cell signaling. Additionally, transgenes and mutant genes can perturb a variety of signaling pathways and cause endoreplication in a population of cells that would have otherwise remained diploid. This section will focus on pathways that have been shown to be involved in regulating the endocycle, including Notch, Insulin/Insulin-like growth factor (IIS) and target of rapamycin (TOR), Hippo, Ras/MAPK and c-Jun N-terminal kinase (JNK).
The Notch Pathway
The Notch signaling pathway is a conserved pathway involved in cell fate determination, differentiation, proliferation and apoptosis [146]. For example, mammalian Notch has been identified as a positive regulator of megakaryocyte specification but inhibits megakaryocyte maturation [147, 148]. In Drosophila, Notch signaling starts by interaction between the Notch receptor and ligands Delta and Serrate found on the surface of neighboring cells. Upon receptor activation by ligands Delta and Serrate, ADAM metalloprotease and γ secretase cleave the Notch receptor between the transmembrane and intracellular domains, releasing the Notch intracellular domain (NICD) into the cytoplasm. NICD is translocated through endosomal vesicles to the nucleus where it forms a complex with DNA binding proteins, Suppressor of Hairless (Su(H)) and Mastermind (Mam). Ultimately, this complex drives the transcription of target genes involved in various cellular processes.
In ovarian follicle cells, Notch signaling triggers differentiation of immature diploid cells to become polyploid by initiating the mitotic cycle to endocycle transition (M/E switch). Activation of the M/E switch takes place during stages 6 and 7 of oogenesis. The germline cells (nurse cells and the oocyte) upregulate the Delta ligand to activate the Notch receptor in the follicle cells, which in turn promotes upregulation of the zinc finger TF Hindsight (Hnt) and downregulation of homeobox TF Cut. Repression of Cut leads to upregulation of Fzr, which enables endocycle entry. Furthermore, it has been shown that Notch downregulates Dap, the Cdk2 inhibitor, and String (Stg), a positive regulator of Cdk1, a mitotic CDK. Due to these changes induced by Notch signaling, follicle cells undergo three rounds of endoreplication to become polyploid [56, 57, 60, 149, 150].
Development of subperineurial glia (SPG) illustrates another example of Notch-regulated ploidy and variant cell cycle determination. SPG cells acquire polyploidy through endocycle and endomitosis. Disrupting Notch signaling increases the ratio of multinuclear to mononuclear SPGs, suggesting that Notch in SPGs is involved in maintaining endocycle and inhibiting endomitosis [151].
The IIS and TOR Pathways
The Insulin/Insulin-like growth factor (IIS) and TOR signaling pathways are conserved pathways that work together to control cellular responses to nutritional stimuli [152]. Regarding dietary restrictions, the inhibitory regulation of IIS and TOR signaling on progression of the endocycle are present across many tissues, including the larval fat body, gut and proventriculus, and salivary gland, where translation of E2F1 has been demonstrated to be impacted via IIS and TOR signaling in response to changing nutrition [79, 153].
In the ovary, ISS and TOR signaling have the potential to alter cellular response to Notch signaling if nutrient availability is poor and are thus directly relevant to follicle cells that are programmed to undergo the M/E switch. Mechanistically, starvation triggers a decrease in insulin-like peptide within the follicle cells, and when Insulin-like receptor (InR) is unbound to its insulin-like peptide, the TF Forkhead box (FoxO) is translocated to the nucleus, where it activates Cut expression, creating a regulatory loop on Notch and pausing activation of the M/E switch. This M/E switch standstill is reversible upon refeeding the flies, thereby causing the follicle cells to enter the endocycle [154]. In the midgut, InR activity is required to promote enterocyte differentiation. Intestinal stem cell clones with null mutations of Pi3K, TOR, or InR are arrested in a diploid state, while overexpression of InR or Rheb in enteroblasts produces enlarged cells with increased ploidy [155].
The Hippo Pathway
The Hippo pathway, first identified in Drosophila, is a conserved pathway in metazoans, and is involved in the control of tissue growth and organ size [156]. The core of this pathway encompasses four components: the Hippo (Hpo), Salvador (Sav), Warts (Wts) and Mob-as-tumor-suppressor (Mats), which collectively form the Hippo kinase complex [157]. This complex negatively regulates the nuclear translocation of Yorkie (Yki), which is the downstream effector of the Hippo signaling cascade that drives the expression of genes related to growth/proliferation such as Myc, ban, diap1, E2f1, and cyclins A, B and E [157].
Yki and its cofactor Multiple Ankyrin repeats Single KH domain (Mask) regulate polyploidy in the sub perineurial glia cells by participating in a double-negative feedback loop with the microRNA miR-285. Mechanistically, miR-285 suppresses Yki activity by direct contact with Mask, resulting in downregulated expression of Yki’s target CycE. miR-285 levels are increased by downregulation of yki or mask, and decreased by yki overexpression. This double-negative feedback is essential to assure proper activity of these components and regulate polyploidy in sub perineurial glia cells. Disturbance of this double-negative feedback loop increases ploidy levels and nuclear sizes, as evidenced by miR-285 knockout or yki overexpression [158].
Upon wounding, loss of tissue can be replaced by cell enlargement through endoreplication. Yki regulates endoreplication in the injured fly abdomen after wounding, and knockdown of yki disrupts wound response [159, 160].
In Mammalia, Hippo has been reported to suppress tumorigenesis and cell ploidy through Skp2 [161]. Tetraploidy caused by cytokinesis failure can activate the Hippo tumor suppressor pathway [162].
The RAS/MAPK Pathway
Drosophila Ras shows substantial amino acid sequence homology (75%) to its mammalian counterpart [163, 164], and participates in the initiation of the MAP kinase cascade, leading to the activation of RAF, MEK and ERK (MAPK), respectively. As the final kinase, ERK translocates to the nucleus where it activates various TFs involved in cell growth and proliferation.
Ras proteins are small GTPases that switch from active to inactive states upon GTP to GDP binding [165]. The GTPase enzymatic domain of Ras is a frequent target for oncogenic mutations because loss of this enzymatic activity confers constitutive activation upon Ras [166]. Generally, knockdown of Ras stalls cells at G1, while activated Ras promotes the G1/S transition in both the cell cycle and endocycle by upregulating CycE post-transcriptionally [167, 168]. Additionally, Ras shortens G1 by upregulating Myc [163]. Interestingly, in the salivary gland, the endocycle can be affected indirectly by ectopic localization of Ras upon knockdown of Rab11, a vesicle trafficking GTPase. These salivary gland cells show reduced DNA content and nuclei size along with decreased transcription levels of the endoreplication regulators CycE, E2f1 and Gem [169].
In Drosophila, oncogenic Ras (RasV12) combined with disruption of the apico-basal polarity gene scribble (scrib) leads to tumor formation in the wing disc. These events are associated with the activation of Yki and JNK pathways, which are known to downregulate CycB, leading to polyploid cell formations in these wing disc tumors [170]. In the pylorous, RasV12 expands the pylorous by cell proliferation in the larval stage but endoreplication in the adult [92].
The JNK Pathway
The JNKs is a conserved signaling pathway involved in cell death, immunity and DNA damage [171]. JNK signaling is activated under cell stress or loss of cell polarity. Upon activation, tumor necrosis factor (TNF) Eiger binds to its receptors Wengen or Grindelwald, and these receptors activate the core kinase cascade that eventually terminates in the final JNK called Basket (Bsk), which activates via phosphorylation various TFs, which upregulate genes often related to caspase-mediated apoptosis, including head involution defective (hid) and reaper (rpr) [172].
JNK signaling has a dual role in polyploid cell development and tumorigenesis that depends on the genetic context of the cell population and crosstalk with other signaling pathways. In imaginal epithelia with peanut knockdown, JNK acts as a tumor suppressor by postranscriptionally suppressing the apoptotic-inhibitor Diap1 in the cells having undergone cytokinesis failure, promoting apoptosis of the tetraploid cells. Additionally, JNK posttranscriptionally represses the M-phase inducer String (Cdc25 homolog), preventing polyploid, error-prone mitosis. Interestingly, these tumor-suppressive effects can be overcome with Yki overexpression, which promotes the transcription of M-phase genes [173]. Notwithstanding the role JNK as a tumor suppressor, JNK signaling can also promote giant polyploid cells via endoreplication in certain neoplastic overgrowths. Some of these tumors include those produced by loss of an endocytic gene rab5, vps25, erupted, or avalanche, and epithelial malignant tumors produced by RasV12/scrib−/− or RasV12/dlg1−/− mutants. Mechanistically, endoreplication is promoted through downrugulation of CycB by JNK signaling and cooperation by Yki causes upregulation of Diap1, preventing cell death [170].
4. Polyploidy in wound repair and tissue homeostasis
4.1. Wound-induced polyploidy
During organismal development, cells are exposed to stress conditions that can result in tissue damage and consequential cell loss. To compensate for the empty space left by cell loss, neighboring surviving cells with limited division capacity can undergo polyploidization to enlarge cell size. This mechanism is called wound-induced polyploidy (WIP) [174], and is conserved between Drosophila and mammals. For example, in mammalian organs with limited division potential such as the heart, liver, cornea, kidney and bladder polyploidization occurs in response to tissue injury [175–179].
WIP can be induced by puncturing the fly abdomen. Quiescent epidermal cells surrounding the puncture wound re-enter S phase and become polyploid. Epidermal cells surrounding the wound undergo WIP via activation of Yki, a downstream effector of the Hippo pathway. Yki activates endocycle genes (Myc and E2f1) and the microRNA bantam (ban). In contrast, AP-1 activation by JNK restricts polyploidization and Yki activation [160]. Interestingly, epithelial cells with stg or fzrRNAi expression that use mitosis as a repair mechanism in lieu of WIP are more prone to accumulate DNA damage and mitotic errors, demonstrating the advantage of WIP as a repair strategy in epithelial tissue [180].
The Drosophila intestine has also been used to study WIP. Though injury to the hindgut pylorus is repaired by mitosis during the larva stage, endoreplication is the preferred repair mechanism during the adult phase. Endoreplication in the adult pylorus is regulated by Fzr, a component of APC/C, which helps with the degradation of mitotic cyclins such as CycA and CycB [92]. The intestinal midgut also demonstrates WIP. Upon damage caused by the enteropathogen Pseudomonas entomophila, midgut stem cells transform into enteroblasts and enterocytes. This transformation is achieved through compensatory endoreplication via EGFR/MAP kinase signaling and results in enterocytes with higher ploidy levels [155].
4.2. Compensatory cellular hypertrophy in tissue homeostasis
Tissue homeostasis corresponds to maintenance of tissue integrity and size upon adverse conditions such as the appearance of mutations that lead to the formation of aberrant cells that are slower-growing or structurally defective. In Drosophila, slower-growing cells can be obtained by loss of the TF Myc or the protooncogene Ras, while structurally defective cells can be induced by loss of apicobasal cell polarity, which can be caused by mutations in genes like discs large (dlg), scribble (scrib) and lethal giant larvae (lgl). Maintaining homeostasis amidst these mutant cells, cell competition is a process used by tissues where less fit cells called the “losers” are eliminated by surrounding neighboring cells, the “winners”. In mitotic epithelia, eliminated loser cells are compensated by neighboring cell proliferation, “compensatory proliferation”. While in post-mitotic tissues, such as endocycling follicle cells, the winner cells perform compensatory cellular hypertrophy, a process in which the winner cells increase their nuclei size two to four times by undergoing extra rounds of endoreplication [104, 181–183].
5. Polyploid mitosis and genomic instability
Genomic instability refers to loss of genome integrity and is characterized by frequent mutations, including chromosomal rearrangements and aneuploidy. Research on genomic instability is of therapeutic value, given that genomic instability is a hallmark of cancer [184, 185]. In Drosophila, tumorigenesis is similarly associated with genomic instability; for example, by disrupting chromosome segregation to promote genomic instability in imaginal disc epithelia, many features of human tumors are recapitulated, including loss of apical-basal polarity, cell delamination, basement membrane degradation, and tumor invasiveness. Substantial evidence from various tissues within the Drosophila model suggests error-prone, polyploid mitosis as a source of genomic instability [48, 90, 186].
Rectal papillae cells can be used to study polyploid mitosis because mitotic division is programmed to follow endoreplication in rectal papillae at metamorphosis. The switch from endoreplicative to mitotic division compromises the genomic stability of rectal papillae, with instability indicated by the presence of broken and acentric chromosomes, lagging chromosomes, and chromatin bridges during an elongated anaphase [48]. Chromosomal abnormalities are also observed in mitotic polyploid ileum cells of the mosquito Culex pipiens, supporting the rationale that genomic instability is due to an inherent imprecision of polyploid mitosis instead of a singular physiological property of Drosophila rectal papillae [48]. Interestingly, rectal papillae accumulate centrioles during the endocycle stages, leading to centrosome amplification and multipolar division at post-endocycle stages. The error-prone nature of multi-polar division causes a high-frequency of aneuploidy [186]. Consequently, rectal papillae possess multiple mechanisms to tolerate and minimize the effects of chromosomal imbalances due to mitotic infidelity. First, unlike the canonical DNA damage response, p53 is not activated to mediate cell cycle arrest or apoptosis in the presence of double-strand breaks in rectal papillae [186]. Tolerance to DNA damage is not unusual for normal development, as many endocycling cells in both flies and humans will also silence cell death genes [15, 187]. Second, rectal papillae activate Blm helicase and Fanconi anemia proteins to coordinate proper alignment and segregation of broken chromosomes and acentric fragments to the poles, helping to prevent micronuclei formation at post-endocycle mitosis [188]. Cytoplasmic sharing has also been investigated as a means whereby specialized gap junctions neutralize the genomic imbalances between nuclei [189].
In contrast to rectal papillae, in most other organs, endoreplication is followed by senescence, but manipulation of the developmental program of the ovary has allowed research on post-endocycle mitosis in ovarian follicle cells, which normally undergo the M/E switch at stages 6 and 7 in a Notch dependent manner [56, 60, 149, 150]. Independent of Notch signaling, cells with CycA knockdown or fzr overexpression can enter precocious endocycles. In either case, endocycle induction protects against apoptosis caused by ionizing gamma radiation [90]. Given relief from heat shock-induced fzr overexpression, cells having undergone precocious endocycles re-enter mitosis – and these cells regain susceptibility to radiation-induced apoptosis. Post-endocycle mitosis in these cells is error-prone, which was evidenced by chromosomal loss and fragmentation, and centrosome amplification [90].
Centrosome amplification has been linked to spindle assembly defects and tumor growth and formation [190]. Sak kinase (PLK4 in humans) is required for centriole duplication, and when overexpressed in larval neuroblasts, centrosome amplification occurs. Neuroblasts showing amplified centrosomes initiate multipolar spindle assembly, but this aberration is frequently resolved at the spindle assembly checkpoint, with only a slight delay as the supernumerary centrosomes cluster together, by which point spindle bipolarity is re-established. Regardless, sak overexpression neuroblasts show subtle differences of spindle alignment in reference to cortical cues. To determine if centrosome amplification can contribute to tumorigenesis, the Sak kinase overexpressed brain tissue was transplanted into wild type hosts. Remarkably, up to 20% of the hosts developed tumors, and several of these tumors metastasized to locations distant from the injection site [74].
Research using the Drosophila model demonstrates several interesting phenomena. First, polyploid cells can become mitotic. This post-endocycle mitosis can lead to genomic instability. Polyploid mitosis can be characterized by aneuploidy and other chromosomal defects. In some but certainly not all contexts, error-prone mitosis caused by extranumerary centrosomes contributes to tumorigenesis; this mechanism is also demonstrated in human cancers [191]. Future investigations on the causal relation between polyploid mitosis and genomic instability, the characterization of multipolar mitosis, as well as the variability by which genomic instability is tolerated and processed by cells sheds light on the basics of cancer biology.
6. Polyploidy in Drosophila tumor models
Polyploid cells can be found in human cancer samples as PGCCs, which are associated with metastasis and anticancer therapy resistance since PGCCs are a source of genomic instability and aneuploidy [22, 23, 192–194]. Moreover, using genomic sequencing of tumor biopsies, the presence of polyploidy has been shown to shape prognosis and progression of advanced cancers [24, 27] In Drosophila, evidence confirms that polyploid cells play important roles in tumorigenesis, tumor growth and invasiveness [133, 170, 173]. This section will address some processes of polyploid cell formation in Drosophila tumors, while elaborating upon the relation and contributions of polyploidy to tumorigenesis and tumor growth.
In the Drosophila eye-antennal disc, activated Ras (RasV12) and simultaneous loss of scrib leads to the formation of tumors with the presence of polyploid cells [170]. Similarly, in the wing and eye discs, polyploid cells can be obtained by loss-of-function mutations in endocytic “neoplastic tumor suppressor” genes rab5, avalanche, erupted or vps25. In rab5−/− and RasV12/scrib−/− mutants, endoreplication is caused by synergic activation of Yki that activates Diap1 and cooperates with JNK to cause downregulation of CycB. In RasV12/scrib−/− tumors, tumor growth is suppressed by eradication of polyploid cells through repression of endoreplication via co-overexpression of CycB or by overexpression of the dominant negative form of the Drosophila JNK, Basket. In addition, co-overexpression of CycB inhibited RasV12/scrib−/− tumor metastatic invasion [170]. Thus, polyploid cells in RasV12/scrib−/− tumors are suggested as an essential factor for tumor growth and metastasis.
A Drosophila salivary gland imaginal ring (ImR) tumor model provides further demonstration of the role of polyploid cells in the tumor initiation and progression [195, 196]. The ImR tumor is formed in a transition zone (TZ) region located between the diploid ImR cells and polyploid salivary gland cells. The TZ region is rich in JNK and Janus kinase (JAK)-signal transducer and activator of transcription (STAT) activity, making this region a tumor hotspot. Because of high activity of JAK-STAT and JNK signaling, Notch induction is sufficient for tumor formation in this region [195]. In this tumor, in addition to polyploid cells re-entering mitosis, continued endoreplication and depolyploidization also contribute to tumor progression. These multiple cell cycle mechanisms ensure tumors always contain a proportion of polyploid cells during tumorigenesis, indicating a role of polyploid cells during tumor progression. Polyploid cell division in the ImR tumor is error-prone since multiple centrosomes, chromosome bridges, lagging chromosomes and asymmetric division were frequently observed, leading to tumors with high chromosome instability. Finally, RNA-seq analysis of ImR tumors showed upregulation of several DNA damage response and repair genes that are also active during meiosis. Knockdown the upregulated DNA damage response and repair genes resulted in depolyploidization defects in the ImR tumors, suggesting a relevant role of DNA damage response and repair pathway during tumor formation [196]. Ploidy reduction also contributes to tumor formation in mammals [26]. Proliferating polyploid cells in liver tissue produce aneuploidy, and increasing ploidy dynamics in liver tissue can raise the risk of liver cancer [197–199].
Tumorigenesis and tumor growth are dependent on cell competition in epithelial cells overexpressing epidermal growth factor receptor (EGFR) combined with microRNA 8 (miR-8) [133]. Interestingly, miR-8 downregulates Peanut, a Septin family protein required for cytokinesis, promoting tumor formation and metastasis when EGFR is overexpressed; however, depletion of Peanut with EGFR overexpression does not lead to tumor formation, raising the possibility that miR-8 has other targets besides Peanut [133]. These epithelial tumors showed metastatic potential and featured giant cells with enlarged nuclei and banding patterns typical of polytene chromosomes. Characteristic to a super-competitor behavior, the giant tumor cells showed elevated Myc levels and induced apoptosis and engulfment of neighboring wild type cells. Using the MARCM method, when Myc was overexpressed in neighboring cells, EGFR + miR-8 clones did not produce giant cells or induce neoplasia. Additionally, reducing Draper (which is involved in engulfment) in tumor cells inhibited cell enlargement and tumorigenesis in EGFR + miR-8 clones. Furthermore, suppressing apoptosis in the EGFR + miR-8 tissue by expression of p35 or Diap-1 also inhibited tumor formation. Taken together, cell competition mediated through induction of apoptosis and engulfment are suggested to be required for tumor formation in this tumor model [133].
Depletion of peanut in the wing disc epithelium leads to cell cytokinesis failure, resulting in tetraploidy and apoptosis. Apoptosis upon cytokinesis failure is mediated through JNK signaling, which suppresses the apoptosis inhibitor Diap1 [173]. Epithelial wing disc tumors are induced when peanut depletion is combined with either Yki overexpression or RasV12. Tumors driven by Yki overexpression and peanut depletion cause cells to demonstrate putative malignant features such as cell and nuclear enlargement, increased centrosome count, increased matrix metalloproteinase 1 (Mmp1) enzyme, invasive potential and polarity disruption. Yki overexpression in these tumors limits cell death through Diap1 expression and through upregulation of Stg which stimulates progression of the cell cycle. Concomitantly, JNK limits Stg expression, demonstrating a tumor suppressive role that is overcome by Yki overexpression [173].
Drosophila tumor models share aspects existent in human solid tumors such as the presence of genomic instability and polyploid cells [23, 192–194]. These Tumor models improve biological understanding of the various factors and mechanisms that involve polyploid cells in tumor formation. These tumor models implicate polyploid cells as key drivers of tumor initiation and growth and dispute the notion that polyploid cells are merely consequential to tumorigenesis.
Conclusion
Polyploid cells are prevalent during Drosophila development, with the functional aspects of polyploidy being multifold and unique across organs. In tissues where mitotic division is no longer an option, polyploidization serves as a response against tissue integrity loss and is inherent to wound repair and tissue homeostasis. The programs leading to polyploidy are subject to a myriad of regulatory factors, providing cell types with endocycles of diverse variation. For example, comparing the ovary and salivary gland, Dap oscillates at different concentrations, possibly driving differential under-replication of euchromatic regions [87]. Nevertheless, some common themes on all endocycles exist. First, cells downregulate mitotic cyclins or decrease mitotic CDK activity, and second, regulated Cdk2-CycE activity and CycE oscillation is necessary to appropriately alternating G and S phases. Furthermore, the endocycle can be affected by internal growth restraints and is responsive to cellular signaling.
The contributions of polyploidy to tumorigenesis and tumor progression have only started to be elucidated. Mounting evidence indicates error-prone, polyploid mitosis as a source of genomic instability, which in various and certain contexts can drive tumor formation and malignancy. Fruit fly research will shed light on the interaction of signaling pathways and various factors involved in the formation of tumorigenic polyploid cells. Rather than having a passive role, in various tumor models, polyploid tumor cells are implicated as key instigators in the progression of tumorigenesis and tumor growth. Considering the wide occurrence of PGCCs in human solid tumors, studies using the Drosophila model that characterize unique facets of polyploid cells are of therapeutic significance.
Figure 2.
Through different Drosophila life cycle stages, polyploid cells promote development and growth in distinct organs.
Figure 3.
Depiction of the endocycle process. Specific expression of cell cycle regulators promotes S and G phase and evasion of M phase.
Figure 4.
Different tumor models in Drosophila containing polyploid-tumor cells. (A) Ectopic expression of oncogenes in combination with tumor suppressor inhibition transforms the wing-disc epithelia into neoplasms containing polyploid giant tumor cells with features that recapitulates certain hallmarks of cancer. (B) Notch hyperactivation promotes neoplastic tumorigenesis in imaginal ring polyploid cells. (C) Polyploid-tumor cells re-enter mitosis and depolyploidization resulting in chromosomal instability.
Acknowledgments
We thank Deeptiman Chatterjee for his assistance in preparing figure legends and relevant critiques. This work was supported by National Institute of Health (GM072562, CA224381, and CA227789) and National Science Foundation (IOS-155790).
Glossary
- C ploidy values
DNA content of a cell
- N ploidy values
the number of chromosome sets
- Polyploidization
process of acquiring polyploidy
- Haploidy
one complete set of chromosomes
- Diploidy
two complete haploid sets of chromosomes
- Hypserploidy
chromosome number slightly greater than an exact multiple of the haploid ploidy value
- Polyploidy
ploidy is greater than two
- Endopolyploidy
polyploidy via endoreplication
- Aneuploidy
a difference of chromosomes within a haploid set
- Polytene chromosome
fusion of multiple chromosomes showing characteristic banding patterns
Footnotes
Conflict of interest
The authors declare that there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shu Z, Row S, and Deng WM, Endoreplication: The Good, the Bad, and the Ugly. Trends Cell Biol, 2018. 28(6): p. 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ovrebo JI and Edgar BA, Polyploidy in tissue homeostasis and regeneration. Development, 2018. 145(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox DT, et al. , Polyploidy: A Biological Force From Cells to Ecosystems. Trends Cell Biol, 2020. 30(9): p. 688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davoli T and de Lange T, The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol, 2011. 27: p. 585610. [DOI] [PubMed] [Google Scholar]
- 5.Ullah Z, Lee CY, and Depamphilis ML, Cip/Kip cyclin-dependent protein kinase inhibitors and the road to polyploidy. Cell Div, 2009. 4: p. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmet J and Ravid K, Polyploidy: occurrence in nature, mechanisms, and significance for the megakaryocyte-platelet system. Exp Hematol, 2000. 28(1): p. 3–16. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka K, et al. , Tetraploidy in cancer and its possible link to aging. Cancer Sci, 2018. 109(9): p. 2632–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreyeva EN, et al. , Regulatory functions and chromatin loading dynamics of linker histone H1 during endoreplication in Drosophila. Genes Dev, 2017. 31(6): p. 603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudkin GT, Replication in polytene chromosomes. Results Probl Cell Differ, 1972. 4: p. 59–85. [DOI] [PubMed] [Google Scholar]
- 10.Hardy K, Winston RM, and Handyside AH, Binucleate blastomeres in preimplantation human embryos in vitro: failure of cytokinesis during early cleavage. J Reprod Fertil, 1993. 98(2): p. 549–58. [DOI] [PubMed] [Google Scholar]
- 11.Iwata K, et al. , Analysis of compaction initiation in human embryos by using time-lapse cinematography. J Assist Reprod Genet, 2014. 31(4): p. 421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kligman I, et al. , The presence of multinucleated blastomeres in human embryos is correlated with chromosomal abnormalities. Hum Reprod, 1996. 11(7): p. 1492–8. [DOI] [PubMed] [Google Scholar]
- 13.Van Royen E, et al. , Multinucleation in cleavage stage embryos. Hum Reprod, 2003. 18(5): p. 1062–9. [DOI] [PubMed] [Google Scholar]
- 14.Cross JC, How to make a placenta: mechanisms of trophoblast cell differentiation in mice--a review. Placenta, 2005. 26 Suppl A: p. S3–9. [DOI] [PubMed] [Google Scholar]
- 15.Ullah Z, et al. , Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev, 2008. 22(21): p. 3024–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauster M, et al. , Factors involved in regulating trophoblast fusion: potential role in the development of preeclampsia. Placenta, 2009. 30 Suppl A: p. S49–54. [DOI] [PubMed] [Google Scholar]
- 17.Paulus JM., DNA metabolism and development of organelles in guinea-pig megakaryocytes: a combined ultrastructural, autoradiographic and cytophotometric study. Blood, 1970. 35(3): p. 298–311. [PubMed] [Google Scholar]
- 18.Ravid K, et al. , Roads to polyploidy: the megakaryocyte example. J Cell Physiol, 2002. 190(1): p. 7–20. [DOI] [PubMed] [Google Scholar]
- 19.Baccini V, et al. , Role of p21(Cip1/Waf1) in cell-cycle exit of endomitotic megakaryocytes. Blood, 2001. 98(12): p. 3274–82. [DOI] [PubMed] [Google Scholar]
- 20.Duncan AW, Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol, 2013. 24(4): p. 347–56. [DOI] [PubMed] [Google Scholar]
- 21.Gentric G and Desdouets C, Polyploidization in liver tissue. Am J Pathol, 2014. 184(2): p. 322–31. [DOI] [PubMed] [Google Scholar]
- 22.Fei F, et al. , Formation of Polyploid Giant Cancer Cells Involves in the Prognostic Value of Neoadjuvant Chemoradiation in Locally Advanced Rectal Cancer. J Oncol, 2019. 2019: p. 2316436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, et al. , Polyploid Giant Cancer Cells (PGCCs): The Evil Roots of Cancer. Curr Cancer Drug Targets, 2019. 19(5): p. 360–367. [DOI] [PubMed] [Google Scholar]
- 24.Zack TI, et al. , Pan-cancer patterns of somatic copy number alteration. Nat Genet, 2013. 45(10): p. 1134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bielski CM, et al. , Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet, 2018. 50(8): p. 1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto T, et al. , Proliferative polyploid cells give rise to tumors via ploidy reduction. Nat Commun, 2021. 12(1): p. 646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinton RJ, et al. , Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature, 2021. 590(7846): p. 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niu N, et al. , Linking genomic reorganization to tumor initiation via the giant cell cycle. Oncogenesis, 2016. 5(12): p. e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith AV and Orr-Weaver TL, The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development, 1991. 112(4): p. 997–1008. [DOI] [PubMed] [Google Scholar]
- 30.Hammond MP and Laird CD, Chromosome structure and DNA replication in nurse and follicle cells of Drosophila melanogaster. Chromosoma, 1985. 91(3–4): p. 267–78. [DOI] [PubMed] [Google Scholar]
- 31.Unhavaithaya Y and Orr-Weaver TL, Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes Dev, 2012. 26(1): p. 31–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nandakumar S, Grushko O, and Buttitta LA, Polyploidy in the adult Drosophila brain. Elife, 2020. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammond MP and Laird CD, Control of DNA replication and spatial distribution of defined DNA sequences in salivary gland cells of Drosophila melanogaster. Chromosoma, 1985. 91(3–4): p. 279–86. [DOI] [PubMed] [Google Scholar]
- 34.Bellen HJ, Tong C, and Tsuda H, 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat Rev Neurosci, 2010. 11(7): p. 514–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuss B, et al. , Control of endoreduplication domains in the Drosophila gut by the knirps and knirps-related genes. Mech Dev, 2001. 100(1): p. 15–23. [DOI] [PubMed] [Google Scholar]
- 36.Stormo BM and Fox DT, Polyteny: still a giant player in chromosome research. Chromosome Res, 2017. 25(3–4): p. 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beyenbach KW, Skaer H, and Dow JA, The developmental, molecular, and transport biology of Malpighian tubules. Annu Rev Entomol, 2010. 55: p. 351–74. [DOI] [PubMed] [Google Scholar]
- 38.Gautam NK, Verma P, and Tapadia MG, Drosophila Malpighian Tubules: A Model for Understanding Kidney Development, Function, and Disease. Results Probl Cell Differ, 2017. 60: p. 3–25. [DOI] [PubMed] [Google Scholar]
- 39.Rodan AR, The Drosophila Malpighian tubule as a model for mammalian tubule function. Curr Opin Nephrol Hypertens, 2019. 28(5): p. 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hochstrasse M., Chromosome structure in four wild-type polytene tissues of Drosophila melanogaster. The 87A and 87C heat shock loci are induced unequally in the midgut in a manner dependent on growth temperature. Chromosoma, 1987. 95(3): p. 197–208. [DOI] [PubMed] [Google Scholar]
- 41.Richards G, The polytene chromosomes in the fat body nuclei of Drosophila melanogaster. Chromosoma, 1980. 79(2): p. 241–50. [DOI] [PubMed] [Google Scholar]
- 42.Lamb MJ, The DNA content of polytene nuclei in midgut and Malpighian tubule cells of adultDrosophila melanogaster. Wilehm Roux Arch Dev Biol, 1982. 191(6): p. 381–384. [DOI] [PubMed] [Google Scholar]
- 43.Mirth C, Truman JW, and Riddiford LM, The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Curr Biol, 2005. 15(20): p. 1796–807. [DOI] [PubMed] [Google Scholar]
- 44.Yamanaka N, Rewitz KF, and O’Connor MB, Ecdysone control of developmental transitions: lessons from Drosophila research. Annu Rev Entomol, 2013. 58: p. 497–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohhara Y, Kobayashi S, and Yamanaka N, Nutrient-Dependent Endocycling in Steroidogenic Tissue Dictates Timing of Metamorphosis in Drosophila melanogaster. PLoS Genet, 2017. 13(1): p. e1006583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu D, et al. , Genetic control of programmed cell death (apoptosis) in Drosophila. Fly (Austin), 2009. 3(1): p. 78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fox DT and Spradling AC, The Drosophila hindgut lacks constitutively active adult stem cells but proliferates in response to tissue damage. Cell Stem Cell, 2009. 5(3): p. 290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fox DT, Gall JG, and Spradling AC, Error-prone polyploid mitosis during normal Drosophila development. Genes Dev, 2010. 24(20): p. 2294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi YJ, Lee G, and Park JH, Programmed cell death mechanisms of identifiable peptidergic neurons in Drosophila melanogaster. Development, 2006. 133(11): p. 2223–32. [DOI] [PubMed] [Google Scholar]
- 50.Hara Y, et al. , Ecdysone-dependent and ecdysone-independent programmed cell death in the developing optic lobe of Drosophila. Dev Biol, 2013. 374(1): p. 127–41. [DOI] [PubMed] [Google Scholar]
- 51.Jiang C, Baehrecke EH, and Thummel CS, Steroid regulated programmed cell death during Drosophila metamorphosis. Development, 1997. 124(22): p. 4673–83. [DOI] [PubMed] [Google Scholar]
- 52.Nicolson S, Denton D, and Kumar S, Ecdysone-mediated programmed cell death in Drosophila. Int J Dev Biol, 2015. 59(1–3): p. 23–32. [DOI] [PubMed] [Google Scholar]
- 53.Kumar S and Cakouros D, Transcriptional control of the core cell-death machinery. Trends Biochem Sci, 2004. 29(4): p. 193–9. [DOI] [PubMed] [Google Scholar]
- 54.Zirin J, et al. , Ecdysone signaling at metamorphosis triggers apoptosis of Drosophila abdominal muscles. Dev Biol, 2013. 383(2): p. 275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salle J, et al. , CycA is involved in the control of endoreplication dynamics in the Drosophila bristle lineage. Development, 2012. 139(3): p. 547–57. [DOI] [PubMed] [Google Scholar]
- 56.Deng WM, Althauser C, and Ruohola-Baker H, Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development, 2001. 128(23): p. 4737–46. [DOI] [PubMed] [Google Scholar]
- 57.Jordan KC, et al. , Notch signaling through tramtrack bypasses the mitosis promoting activity of the JNK pathway in the mitotic-to-endocycle transition of Drosophila follicle cells. BMC Dev Biol, 2006. 6: p. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knapp EM, Li W, and Sun J, Downregulation of homeodomain protein Cut is essential for Drosophila follicle maturation and ovulation. Development, 2019. 146(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaeffer V., et al., Notch-dependent Fizzy-related/Hec1/Cdh1 expression is required for the mitotic-to-endocycle transition in Drosophila follicle cells. Curr Biol, 2004. 14(7): p. 630–6. [DOI] [PubMed] [Google Scholar]
- 60.Sun J, et al. , Regulation of the endocycle/gene amplification switch by Notch and ecdysone signaling. J Cell Biol, 2008. 182(5): p. 885–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng MH, et al. , The Drosophila LIN54 homolog Mip120 controls two aspects of oogenesis. Biol Open, 2017. 6(7): p. 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zielke N, Edgar BA, and DePamphilis ML, Endoreplication. Cold Spring Harb Perspect Biol, 2013. 5(1): p. a012948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dapples CC and King RC, The development of the nucleolus of the ovarian nurse cell of Drosophila melanogaster. Z Zellforsch Mikrosk Anat, 1970. 103(1): p. 34–47. [DOI] [PubMed] [Google Scholar]
- 64.Pepling ME, DEVELOPMENT. Nursing the oocyte. Science, 2016. 352(6281): p. 35–6. [DOI] [PubMed] [Google Scholar]
- 65.Shen W and Sun J, Dynamic Notch Signaling Specifies Each Cell Fate in Drosophila Spermathecal Lineage. G3 (Bethesda), 2017. 7(5): p. 1417–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayhew ML and Merritt DJ, The morphogenesis of spermathecae and spermathecal glands in Drosophila melanogaster. Arthropod Struct Dev, 2013. 42(5): p. 385–93. [DOI] [PubMed] [Google Scholar]
- 67.Sun J and Spradling AC, Ovulation in Drosophila is controlled by secretory cells of the female reproductive tract. Elife, 2013. 2: p. e00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Micchelli CA and Perrimon N, Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature, 2006. 439(7075): p. 475–9. [DOI] [PubMed] [Google Scholar]
- 69.Ohlstein B and Spradling A, The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature, 2006. 439(7075): p. 470–4. [DOI] [PubMed] [Google Scholar]
- 70.Malumbres M, Cyclin-dependent kinases. Genome Biol, 2014. 15(6): p. 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lim S and Kaldis P, Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development, 2013. 140(15): p. 3079–93. [DOI] [PubMed] [Google Scholar]
- 72.Barnum KJ and O’Connell MJ, Cell cycle regulation by checkpoints. Methods Mol Biol, 2014. 1170: p. 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sigrist SJ and Lehner CF, Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell, 1997. 90(4): p. 671–81. [DOI] [PubMed] [Google Scholar]
- 74.Narbonne-Reveau K, et al. , APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development, 2008. 135(8): p. 1451–61. [DOI] [PubMed] [Google Scholar]
- 75.Lilly MA and Duronio RJ, New insights into cell cycle control from the Drosophila endocycle. Oncogene, 2005. 24(17): p. 2765–75. [DOI] [PubMed] [Google Scholar]
- 76.Follette PJ, Duronio RJ, and O’Farrell PH, Fluctuations in cyclin E levels are required for multiple rounds of endocycle S phase in Drosophila. Curr Biol, 1998. 8(4): p. 235–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weiss A, et al. , Continuous Cyclin E expression inhibits progression through endoreduplication cycles in Drosophila. Curr Biol, 1998. 8(4): p. 239–42. [DOI] [PubMed] [Google Scholar]
- 78.Zielke N, et al. , The anaphase-promoting complex/cyclosome (APC/C) is required for rereplication control in endoreplication cycles. Genes Dev, 2008. 22(12): p. 1690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zielke N, et al. , Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature, 2011. 480(7375): p. 123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knoblich JA, et al. , Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell, 1994. 77(1): p. 107–20. [DOI] [PubMed] [Google Scholar]
- 81.Sauer K, et al. , Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev, 1995. 9(11): p. 1327–39. [DOI] [PubMed] [Google Scholar]
- 82.Lilly MA and Spradling AC, The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev, 1996. 10(19): p. 2514–26. [DOI] [PubMed] [Google Scholar]
- 83.Wang XF, et al. , Evolutionarily Conserved Roles for Apontic in Induction and Subsequent Decline of Cyclin E Expression. iScience, 2020. 23(8): p. 101369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moberg KH, et al. , Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature, 2001. 413(6853): p. 311–6. [DOI] [PubMed] [Google Scholar]
- 85.Shcherbata HR, et al. , The mitotic-to-endocycle switch in Drosophila follicle cells is executed by Notch-dependent regulation of G1/S, G2/M and M/G1 cell-cycle transitions. Development, 2004. 131(13): p. 3169–81. [DOI] [PubMed] [Google Scholar]
- 86.Hong A., et al., The cyclin-dependent kinase inhibitor Dacapo promotes replication licensing during Drosophila endocycles. EMBO J, 2007. 26(8): p. 2071–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Swanson CI, et al. , Expression of an S phase-stabilized version of the CDK inhibitor Dacapo can alter endoreplication. Development, 2015. 142(24): p. 4288–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lehner CF and O’Farrell PH, The roles of Drosophila cyclins A and B in mitotic control. Cell, 1990. 61(3): p. 535–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rotelli MD, et al. , A Cyclin A-Myb-MuvB-Aurora B network regulates the choice between mitotic cycles and polyploid endoreplication cycles. PLoS Genet, 2019. 15(7): p. e1008253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hassel C, et al. , Induction of endocycles represses apoptosis independently of differentiation and predisposes cells to genome instability. Development, 2014. 141(1): p. 112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pesin JA and Orr-Weaver TL, Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol, 2008. 24: p. 475–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cohen E, et al. , Fizzy-Related dictates A cell cycle switch during organ repair and tissue growth responses in the Drosophila hindgut. Elife, 2018. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Nooij JC, Letendre MA, and Hariharan IK, A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell, 1996. 87(7): p. 1237–47. [DOI] [PubMed] [Google Scholar]
- 94.Lane ME, et al. , Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell, 1996. 87(7): p. 1225–35. [DOI] [PubMed] [Google Scholar]
- 95.Warbrick E, et al. , PCNA binding proteins in Drosophila melanogaster : the analysis of a conserved PCNA binding domain. Nucleic Acids Res, 1998. 26(17): p. 3925–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Borlado LR and Mendez J, CDC6: from DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis, 2008. 29(2): p. 237–43. [DOI] [PubMed] [Google Scholar]
- 97.Wu Z, et al. , Juvenile Hormone Activates the Transcription of Cell-division-cycle 6 (Cdc6) for Polyploidy-dependent Insect Vitellogenesis and Oogenesis. J Biol Chem, 2016. 291(10): p. 5418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Drury LS, Perkins G, and Diffley JF, The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J, 1997. 16(19): p. 5966–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ding Q and MacAlpine DM, Preferential re-replication of Drosophila heterochromatin in the absence of geminin. PLoS Genet, 2010. 6(9): p. e1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.May NR, et al. , Levels of the origin-binding protein Double parked and its inhibitor Geminin increase in response to replication stress. J Cell Sci, 2005. 118(Pt 18): p. 4207–17. [DOI] [PubMed] [Google Scholar]
- 101.Yeh JE, Toniolo PA, and Frank DA, Targeting transcription factors: promising new strategies for cancer therapy. Curr Opin Oncol, 2013. 25(6): p. 652–8. [DOI] [PubMed] [Google Scholar]
- 102.Papavassiliou KA and Papavassiliou AG, Transcription Factor Drug Targets. J Cell Biochem, 2016. 117(12): p. 2693–2696. [DOI] [PubMed] [Google Scholar]
- 103.Gatti G, et al. , MYC prevents apoptosis and enhances endoreduplication induced by paclitaxel. PLoS One, 2009. 4(5): p. e5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tamori Y and Deng WM, Tissue repair through cell competition and compensatory cellular hypertrophy in postmitotic epithelia. Dev Cell, 2013. 25(4): p. 350–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Edgar BA and Orr-Weaver TL, Endoreplication cell cycles: more for less. Cell, 2001. 105(3): p. 297–306. [DOI] [PubMed] [Google Scholar]
- 106.van den Heuvel S and Dyson NJ, Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol, 2008. 9(9): p. 713–24. [DOI] [PubMed] [Google Scholar]
- 107.Duronio RJ and O’Farrell PH, Developmental control of the G1 to S transition in Drosophila: cyclin Eis a limiting downstream target of E2F. Genes Dev, 1995. 9(12): p. 1456–68. [DOI] [PubMed] [Google Scholar]
- 108.Duronio RJ., Bonnett PCe, and O’Farrel PHl, Mutations of the Drosophila dDP, dE2F, and cyclin E genes reveal distinct roles for the E2F-DP transcription factor and cyclin E during the G1-S transition. Mol Cell Biol, 1998. 18(1): p. 141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Royzman I, Whittaker AJ, and Orr-Weaver TL, Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes Dev, 1997. 11(15): p. 1999–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim M, Delos Santos K, and Moon NS, Proper CycE-Cdk2 activity in endocycling tissues requires regulation of the cyclin-dependent kinase inhibitor Dacapo by dE2F1b in Drosophila. Genetics, 2021. 217(1): p. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shibutani ST, et al. , Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell, 2008. 15(6): p. 890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eulenberg KG and Schuh R, The tracheae defective gene encodes a bZIP protein that controls tracheal cell movement during Drosophila embryogenesis. EMBO J, 1997. 16(23): p. 7156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gellon G, et al. , A genetic screen for modifiers of Deformed homeotic function identifies novel genes required for head development. Development, 1997. 124(17): p. 3321–31. [DOI] [PubMed] [Google Scholar]
- 114.Liu QX, et al. , Drosophila MBF1 is a co-activator for Tracheae Defective and contributes to the formation of tracheal and nervous systems. Development, 2003. 130(4): p. 719–28. [DOI] [PubMed] [Google Scholar]
- 115.Liu QX, et al. , Evolutionarily conserved transcription factor Apontic controls the G1/S progression by inducing cyclin E during eye development. Proc Natl Acad Sci U S A, 2014. 111(26): p. 9497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shen Y, et al. , The transcriptional factor Apt regulates neuroblast differentiation through activating CycE expression. Biochem Biophys Res Commun, 2018. 499(4): p. 889–894. [DOI] [PubMed] [Google Scholar]
- 117.Su MT, et al. , The pioneer gene, apontic, is required for morphogenesis and function of the Drosophila heart. Mech Dev, 1999. 80(2): p. 125–32. [DOI] [PubMed] [Google Scholar]
- 118.Wang XF, et al. , Apontic directly activates hedgehog and cyclin E for proper organ growth and patterning. Sci Rep, 2017. 7(1): p. 12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brehm A, et al. , Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 1998. 391(6667): p. 597–601. [DOI] [PubMed] [Google Scholar]
- 120.Nielsen SJ, et al. , Rb targets histone H3 methylation and HP1 to promoters. Nature, 2001. 412(6846): p. 561–5. [DOI] [PubMed] [Google Scholar]
- 121.Talluri S and Dick FA, Regulation of transcription and chromatin structure by pRB: here, there and everywhere. Cell Cycle, 2012. 11(17): p. 3189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kosman D, et al. , Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science, 1991. 254(5028): p. 118–22. [DOI] [PubMed] [Google Scholar]
- 123.Boulay JL, Dennefeld C, and Alberga A, The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. Nature, 1987. 330(6146): p. 395–8. [DOI] [PubMed] [Google Scholar]
- 124.Alberga A, et al. , The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development, 1991. 111(4): p. 983–92. [DOI] [PubMed] [Google Scholar]
- 125.Leptin M, twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev, 1991. 5(9): p. 1568–76. [DOI] [PubMed] [Google Scholar]
- 126.Hemavathy K, Meng X, and Ip YT, Differential regulation of gastrulation and neuroectodermal gene expression by Snail in the Drosophila embryo. Development, 1997. 124(19): p. 3683–91. [DOI] [PubMed] [Google Scholar]
- 127.Zeng J, et al. , Snail synchronizes endocycling in a TOR-dependent manner to coordinate entry and escape from endoreplication pausing during the Drosophila critical weight checkpoint. PLoS Biol, 2020. 18(2): p. e3000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sadasivam S. and DeCaprio JA, The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer, 2013. 13(8): p. 585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fung SM, Ramsay G, and Katzen AL, Mutations in Drosophila myb lead to centrosome amplification and genomic instability. Development, 2002. 129(2): p. 347–59. [DOI] [PubMed] [Google Scholar]
- 130.Bjorum SM, et al. , The Drosophila BTB domain protein Jim Lovell has roles in multiple larval and adult behaviors. PLoS One, 2013. 8(4): p. e61270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhou F, Qiang KM, and Beckingham KM, Failure to Burrow and Tunnel Reveals Roles for jim lovell in the Growth and Endoreplication of the Drosophila Larval Tracheae. PLoS One, 2016. 11(8): p. e0160233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhou F, et al. , The roles of jim lovell and uninflatable in different endopolyploid larval tissues of Drosophila melanogaster. PLoS One, 2020. 15(8): p. e0237662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Eichenlaub T, Cohen SM, and Herranz H, Cell Competition Drives the Formation of Metastatic Tumors in a Drosophila Model of Epithelial Tumor Formation. Curr Biol, 2016. 26(4): p. 419–27. [DOI] [PubMed] [Google Scholar]
- 134.Pierce SB, et al. , dMyc is required for larval growth and endoreplication in Drosophila. Development, 2004. 131(10): p. 2317–27. [DOI] [PubMed] [Google Scholar]
- 135.Schaub FX and Cleveland JL, Tipping the MYC-MIZ1 balance: targeting the HUWE1 ubiquitin ligase selectively blocks MYC-activated genes. EMBO Mol Med, 2014. 6(12): p. 1509–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yanku Y, et al. , Drosophila HUWE1 Ubiquitin Ligase Regulates Endoreplication and Antagonizes JNK Signaling During Salivary Gland Development. Cells, 2018. 7(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Aughey GN, Grice SJ, and Liu JL, The Interplay between Myc and CTP Synthase in Drosophila. PLoS Genet, 2016. 12(2): p. e1005867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kitagawa M and Lee SH, The chromosomal passenger complex (CPC) as a key orchestrator of orderly mitotic exit and cytokinesis. Front Cell Dev Biol, 2015. 3: p. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Giet R and Glover DM, Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol, 2001. 152(4): p. 669–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lordier L, et al. , Aurora B is dispensable for megakaryocyte polyploidization, but contributes to the endomitotic process. Blood, 2010. 116(13): p. 2345–55. [DOI] [PubMed] [Google Scholar]
- 141.Graziadio L, et al. , Phenotypic characterization of diamond (dind), a Drosophila gene required for multiple aspects of cell division. Chromosoma, 2018. 127(4): p. 489–504. [DOI] [PubMed] [Google Scholar]
- 142.Armstrong RL, et al. , H3K9 Promotes Under-Replication of Pericentromeric Heterochromatin in Drosophila Salivary Gland Polytene Chromosomes. Genes (Basel), 2019. 10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Drelon C, et al. , The histone demethylase KDM5 controls developmental timing in Drosophila by promoting prothoracic gland endocycles. Development, 2019. 146(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Singh A, et al. , Pleiotropic Functions of the Chromodomain-Containing Protein Hat-trick During Oogenesis in Drosophila melanogaster. G3 (Bethesda), 2018. 8(3): p. 1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang S, et al. , Mechanotransduction via the LINC complex regulates DNA replication in myonuclei. J Cell Biol, 2018. 217(6): p. 2005–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guruharsha KG, Kankel MW, and Artavanis-Tsakonas S, The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet, 2012. 13(9): p. 654–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mercher T, et al. , Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell Stem Cell, 2008. 3(3): p. 314–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Poirault-Chassac S, et al. , Notch/Delta4 signaling inhibits human megakaryocytic terminal differentiation. Blood, 2010. 116(25): p. 5670–8. [DOI] [PubMed] [Google Scholar]
- 149.Sun J. and Deng WM, Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells. Development, 2005. 132(19): p. 4299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sun J and Deng WM, Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation. Dev Cell, 2007. 12(3): p. 431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Von Stetina JR, et al. , Variant cell cycles regulated by Notch signaling control cell size and ensure a functional blood-brain barrier. Development, 2018. 145(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lin X and Smagghe G, Roles of the insulin signaling pathway in insect development and organ growth. Peptides, 2019. 122: p. 169923. [DOI] [PubMed] [Google Scholar]
- 153.Saucedo LJ, et al. , Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol, 2003. 5(6): p. 566–71. [DOI] [PubMed] [Google Scholar]
- 154.Jouandin P, Ghiglione C, and Noselli S, Starvation induces FoxOdependent mitotic-to-endocycle switch pausing during Drosophila oogenesis. Development, 2014. 141(15): p. 3013–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Xiang J, et al. , EGFR-dependent TOR-independent endocycles support Drosophila gut epithelial regeneration. Nat Commun, 2017. 8: p. 15125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zheng Y and Pan D, The Hippo Signaling Pathway in Development and Disease. Dev Cell, 2019. 50(3): p. 264–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Staley BK and Irvine KD, Hippo signaling in Drosophila: recent advances and insights. Dev Dyn, 2012. 241(1): p. 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li D, et al. , miR-285-Yki/Mask double-negative feedback loop mediates blood-brain barrier integrity in Drosophila. Proc Natl Acad Sci U S A, 2017. 114(12): p. E2365-E2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Losick VP, Fox DT, and Spradling AC, Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr Biol, 2013. 23(22): p. 2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Losick VP, Jun AS, and Spradling AC, Wound-Induced Polyploidization: Regulation by Hippo and JNK Signaling and Conservation in Mammals. PLoS One, 2016. 11(3): p. e0151251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang S, et al. , Hippo Signaling Suppresses Cell Ploidy and Tumorigenesis through Skp2. Cancer Cell, 2017. 31(5): p. 669–684 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ganem NJ, et al. , Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell, 2014. 158(4): p. 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Prober DA and Edgar BA, Ras1 promotes cellular growth in the Drosophila wing. Cell, 2000. 100(4): p. 435–46. [DOI] [PubMed] [Google Scholar]
- 164.Neuman-Silberberg FS, et al. , The Drosophila ras oncogenes: structure and nucleotide sequence. Cell, 1984. 37(3): p. 1027–33. [DOI] [PubMed] [Google Scholar]
- 165.Wennerberg K, Rossman KL, and Der CJ, The Ras superfamily at a glance. J Cell Sci, 2005. 118(Pt 5): p. 843–6. [DOI] [PubMed] [Google Scholar]
- 166.Schubbert S, Shannon K, and Bollag G, Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer, 2007. 7(4): p. 295–308. [DOI] [PubMed] [Google Scholar]
- 167.Coleman ML, Marshall CJ, and Olson MF, RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat Rev Mol Cell Biol, 2004. 5(5): p. 355–66. [DOI] [PubMed] [Google Scholar]
- 168.Leone G, et al. , Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature, 1997. 387(6631): p. 422–6. [DOI] [PubMed] [Google Scholar]
- 169.Choubey PK and Roy JK, Rab11, a vesicular trafficking protein, affects endoreplication through Ras-mediated pathway in Drosophila melanogaster. Cell Tissue Res, 2017. 367(2): p. 269–282. [DOI] [PubMed] [Google Scholar]
- 170.Cong B., Ohsaw Sa, and Igak Ti, JNK and Yorkie drive tumor progression by generating polyploid giant cells in Drosophila. Oncogene, 2018. 37(23): p. 3088–3097. [DOI] [PubMed] [Google Scholar]
- 171.Tafesh-Edwards G and Eleftherianos I, JNK signaling in Drosophila immunity and homeostasis. Immunol Lett, 2020. 226: p. 7–11. [DOI] [PubMed] [Google Scholar]
- 172.La Marca JE and Richardson HE, Two-Faced: Roles of JNK Signalling During Tumourigenesis in the Drosophila Model. Front Cell Dev Biol, 2020. 8: p. 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gerlach SU, Eichenlaub T, and Herranz H, Yorkie and JNK Control Tumorigenesis in Drosophila Cells with Cytokinesis Failure. Cell Rep, 2018. 23(5): p. 1491–1503. [DOI] [PubMed] [Google Scholar]
- 174.Losick VP, Wound-Induced Polyploidy Is Required for Tissue Repair. Adv Wound Care (New Rochelle), 2016. 5(6): p. 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ikebe H, et al. , Changes in nuclear DNA content and cell size of injured human corneal endothelium. Exp Eye Res, 1988. 47(2): p. 205–15. [DOI] [PubMed] [Google Scholar]
- 176.Laflamme MA and Murry CE, Heart regeneration. Nature, 2011. 473(7347): p. 326–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Miyaoka Y, et al. , Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol, 2012. 22(13): p. 1166–75. [DOI] [PubMed] [Google Scholar]
- 178.Wang J, et al. , Polyploid Superficial Cells that Maintain the Urothelial Barrier Are Produced via Incomplete Cytokinesis and Endoreplication. Cell Rep, 2018. 25(2): p. 464–477 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lazzeri E, et al. , Endocycle-related tubular cell hypertrophy and progenitor proliferation recover renal function after acute kidney injury. Nat Commun, 2018. 9(1): p. 1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Grendler J, et al. , Wound-induced polyploidization is driven by Myc and supports tissue repair in the presence of DNA damage. Development, 2019. 146(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Amoyel M and Bach EA, Cell competition: how to eliminate your neighbours. Development, 2014. 141(5): p. 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tamori Y and Deng WM, Cell competition and its implications for development and cancer. J Genet Genomics, 2011. 38(10): p. 483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tamori Y and Deng WM, Compensatory cellular hypertrophy: the other strategy for tissue homeostasis. Trends Cell Biol, 2014. 24(4): p. 230–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Negrini S, Gorgoulis VG, and Halazonetis TD, Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol, 2010. 11(3): p. 220–8. [DOI] [PubMed] [Google Scholar]
- 185.Dekanty A, et al. , Aneuploidy-induced delaminating cells drive tumorigenesis in Drosophila epithelia. Proc Natl Acad Sci U S A, 2012. 109(50): p. 20549–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Schoenfelder KP, et al. , Indispensable pre-mitotic endocycles promote aneuploidy in the Drosophila rectum. Development, 2014. 141(18): p. 3551–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mehrotra S, et al. , Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev, 2008. 22(22): p. 3158–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bretscher HS. and Fox DT, Proliferation of Double-Strand Break-Resistant Polyploid Cells Requires Drosophila FANCD2. Dsev Cell, 2016. 37(5): p. 444–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Peterson NG, et al. , Cytoplasmic sharing through apical membrane remodeling. Elife, 2020. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Basto R, et al. , Centrosome amplification can initiate tumorigenesis in flies. Cell, 2008. 133(6): p. 1032–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ganem NJ, Godinho SA, and Pellman D, A mechanism linking extra centrosomes to chromosomal instability. Nature, 2009. 460(7252): p. 278–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Amend SR, et al. , Polyploid giant cancer cells: Unrecognized actuators of tumorigenesis, metastasis, and resistance. Prostate, 2019. 79(13): p. 1489–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bou-Nader M, et al. , Polyploidy spectrum: a new marker in HCC classification. Gut, 2020. 69(2): p. 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mirzayans R, Andrais B, and Murray D, Roles of Polyploid/Multinucleated Giant Cancer Cells in Metastasis and Disease Relapse Following Anticancer Treatment. Cancers (Basel), 2018. 10(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yang SA, et al. , Oncogenic Notch Triggers Neoplastic Tumorigenesis in a Transition-Zone-like Tissue Microenvironment. Dev Cell, 2019. 49(3): p. 461–472 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang XF, et al. , Polyploid mitosis and depolyploidization promote chromosomal instability and tumor progression in a Notch-induced tumor model. Dev Cell, 2021. 56(13): p. 1976–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Duncan AW, et al. , The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature, 2010. 467(7316): p. 707–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Duncan AW, et al. , Frequent aneuploidy among normal human hepatocytes. Gastroenterology, 2012. 142(1): p. 25–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Duncan AW, et al. , Aneuploidy as a mechanism for stress-sinduced liver adaptation. J Clin Invest, 2012. 122(9): p. 3307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]