Abstract
The strictly regulated bone remodeling process ensures that osteoblastic bone formation is coupled to osteoclastic bone resorption. This coupling is regulated by a panel of coupling factors, including clastokines promoting the recruitment, expansion, and differentiation of osteoprogenitor cells within the eroded cavity. The osteoprogenitor cells on eroded surfaces are called reversal cells. They are intermixed with osteoclasts and become bone-forming osteoblast when reaching a critical density and maturity. Several coupling factors have been proposed in the literature, but their effects and expression pattern vary between studies depending on species and experimental setup. In this study, we investigated the mRNA levels of proposed secreted and membrane-bound coupling factors and their receptors in cortical bone remodeling events within the femur of healthy adolescent human controls using high-sensitivity RNA in situ hybridization. Of the proposed coupling factors, human osteoclasts showed mRNA-presence of LIF, PDGFB, SEMA4D, but no presence of EFNB2, and OSM. On the other hand, the osteoblastic reversal cells proximate to osteoclasts presented with LIFR, PDGFRA and PLXNB1, but not PDGFRB, which are all known receptors of the proposed coupling factors. Although EFNB2 was not present in mature osteoclasts, the mRNA of the ligand-receptor pair EFNB2:EPHB4 were abundant near the central blood vessels within intracortical pores with active remodeling. EPHB4 and SEMA4D were also abundant in mature bone-forming osteoblasts. This study highlights that especially LIF:LIFR, PDGFB:PDGFRA, SEMA4D:PLXNB1 may play a critical role in the osteoclast-osteoblast coupling in human remodeling events, as they are expressed within the critical cells.
Keywords: bone remodeling, osteoclast (OC), coupling, osteoblast (OB), bone cells interaction
Introduction
Bone remodeling is responsible for maintenance of the adult human skeleton. Imbalances in bone resorption and formation during the bone remodeling process causes either a gain or loss of bone (Delaisse et al., 2020). Such imbalance may be due to uncoupled resorption and formation, as characteristic of ageing, osteoporosis and multiple myeloma (Andersen et al., 2010, 2013; Jensen et al., 2014; Andreasen et al., 2020). The bone remodeling process includes three different phases: First, the resorptive phase where osteoclasts start resorption of old bone. Second, the reversal-resorption phase, where osteoclasts expand the resorbed area. In the reversal-resorption phase, osteoclasts are intermixed with osteoprogenitors recruited to the eroded bone surface. These osteoprogenitors, known as reversal cells, prepare the bone surface for bone formation, while gradually undergoing differentiation into mature bone forming osteoblasts. Third, the bone formation phase (Andersen et al., 2013; Lassen et al., 2017; Delaisse et al., 2020; Sims and Martin, 2020).
The proximity between osteoclasts and osteoblastic reversal cells during the reversal-resorption phase allows active interaction and signaling between these cell types. Furthermore, reversal cells comprise a possible target cell available for osteoclastic coupling factors. Osteoclastic coupling factors include: 1) matrix-derived factors released during resorption, 2) factors secreted by osteoclasts, 3) membrane-bound factors on osteoclasts, and 4) factors packed in exosomes released by osteoclasts (Charles and Aliprantis, 2014; Sims and Martin, 2020).
The discovery of reversal cells vacating eroded bone surfaces near osteoclasts have provided a potential bridge in the communication between osteoclasts and osteoblastic cells during bone remodeling, as bone-resorbing osteoclasts are rarely observed near bone-forming osteoblasts (Eriksen, Melsen and Mosekilde, 1984; Andersen et al., 2009; Lassen et al., 2017). Initially, reversal cells were described as mononucleated cells, covering approximately 80% of eroded surfaces in trabecular bone and proposed to be pre-osteoclasts due to the presence of TRAcP (Baron et al., 1983; Eriksen et al., 1984; Eriksen et al., 1984; Bianco et al., 1988; Mocetti et al., 2000). We now know that they are osteoblast-lineage cells, expressing markers specific for early osteogenic commitment (Andersen et al., 2009; Abdelgawad et al., 2016; Abdallah et al., 2017; Jafari et al., 2017; Lassen et al., 2017; Chen et al., 2019). Furthermore, we have shown that early reversal cells have direct cell-cell interactions with osteoclasts, they take up Tartrate-resistant acid phosphatase (TRAcP) released by osteoclasts, and they decompose resorption debris left by the osteoclast (Everts et al., 2002; Abdelgawad et al., 2016). Collectively, this supports the concept that osteoblastic reversal cells are a key recipient of osteoclastic coupling factors (Charles and Aliprantis, 2014; Delaisse et al., 2020).
The proposed osteoclastic coupling factors include secreted and membrane-bound coupling factors. Potential secreted coupling factors (clastokines) includes Leukemia Inhibitory Factor (LIF), Cardiotrophin-1 (CTF1) and Oncostatin M (OSM) from the IL-6 family of cytokines. These cytokines have been suggested to play a role in bone metabolism (Sims, 2009, 2021). An interesting feature of these cytokines is their dependency of the glycoprotein 130 subunit during signaling, and their ability to react with other receptors within this group of cytokines (Kishimoto et al., 1995). LIF has been associated with metabolic and immunological processes and especially with growth and bone metabolism (Ware et al., 1995; Jones and Jenkins, 2018). The receptor of LIF (LIFR) is expressed by murine osteoblastic cells in vitro (Allan et al., 1990; Reid et al., 1990; Bellido et al., 1997; Walker et al., 2010). The amino acid sequence of CTF1 is similar to LIF and able to bind and activate LIFR(Pennica et al., 1995). In primary murine osteoblastic cells, Ctf1 expression increases with differentiation (Liu, Aubin and Malaval, 2002) whereas CTF1 protein has been reported in mature murine osteoclasts (Walker et al., 2008). OSM also has the ability to bind and activate LIFR (Rose and Bruce, 1991; Liu et al., 1992) besides the specific OSM receptor (OSMR) (Thoma et al., 1994). Murine osteoblastic cells express both Lifr and Osmr, but their expression levels differ throughout differentiation (Bellido et al., 1996). However, knowledge on how LIF, CTF1, OSM, OSMR and LIFR are expressed in human bone is scarce.
Platelet-Derived Growth Factor (PDGF) has also attracted attention as a possible secreted coupling factor regulating bone formation (Horner et al., 1996). PDGFs are dimeric proteins of two polypeptide chains, forming either homodimers (AA, BB) or heterodimers (AB). Likewise, PDGF receptors are dimeric and either homodimers or heterodimers (PDGFRA, PDGFRB or PDGFRAB). PDGF-BB is considered the universal PDGF with binding affinity for all PDGF receptors (Horner et al., 1996; Alvarez, Kantarjian and Cortes, 2006). In human trabecular bone, PDGFB expression was recently observed in osteoclasts, while its receptors PDGFRA and PDGFRB were expressed in proximate reversal cells and osteoblastic canopy cells, separating bone surface cells from the marrow cavity (Brun et al., 2020).
Proposed membrane-bound coupling factors include semaphorin 4D (SEMA4D), a transmembrane glycoprotein with high affinity to PlexinB1 (PLXNB1) (Kang and Kumanogoh, 2013). SEMA4D is believed to be a repressor of bone formation, as knockdown in mice leads to a high bone-mass phenotype with no effect on bone resorption (Negishi-Koga et al., 2011). In humans, high serum levels of SEMA4D has been associated with low BMD and decreased markers of bone formation (Zhang et al., 2015). However, not much is known about the spatial expression of SEMA4D and PLXNB1 within the bone environment. EphrinB2 (EFNB2) is yet another proposed membrane-bound coupling factor. EFNB2 is a transmembrane ligand of the receptor tyrosine kinase EPHB4. Activation of receptor tyrosine kinases initiates bidirectional signaling, forward through the receptor and reverse through the ligand (Pasquale, 2010; Taylor, Campbell and Nobes, 2017). Expression of Efnb2 has been shown in osteoclasts, osteoblasts and osteocytes of mice, whereas Ephb4 expression has only been shown in osteoblasts (Arthur et al., 2011, 2018; Wang et al., 2014).
In this study we investigated spatial mRNA localization of several suggested coupling factors, secreted or membrane-bound in osteoclasts, and their receptors in osteoblastic reversal cells and osteoblasts within human intracortical bone remodeling events.
Materials and Methods
Human bone specimens were collected from the proximal femur of nine adolescent patients aged 6–15 years undergoing corrective surgery for Coxa Valga. Collected specimens were fixated in 4% paraformaldehyde for 2 days and subsequently decalcified for 30 days in 0.5 M EDTA containing 0.4% paraformaldehyde. Decalcified specimens were dehydrated, paraffin-embedded and cut in series of 3.5-µm-thick adjacent sections. Every fifth section was Masson Trichrome stained to select samples with active bone remodeling (identified as erosion or formation in cortical pores). Selected sections were stained with in situ hybridization combined with TRAcP. Spatial localization of each mRNA was validated in at least three different individuals. The study was approved by the Danish National Committee on Biomedical Research Ethics (Project-ID: S-2012-0193).
In situ Hybridization Combined With Immunostaining
Sections adjacent to Masson Trichrome stained sections were in situ hybridized for the mRNA abundance of proposed coupling factors LIF, CTF1, OSM, SEMA4D, EPHB4 and PDGFA, as well as their receptors LIFR, OSMR, PLXNB1, EFNB2, PDGFRA and PDGFRB. In situ hybridization was performed using a modified RNAscope 2.5 high-definition procedure (R2283, Sigma-Aldrich). After deparaffinization and rehydration, sections were treated with 1.5% hydrogen peroxidase for 30 min at room temperature to inactivate endogenous peroxidases. Subsequently, sections were pretreated with RNAscope Target Retrieval for 15 min at 90°C and pepsin (322300, ACD Bioscience) for 20 min at 40°C. After pretreatment, sections were hybridized in a HybEZ™ hybridization oven at 40°C overnight with 20- probe-pairs for human LIF (cat. No: 445721, binding nt 839-1780 of NM_002309.4), CTF1 (Cat. No. 895601, binding nt 40-1222 of NM_001330.5), OSM (Cat. No. 456381, binding nt 32-1175 of NM_020530.4), SEMA4D (Cat. No, 430711, binding nt 611-1623 of NM_006378.3), EPH receptor panel with high affinity for EPHB4 and affinity for EPHB1/EPHB2/EPHB3 (Cat. No. 516401, binding nt 2019-2577 of NM_004444.4), PDGFB (Cat. No. 406701, binding nt 665-2037 of NM_033016.2), LIFR (Cat. No. 441021, binding nt 2411-3421 of NM_001127671.1), OSMR (Cat. No. 537121, binding nt 307-1357 of NM_001323505.1), PLXNB1 (Cat. No. 430681, binding nt 1208-2101 of NM_002673.5), EFNB2 (Cat. No. 430651, binding nt 2-919 of NM_004093.3), PDGFRA (Cat. No. 604481, binding nt 844-1774 of NM_006206.4) and PDGFRB (Cat. No. 548991, binding 523-2984 of NM_002609.3) from ACD Bioscience. The probes were diluted 1:1 in probe diluent (449819, ACD Bioscience) and negative controls were with only probe diluent. Each probe was validated on a tissue array with 36 different anonymized tissue-samples. Hybridized probes were branch amplified through six steps in the HybEZ™ hybridization oven according to manufactures instructions, and further enhanced with digoxigenin-conjugated tyramide (NEL748001KT, PerkinElmer) detected with alkaline-phosphatase conjugated sheep anti-digoxigenin Fab fragments (11093274910, Roche) and visualized using Liquid Permanent Red (Agiliant). After the in situ procedure, osteoclasts were immunostained with mouse-anti-TRAcP IgG2B antibody (clone 9C5, MABF96, Merck Millipore) detected with horseradish peroxidase-conjugated anti-mouse IgG polymers (BrightVision, Immunologic, Duiven, Holland) and visualized using Deep Space Black (Biocare Medical Concord, CA, United States). Finally, sections were counterstained with Mayer’s hematoxylin.
Microscopy
The stained sections were imaged on a VS200 slide scanner (Olympus) using Z-stack condensed into a single plane with optimal focus, which were investigated using the Olivia software (Olympus).
Results
In this observational study, we examined the presence of mRNA encoding secreted and membrane-bound coupling factors proposed in the literature and their receptors in human cortical bone remodeling events. Here, we focused particularly on reversal cells situated adjacent to mature bone resorbing osteoclasts. We examined femur cortical bone specimens from nine adolescents and each mRNA was evaluated in at least three different individuals.
IL-6 Family Cytokines and Their Receptors are Present in Human Bone Remodeling Events
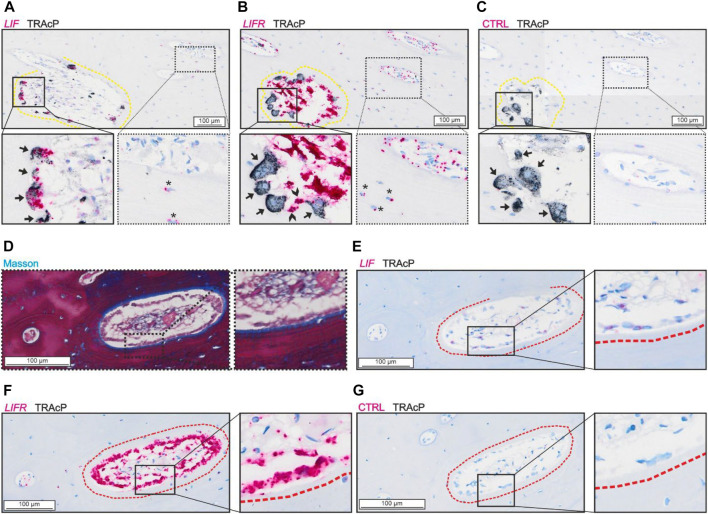
Analysis of the spatial mRNA localization of LIF and LIFR revealed a high abundance of LIF in mature bone-resorbing osteoclasts and a lower presence in osteocytes (Figure 1A). On the other hand, the LIFR mRNA was not detected in mature bone-resorbing osteoclasts (Figure 1B). Instead, LIFR was highly abundant in reversal cells near osteoclasts on the eroded surfaces, and in mononucleated cells within the pore lumen, which to a great extend reflect osteoprogenitors being recruited to the eroded surfaces as reversal cells (Lassen et al., 2017) (Figure 1B). LIFR was also abundant in mature bone-forming osteoblasts on osteoid surfaces and only weakly present in some osteocytes (Figures 1D,F). LIF was only weakly present in mature bone-forming osteoblasts (Figure 1E).
FIGURE 1.
LIF is present in osteoclasts and LIFR in osteoblasts and mononucleated cells in the lumen of adjacent sections of intracortical canals. (A–C) Adjacent sections of eroded pore with in situ hybridization (red) of LIF (A), LIFR (B) and negative control (C) combined with immunostaining of TRAcP (black). (D–G) Adjacent sections of formative pore with masson (D) and in situ hybridization of LIF (E), LIFR (F) or negative control (G) combined with immunostaining of TRAcP (black). Eroded surfaces are marked by a yellow dashed line, formative bone surfaces are marked by a red dashed line. Osteoclasts are indicated by black arrows in the zooms, Reversal cells are indicated by arrowheads and osteocytes with signal from in situ hybridization are shown with*.
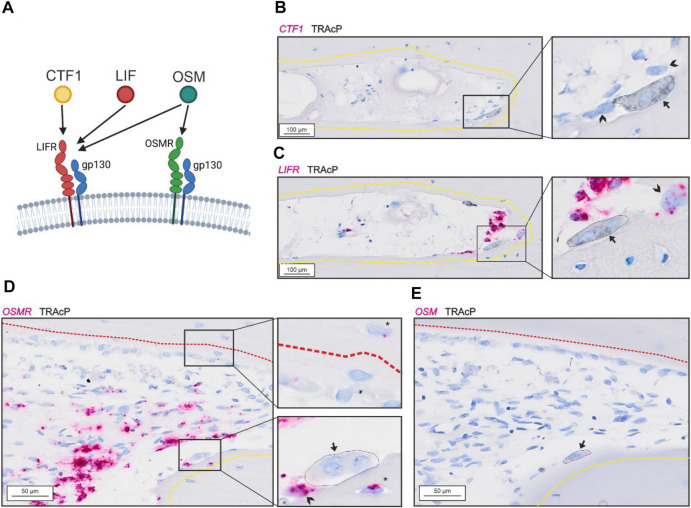
Bone-resorbing osteoclasts showed no presence of CTF1 (Figure 2B) despite presence of LIFR in proximate reversal cells and mononucleated cells within the lumen (potential osteoprogenitors) (Figure 2C). OSMR was abundant in reversal cells and proximate mononucleated cells within the lumen (potential osteoprogenitors), as well as to some extend in osteocytes. In contrast to LIFR, OSMR was not notably present in bone-forming osteoblasts (Figure 2D). Surprisingly, bone-resorbing osteoclasts showed no evidence of OSM mRNA (Figure 2E), as the case for CTF1. Levels of OSM and CFT1 was generally low and restricted to a few mononucleated cells within the intracortical pores. Both OSM and CTF1 were detected in different tissues in the control tissue array (Suppl. 1).
FIGURE 2.
Gp130-associating receptors are present within intracortical pores with active bone remodeling. (A) schematic illustration of the two gp130-associating receptors (LIFR and OSMR) and their affinity for CTF-1, LIF and OSM. (A–B) Adjacent sections of eroded pore with in situ hybridization (red) of CTF1 (A) and LIFR (B) combined with immunostaining of TRAcP (black). (D–E) Adjacent sections of pore with resorption and formation with in situ hybridization (red) of OSMR (D) and OSM (E) combined with immunostaining of TRAcP (black). Eroded surfaces are marked by a yellow dashed line and formative surfaces are marked by a red dashed line. Osteoclasts are marked by a black arrow and outlined in the zooms, reversal cells are marked by arrowheads.
PDGF and its Receptors are Present in Human Bone Remodeling Events
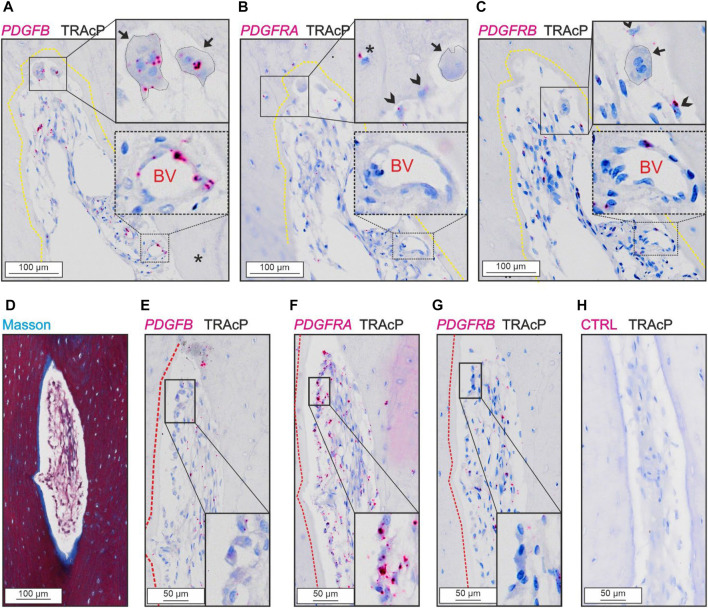
PDGFB was detected in osteoclasts and in cells near the vascular structures, not in reversal cells (Figures 3A,E). The two receptors were present at different levels in the tissue.
FIGURE 3.
Levels of PDGFRA and PDGFRB differ in cortical bone. (A–C) Adjacent sections of eroded pore with immunostaining of TRAcP (black) and in situ hybridization (red) of PDGFB (A), PDGFRA (B) and PDGFRB (C). (D–H) Adjacent sections of pore with bone formation with Masson Trichrome (D) or in situ hybridization of PDGFB (E), PDGFRA (F), PDGFRB (G) or negative control (H) combined with immunostaining of TRAcP (black). Eroded surfaces are marked by a yellow dotted line, formative surfaces are marked by a red dotted line. Osteoclasts are outlined and marked with black arrows, reversal cells with arrowheads and osteocytes with in situ signal are marked with (*), blood vessels are indicated with “BV”.
PDGFRA and PDGFRB were present in reversal cells on eroded surfaces (Figures 3B, C). Furthermore, PDGFRA was present in osteocytes and osteoblasts (Figures 3B,F), whereas PDGFRB was primarily located near vascular structures within the lumen of intracortical pores and not in bone forming osteoblasts or osteocytes (Figures 3C,G).
SEMA4D and PLXNB1 are Present in Human Bone Remodeling Events
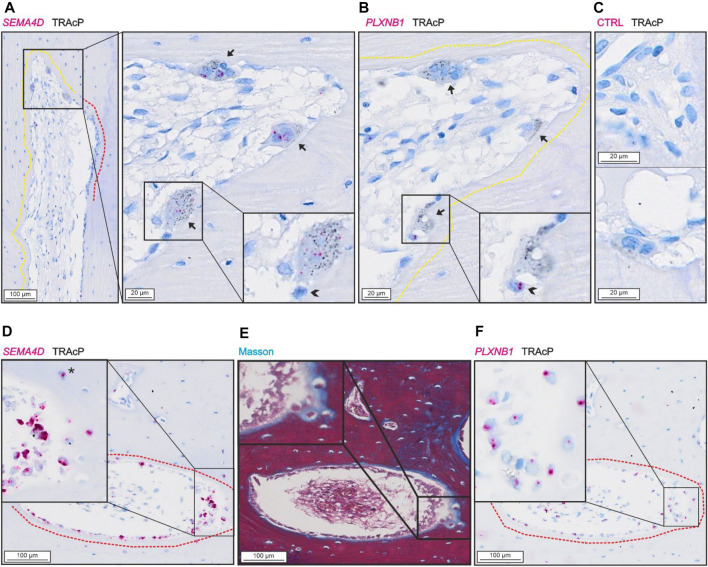
SEMA4D was present in mature bone-resorbing osteoclasts (Figure 4A) and in bone-forming osteoblasts on osteoid surfaces and in some osteocytes (Figures 4D,E). The few mononucleated cells within the lumen showing low levels of SEMA4D, appeared morphologically like endothelial cells instead of potential osteoprogenitors. Discrete levels of PLXNB1 (receptor of SEMA4D) were observed in reversal cells next to SEMA4D-positive osteoclasts (Figure 4B), and in bone-forming osteoblasts on osteoid surfaces and in some osteocytes (Figures 4E,F). No PLXNB1 was observed in mature bone-resorbing osteoclasts (Figure 4B).
FIGURE 4.
SEMA4D is present in osteoclasts and mature osteoblasts and PLXNB1 is present in osteoblasts. (A–C) Adjacent sections of pore with resorption with in situ hybridization (red) of SEMA4D (A), PLXNB1 (B) or negative control (C) combined with immunostaining of TRAcP (black). (D–F) Adjacent sections of pore with bone formation with masson trichrome staining (E) or in situ hybridization (red) of SEMA4D (D) or PLXNB1 (F) combined with immunostaining of TRAcP (black). Eroded surfaces are marked by a yellow dotted line, formative surfaces are marked by a red dotted line. Osteoclasts are outlined and marked with black arrows, reversal cells with arrowheads (*).
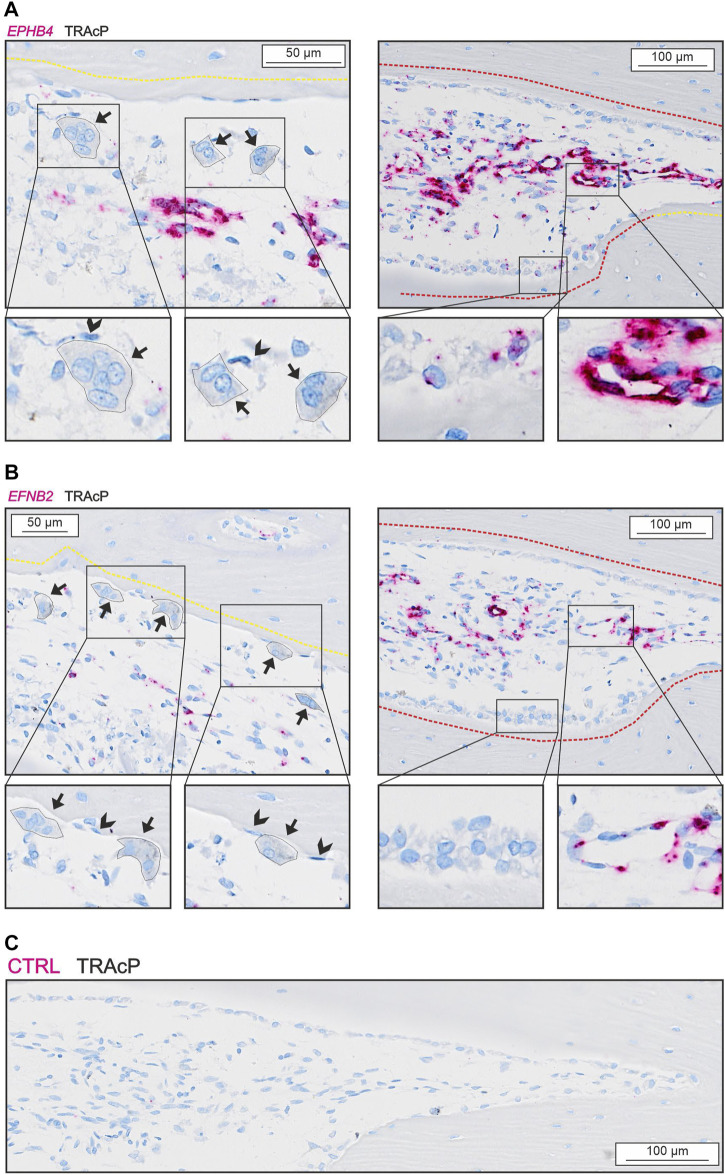
Vascular Structures Express EFNB2 and EPHB4 in Human Bone Remodeling Events
We observed no presence of either EPHB4 or EFNB2 in osteoclasts, reversal cells or osteocytes (Figure 5), but some mature bone-forming osteoblasts contained EPHB4 mRNA (Figure 5A). In contrast, both EPHB4 and EFNB2 were highly present in vascular structures within the intracortical pores (Figure 5).
FIGURE 5.
EFNB2 and EPHB4 are mainly present near vascular structures. (A) In situ hybridization of EPHB4 (red) and immunohistochemical staining of TRAcP (black). (B) In situ hybridization of EFNB2 (red) and immunohistochemical staining of TRAcP (black). Eroded surfaces are marked by a yellow dotted line, formative surfaces are marked by a red dotted line. Osteoclasts are outlined and marked with black arrows, reversal cells with arrowheads.
Discussion
The elusive coupling of bone formation to osteoclastic bone resorption is a critical step in the bone remodeling process, which we are only starting to understand (Delaisse et al., 2020). Osteoclastic coupling factors play a central role in the osteoclast-osteoblast coupling, ensuring the initiation of bone formation within the vacated resorption cavities (Sims and Martin, 2020). The present study examines in situ mRNA localization of potential membrane-bound and secreted osteoclastic coupling factors and their respective receptors in human cortical remodeling events. The study demonstrates that mRNA of several coupling factors are present in osteoclasts, while their receptors were present in neighboring osteoblastic reversal cells (e.g., osteoprogenitors) during the reversal-resorption phase. This supports the notion that interactions between osteoclasts and osteoprogenitors within the reversal-resorption phase play a key role in the coupling mechanism, potentially involving a dedicated panel of secreted and membrane-bound coupling factors.
Secreted Osteoclastic Coupling Factors and Their Receptors in Human Bone Remodeling
In human cortical remodeling events, the osteoclastic levels of LIF and high levels of LIFR in neighboring reversal cells and potential osteoprogenitors within the lumen, support that LIF:LIFR signaling may likely have a functional role in the coupling during human bone remodeling. This supports previous studies in genetic mice models, suggesting a pro-osteogenic effect of LIF:LIFR signaling during bone remodeling. These mice studies showed an increased bone volume when Lif was overexpressed (Metcalf and Gearing, 1989), and decreased bone volume and increased number of osteoclasts in Lif knockout mice (Bozec et al., 2008) and Lifr knockout mice (Ware et al., 1995). This skeletal effect is partly transferable to humans, where mutations in the LIFR gene cause Stüve-Wiedemann syndrome (OMIM #610559), characterized by bowing and thickening in the lower limbs and abnormal trabecular bone structure (Cormier-Daire et al., 1998; Dagoneau et al., 2004). Stüve-Wiedemann syndrome is often fatal and associated with early lethality, as also observed in Lif and Lifr knockout mice (Cormier-Daire et al., 1998; Sims, 2009). The early lethality makes the effects of LIF:LIFR signaling on remodeling versus modeling and growth hard to interpret, and differing roles of LIF signaling in bone development and remodeling has been reported (Poulton et al., 2012). Our findings of LIF and LIFR in interacting osteoclasts and osteoprogenitors, support that LIF:LIFR signaling plays a role in the osteoclast-osteoblast coupling within the reversal-resorption phase.
Importantly, LIFR signaling can also be activated by several other ligands of the IL-6 family cytokines (Kishimoto et al., 1995). CTF1 and OSM are two alternative ligands of LIFR, which have been suggested to play a regulatory role in bone remodeling. Like LIF, CTF1 might possess different roles in modeling versus remodeling events. Studies on Ctf1 knockout mice have shown that they are osteopenic at birth but had a high bone mass phenotype at 10- and 26-weeks of age (Walker et al., 2008; Poulton et al., 2012). In the same study, CTF1 protein was observed in murine osteoclasts. We did not observe any notable levels of CTF1 mRNA in osteoclasts, reversal cells or osteoblasts in human cortical bone remodeling events, questioning its importance in human bone remodeling. OSM is an alternative ligand of LIFR, which has been extensively studied. Studies treating mice with OSM has indicated both pro-osteogenic effects (Jay et al., 1996; Bellido et al., 1997; Walker et al., 2010), as well as an increased osteoclast formation and activity (Tamura et al., 1993; Palmqvist et al., 2002). Recently, it was suggested that OSM signaling through LIFR stimulates bone formation (Walker et al., 2010), consistent with a high bone mass phenotype observed in mice overexpressing bovine Osm (Malik et al., 1995). Conversely, OSM signaling through OSMR is suggested to induce osteoclastogenesis indirectly by upregulating RANKL expression (Walker et al., 2010). However, we did not observe any notable presence in osteoclasts, reversal cells or osteoblasts in human cortical bone remodeling events, questioning its importance in human bone remodeling. On the other hand, we did observe OSMR mRNA in reversal cells and potential osteoprogenitors within the lumen of intracortical pores, which may respond to an alternative unknown ligand.
Another potential secreted osteoclastic coupling factor is PDGF homodimers or heterodimers, which have attracted attention as regulators of bone remodeling. This attention originates from clinical studies with the tyrosine kinase inhibitors Imatinib and Nilutinib observed to increase serum markers of bone formation, but not resorption (Grey et al., 2006). Subsequently, in vitro studies ascribed this effect of Imatinib and Nilutinib treatment to PDGFR-β signaling causing increased Opg expression (O’Sullivan et al., 2007, 2011, 2016). Treatment with PDGF-BB has also been shown to increase mesenchymal cell proliferation and osteoblast differentiation in vitro, but also the expression of pro-resorptive factors, such as Csf1 and Rankl (Chen et al., 2015). In human trabecular bone, PDGFB was expressed by osteoclasts whereas both PDGF receptors (PDGFRA and PDGFRB) were expressed by osteoblastic canopy cells and reversal cells (Brun et al., 2020). In the present study, we observed expression of both receptors in reversal cells but differing expression pattern in other cells. Besides in reversal cells, PDGFRA was expressed by osteocytes and osteoblasts, whereas PDGFRB was expressed near vascular structures within intracortical pores.
Membrane-Bound Osteoclastic Coupling Factor and Their Receptors in Human Bone Remodeling
In human cortical bone remodeling events, SEMA4D was present in osteoclasts and PLXNB1 was observed in reversal cells, supporting that SEMA4D:PLXNB1 binding may play a role in their communication. This is in line with murine studies, showing Sema4d expression in osteoclasts and osteoclast progenitors, and increased Plxnb1 expression during osteoblast differentiation (Negishi-Koga et al., 2011). Functional studies in mice, suggest that Sema4D is a suppressor of bone formation with knockdown leading to a higher bone mass. However, the cause of high bone mass in knockout mice does not concur between studies. Negishi-Koga and colleagues reported increased bone formation without osteoclastic effect (Negishi-Koga et al., 2011) whereas Dacquin and colleagues observed reduced resorptive activity (Dacquin et al., 2011). In a clinical study, serum levels of SEMA4D positively correlated with serum markers of resorption in patients with multiple myeloma (Zhang et al., 2015; Terpos et al., 2018). Later, SEMA4D secreted from a human lung cancer cell line were shown to inhibit osteoblast differentiation in vitro (Chen et al., 2019). In contrast to this study, treatment of osteoporotic postmenopausal women with the antiresorptive Denosumab have been shown to increase serum levels of SEMA4D compared to controls (Anastasilakis et al., 2015), suggesting that SEMA4D is produced by other sources than osteoclasts. This study suggests that SEM4D originate from mature bone-forming osteoblasts, showing presence of SEMA4D mRNA at human bone remodeling sites.
EFNB2:EPHB4 signaling has also been proposed as a coupling pathway requiring cell-cell contact. Efnb2 and Ephb4 have been reported in several bone cells (Arthur et al., 2011, 2018; Wang et al., 2014) and EFNB2:EPHB4 signaling within the osteoblast lineage is believed to promote osteoblast differentiation (Takyar et al., 2013; Tonna et al., 2014). Nevertheless, we were unable to observe any notable presence of EFNB2 and EPHB4 in human osteoclasts and reversal cells questioning its direct importance in the osteoclast-osteoblasts coupling mechanism of human bone remodeling. On the other hand, EFNB2 and EPHB4 are highly expressed in the vascular structures within the lumen of intracortical pores, consistent with a role in the local vascularization and angiogenesis as shown in other studies (Wang et al., 2010). Vascularization is essential for osteoprogenitor recruitment and thereby indirectly the activation of bone formation on eroded bone surfaces vacated by the osteoclasts.
In this study, we qualitatively investigated the spatial in situ mRNA localization of proposed coupling factors and their receptors using bone specimens from adolescents undergoing corrective surgery for Coxa Valga. Therefore, we consider the analyzed cortical bone as healthy. By investigating intracortical pores, we ensure that well-defined remodeling processes were examined, despite the young age of patients. Our investigations are limited to the in situ cellular mRNA-levels, which are affected by expression and stability of each individual mRNA. Despite the use of a tissue array to validate probes, stability and retention time within bone may vary from other tissues. The study does not investigate the distribution of proteins or functional analyses of included coupling factors. In the applied mRNA detection-procedure we used probe pairs designed by ACD Bioscience. Each set of probe pairs included 20 different probe pairs targeting a specific region within the gene of interest. Levels of mRNA detected were described af high/low when compared to other probes or differing levels between cell types.
Further investigation of the mRNA and protein abundance, as well as functional significance of these coupling factors are needed in human bone remodeling.
Conclusion
Our mRNA analysis of human cortical bone remodeling events revealed presence of proposed coupling factors LIF, SEMA4D and PDGFB mRNA in mature bone-resorbing osteoclasts and presence of their respective receptors LIFR, PLXNB1, PDGFRA and PDGFRB mRNA in neighboring reversal cells. These results are complementary to previous functional studies, supporting a functional role in the coupling mechanism of human bone remodeling. Conversely, we did not observe presence of CTF1 or OSM mRNA in mature osteoclasts, despite the presence of OSMR mRNA in neighboring reversal cells (Figure 6). Finally, presence of EFNB2 and EPHB4 mRNA was restricted to vascular structures within intracortical pores, with no indications of presence within osteoclasts nor reversal cells.
FIGURE 6.
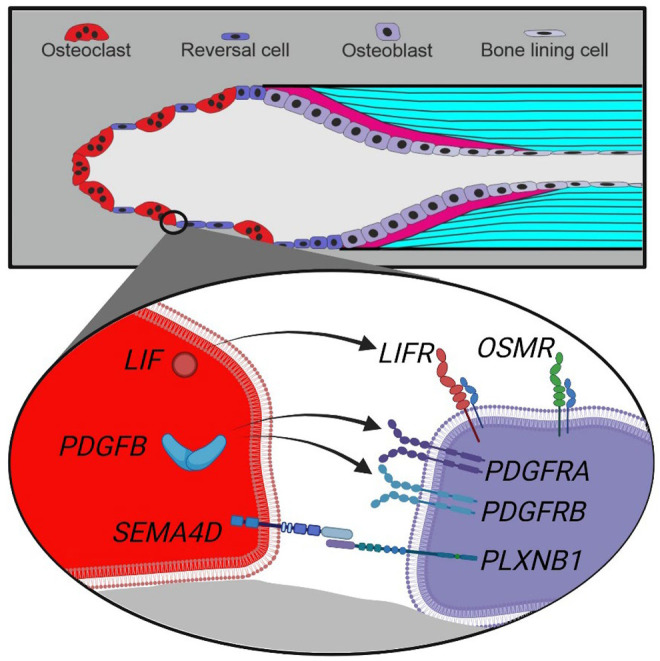
Schematic presentation of a cutting cone with observed coupling factors and receptors in osteoclasts and reversal cells in human bone remodeling. Top, a cutting cone with initial resorption (left), reversal-resorption phase with osteoclasts intermixed with reversal cells (middle) followed by bone forming osteoblasts laying down osteoid (right). Bottom, zoom on the cell-cell interface between osteoclasts (red) and reversal cells (purple) with observed expression of coupling factors and receptors.
Acknowledgments
We acknowledge Dr Søren Harving, Aalborg University Hospital for collection of specimens.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Danish National Committee on Biomedical Research Ethics. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. (Project-ID: S-2012-0193).
Author Contributions
Authorship contribution statement: Conceptualization and design; TA, J-MD, and CA Acquisition of data: XB and MN Investigation and methodology: XB, MN, TA, and CA Analysis and interpretation of data: all authors Funding acquisition, project administration and supervision: TA, J-MD, CA, and XB Drafting: XB and TA Final approval: all authors.
Funding
Salary for PhD student Xenia Borggaard was covered by The Velux Foundation (Grant no. 25723) and Department of Clinical Research, University of Southern Denmark. Laboratory costs were covered by The Velux Foundation (Grant no. 25723), Aase og Ejnar Danielsens fond (Grant no. 18-10-0473) and The Danish National Association for Osteoporosis (Osteoporoseforeningen). Schematic figures in this publication were created with www.biorender.com.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abdallah B. M., Figeac F., Larsen K. H., Ditzel N., Keshari P., Isa A., et al. (2017). CRMP4 Inhibits Bone Formation by Negatively Regulating BMP and RhoA Signaling. J. Bone Min. Res. 32 (5), 913–926. 10.1002/jbmr.3069 [DOI] [PubMed] [Google Scholar]
- Abdelgawad M. E., Delaisse J.-M., Hinge M., Jensen P. R., Alnaimi R. W., Rolighed L., et al. (2016). Early Reversal Cells in Adult Human Bone Remodeling: Osteoblastic Nature, Catabolic Functions and Interactions with Osteoclasts. Histochem Cell Biol. 145 (6), 603–615. 10.1007/s00418-016-1414-y [DOI] [PubMed] [Google Scholar]
- Allan E. H., Hilton D. J., Brown M. A., Evely R. S., Yumita S., Metcalf D., et al. (1990). Osteoblasts Display Receptors for and Responses to Leukemia-Inhibitory Factor. J. Cell. Physiol. 145 (1), 110–119. 10.1002/jcp.1041450116 [DOI] [PubMed] [Google Scholar]
- Alvarez R. H., Kantarjian H. M., Cortes J. E. (2006). Biology of Platelet-Derived Growth Factor and its Involvement in Disease. Mayo Clin. Proc. 81 (9), 1241–1257. 10.4065/81.9.1241 [DOI] [PubMed] [Google Scholar]
- Anastasilakis A. D., Polyzos S. A., Makras P., Gkiomisi A., Sakellariou G., Savvidis M., et al. (2015). Circulating semaphorin-4D and Plexin-B1 Levels in Postmenopausal Women with Low Bone Mass: The 3-month Effect of Zoledronic Acid, Denosumab or Teriparatide Treatment. Expert Opin. Ther. Targets 19 (3), 299–306. 10.1517/14728222.2014.983078 [DOI] [PubMed] [Google Scholar]
- Andersen T. L., Sondergaard T. E., Skorzynska K. E., Dagnaes-Hansen F., Plesner T. L., Hauge E. M., et al. (2009). A Physical Mechanism for Coupling Bone Resorption and Formation in Adult Human Bone. Am. J. Pathology 174 (1), 239–247. 10.2353/ajpath.2009.080627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen T. L., Søe K., Sondergaard T. E., Plesner T., Delaisse J.-M. (2010). Myeloma Cell-Induced Disruption of Bone Remodelling Compartments Leads to Osteolytic Lesions and Generation of Osteoclast-Myeloma Hybrid Cells. Br. J. Haematol. 148 (4), 551–561. 10.1111/j.1365-2141.2009.07980.x [DOI] [PubMed] [Google Scholar]
- Andersen T. L., Abdelgawad M. E., Kristensen H. B., Hauge E. M., Rolighed L., Bollerslev J., et al. (2013). Understanding Coupling between Bone Resorption and Formation: Are Reversal Cells the Missing Link?. Am. J. Pathology 183 (1), 235–246. 10.1016/j.ajpath.2013.03.006 [DOI] [PubMed] [Google Scholar]
- Andreasen C. M., Bakalova L. P., Brüel A., Hauge E. M., Kiil B. J., Delaisse J.-M., et al. (2020). The Generation of Enlarged Eroded Pores upon Existing Intracortical Canals Is a Major Contributor to Endocortical Trabecularization. Bone 130, 115127. 10.1016/j.bone.2019.115127 [DOI] [PubMed] [Google Scholar]
- Arthur A., Zannettino A., Panagopoulos R., Koblar S. A., Sims N. A., Stylianou C., et al. (2011). EphB/ephrin-B Interactions Mediate Human MSC Attachment, Migration and Osteochondral Differentiation. Bone 48 (3), 533–542. 10.1016/j.bone.2010.10.180 [DOI] [PubMed] [Google Scholar]
- Arthur A., Nguyen T. M., Paton S., Klisuric A., Zannettino A. C. W., Gronthos S. (2018). The Osteoprogenitor-specific Loss of ephrinB1 Results in an Osteoporotic Phenotype Affecting the Balance between Bone Formation and Resorption. Sci. Rep. 8 (1), 1–12. 10.1038/s41598-018-31190-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron R., Silverglate A., Broadus A., Lang R. (1983). Estimation of Trabecular Bone Resorption by Histomorphometry: Evidence for a Prolonged Reversal Phase with Normal Resorption in Post-menopausal Osteoporosis and Coupled Increased Resorption in Primary Hyperparathyroidism. Clin. Disord. Bone Min. Metab. 1 191–195. [Google Scholar]
- Bellido T., Stahl N., Farruggella T. J., Borba V., Yancopoulos G. D., Manolagas S. C. (1996). Detection of Receptors for Interleukin-6, Interleukin-11, Leukemia Inhibitory Factor, Oncostatin M, and Ciliary Neurotrophic Factor in Bone Marrow Stromal/osteoblastic Cells. J. Clin. Invest. 97 (2), 431–437. 10.1172/JCI118432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellido T., Borba V. Z. C., Roberson P., Manolagas S. C. (1997). Activation of the Janus Kinase/STAT (Signal Transducer and Activator of Transcription) Signal Transduction Pathway by Interleukin-6-type Cytokines Promotes Osteoblast Differentiation*. Endocrinology 138 (9), 3666–3676. 10.1210/endo.138.9.5364 [DOI] [PubMed] [Google Scholar]
- Bianco P., Ballanti P., Bonucci E. (1988). Tartrate-resistant Acid Phosphatase Activity in Rat Osteoblasts and Osteocytes. Calcif. Tissue Int. 43 (3), 167–171. 10.1007/BF02571315 [DOI] [PubMed] [Google Scholar]
- Bozec A., Bakiri L., Hoebertz A., Eferl R., Schilling A. F., Komnenovic V., et al. (2008). Osteoclast Size Is Controlled by Fra-2 through LIF/LIF-receptor Signalling and Hypoxia. Nature 454 (7201), 221–225. 10.1038/nature07019 [DOI] [PubMed] [Google Scholar]
- Brun J., Andreasen C. M., Ejersted C., Andersen T. L., Caverzasio J., Thouverey C. (2020). PDGF Receptor Signaling in Osteoblast Lineage Cells Controls Bone Resorption through Upregulation of Csf1 Expression. J. Bone Min. Res. 35 (12), 2458–2469. 10.1002/jbmr.4150 [DOI] [PubMed] [Google Scholar]
- Charles J. F., Aliprantis A. O. (2014). Osteoclasts: More Than 'bone Eaters'. Trends Mol. Med. 20 (8), 449–459. 10.1002/nme.211010.1016/j.molmed.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Baylink D. J., Brier-Jones J., Neises A., Kiroyan J. B., Rundle C. H., et al. (2015). PDGFB-based Stem Cell Gene Therapy Increases Bone Strength in the Mouse. Proc. Natl. Acad. Sci. U.S.A. 112 (29), E3893–E3900. 10.1073/pnas.1501759112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Shi K., Andersen T. L., Qiu W., Kassem M. (2019). KIAA1199 is a Secreted Molecule that Enhances Osteoblastic Stem Cell Migration and Recruitment. Cell Death Dis. 10 (2). 126. 10.1038/s41419-018-1202-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-g., Sun J., Shen W.-w., Yang S.-z., Zhang Y., Hu X., et al. (2019). Sema4D Expression and Secretion Are Increased by HIF-1α and Inhibit Osteogenesis in Bone Metastases of Lung Cancer. Clin. Exp. Metastasis 36 (1), 39–56. 10.1007/s10585-018-9951-5 [DOI] [PubMed] [Google Scholar]
- Cormier-Daire V., Munnich A., Lyonnet S., Rustin P., Delezoide A.-L., Maroteaux P., et al. (1998). Presentation of Six Cases of Stüve-Wiedemann Syndrome. Pediatr. Radiol. 28 (10), 776–780. 10.1007/s002470050464 [DOI] [PubMed] [Google Scholar]
- Dacquin R., Domenget C., Kumanogoh A., Kikutani H., Jurdic P., Machuca-Gayet I. (2011). Control of Bone Resorption by Semaphorin 4D Is Dependent on Ovarian Function. PLoS ONE 6 (10), e26627. 10.1371/journal.pone.0026627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagoneau N., Scheffer D., Huber C., Al-Gazali L. I., Di Rocco M., Godard A., et al. (2004). Null Leukemia Inhibitory Factor Receptor (LIFR) Mutations in Stüve-Wiedemann/Schwartz-Jampel Type 2 Syndrome. Am. J. Hum. Genet. 74 (2), 298–305. 10.1086/381715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaisse J.-M., Andersen T. L., Kristensen H. B., Jensen P. R., Andreasen C. M., et al. (2020). Re-thinking the Bone Remodeling Cycle Mechanism and the Origin of Bone Loss. Bone 723 138028. 10.1016/j.scitotenv.2020.138028 [DOI] [PubMed] [Google Scholar]
- Eriksen E. F., Gundersen H. J. G., Melsen F., Mosekilde L. (1984). Reconstruction of the Formative Site in Iliac Trabecular Bone in 20 Normal Individuals Employing a Kinetic Model for Matrix and Mineral Apposition. Metabolic Bone Dis. Relat. Res. 5 (5), 243–252. 10.1016/0221-8747(84)90066-3 [DOI] [PubMed] [Google Scholar]
- Eriksen E. F., Melsen F., Mosekilde L. (1984). Reconstruction of the Resorptive Site in Iliac Trabecular Bone: A Kinetic Model for Bone Resorption in 20 Normal Individuals. Metabolic Bone Dis. Relat. Res. 5 (5), 235–242. 10.1016/0221-8747(84)90065-1 [DOI] [PubMed] [Google Scholar]
- Everts V., Delaissé J. M., Korper W., Jansen D. C., Tigchelaar-Gutter W., Saftig P., et al. (2002). The Bone Lining Cell: Its Role in Cleaning Howship's Lacunae and Initiating Bone Formation. J. Bone Min. Res. 17 (1), 77–90. 10.1359/jbmr.2002.17.1.77 [DOI] [PubMed] [Google Scholar]
- Grey A., O'Sullivan S., Reid I. R., Browett P. (2006). Imatinib Mesylate, Increased Bone Formation, and Secondary Hyperparathyroidism. N. Engl. J. Med. 355 (23), 2494–2495. 10.1056/nejmc062388 [DOI] [PubMed] [Google Scholar]
- Horner A., Bord S., Kemp P., Grainger D., Compston J. E. (1996). Distribution of Platelet-Derived Growth Factor (PDGF) a Chain mRNA, Protein, and PDGF-α Receptor in Rapidly Forming Human Bone. Bone 19 (4), 353–362. 10.1016/S8756-3282(96)00217-7 [DOI] [PubMed] [Google Scholar]
- Jafari A., Qanie D., Andersen T. L., Zhang Y., Chen L., Postert B., et al. (2017). Legumain Regulates Differentiation Fate of Human Bone Marrow Stromal Cells and Is Altered in Postmenopausal Osteoporosis. Stem Cell Rep. 8 (2), 373–386. 10.1016/j.stemcr.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay P. R., Centrella M., Lorenzo J., Bruce A. G., Horowitz M. C. (1996). Oncostatin-M: a New Bone Active Cytokine that Activates Osteoblasts and Inhibits Bone Resorption. Endocrinology 137 (4), 1151–1158. 10.1210/endo.137.4.8625883 [DOI] [PubMed] [Google Scholar]
- Jensen P. R., Andersen T. L., Hauge E.-M., Bollerslev J., Delaissé J.-M. (2014). A Joined Role of Canopy and Reversal Cells in Bone Remodeling - Lessons from Glucocorticoid-Induced Osteoporosis. Bone 73, 16–23. 10.1016/j.bone.2014.12.004 [DOI] [PubMed] [Google Scholar]
- Jones S. A., Jenkins B. J. (2018). Recent Insights into Targeting the IL-6 Cytokine Family in Inflammatory Diseases and Cancer. Nat. Rev. Immunol. 18 (12), 773–789. 10.1038/s41577-018-0066-7 [DOI] [PubMed] [Google Scholar]
- Kang S., Kumanogoh A. (2013). Semaphorins in Bone Development, Homeostasis, and Disease. Seminars Cell & Dev. Biol. 24 (3), 163–171. 10.1016/j.semcdb.2012.09.008 [DOI] [PubMed] [Google Scholar]
- Kishimoto T., Akira S., Narazaki M., Taga T. (1995). Interleukin-6 Family of Cytokines and Gp130. Blood 86 (4), 1243–1254. 10.1182/blood.v86.4.1243.bloodjournal8641243 [DOI] [PubMed] [Google Scholar]
- Lassen N. E., Andersen T. L., Pløen G. G., Søe K., Hauge E. M., Harving S., et al. (2017). Coupling of Bone Resorption and Formation in Real Time: New Knowledge Gained from Human Haversian BMUs. J. Bone Min. Res. 32 (7), 1395–1405. 10.1002/jbmr.316910.1002/jbmr.3091 [DOI] [PubMed] [Google Scholar]
- Liu J., Modrell B., Aruffo A., Marken J. S., Taga T., Yasukawa K., et al. (1992). Interleukin-6 Signal Transducer Gp130 Mediates Oncostatin M Signaling. J. Biol. Chem. 267 (24), 16763–16766. 10.1016/s0021-9258(18)41845-5 [DOI] [PubMed] [Google Scholar]
- Liu F., Aubin J. E., Malaval L. (2002). Expression of Leukemia Inhibitory Factor (LIF)/interleukin-6 Family Cytokines and Receptors during In Vitro Osteogenesis: Differential Regulation by Dexamethasone and LIF. Bone 31 (1), 212–219. 10.1016/S8756-3282(02)00806-2 [DOI] [PubMed] [Google Scholar]
- Malik N., Haugen H. S., Modrell B., Shoyab M., Clegg C. H. (1995). Developmental Abnormalities in Mice Transgenic for Bovine Oncostatin M. Mol. Cell Biol. 15 (5), 2349–2358. 10.1128/mcb.15.5.2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf D., Gearing D. P. (1989). Fatal Syndrome in Mice Engrafted with Cells Producing High Levels of the Leukemia Inhibitory Factor. Proc. Natl. Acad. Sci. U.S.A. 86 (15), 5948–5952. 10.1073/pnas.86.15.5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocetti P., Ballanti P., Zalzal S., Silvestrini G., Bonucci E., Nanci A. (2000). A Histomorphometric, Structural, and Immunocytochemical Study of the Effects of Diet-Induced Hypocalcemia on Bone in Growing Rats. J. Histochem Cytochem. 48 (8), 1059–1077. 10.1177/002215540004800804 [DOI] [PubMed] [Google Scholar]
- Negishi-Koga T., Shinohara M., Komatsu N., Bito H., Kodama T., Friedel R. H., et al. (2011). Suppression of Bone Formation by Osteoclastic Expression of Semaphorin 4D. Nat. Med. 17 (11), 1473–1480. 10.1038/nm.2489 [DOI] [PubMed] [Google Scholar]
- O'Sullivan S., Naot D., Callon K., Porteous F., Horne A., Wattie D., et al. (2007). Imatinib Promotes Osteoblast Differentiation by Inhibiting PDGFR Signaling and Inhibits Osteoclastogenesis by Both Direct and Stromal Cell-dependent Mechanisms. J. Bone Min. Res. 22 (11), 1679–1689. 10.1359/jbmr.070719 [DOI] [PubMed] [Google Scholar]
- O'Sullivan S., Lin J.-M., Watson M., Callon K., Tong P. C., Naot D., et al. (2011). The Skeletal Effects of the Tyrosine Kinase Inhibitor Nilotinib. Bone 49 (2), 281–289. 10.1016/j.bone.2011.04.014 [DOI] [PubMed] [Google Scholar]
- O’Sullivan S., Tay M. L., Lin J.-M., Bava U., Callon K., Cornish J., et al. (2016). Tyrosine Kinase Inhibitors Regulate OPG through Inhibition of PDGFRβ. PLoS ONE 11 (10), e0164727–13. 10.1371/journal.pone.0164727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist P., Persson E., Conaway H. H., Lerner U. H. (2002). IL-6, Leukemia Inhibitory Factor, and Oncostatin M Stimulate Bone Resorption and Regulate the Expression of Receptor Activator of NF-Κb Ligand, Osteoprotegerin, and Receptor Activator of NF-Κb in Mouse Calvariae. J. Immunol. 169 (6), 3353–3362. 10.4049/jimmunol.169.6.3353 [DOI] [PubMed] [Google Scholar]
- Pasquale E. B. (2010). Eph Receptors and Ephrins in Cancer: Bidirectional Signalling and beyond. Nat. Rev. Cancer 10 (3), 165–180. 10.1038/nrc2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennica D., King K. L., Shaw K. J., Luis E., Rullamas J., Luoh S. M., et al. (1995). Expression Cloning of Cardiotrophin 1, a Cytokine that Induces Cardiac Myocyte Hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 92 (4), 1142–1146. 10.1073/pnas.92.4.1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton I. J., McGregor N. E., Pompolo S., Walker E. C., Sims N. A. (2012). Contrasting Roles of Leukemia Inhibitory Factor in Murine Bone Development and Remodeling Involve Region-specific Changes in Vascularization. J. Bone Min. Res. 27 (3), 586–595. 10.1002/jbmr.1485 [DOI] [PubMed] [Google Scholar]
- Reid I. R., Lowe C., Cornish J., Skinner S. J. M., Hilton D. J., Willson T. A., et al. (1990). Leukemia Inhibitory Factor: A Novel Bone-Active Cytokine*. Endocrinology 126 (3), 1416–1420. 10.1210/endo-126-3-1416 [DOI] [PubMed] [Google Scholar]
- Rose T. M., Bruce A. G. (1991). Oncostatin M Is a Member of a Cytokine Family that Includes Leukemia-Inhibitory Factor, Granulocyte Colony-Stimulating Factor, and Interleukin 6. Proc. Natl. Acad. Sci. U.S.A. 88 (19), 8641–8645. 10.1073/pnas.88.19.8641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims N. A., Martin T. J. (2020). Osteoclasts Provide Coupling Signals to Osteoblast Lineage Cells through Multiple Mechanisms. Annu. Rev. Physiol. 82 (1), 507–529. 10.1146/annurev-physiol-021119-034425 [DOI] [PubMed] [Google Scholar]
- Sims N. A. (2009). gp130 Signaling in Bone Cell Biology: Multiple Roles Revealed by Analysis of Genetically Altered Mice. Mol. Cell. Endocrinol. 310 (1–2), 30–39. 10.1016/j.mce.2008.08.025 [DOI] [PubMed] [Google Scholar]
- Sims N. A. (2021). Influences of the IL-6 Cytokine Family on Bone Structure and Function. Cytokine 146, 155655. 10.1016/j.cyto.2021.155655 [DOI] [PubMed] [Google Scholar]
- Takyar F. M., Tonna S., Ho P. W., Crimeen-Irwin B., Baker E. K., Martin T. J., et al. (2013). EphrinB2/EphB4 Inhibition in the Osteoblast Lineage Modifies the Anabolic Response to Parathyroid Hormone. J. Bone Min. Res. 28 (4), 912–925. 10.1002/jbmr.1820 [DOI] [PubMed] [Google Scholar]
- Tamura T., Udagawa N., Takahashi N., Miyaura C., Tanaka S., Yamada Y., et al. (1993). Soluble Interleukin-6 Receptor Triggers Osteoclast Formation by Interleukin 6. Proc. Natl. Acad. Sci. U.S.A. 90 (24), 11924–11928. 10.1073/pnas.90.24.11924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor H., Campbell J., Nobes C. D. (2017). Ephs and Ephrins. Curr. Biol. 27 (3), R90–R95. 10.1016/j.cub.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Terpos E., Ntanasis-Stathopoulos I., Christoulas D., Bagratuni T., Bakogeorgos M., Gavriatopoulou M., et al. (2018). Semaphorin 4D Correlates with Increased Bone Resorption, Hypercalcemia, and Disease Stage in Newly Diagnosed Patients with Multiple Myeloma. Blood Cancer J. 8 (5). 42. 10.1038/s41408-018-0075-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma B., Bird T. A., Friend D. J., Gearing D. P., Dower S. K. (1994). Oncostatin M and Leukemia Inhibitory Factor Trigger Overlapping and Different Signals through Partially Shared Receptor Complexes. J. Biol. Chem. 269 (8), 6215–6222. 10.1016/s0021-9258(17)37590-7 [DOI] [PubMed] [Google Scholar]
- Tonna S., Takyar F. M., Vrahnas C., Crimeen‐Irwin B., Ho P. W. M., Poulton I. J., et al. (2014). EphrinB2 Signaling in Osteoblasts Promotes Bone Mineralization by Preventing Apoptosis. FASEB J. 28 (10), 4482–4496. 10.1096/fj.14-254300 [DOI] [PubMed] [Google Scholar]
- Walker E. C., McGregor N. E., Poulton I. J., Pompolo S., Allan E. H., Quinn J. M., et al. (2008). Cardiotrophin-1 Is an Osteoclast-Derived Stimulus of Bone Formation Required for Normal Bone Remodeling. J. Bone Mineral Res. 23 (12), 2025–2032. 10.1359/jbmr.080706 [DOI] [PubMed] [Google Scholar]
- Walker E. C., McGregor N. E., Poulton I. J., Solano M., Pompolo S., Fernandes T. J., et al. (2010). Oncostatin M Promotes Bone Formation Independently of Resorption when Signaling through Leukemia Inhibitory Factor Receptor in Mice. J. Clin. Invest. 120 (2), 582–592. 10.1172/JCI40568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Nakayama M., Pitulescu M. E., Schmidt T. S., Bochenek M. L., Sakakibara A., et al. (2010). Ephrin-B2 Controls VEGF-Induced Angiogenesis and Lymphangiogenesis. Nature 465 (7297), 483–486. 10.1038/nature09002 [DOI] [PubMed] [Google Scholar]
- Wang Y., Menendez A., Fong C., ElAlieh H. Z., Chang W., Bikle D. D. (2014). Ephrin B2/EphB4 Mediates the Actions of IGF-I Signaling in Regulating Endochondral Bone Formation. J. Bone Min. Res. 29 (8), 1900–1913. 10.1002/jbmr.2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware C. B., Horowitz M. C., Renshaw B. R., Hunt J. S., Liggitt D., Koblar S. A., et al. (1995). Targeted Disruption of the Low-Affinity Leukemia Inhibitory Factor Receptor Gene Causes Placental, Skeletal, Neural and Metabolic Defects and Results in Perinatal Death. Development 121 (5), 1283–1299. 10.1242/dev.121.5.1283 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng E., Xu Y., Wang W., Zhang T., Xiao L., et al. (2015). Serum Sema4D Levels Are Associated with Lumbar Spine Bone Mineral Density and Bone Turnover Markers in Patients with Postmenopausal Osteoporosis. Int. J. Clin. Exp. Med. 8 (9), 16352–16357. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.