Abstract
A Saccharomyces cerevisiae strain, KK-211, isolated by the long-term bioprocess of stereoselective reduction in isooctane, showed extremely high tolerance to the solvent, which is toxic to yeast cells, but, in comparison with its wild-type parent, DY-1, showed low tolerance to hydrophilic organic solvents, such as dimethyl sulfoxide and ethanol. In order to detect the isooctane tolerance-associated genes, mRNA differential display (DD) was employed using mRNAs isolated from strains DY-1 and KK-211 cultivated without isooctane, and from strain KK-211 cultivated with isooctane. Thirty genes were identified as being differentially expressed in these three types of cells and were classified into three groups according to their expression patterns. These patterns were further confirmed and quantified by Northern blot analysis. On the DD fingerprints, the expression of 14 genes, including MUQ1, PRY2, HAC1, AGT1, GAC1, and ICT1 (YLR099c) was induced, while the expression of the remaining 16 genes, including JEN1, PRY1, PRY3, and KRE1, was decreased, in strain KK-211 cultivated with isooctane. The genes represented by HAC1, PRY1, and ICT1 have been reported to be associated with cell stress, and AGT1 and GAC1 have been reported to be involved in the uptake of trehalose and the production of glycogen, respectively. MUQ1 and KRE1, encoding proteins associated with cell surface maintenance, were also detected. Based on these results, we concluded that alteration of expression levels of multiple genes, not of a single gene, might be the critical determinant for isooctane tolerance in strain KK-211.
The toxicity of organic solvents is a serious problem in utilizing living microorganisms for stereoselective biochemical reactions, which are usually performed in organic solvents or water–organic-solvent two-phase systems (13). Although the detailed mechanism of their toxicity to microorganisms has not been extensively studied, it is thought that highly toxic organic solvents, such as toluene and hexane, destroy the integrity of cell membranes by accumulating in the lipid bilayer of plasma membranes (34, 35, 40). The toxicity of an organic solvent correlates inversely with the logarithm of its partition coefficient between octanol and water (logPow) (15). Organic solvents with lower logPow values are more toxic than those with higher logPow values. The organic solvent with the lowest logPow in which target microorganisms can grow is called the index solvent, and the logPow value of the index solvent is called the index value.
Recently, some strains of Pseudomonas aeruginosa, Pseudomonas fluorescens, Pseudomonas putida, and Escherichia coli have been found to be tolerant to toxic organic solvents (1, 2, 17, 27), and mechanisms of tolerance in these bacteria have been proposed (20). Different mechanisms, such as the presence of an efflux system localized in the outer membrane (18, 33, 39), which actively decreases the amount of solvent in the cell and alters the composition of the outer membrane (16, 29, 38), were also considered to contribute to organic-solvent tolerance in these strains. But until now, there have been no reports on eukaryotic microorganisms with organic-solvent tolerance.
An isooctane (2,2,4-trimethylpentane)-tolerant Saccharomyces cerevisiae mutant, strain KK-211, was first isolated from commercial dry yeast which was utilized as an immobilized biocatalyst in the double entrapment of calcium alginate and polyurethane for long-term stereoselective microbial reduction in isooctane (22). Strain KK-211 could grow in the medium overlaid with isooctane, which was previously believed to be lethal to S. cerevisiae. Since the index solvent of S. cerevisiae is usually dodecane (14), this strain will provide some important information about organic-solvent tolerance in eukaryotes, which may lead to a much wider application of yeast in industrial bioprocesses in general.
To identify the genes involved in the isooctane tolerance of strain KK-211, mRNA differential display (DD) was used. DD is a powerful method for detecting genes which are differentially expressed among different cells or among cells under altered conditions (25). It was reported that the expression levels of some genes in organic-solvent-tolerant bacteria changed compared with those of their wild-type parents (5). It is also known that genes encoding efflux pumps of the transmembrane type are expressed at high levels in multidrug-resistant S. cerevisiae (6, 11); such genes sometimes correlate to organic-solvent tolerance in some bacteria (3, 19, 24). It is, therefore, reasonable and effective to use DD for detecting the genes associated with organic-solvent tolerance in strain KK-211.
In this work, the genes identified are classified into three groups according to their expression patterns, and a possible mechanism for the organic-solvent tolerance in strain KK-221 is proposed.
MATERIALS AND METHODS
Strains and media.
Baker's yeast (S. cerevisiae [no sporulation, unknown ploidy]; purchased from Oriental Yeast Co., Tokyo, Japan) was precultivated in 10 ml of YPD liquid medium (1% [wt/vol] Bacto Yeast Extract [Difco Laboratories, Detroit, Mich.], 2% [wt/vol] Bacto Peptone [Difco], 2% [wt/vol] glucose) overnight at 30°C. Precultivated cells were streaked on YPD plate medium and incubated for 2 days at 30°C; then single colonies were picked up and incubated on a YPD plate overlaid with isooctane to a thickness of 5 mm for 3 days at 30°C. During incubation, the plates were sealed with Parafilm to prevent evaporation of isooctane. Strain DY-1 was one of those that could not form any colonies under such conditions and was used in this study as a control strain. Strain KK-211 was one of six colonies spontaneously isolated. These strains, obtained from baker's yeast, proliferated and leaked from the immobilized support by entrapment into the medium during the intermittent cultivation for repeated reduction of n-butyl 3-oxobutanoate in isooctane in our laboratory (22), and showed the highest tolerance in the medium containing isooctane. The strains might appear during long-term adaptation (50 days of repeated use) in the organic solvent without treatment by mutagens. The properties of the strains, including morphology, were the same as those of the parental baker's yeast. The difference between the strains was confirmed to be only the isooctane tolerance phenotype. Strain KK-211 could form many colonies within 2 days on a YPD plate overlaid with isooctane. In DNA subcloning, E. coli DH5α [F− endA1 hsdR17(rK− mK+) supE44 thi-1 λ− recA1 gyrA96 ΔlacU169 (φ80dlacZΔM15)] was used as the host for transformation. E. coli was grown in Luria-Bertani (LB) medium (1% [wt/vol] tryptone peptone (Difco), 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] sodium chloride).
Growth in organic-solvent-containing medium.
Yeast cells grown overnight in 10 ml of YPD medium were harvested, washed with sterilized water, and transferred to 50 ml of fresh YPD medium that was either overlaid with 50 ml of hydrophobic organic solvents (n-dodecane, n-decane, n-nonane, isooctane, cyclooctane, diphenyl ether, and n-hexane) or mixed with 5 and 10% (vol/vol) hydrophilic organic solvents (dimethyl sulfoxide [DMSO] and ethanol, respectively) to give a final optical density at 600 nm (OD600) of 0.1, and the yeast cells were further incubated with shaking at 30°C. When a synthetic medium was used, 2% (vol/vol) isooctane was added instead of glucose as the sole carbon source. Cell growth was monitored by measuring the turbidity (OD600) of the cultures.
Adhesion of cells to isooctane droplets.
Yeast cells cultivated overnight from 10 ml of culture broth were centrifuged at 3,000 × g for 1 min, rinsed with sterilized water, and resuspended in 3 ml of sterilized water. The cell suspension (100 μl) mixed with sterilized water (700 μl) was vigorously mixed with 200 μl of isooctane for 1 min and immediately observed under a microscope.
RNA preparation.
Total RNA was isolated according to the method of Kaiser et al. (21) from baker's yeast cultivated without isooctane (DY-1/N) and from strain KK-211 cultivated without isooctane (KK-211/N) and with isooctane (KK-211/I). The final pellet was resuspended in 0.1% diethylpyrocarbonate-treated H2O to make 1-μg/μl total-RNA solutions. To avoid contamination by genomic DNA, total-RNA solutions were treated with RNase-free DNase I (Gibco BRL, Gaithersburg, Md.) in the presence of recombinant RNase inhibitor (TOYOBO, Osaka, Japan). Degradation of genomic DNA in the total-RNA solution was confirmed by PCR (data not shown). The DNA-free total-RNA solutions were used for reverse transcription. All solutions and equipment were treated to ensure that they were RNase free.
Reverse transcription of RNA.
The total RNAs from the DY-1/N, KK-211/N, and KK-211/I strains were reverse transcribed using SuperScriptII RNase H− reverse transcriptase (GIBCO-BRL) and anchor primers as shown in the manufacturer's manual, except that 30 pmol of the anchor primer was used. Altogether, 30 distinct cDNA pools were synthesized by 10 types of anchor primers (T12AM, T12GA, T12GG, T12CG, and GT15N, where M represents A, T, G, or C and N represents A, G, or C) for each RNA sample, and these cDNA pools were directly used as templates for differential display PCR (DD-PCR).
mRNA DD and DNA subcloning.
DD-PCRs were individually performed with 117 sets of anchor primers and arbitrary primers in the presence of 83 μM [α-32P]dCTP (6,000 Ci/mmol) (Amersham Pharmacia Biotech, Uppsala, Sweden) and 1 U of AmpliTaq DNA polymerase (Perkin-Elmer, Branchburg, N.J.) for 40 cycles (94°C for 30 s, 40°C for 2 min, and 72°C for 30 s) followed by an additional extension at 72°C for 5 min as previously described by Liang and colleagues (25, 26). In every set of primers, one was from 10 3′ anchor primers used for the reverse transcription and another was from 21 5′ arbitrary primers as shown in Table 1. The radiolabeled PCR products were separated on a 6% polyacrylamide gel containing 16.7 M urea and were visualized by autoradiography. To verify the reproducibility of the bands, the analysis was repeated on newly isolated RNA using those primer sets that generated differentially expressed cDNA patterns in the primary analysis.
TABLE 1.
Sequences of the 21 arbitrary primers used in DD
| Primer name | Sequence |
|---|---|
| AP2 | 5′-CTTCGACTGT-3′ |
| AP3 | 5′-CTTTGGTCAG-3′ |
| AP4 | 5′-AGTATTCCAC-3′ |
| AP11 | 5′-CTTCGGGTAA-3′ |
| AP14 | 5′-AATGGGCTGA-3′ |
| ARP01 | 5′-TCGAATACGC-3′ |
| ARP02 | 5′-AATATCCTGG-3′ |
| ARP03 | 5′-CCGGACAAGT-3′ |
| ARP04 | 5′-TGCGAGTAAT-3′ |
| ARP05 | 5′-ATGAAACTCC-3′ |
| ARP06 | 5′-GACCATATCG-3′ |
| Ltk3 | 5′-CTTGATTGCC-3′ |
| Ldd1 | 5′-CTGATCCATG-3′ |
| D5 | 5′-GGAACCAATC-3′ |
| D13 | 5′-GTTTTCGCAG-3′ |
| UTR11 | 5′-GTACCTGCATTTA-3′ |
| UTR12 | 5′-CATTGTTATATTA-3′ |
| OPA16 | 5′-AGCCAGCGAA-3′ |
| OPA17 | 5′-GACCGCTTGT-3′ |
| OPA19 | 5′-CAAACGTCGG-3′ |
| OPA20 | 5′-GTTGCGATCC-3′ |
After the X-ray film (Fuji Film Co., Tokyo, Japan) was developed, the cDNA bands of interest were cut off and cDNAs were recovered from the dried gel. After the bands that had been cut off were soaked in 100 μl of H2O at room temperature for 10 min, they were boiled for 15 min. The solution was centrifuged and, from the supernatant obtained, cDNAs were recovered as precipitates by using ethanol. The cDNAs recovered were further reamplified in 40 μl of reaction mixture with the same primer set and PCR conditions used in the mRNA DD, except that 50 μM deoxynucleoside triphosphates were added instead of the labeled compound. The reamplified cDNA fragments were subcloned into the pT7Blue2 T-vector (Novagen, Madison, Wis.). DNA sequencing of the subcloned cDNA fragments was carried out by a model 373A DNA sequencing system (Perkin-Elmer Applied Biosystems, Foster City, Calif.) with the universal primer set. Homology searches against databases were performed using the BLAST program with the S. cerevisiae genome database.
Northern blot analysis and quantitation.
Northern blot analysis was carried out with 20 μg of total RNA used in DD. Total RNA (20 μg) was separated by electrophoresis on a formaldehyde-denatured 1.2% agarose gel and blotted onto a positively charged nylon membrane (Boehringer GmbH, Mannheim, Germany). Cross-linking of mRNA to the membrane was achieved with a GS GENE LINKER (Bio-Rad, Richmond, Calif.) at 150 mJ. mRNA was detected with an Alkphos Direct Labelling and Detection Kit (Amersham Pharmacia Biotech) using CDP-Star as the substrate. About 400 bp of the coding region, amplified from the genomic DNA of strain KK-211 by PCR, was used as the probe. The band intensities on blots, corresponding to the mRNA levels, were quantified using NIH Image 1.61. Data obtained by Northern blot analyses were captured into a Macintosh computer (Apple Computers, Cupertino, Calif.) by a ScanJetII scanner (Hewlett-Packard, Palo Alto, Calif.) and analyzed by the public-domain NIH Image program (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/). The resulting values were normalized against that of ACT1. Since the levels of ACT1 mRNA were almost the same under each condition used (data not shown), the band intensity of internal ACT1 mRNA in each cell was used for normalization of the levels of various mRNAs in the cells under different conditions.
RESULTS
Growth of strain KK-211 in hydrophobic and hydrophilic organic-solvent-containing media.
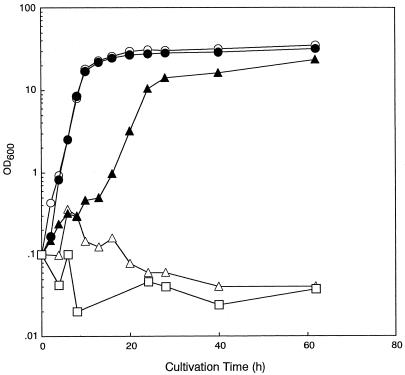
The organic-solvent tolerance of yeast strains DY-1 and KK-211 was investigated by measuring their growth in organic-solvent-containing media after precultivation. n-Dodecane (logPow = 7.0), n-decane (logPow = 6.0), n-nonane (logPow = 5.5), isooctane (logPow = 4.8), cyclooctane (logPow = 4.5), diphenyl ether (logPow = 4.2), and n-hexane (logPow = 3.9) were selected as the hydrophobic organic solvents, and DMSO and ethanol were selected as the hydrophilic organic solvents. Strain DY-1 could not grow in the n-nonane (Table 2)- or isooctane (Fig. 1)-containing medium but showed very slow growth in the presence of n-decane (Table 2). Thus, the index solvent of this strain seemed to be n-decane, and therefore, the DY-1 strain showed slightly higher organic-solvent tolerance than some experimental haploid strains, for which the index solvent was reported to be n-dodecane (logPow = 7.0) (14). On the other hand, strain KK-211 could grow in the presence of isooctane, although the growth was slow (Fig. 1). The index solvent of strain KK-211 seemed to be isooctane, because growth of strain KK-211 in the present of diphenyl ether and cyclooctane was extremely interrupted. The question of whether or not strain KK-211 could assimilate isooctane as the carbon source was also examined. Strain KK-211 could not grow in the synthetic medium containing isooctane as the sole carbon source (Fig. 1), confirming that strain KK-211 was not an isooctane-assimilating strain but an isooctane-tolerant strain.
TABLE 2.
Growth of strains DY-1 and KK-211 in YPD medium containing organic solvents
| Strain and organic solvent | Growth ratea (OD600/h) | Growth yieldb (OD600) |
|---|---|---|
| DY-1 | ||
| None | 2.0 | 30 |
| 50% (vol/vol) | ||
| n-Dodecane | 1.0 | 20 |
| n-Decane | 0.001 | 1 |
| n-Nonane | 0 | 0 |
| 10% (vol/vol) | ||
| DMSO | 0.1 | 4 |
| Ethanol | 0.02 | 4 |
| KK-211 | ||
| None | 2.0 | 30 |
| 50% (vol/vol) | ||
| n-Nonane | 0.5 | 10 |
| Diphenyl ether | 0 | 0 |
| Cyclooctane | 0 | 0 |
| n-Hexane | 0 | 0 |
| 10% (vol/vol) | ||
| DMSO | 0.03 | 2 |
| Ethanol | 0 | 0 |
At the mid-exponential phase.
At the stationary phase.
FIG. 1.
Growth of strains DY-1 and KK-211 cultivated in YPD liquid medium containing isooctane. DY-1 (○ and ▵) and KK-211 (●, ▴, and □) were precultured overnight in YPD medium and inoculated into YPD liquid medium both without isooctane (○ and ●) and with isooctane (▵ and ▴), and into a synthetic medium which used isooctane as the sole carbon source (□). The initial OD600 was 0.1.
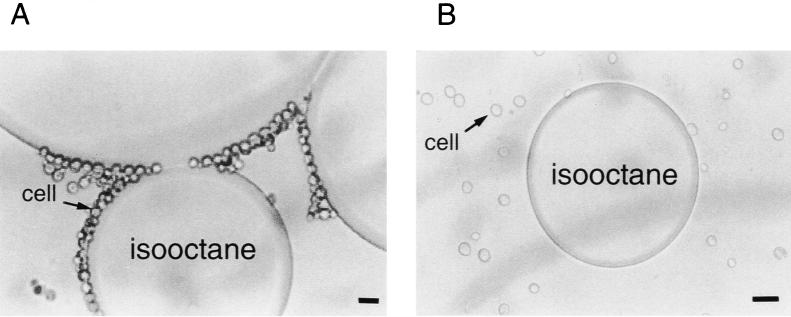
Observation of cell growth in the presence of hydrophilic organic solvents, DMSO or ethanol, would give us valuable information. Both of these solvents inhibited the growth of yeast strains in a concentration-dependent manner (data not shown). Surprisingly, the tolerance of strain DY-1 to DMSO or ethanol was apparently higher than that of strain KK-211, although the tolerance of strain DY-1 to hydrophobic organic solvents was lower than that of strain KK-211. These results were unexpected, suggesting that cell surface properties might be different in these two yeast strains. To verify this prediction, cell surface affinity to the hydrophobic organic solvent isooctane was investigated by observing the adherence of the cells to isooctane droplets in the aqueous phase as previously performed by Aono and Kobayashi (4). Isooctane was emulsified by vigorous mixing in a suspension of yeast cells. Most of the DY-1 cells adhered to isooctane droplets (Fig. 2), while the KK-211 cells rarely adhered. These results, in addition to the growth characteristics in hydrophilic organic solvents like DMSO and ethanol, suggested that the cell surface of strain KK-211 is more hydrophilic than that of strain DY-1, so that isooctane cannot penetrate through the cellular membrane or into the intracellular space, although there is some possibility that changes in cell surface structures or the functions of certain cell surface proteins might contribute to isooctane tolerance in either of the two strains.
FIG. 2.
Adhesion of DY-1 (A) and KK-211 (B) cells to isooctane droplets. Overnight-cultured cells were added to sterilized water containing isooctane, then agitated vigorously with a vortex mixer and observed with a microscope. Bar, 10 μm.
Identification of the specific genes for organic-solvent tolerance by the DD method.
Strain KK-211 was incapable of sporulation, and its ploidy was not apparent, which made it difficult for us to identify the organic-solvent tolerance-associated genes by genetic methods. In this study, the mRNA DD technique was used in an attempt to isolate genes that confer higher organic-solvent tolerance on normal yeast strains.
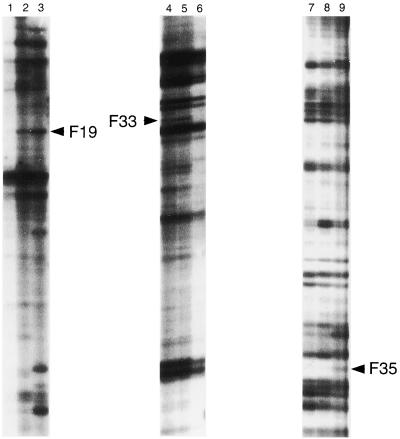
Total RNAs were isolated from three types of cells, strain KK-211 cultivated in YPD medium without isooctane (KK-211/N) and with 50% (vol/vol) isooctane (KK-211/I) and, as a control, strain DY-1 cultivated in YPD medium without isooctane (DY-1/N) at 30°C. Since genes responsible for or associated with organic-solvent stress might be considered to be induced at early-exponential phase, the three types of total RNAs used in the DD method were obtained from cells cultivated for 20 h, because strain KK-211 began to grow at this cultivation time in isooctane-containing medium, although strain DY-1 showed no growth (Fig. 1). We used 10 anchor primers to increase the variety of the cDNA pool. DD-PCR was performed using 16 arbitrary primers as shown in Table 1. DD-PCRs were performed with 117 combinations of anchor primers and arbitrary primers on three types of cDNA templates (Fig. 3). The fingerprints obtained exhibited 30 to 50 bands in each lane (DY-1/N, KK-211/N, and KK-211/I). When band patterns in the three lanes were compared, almost all of the bands were found to be constitutively expressed in each lane, while several bands showed different expression patterns. These different patterns were classified into three groups. Bands which appeared preferentially and strongly in KK-211/I were classified as belonging to group I, and those which appeared in both KK-211/N and KK-211/I were assigned to group II. Group III contained bands which showed other patterns, for example, bands which appeared only in DY-1/N or in both DY-1/N and KK-211/N. After 351 DD-PCRs, 38 differentially displayed bands were detected. Among these, 11 bands were classified as belonging to group I, 8 bands as belonging to group II, and 19 bands as belonging to group III.
FIG. 3.
Representative mRNA DD fingerprints generated from PCR amplification with different primer sets as shown in Table 1. Total RNAs were isolated from DY-1/N (lanes 1, 4, and 7), KK-211/N (lanes 2, 5, and 8), and KK-211/I (lanes 3, 6, and 9) cells cultivated for 20 h at 30°C and were then analyzed by the DD method. Samples were loaded in sets of three according to the three cell types (DY-1, KK-211/N, and KK-211/I) for each primer set. cDNA bands that appeared differentially among the three samples and were subsequently isolated (F19, F33, and F35) are indicated by arrowheads.
Sequencing of subcloned gene fragments and their identification by the S. cerevisiae genome database revealed that 30 genes might be expressed differentially between KK-211 and DY-1. As shown in Table 3, some stress-responsive genes were involved. For example, GAC1, which is induced by osmotic stress, has an STRE element in its promoter region. PRY1 and PRY3, belonging to group III, and PRY2, belonging to group II, are also interesting because, although the expression patterns of these genes are different, the amino acid sequences of proteins encoded by PRY1, PRY2, and PRY3 are found to be 60% identical. KRE1 and MUQ1 encode proteins that have roles in the maintenance of the cell wall and plasma membrane, respectively.
TABLE 3.
Sequence analysis of cDNAs differentially expressed among three types of cells
| Clone name | Primer set (anchor/arbitrary) | ORFa | Gene productb | SGDc accession no. | Length of cDNA obtained (bp) |
|---|---|---|---|---|---|
| Group I | |||||
| F2 | GT15A/ARP04 | GAC1 | Regulatory subunit of glycogen synthesis | S0005704 | 191 |
| F7 | GT15A/ARP04 | SPA2 | Spindle pole antigen | S0003944 | 150 |
| F8 | GT15A/ARP04 | NFS1 | Unknown | S0000522 | 257 |
| F16 | GT15A/ARP03 | PRY2 | Unknown | S0001721 | 129 |
| F17 | T12AG/D13 | YOL027c | Unknown | S0005387 | 150 |
| F27 | GT15C/UTR12 | HHT2 | Histone H3 | S0004976 | 477 |
| F60 | T12AA/D5 | MUQ1 | Choline phosphate cytidylyltransferase | S0003239 | 415 |
| F68 | GT15G/Ltk3 | AGT1 | α-Glucoside transporter | S0003521 | 617 |
| Group II | |||||
| F1 | GT15A/ARP04 | YNL190w | Unknown | S0005134 | 300 |
| F9 | GT15A/ARP03 | HAC1 | Basic-leucine zipper protein | S0001863 | 542 |
| F19 | T12AG/ARP01 | ICT1 | Unknown | S0004089 | 505 |
| F35 | T12GA/Ltk3 | YER041w | Unknown | S0000843 | 307 |
| F57 | GT15G/AP14 | FCY21 | Purine-cytosine permease | S0000862 | 200 |
| F58 | GT15G/AP14 | CHD1 | Transcriptional regulator | S0000966 | 200 |
| Group III | |||||
| F24 | T12GA/ARP02 | GCN4 | Transcriptional activator of amino acid biosynthetic genes | S0000735 | 479 |
| F26 | T12GA/ARP02 | KAP123 | Karyopherin 4 | S0000912 | 419 |
| F28 | GT15G/Ltk3 | UBP13 | Ubiquitin carboxyl-terminal hydrolase | S0000163 | 173 |
| F29 | GT15G/OPA20 | SDS24 | Unknown | S0000418 | 235 |
| F31 | GT15G/OPA20 | FLR1 | Major facilitator transporter | S0000212 | 138 |
| F32 | GT15G/OPA20 | PET8 | Member of family of mitochondrial carrier proteins | S0004948 | 138 |
| F33 | T12GA/ARP06 | KRE1 | Unknown | S0005266 | 439 |
| F34 | T12GA/ARP06 | VPS8 | Regulatory subunit in vesicular transport | S0000002 | 182 |
| F40 | GT15A/ARP04 | SAC6 | Fibrin homolog (actin-filament bundling protein) | S0002536 | 207 |
| F42 | GT15A/ARP04 | GLN1 | Glutamine synthetase | S0006239 | 144 |
| F45 | GT15A/ARP03 | ATP9 | F0-ATP synthetase subunit 9 | S0002981 | 271 |
| F46 | T12AG/ARP01 | PCK1 | Phosphoenolpyruvate carboxylkinase | S0001805 | 434 |
| F48 | T12AG/ARP01 | PRY3 | Unknown | S0003614 | 180 |
| F50 | GT15A/ARP04 | YBR077c | Unknown | S0000281 | 383 |
| F51 | T12AC/UTR11 | JEN1 | Carboxylic acid transporter protein homolog | S0001700 | 370 |
| F69 | T12AC/ARP01 | PRY1 | Unknown | S0003615 | 299 |
ORF, open reading frame. ORFs were determined by comparison with the S. cerevisiae genome database. The sequences of cDNAs obtained exhibit more than 90% nucleotide sequence identity with those of ORFs in the SGD.
Gene products are listed according to the SGD and the yeast protein database (YPD).
SGD, Saccharomyces cerevisiae genome database.
Confirmation of gene expression patterns by Northern blot analysis.
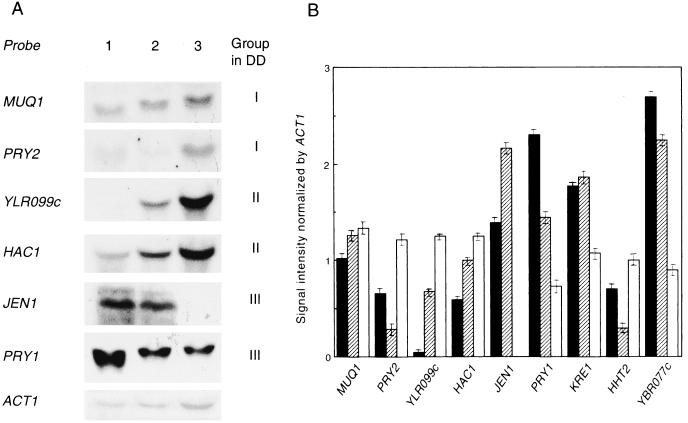
The DD method is a convenient method for detecting the genes that are expressed differently under different conditions, but the accuracy for its quantification is not sufficient. Northern blot analysis was therefore performed to quantitatively confirm the expression patterns of the genes screened by the DD method (Fig. 4). By Northern blot analysis using the same lot of total RNA used in the DD method, the expression patterns of nine genes were the same as those detected in the DD fingerprint. For example, the expression of PRY1, KRE1, and JEN1 was reduced to 30, 60, and 0%, respectively, in KK-211/I. In contrast, the expression of HAC1, PRY2, and ICT1 (YLR099c) in KK-211/I was 2.1-, 1.8-, and 24.5-fold higher, respectively, than that in DY-1/N. Thus, it seems that the isooctane-tolerant phenotype of strain KK-211 may result from the alteration of the expression of several genes due to the loss of their proper regulation.
FIG. 4.
Confirmation of organic-solvent-responsive gene expression. (A) Representative results of Northern blot analysis. Total RNA used in the DD analysis was subjected to denaturing gel electrophoresis. Northern blotting was carried out by hybridization with phosphatase-conjugated probes, and bands were detected by luminescence reaction. ACT1 mRNA hybridized with the ACT1 probe was used as the internal control in each cell. Lanes: 1, DY-1/N; 2, KK-211/N; 3, KK-211/I. (B) Quantification of signal intensity. Quantification of the mRNA levels and normalization using the ACT1 signal were carried out as described in Materials and Methods. Three independent experiments were performed, and the data obtained are shown as means ± standard errors (n = 3). Solid bar, DY-1/N; striped bar, KK-211/N; open bar, KK-211/I.
DISCUSSION
The isooctane-tolerant S. cerevisiae strain KK-211 was isolated from commercial dry yeast that was utilized in isooctane as an immobilized living biocatalyst for long-term (more than 50 days) stereoselective reduction of ethyl 3-oxobutanoate to ethyl (S)-3-hydroxybutanoate. Strain KK-211 is the first isooctane-tolerant strain of yeast. Recently, highly organic-solvent-tolerant bacteria, E. coli strain K-12, P. aeruginosa, and P. putida, have been studied extensively to reveal the tolerance mechanism. Many kinds of phenomena have been reported as the tolerance mechanisms in these bacteria, for example, the existence of some cell surface proteins that act as an efflux pump of hydrophobic compounds (18), modification of lipopolysaccharides (29), an increased proportion of saturated fatty acids in the plasma membrane (16, 29), and a lack of OprF porin (23). Almost all of these seem to be related to different expressions of some genes. Thus, we predicted that genes encoding the proteins associated with organic-solvent stress in strain KK-211 could be detected by the DD method, both in cases where such genes are overexpressed and in cases where they are defective in strain KK-211.
In this study, DD-PCRs were performed on DY-1/N, KK-211/N, and KK-211/I using 117 combinations of anchor primers and arbitrary primers, and the resulting 38 bands appeared differentially. These bands were classified into three groups according to their band patterns. Groups I and II contained the genes strongly expressed in strain KK-211 in the presence or absence of isooctane, respectively. The expression of genes belonging to group III was repressed in strain KK-211. It was predictable that many bands were classified into group III, because strain KK-211 was isolated from dry yeast that was used for long-term bioreaction in isooctane and might have some mutations to repress the expression of several genes which were not associated with isooctane tolerance. In contrast, group I possibly comprises genes that were induced only by organic solvents or by the associated stress.
As shown in Table 3, GAC1 and AGT1 were classified as group I genes. GAC1 encodes a protein phosphatase that forms a holoenzyme with Glc7p to control glycogen accumulation by dephosphorylation of glycogen synthase (Gsy1p or Gsy2p) (32) and is induced by heat shock and osmotic stress (28). AGT1 encodes an α-glucoside transporter of plasma membrane. Agt1p can transport many kinds of hexose substrates (maltose, turanose, isomaltose, trehalose, etc.), especially trehalose, with high affinity, across the plasma membrane (30, 37). These two compounds, glycogen and trehalose, are known to protect the cell from heat shock stress, osmotic stress, and salt stress, and they are called compatible solutes (31, 36). Therefore, these compatible solutes, glycogen and trehalose, may play important roles in maintaining the functions of enzymes against organic-solvent stress or by increasing the hydrophilicity of the inner side of the plasma membrane by localizing along the plasma membrane. HAC1 belongs to group II and encodes a basic-leucine zipper transcriptional activator of KAR2 in the unfolded protein response (UPR) pathway (8), which is indirectly required to protect cells from stress in the endoplasmic reticulum (9). When organic solvents penetrate into the intracellular space, it may affect the proper function of enzymes or destroy the conformation of functional enzymes by hydrophobic interactions. Thus, Hac1p may be necessary for the maintenance of protein function against the invading organic solvent.
Considering the different cell surface properties in strains DY-1 and KK-211 as shown in Fig. 2, MUQ1, SDS24, ICT1 (YLR099c), and KRE1 are interesting. The expression of MUQ1, a group I gene, was increased in KK-211/I. Muq1p is a transferase that converts ethanolamine phosphate to CDP-ethanolamine in the second step of the ethanolamine branch of the Kennedy pathway for synthesis of phosphatidylethanolamine (PtdEtn) (10). Overproduction of this protein may lead to an increase in the rate of PtdEtn in the phospholipid of the plasma membrane, which may in turn contribute to a more hydrophilic cell surface in strain KK-211 (as observed in its reduced adhesion to isooctane droplets [Fig. 2]) and, as a result, decrease the permeability of the cell membrane toward organic solvents. SDS24, the expression of which is repressed in KK-211/I, is also noteworthy. Sds24p, which has an unknown function, is homologous to stearoyl-coenzyme A desaturase (delta-9 fatty acid desaturase) of S. cerevisiae, which is required for synthesis of unsaturated fatty acids (Ole1p) (7). There is some possibility that the decreased expression of this gene causes the increased ratio of saturated to unsaturated fatty acids. This prediction is sustained by the fact that the ratio of saturated fatty acids in the phospholipid bilayer of KK-211/I cells was increased compared with that in DY-1/N cells (data not shown). It is therefore thought that the increased ratio of saturated fatty acids in the plasma membrane resulted in the stabilization of the cell membrane and could compensate for the effect caused by organic solvents, as reported for organic-solvent-tolerant bacteria (16, 38). ICT1 (YLR099c) belongs to group II, and Northern blot analysis confirmed that it was 24.5-fold induced in KK-211/I cells. This gene also encodes a protein of unknown function that has a hypothetical serine active site, such as is found in lipase or serine protease. Disruption of the gene results in elevated sensitivity to the cell surface-perturbing reagent calcofluor white (12). This suggests that the gene product of YLR099c particularly affects the cell surface properties of strain KK-211. From the results of both DD and Northern blot analysis, changes in the expression levels of some genes seem to be important factors for organic-solvent tolerance in strain KK-211. It seems that the altered expression levels of multiple genes, not a single gene, correlate to organic-solvent tolerance in strain KK-211.
Until strain KK-211 was isolated, no organic-solvent-tolerant yeast strain was reported. This strain will thus provide a considerable amount of information that may help in understanding the stress response mechanism in eukaryotic microorganisms, which may be different from that in organic-solvent-tolerant bacteria. Moreover, elucidation of the tolerance mechanism in this strain will contribute to the extension of the use of yeast in bioprocesses for the production of fine chemicals, agricultural chemicals, and pharmaceuticals, since living yeast has been utilized only in the water phase.
REFERENCES
- 1.Aono R, Aibe K, Inoue A, Horikoshi K. Preparation of organic solvent-tolerant mutants from Escherichia coli K-12. Agric Biol Chem. 1991;55:1935–1938. [Google Scholar]
- 2.Aono R, Ito M, Horikoshi K. Isolation of novel toluene-tolerant strain of Pseudomonas aeruginosa. Biosci Biotechnol Biochem. 1992;56:145–146. [Google Scholar]
- 3.Aono R, Kobayashi M, Nakajima H, Kobayashi H. A close correlation between improvement of organic solvent tolerance levels and alteration of resistance toward low levels of multiple antibiotics in Escherichia coli. Biosci Biotechnol Biochem. 1995;59:213–218. doi: 10.1271/bbb.59.213. [DOI] [PubMed] [Google Scholar]
- 4.Aono R, Kobayashi H. Cell surface properties of organic solvent-tolerant mutants of Escherichia coli K-12. Appl Environ Microbiol. 1997;63:3637–3642. doi: 10.1128/aem.63.9.3637-3642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asako H, Nakajima H, Kobayashi K, Kobayashi M, Aono R. Organic solvent tolerance and antibiotic resistance increased by overexpression of marA in Escherichia coli. Appl Environ Microbiol. 1997;63:1428–1433. doi: 10.1128/aem.63.4.1428-1433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balzi E, Goffeau A. Yeast multidrug resistance: the PDR network. J Bioenerg Biomembr. 1995;27:71–76. doi: 10.1007/BF02110333. [DOI] [PubMed] [Google Scholar]
- 7.Carratu L, Franceschelli S, Pardini C L, Kobayashi G S, Horvath I, Vigh L, Maresca B. Membrane lipid perturbation modifies the set point of the temperature of heat shock response in yeast. Proc Natl Acad Sci USA. 1996;93:3870–3875. doi: 10.1073/pnas.93.9.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman R E, Walter P. Translational attenuation mediated by an mRNA intron. Curr Biol. 1997;7:850–859. doi: 10.1016/s0960-9822(06)00373-3. [DOI] [PubMed] [Google Scholar]
- 9.Cox J S, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 10.Daum G, Lees N D, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 11.Decottignies A, Grant A M, Nichols J W, de Wet H, McIntosh D B, Goffeau A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J Biol Chem. 1998;273:12612–12622. doi: 10.1074/jbc.273.20.12612. [DOI] [PubMed] [Google Scholar]
- 12.Entian K D, Schuster T, Hegemann J H, Becher D, Feldmann H, Guldener U, Gotz R, Hansen M, Hollenberg C P, Jansen G, Kramer W, Klein S, Kotter P, Kricke J, Launhardt H, Mannhaupt G, Maierl A, Meyer P, Mewes W, Munder T, Niedenthal R K, Ramezani R M, Rohmer A, Romer A, Hinnen A. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol Gen Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- 13.Favre-Bulle O, Schouten T, Kingma J, Witholt B. Bioconversion of n-octane to octanoic acid by a recombinant Escherichia coli cultured in a two-liquid phase bioreactor. Bio/Technology. 1991;9:367–371. doi: 10.1038/nbt0491-367. [DOI] [PubMed] [Google Scholar]
- 14.Fukumaki T, Inoue A, Moriyama K, Horikoshi K. Isolation of a marine yeast that degrades hydrocarbon in the presence of organic solvent. Biosci Biotechnol Biochem. 1994;58:1784–1788. [Google Scholar]
- 15.Hansch C, Anderson S M. The effect of intramolecular hydrophobic bonding on partition coefficients. J Org Chem. 1967;32:2583–2586. [Google Scholar]
- 16.Ingram L O. Changes in lipid composition of Escherichia coli resulting from growth with organic solvents and with food additives. Appl Environ Microbiol. 1977;33:1233–1236. doi: 10.1128/aem.33.5.1233-1236.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue A, Horikoshi K. A Pseudomonas thrives in high concentration of toluene. Nature (London) 1989;338:264–265. [Google Scholar]
- 18.Isken S, de Bont J A M. Active efflux of toluene in a solvent-resistant bacterium. J Bacteriol. 1996;178:6056–6058. doi: 10.1128/jb.178.20.6056-6058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isken S, Santos P M A C, de Bont J A M. Effect of solvent adaptation on the antibiotic resistance in Pseudomonas putida S12. Appl Microbiol Biotechnol. 1997;48:642–647. [Google Scholar]
- 20.Isken S, de Bont J A M. Bacteria tolerant to organic solvent. Extremophiles. 1998;2:229–238. doi: 10.1007/s007920050065. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser C, Michaelis S, Mitchell A, editors. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1994. [Google Scholar]
- 22.Kanda T, Miyata N, Fukui T, Kawamoto T, Tanaka A. Doubly entrapped baker's yeast survives during the long-term stereoselective reduction of ethyl 3-oxobutanoate in an organic solvent. Appl Microbiol Biotechnol. 1998;49:377–381. doi: 10.1007/s002530051185. [DOI] [PubMed] [Google Scholar]
- 23.Li L, Komatsu T, Inoue A, Horikoshi K. A toluene-tolerant mutant of Pseudomonas aeruginosa lacking the outer membrane protein F. Biosci Biotechnol Biochem. 1995;59:2358–2359. doi: 10.1271/bbb.59.2358. [DOI] [PubMed] [Google Scholar]
- 24.Li X-Z, Poole K. Organic solvent-tolerant mutants of Pseudomonas aeruginosa display multiple antibiotic resistance. Can J Microbiol. 1999;45:18–22. doi: 10.1139/cjm-45-1-18. [DOI] [PubMed] [Google Scholar]
- 25.Liang P, Pardee A B. Differential display of eukaryotic mRNA by means of the polymerase chain reaction. Science (Washington, DC) 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 26.Liang P, Averboukh L, Pardee A B. Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res. 1993;21:3269–3275. doi: 10.1093/nar/21.14.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima H, Kobayashi H, Aono R, Horikoshi K. Effective isolation and identification of toluene-tolerant Pseudomonas strains. Biosci Biotechnol Biochem. 1992;56:1872–1873. [Google Scholar]
- 28.Parrou J L, Teste M A, Francois J. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology. 1997;143:1891–1900. doi: 10.1099/00221287-143-6-1891. [DOI] [PubMed] [Google Scholar]
- 29.Pinkart H C, Wolfram J W, Rogers R, White D C. Cell envelope changes in solvent-tolerant and solvent-sensitive Pseudomonas putida strain following exposure to o-xylene. Appl Environ Microbiol. 1996;62:1129–1132. doi: 10.1128/aem.62.3.1129-1132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plourde-Owobi L, Durner S, Parrou J L, Wieczorke R, Goma G, Francois J. AGT1, encoding an α-glucoside transporter involved in uptake and intracellular accumulation of trehalose in Saccharomyces cerevisiae. J Bacteriol. 1999;181:3830–3832. doi: 10.1128/jb.181.12.3830-3832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poolman B, Glaasker E. Regulation of compatible solute accumulation in bacteria. Mol Microbiol. 1998;29:397–407. doi: 10.1046/j.1365-2958.1998.00875.x. [DOI] [PubMed] [Google Scholar]
- 32.Ramaswamy N T, Li L, Khalil M, Cannon J F. Regulation of yeast glycogen metabolism and sporulation by Glc7p protein phosphatase. Genetics. 1998;149:57–72. doi: 10.1093/genetics/149.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramos J L, Duque E, Rodriguez-Herva J J, Godoy P, Haidour A, Reyes F, Fernandez-Barrero A. Mechanism for solvent tolerance in bacteria. J Biol Chem. 1997;272:3887–3890. doi: 10.1074/jbc.272.7.3887. [DOI] [PubMed] [Google Scholar]
- 34.Sikkema J, de Bont J A M, Poolman B. Interactions of cyclic hydrocarbons with biological membranes. J Biol Chem. 1994;269:8022–8028. [PubMed] [Google Scholar]
- 35.Sikkema J, de Bont J A M, Poolman B. Mechanism of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer M A, Lindquist S. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol Cell. 1998;1:639–648. doi: 10.1016/s1097-2765(00)80064-7. [DOI] [PubMed] [Google Scholar]
- 37.Stambuk B U, da Silva M A, Panek A D, de Araujo P S. Active α-glucoside transport in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1999;170:105–110. doi: 10.1111/j.1574-6968.1999.tb13361.x. [DOI] [PubMed] [Google Scholar]
- 38.Weber F J, Isken S, de Bont J A M. cis/trans isomerization of fatty acids as a defence mechanism of Pseudomonas putida strains to toxic concentrations of toluene. Microbiology. 1994;140:2013–2017. doi: 10.1099/13500872-140-8-2013. [DOI] [PubMed] [Google Scholar]
- 39.White D G, Goldman J D, Demple B, Levy S B. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woldringh C L. Effect of toluene and phenethyl alcohol on the ultrastructure of Escherichia coli. J Bacteriol. 1973;114:1359–1361. doi: 10.1128/jb.114.3.1359-1361.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]