Abstract
The objective of this study was to determine the effect of high pressure (HP) on the inactivation of microbial contaminants in Cheddar cheese (Escherichia coli K-12, Staphylococcus aureus ATCC 6538, and Penicillium roqueforti IMI 297987). Initially, cheese slurries inoculated with E. coli, S. aureus, and P. roqueforti were used as a convenient means to define the effects of a range of pressures and temperatures on the viability of these microorganisms. Cheese slurries were subjected to pressures of 50 to 800 MPa for 20 min at temperatures of 10, 20, and 30°C. At 400 MPa, the viability of P. roqueforti in cheese slurry decreased by >2-log-unit cycles at 10°C and by 6-log-unit cycles at temperatures of 20 and 30°C. S. aureus and E. coli were not detected after HP treatments in cheese slurry of >600 MPa at 20°C and >400 MPa at 30°C, respectively. In addition to cell death, the presence of sublethally injured cells in HP-treated slurries was demonstrated by differential plating using nonselective agar incorporating salt or glucose. Kinetic experiments of HP inactivation demonstrated that increasing the pressure from 300 to 400 MPa resulted in a higher degree of inactivation than increasing the pressurization time from 0 to 60 min, indicating a greater antimicrobial impact of pressure. Selected conditions were subsequently tested on Cheddar cheese by adding the isolates to cheese milk and pressure treating the resultant cheeses at 100 to 500 MPa for 20 min at 20°C. The relative sensitivities of the isolates to HP in Cheddar cheese were similar to those observed in the cheese slurry, i.e., P. roqueforti was more sensitive than E. coli, which was more sensitive than S. aureus. The organisms were more sensitive to pressure in cheese than slurry, especially with E. coli. On comparison of the sensitivities of the microorganisms in a pH 5.3 phosphate buffer, cheese slurry, and Cheddar cheese, greatest sensitivity to HP was shown in the pH 5.3 phosphate buffer by S. aureus and P. roqueforti while greatest sensitivity to HP by E. coli was exhibited in Cheddar cheese. Therefore, the medium in which the microorganisms are treated is an important determinant of the level of inactivation observed.
Research into the application of high-pressure (HP) processing for food preservation began more than a century ago, when Hite (20) demonstrated that the shelf life of milk, fruit, and vegetables could be extended by HP treatment (20, 21). In contrast to thermal processing, the application of HP to food does not affect its taste, color, or flavor (18, 22). The pressures required to achieve microbial inactivation are high, usually in the range of 300 to 700 MPa, but as the pressure applied is uniform in all directions, the treated product remains intact. Consequently, products such as cheese can be treated with minimal effect on texture and consistency. The first commercial HP-treated products appeared on the market in 1991 in Japan (24), where HP processing is now used commercially for products such as jams, sauces, fruit juices, rice cakes, and desserts. Progress in the design of HP equipment has facilitated commercial semicontinuous HP processing of liquid and solid foods (10).
The mechanisms by which HP-induced inactivation of microorganisms takes place have not yet been fully elucidated, but HP is known to cause morphological, biochemical, and genetic alterations in vegetative microorganisms (9, 22). Generally, yeasts and molds are most sensitive to HP, gram-positive microorganisms are most resistant, possibly due to their cell wall structure, and gram-negative microorganisms are moderately sensitive (9). Bacterial spores are extremely resistant to HP and can survive pressures in excess of 1,000 MPa (9). The stage of growth of microorganisms is also important in determining sensitivity to HP, as log-phase cells are more sensitive than stationary-phase cells (9). Important parameters affecting HP inactivation of microorganisms in food include the constituents and physical conditions within the food, which may induce a protective effect against HP (8). Therefore, a direct extrapolation of data for microbial inactivation by HP obtained with buffer or physiological solutions to predict levels of inactivation in foodstuffs may give misleading results.
HP treatment of microorganisms may result in either no loss of viability, sublethal injury, or cell death, although the last may or may not be accompanied by cell lysis. Sublethal injury may occur at pressures lower than those at which cell death occurs (29). After treating E. coli in phosphate buffer at 270 MPa, Hauben et al. (17) found that 99.58% of the surviving population was comprised of injured cells.
The present study defines the effects of a range of pressures and temperatures on the viability of selected contaminants of Cheddar cheese and demonstrates the potential application of HP for controlling undesirable microorganisms in this product.
MATERIALS AND METHODS
Stock cultures.
The cultures used in this study were Escherichia coli K-12 (ATCC 29425; American Type Culture Collection, Rockville, Md.), Staphylococcus aureus ATCC 6538 (American Type Culture Collection), and Penicillium roqueforti IMI 297987 (International Mycological Institute, Egham, Surrey, England). These cultures were selected because they are well-defined typed strains which have been used in many previous laboratory investigations, including HP studies (17, 26).
Mold spores of P. roqueforti IMI 297987 were harvested following growth for 5 days at 25°C on malt extract agar (Oxide, Unipath Ltd., Basingstoke, Hampshire, England) with 20% sucrose in a 140-mm-diameter petri dish. Spores were released by washing cultures with sterile deionized water containing 0.05% Tween 20 (37), and the suspension was filtered through sterile glass wool to remove mycelial fragments (36). The spores were recovered by centrifugation at 10,120 × g for 15 min at 4°C. The spore pellet was suspended in sterile deionized water and stored at 4°C until required.
Preparation of cultures for addition to cheese curd.
Tryptone yeast phosphate broth was inoculated with E. coli K-12 and incubated at 30°C overnight. S. aureus ATCC 6538 was grown in Trypticase soy broth (Becton Dickinson and Co.) containing 0.6% yeast extract at 37°C overnight. Both cultures (3 ml) were centrifuged at 20,800 × g for 1 min, the supernatants were discarded, and the pellets were washed with maximum-recovery diluent (MRD; Oxoid). The washed pellets were resuspended in 2 ml of MRD. Four milliliters of the P. roqueforti spore suspension was centrifuged at 20,800 × g, and the pellet was resuspended in 4 ml of MRD.
Preparation of Cheddar cheese slurries.
Cheddar cheese curd chips prior to being salted were obtained from Wexford Creamery (Wexford, Ireland) and stored at −20°C until required. Starter bacteria, lactobacilli, and coliforms were enumerated as described by Ryan et al. (30) in the cheese curd chips. S. aureus organisms were enumerated on Baird Parker medium (Merck) plus egg yolk tellurite (Merck) after incubation at 37°C for 48 h, E. coli organisms were enumerated on violet red bile agar (VRBA) after incubation at 30°C for 24 h, and yeasts and molds were enumerated on yeast extract-glucose-chloramphenicol (Merck) agar after incubation at 25°C for 5 days.
Cheese slurries (pH 5.2 to 5.4) were prepared with 100 g of Cheddar cheese curd chips and 42 g of 10% (wt/vol) sterile saline solution (to achieve a salt-in-moisture content of 4.5% in the slurry). All three organisms were added together to the cheese slurry to expedite HP treatment and subsequent microbial assay. E. coli K-12 and S. aureus ATCC 6538 were added to achieve 107 CFU/g, and P. roqueforti IMI 297987 was added to achieve 106 CFU/g. Inoculated slurries were blended for 5 min in a sterile homogenizer (Waring Commercial, New Hartford, Conn.). Portions of cheese slurry (10 g) were then packed into vacuum pouches (20 by 30 cm; G. B. Miller and Sons, Bray, County Wicklow, Republic of Ireland) and vacuum packed. These were then placed in a second vacuum pouch and vacuum sealed to prevent contact between the hydrostatic pressurization fluid and the cheese. The slurry samples were stored at 4°C until they were treated with HP (less than 4 h was required for HP treatment of a full batch of samples).
HP treatment of inoculated Cheddar cheese slurries.
Inoculated Cheddar cheese slurry samples were pressurized in an HP rig (Stansted Fluid Power Ltd., Stansted, England), which has a 300-mm-deep chamber with a diameter of 37 mm. The pressurization medium used was 15% (vol/vol) castor oil in ethanol. Pressurization was carried out at pressures of 50, 100, 200, 300, 400, 500, 600, 700, and 800 MPa for 20 min at temperatures of 10, 20, and 30°C on two independent replicate slurry samples. Duplicate unpressurized control slurry samples were maintained at ambient temperature, as there would be little difference in the levels of growth detected following the short incubation time (20 min).
A range of pressures characteristic of commercial HP rigs was evaluated, and the narrow range of temperatures used was necessary to avoid undesirable changes in cheese structure and texture due to extremes of temperature; thus, temperatures applicable to cheese processing were used. Kinetic experiments to determine inactivation indicated a greater effect of pressure than of time, and thus a holding time of 20 min was used in all experiments.
Microbiological assay of the pressurized product.
Immediately following decompression, serial dilutions of the cheese slurry samples were prepared. E. coli K-12, S. aureus ATCC 6538, and P. roqueforti IMI 297987 organisms were enumerated as described above. The species-selective media used were determined to be suitable for enumerating individual species in the mixed microbial population and were effective in suppressing the background microflora present in the cheese curd chips. Each replicate cheese slurry sample was plated in duplicate.
Kinetic experiments to determine microbial inactivation by HP.
The decimal reduction time (D value) is the time required at a specified pressure to reduce the number of cells by 90%, and the Z value is the increase in pressure (in megapascals) needed to change the D value by 90%.
Pressure treatments were carried out at 300, 350, and 400 MPa for 0, 10, 20, 30, 40, 50, and 60 min at 20°C on two independent replicate slurry samples as described above. Microorganisms surviving HP treatment were enumerated as described above. The D values were calculated from the absolute values of the inverses of the slopes of the linear-regression equations by plotting log numbers of survivors versus time (3) at 300, 350, and 400 MPa at 20°C. The Z values were calculated from the absolute values of the inverses of the slopes of the linear-regression equations relating log units (D values) to pressure (4).
Strain-to-strain variation in HP sensitivity.
Variation in strain sensitivity to HP was examined by subjecting a range of strains of S. aureus, E. coli, and mold spores to HP treatment at 100 to 400 MPa in cheese slurries for 20 min at 20°C as described above. The extra strains of S. aureus and E. coli tested were isolated from commercial-smear-ripened cheeses, and the species were confirmed. Mold strains were isolated either from a commercial-smear-ripened cheese or from Cheddar cheese made at pilot scale. These molds were not confirmed as being P. roqueforti but were typical mold contaminants of cheese.
Effect of HP treatment on log-phase cells.
As the inactivation experiments described above used stationary-phase cells, log-phase cells of E. coli and S. aureus were also added to cheese slurries to compare inactivation rates of log- and stationary-phase cells. The cultures were grown to log phase, as determined by optical density measurement, inoculated into the slurries, and HP treated at 100 to 400 MPa for 20 min at 20°C as described previously.
Detection of sublethal injury.
In order to determine the level of sublethal injury, a differential plating technique using nonselective agar incorporating salt or glucose was employed. These are injury-selective media, as opposed to the species-selective media described above. Tryticase soy agar yeast extract (TSAYE) and TSAYE with salt incorporated were used for detection of unstressed and stressed E. coli (5% salt) and S. aureus (7% salt) (23). Yeast glucose [YG; yeast extract at 5.0 g/liter and d-(+)-glucose at 20.0 g/liter] agar and YG agar with the d-(+)-glucose level increased to 30% (2) were used to elucidate the stress response in P. roqueforti. Both noninjured and injured cells were able to form colonies on TSAYE and YG agar, whereas only noninjured cells formed colonies in the presence of salt or glucose. To enable the use of nonselective media, the slurries were pasteurized at 63.5°C for 30 min prior to the addition of the individual cultures to inactivate residual starter microflora, whose growth was suppressed on the species-selective media. In these experiments, individual addition of the microorganisms to the slurry was necessitated by the use of the nonselective differential plating media used for injury detection. Two independent replicate slurry samples of E. coli and P. roqueforti were HP treated (20°C for 20 min) at 300, 400, and 500 MPa, and those containing S. aureus were treated at 400, 500, 600, and 700 MPa. Following HP treatment, slurries were incubated at 8°C, at which temperature growth did not occur, and any increase in cell numbers could thus be attributed to recovery from sublethal injury. Following incubation at 8°C for 0, 24, and 72 h, HP-treated and control slurries were sampled and the numbers of injured cells which recovered were determined by using the nonselective differential plating technique. Samples were also plated on the species-selective media used previously to determine the influence of these media on the growth of sublethally injured cells.
Manufacture of cheese.
Miniature cheeses were produced by using a modification of the protocol of Shakeel-Ur-Rehman et al. (32). Pasteurized milk (800 ml) was added to six plastic centrifuge bottles (1 liter) and cooled to 31°C. The starter culture (1% inoculum of Lactococcus lactis subsp. cremoris 303) and the three microbial species under study were then added to the milk, which was ripened for 30 min. Following renneting and coagulum formation, the curd was cut and stirred. Curds and whey were then centrifuged at room temperature for 60 min at 5,442 × g in a Mistral 6,000 centrifuge (MSE Scientific Instruments, Sussex, England). The whey was drained, and the curds were held in the centrifuge bottles at 36°C until the pH decreased to 5.2 to 5.3, whereupon cheeses were inverted and recentrifuged at 5,442 × g for 20 min at room temperature. The remaining whey was then drained, and cheeses were brine salted (20% NaCl, 0.05% CaCl2 · 2H2O) for 100 min at room temperature, vacuum packed, and stored at 8°C.
Compositional analysis (pH, salt, and moisture) of the cheeses, determined the day after manufacture as described by Guinee et al. (16), gave results typical for Cheddar cheese (15). At least two independent replicate cheese trials (six cheeses per trial), produced on separate occasions, including the test microorganisms were carried out. Duplicate samples of cheese from each trial were HP treated at 100 to 500 MPa for 20 min at 20°C. Species-selective media, as described earlier, were used to enumerate the target organisms immediately after HP treatment in duplicate.
Microbial inactivation in buffer systems.
The test microorganisms were suspended in 0.06 M phosphate buffer (pH 5.3), and two independent replicates were treated at 100 to 500 MPa for 20 min at 20°C in order to compare inactivation ratios (the log of the number of cells detected following pressurization treatment to the total number of CFU in the control) with those obtained in the model cheese slurry system and in the miniature cheese.
RESULTS
Inactivation experiments with cheese slurry.
Analyses of the background microflora of the Cheddar cheese curd chips showed that starter bacteria and nonstarter lactic acid bacteria were present at 1.14 × 109 and 2.90 × 102 CFU/g, respectively, while yeasts, molds, coliforms, and S. aureus were not detected.
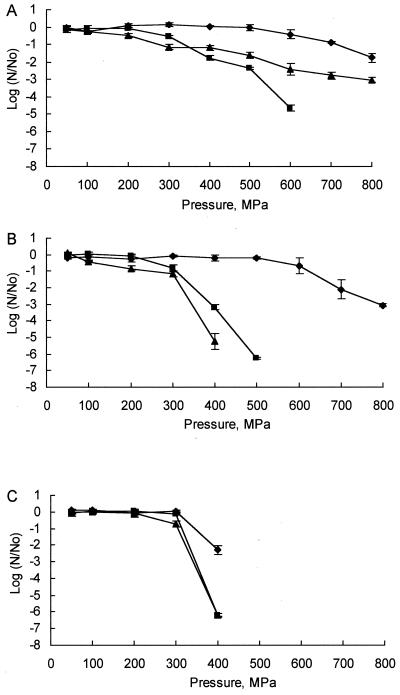
Results obtained for the inactivation of E. coli K-12, S. aureus ATCC 6538, and P. roqueforti IMI 297987 in the cheese slurry after HP treatment are shown in Fig. 1. The tested strains of both E. coli and S. aureus were remarkably resistant to pressures up to 500 MPa at 10°C. However, S. aureus showed a greater resistance, with a 1.5-log-unit cycle reduction in numbers at 800 MPa compared to a 3-log-unit cycle reduction in numbers of E. coli organisms at this pressure. Under these conditions, the mold species was more sensitive, with a >2-log-unit cycle reduction at 400 MPa and a >6-log-unit cycle reduction at 500 MPa. At 20°C, inactivation of E. coli and S. aureus became evident at 300 MPa, with total viable numbers of the former being reduced by >6-log-unit cycles at 500 MPa and of the latter being reduced by >4-log-unit cycles at 600 MPa. Under these conditions, numbers of the mold spores were reduced by 6-log-unit cycles at 400 MPa. At 30°C, the sensitivity of E. coli to HP increased, with inactivation becoming evident at 100 MPa and a 5-log-unit cycle reduction occurring at 400 MPa. At 30°C, however, S. aureus exhibited anomalous behavior, with an inactivation rate somewhere between that obtained at 10 and 20°C. An almost linear decline in cell numbers at pressures above 100 MPa culminated in a 3-log-unit cycle reduction at 800 MPa, indicating much greater resistance to HP by this organism at 30°C than at 20°C. P. roqueforti exhibited similar sensitivity to HP at 20 and 30°C.
FIG. 1.
Inactivation of S. aureus ATCC 6538 (A), E. coli K-12 (B), and P. roqueforti IMI 297987 (C) in Cheddar cheese slurry by HP treatment at 10°C (⧫), 20°C (■), and 30°C (▴) for 20 min (No is the total number of CFU in the control, and N is the number of cells detected following pressurization treatments). Each curve shows the mean values obtained from results from two independent replicates, and error bars represent standard deviations.
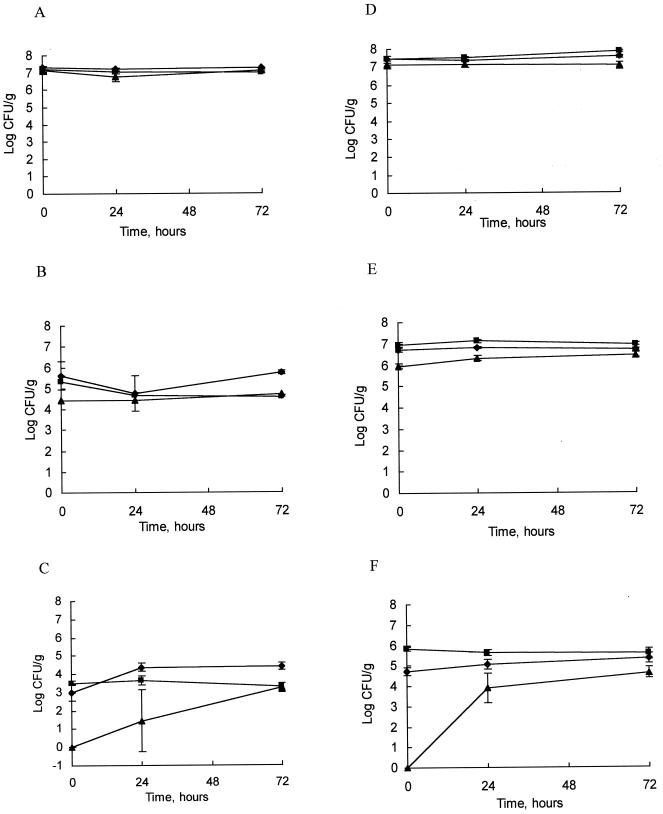
Sublethal injury and recovery of microorganisms after HP treatment.
Sublethal injury and recovery of S. aureus ATCC 6538 (Fig. 2A to C), E. coli K-12 (Fig. 2D to F), and P. roqueforti IMI 297989 (data not shown) after HP treatment at 20°C were investigated using injury-selective media. For control S. aureus samples, to which no pressure was applied, no difference was evident between growth on nonselective and growth on selective media (Fig. 2A). At 400 MPa, a >1-log-unit cycle of kill (indicated by the difference in levels obtained in Fig. 2A and B) and a further 1-log-unit cycle of injury (indicated by the difference in counts obtained on TSAYE and TSAYE plus salt) occurred, and slow recovery of injured cells was seen following incubation for 72 h (Fig. 2B). At 500 MPa there was a 3- to 4-log-unit cycle of kill, accompanied by 3-log-unit cycles of injury, with complete recovery following 72 h of incubation at 8°C (Fig. 2C). At 600 MPa there was a 7-log-unit cycle of kill on TSAYE, from which S. aureus did not recover following 72 h of incubation at 8°C, and on Baird Parker agar there was a 6-log-unit cycle of kill and a 1-log-unit cycle of injury, from which the organism did not recover after 72 h at 8°C (not shown). At 700 MPa the entire culture was killed, as no recovery was evident following incubation at 8°C (not shown). These results suggest that Baird Parker medium supported growth of noninjured and injured cells equally well; thus, all inactivation results obtained in the model cheese slurry system for S. aureus plated on this medium reflect actual kill.
FIG. 2.
Sublethal injury in Cheddar cheese slurry of S. aureus ATCC 6538 without pressurization (control) (A), at 400 MPa (B), and at 500 MPa (C) plated on Baird Parker agar (⧫), TSAYE (■), and TSAYE plus 7% salt (▴) and of E. coli K-12 without pressurization (D), at 300 MPa (E), and at 400 MPa (F) plated on VRBA (⧫), TSAYE (■), and TSAYE plus 5% salt (▴) (pressure treatments were carried out for 20 min at 20°C). Each curve shows the mean values obtained from results from two independent replicates, and error bars represent standard deviations.
Again, with E. coli control samples, there was no difference detected between growth on nonselective media and growth on selective media (Fig. 2D). At 300 MPa there were 0.5-log-unit cycles of kill accompanied by a 1-log-unit cycle of injured cells, which exhibited almost complete recovery following 72 h of incubation at 8°C (Fig. 2E). At 400 MPa there was a >1-log-unit cycle of kill accompanied by 6-log-unit cycles of injury, from which E. coli showed almost complete recovery after 24 h (Fig. 2F). At 500 MPa a >4-log-unit cycle of kill was evident, accompanied by 3-log-unit cycles of injury (not shown). Recovery in this case took longer but was almost complete after 72 h, although no recovery was evident on VRBA. These results suggest that the inactivation data generated for E. coli plated on the VRBA medium reflect killing as well as an element of sublethal injury.
P. roqueforti samples showed no difference in levels of growth on the three media used. There was no decline in cell numbers at 300 MPa, but viability decreased by 5-log-unit cycles at 400 MPa with 2-log-unit cycles of spore injury, from which the organism almost completely recovered following incubation at 8°C for 72 h (data not shown). At 500 MPa P. roqueforti was completely inactivated and did not recover following incubation at 8°C.
Kinetics of inactivation.
The rates of inactivation of the three test organisms at 20°C in Cheddar cheese slurry were studied (Table 1). S. aureus exhibited a linear rate of inactivation at the three pressures tested. Increasing the pressure from 300 to 400 MPa had a greater effect on inactivation than extending the pressurization time (not shown in Table 1), with the D values decreasing from 38 to 20 min over this pressure range. With the mold species, results obtained at the lower pressure (300 MPa) were very similar to those obtained with S. aureus, in that little effect of increasing pressurization time was evident (not shown), and a D value of 56 min was obtained at this pressure. However, at 350 and 400 MPa the effects of extending the pressurization time became significant and even greater in magnitude than were observed for S. aureus. The D value was reduced to 5 min after pressurization of P. roqueforti at 400 MPa. With E. coli at 300 and 350 MPa, the D values were 22 and 19 min, respectively, although the D value could not be calculated at 400 MPa as the inactivation curve was not linear at this pressure.
TABLE 1.
D values and Z values for S. aureus ATCC 6538, E. coli K-12, and P. roqueforti IMI 297987 following pressurization at 300, 350, and 400 MPa at 20°C
| Microorganism | D valuea after pressurization at:
|
Z valueb | ||
|---|---|---|---|---|
| 300 MPa | 350 MPa | 400 MPa | ||
| S. aureus ATCC 6538 | 38 | 33 | 20 | 359 |
| E. coli K-12 | 22 | 19 | NVc | NV |
| P. roqueforti IMI 297987 | 56 | 15 | 5 | 95 |
D values (in minutes) calculated from the absolute values of the inverse slopes from linear-regression equations between log-unit CFU/gram and time (3). Values shown were calculated from the means of results from two independent replicates.
Z values (in megapascals) calculated from the absolute values of the inverse slopes from linear-regression equations relating log-unit D value and pressure (4).
NV denotes no value obtained due to a nonlinear inactivation curve.
Comparison of the Z values for the mold and S. aureus species demonstrated much greater sensitivity of the former to HP.
Influence of strain variation on inactivation by HP treatment.
Pressure treatment at 100 to 200 MPa caused negligible inactivation of any of the three strains of S. aureus, E. coli, or mold species studied (data not shown). Inactivation increased for all strains at 300 MPa, where the greatest degree of interstrain variation was seen for the mold species (2-log-unit cycle difference between strains). However, as the viability of the mold spores was greatly reduced at 400 MPa, the degree of strain variation at this pressure was small (<1-log-unit cycle). The range of pressure sensitivity within strains of S. aureus and E. coli increased to a 2-log-unit cycle difference at 400 MPa.
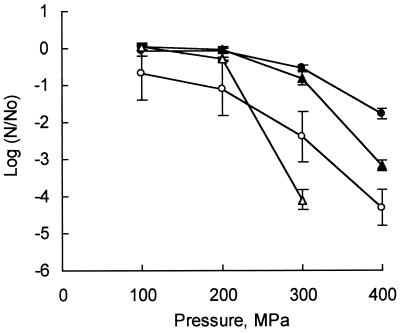
Effect of HP treatment on log-phase and stationary-phase cells.
Both S. aureus ATCC 6538 and E. coli K-12 cells in the log phase of growth were more sensitive to HP treatment in Cheddar cheese slurry than they were in stationary phase (Fig. 3). Log-phase S. aureus cells were more resistant than log-phase E. coli cells.
FIG. 3.
Inactivation of stationary-phase S. aureus ATCC 6538 (●) and E. coli K-12 (▴) cells and log-phase S. aureus ATCC 6538 (○) and E. coli K-12 (▵) cells in Cheddar cheese slurry by HP treatment at 20°C for 20 min (No is the total number of CFU in the control, and N is the number of cells detected following pressurization treatments). Each curve shows the mean values obtained from results from two independent replicates, and error bars represent standard deviations.
Inactivation of microorganisms in cheese by HP treatment.
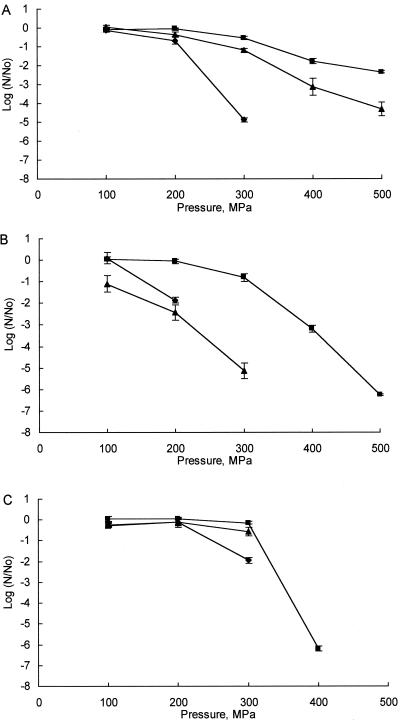
Relative sensitivities of the three microbial species to HP in the miniature cheeses assayed immediately after pressurization were as found for the cheese slurry system, i.e., P. roqueforti was more sensitive than E. coli, which was more sensitive than S. aureus (Fig. 4). However, there were substantial differences between media, particularly with E. coli; e.g., organisms in cheese were much more sensitive to HP than they were in the cheese slurry system (Fig. 4B). The same trend was evident for S. aureus and P. roqueforti, although the effect was less dramatic.
FIG. 4.
Inactivation of S. aureus ATCC 6538 (A), E. coli K-12 (B), and P. roqueforti IMI 297987 (C) in pH 5.3 buffer (⧫), Cheddar cheese slurry (■), and Cheddar cheese (▴) by HP treatment at 20°C for 20 min (No is the total number of CFU in the control, and N is the number of cells detected following pressurization treatments). Each curve shows the mean values obtained from results of at least two independent experiments, and error bars represent standard deviations.
Comparisons with the buffer system indicated that HP treatment of microorganisms in this medium achieved the greatest inhibitory effect (Fig. 4). However, it is interesting that in the case of E. coli at relatively low pressures, inactivation was greater in the Cheddar cheese product.
DISCUSSION
The purpose of this study was to determine the effect of HP treatment on microbial contaminants in Cheddar cheese and to provide information, not previously available, on the influence of the Cheddar cheese environment on HP inactivation.
HP inactivation was studied in three different systems: cheese slurry, Cheddar cheese, and a buffer system. Cheese slurries are recognized as an acceptable experimental model for cheese conditions (12, 13) and were used to develop a matrix relating microbial inactivation to conditions of pressure and temperature. The data from the matrix were subsequently selectively applied to laboratory-manufactured Cheddar cheese deliberately contaminated with the microorganisms of interest. Inactivation in buffer was studied in order to elucidate the degree of protection provided by the cheese against HP inactivation.
The greater sensitivity of the microorganisms in the Cheddar cheese than in the cheese slurry system may be explained by acid injury to the bacteria during fermentation. Also, some cells may have been in the log phase of growth, as overnight cultures were added directly to the milk, which underwent a 5-h cheese-making process. This circumstance may have resulted in greater inactivation, as the HP sensitivity of log-phase cells is greater than that of stationary-phase cells (9), as demonstrated also in the present study (Fig. 3). It has been shown that water availability can markedly affect the response of microorganisms to HP (28, 31, 33), with microorganisms in dehydrated foods being very pressure resistant (9). It is likely that differences in water availability explain the greater sensitivity of microorganisms to HP inactivation in the buffer system than in cheese.
The importance of the interaction between pressure and temperature (even over the range of moderate temperatures used here) in determining the degree of inactivation of microbial species was evident from the results obtained in the cheese slurry experiment. Adjustment of temperature can significantly affect the pressure necessary to achieve inactivation. Results obtained by others (6, 7, 34, 35) working over a greater range of temperatures indicate an even greater response to pressure-temperature interactions. However, temperatures higher than those used in this study cannot be realistically applied in a commercial Cheddar cheese process.
Yeasts and mold spores have been previously shown to be readily inactivated at 400 MPa (33), while Ogawa et al. (27) reported a >5-log-unit reduction when nine species of yeasts and molds in fruit juice were treated at 350 MPa for 30 min or 400 MPa for 5 min. These results are in agreement with results obtained in this study for P. roqueforti in cheese slurry at temperatures of 20 and 30°C. The pattern of inactivation by HP observed for the mold spores was different from that obtained with the bacterial species. However, it has been demonstrated that eucaryotic microorganisms are generally more sensitive to pressure than procaryotic microorganisms (22). Inactivation of molds was not evident until a pressure of 300 MPa was reached, after which cell numbers decreased rapidly.
Sublethal injury experiments indicated that, at the higher pressures, the injured population was more severely damaged and took longer to recover. Hauben et al. (17) also found that increasing the pressure from 180 to 270 MPa resulted in an increased percentage of sublethally injured cells. Those authors studied the effect of 320 MPa for 15 min at room temperature on E. coli K-12 in buffer at pH 7 and found approximately a 4-log-unit cycle reduction in numbers, of which 2 log units of organisms were sublethally injured.
Previous studies of HP inactivation of microorganisms have reported either a first-order kinetic reaction or a sigmoidal response, indicating the presence of a pressure-resistant subpopulation (11, 19, 22, 33). Generally, S. aureus displays first-order inactivation kinetics (5, 14), as found in this study with Cheddar cheese slurries. E. coli is reported to demonstrate either first-order inactivation kinetics (25, 34) or a two-phase tailing effect (1, 9), depending on the strain and treatment conditions, both of which effects were observed for E. coli in the present study.
Strain-to-strain variation is a significant problem which arises when the extent of HP inactivation is being defined for a particular species. This study demonstrated a strain response to pressure, with values varying by ∼2-log-unit cycles for both the mold and bacterial species. Other studies have found greater degrees of variation. Benito et al. (1) found a 5-log-unit cycle difference in pressure sensitivity between stationary-phase cells of strains of E. coli O157 treated at 500 MPa for 20 min in pH 7.0 buffer. Food isolates are generally reported to be more pressure resistant than strains from culture collections (9). In this study the food isolates of S. aureus were more pressure resistant than S. aureus ATCC 6538 pressure treated at 400 MPa at 20°C for 20 min. However, E. coli K-12 and P. roqueforti IMI 297987 spores were more pressure resistant than the food isolates under the conditions studied.
The results from this study underline the importance of taking the suspending medium and the effects of processing into account when assessing the impact of HP on the viability of microorganisms. Application of HP substantially reduces the microbial load in Cheddar cheese, with 400 MPa for 20 min at 20°C being sufficient to reduce the numbers of viable E. coli and P. roqueforti by 7- and 6-log-unit cycles, respectively, and to reduce the levels of S. aureus by 3-log-unit cycles. The technology could be applied to control post pasteurization contaminants in cheese where heat treatment cannot be used. However, the HPs required to inactivate microorganisms combined with the high cost of HP technology are factors currently limiting the use of HP processing in industrial applications. Nevertheless, there is increasing interest in combining HP with other technologies, such as the use of bacteriocins as demonstrated by Morgan et al. (26), which may lead to microbial inactivation at reduced pressures. These results thus favor the development of commercial processes for HP treatment of dairy products such as cheese.
ACKNOWLEDGMENTS
This project was funded by the Commission of the European Communities through the Agricultural and Fisheries (FAIR) RTD program (grant CT96-1113). Ciara O'Reilly was funded by a Teagasc Walsh Fellowship.
We thank J. Costello and J. Molloy (Wexford Creamery).
REFERENCES
- 1.Benito A, Ventoura G, Casadei M, Robinson T, Mackey B. Variation in resistance of natural isolates of Escherichia coli O157 to high hydrostatic pressure, mild heat, and other stresses. Appl Environ Microbiol. 1999;65:1564–1569. doi: 10.1128/aem.65.4.1564-1569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beuchat L R, Pitt J I. Influence of solute, pH, and incubation temperature on recovery of heat-stressed Wallemia sebi conidid. Appl Environ Microbiol. 1990;56:2545–2550. doi: 10.1128/aem.56.8.2545-2550.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradshaw J G, Peeler J T, Corwin J J, Barnett J E, Twedt R M. Thermal resistance of disease-associated Salmonella typhimurium in milk. J Food Prot. 1987;50:95–96. doi: 10.4315/0362-028X-50.2.95. [DOI] [PubMed] [Google Scholar]
- 4.Bradshaw J G, Shah D B, Wehby A J, Peeler J T, Twedt R M. Thermal inactivation of the Kanagawa hemolysin of Vibrio parahaemolyticus in buffer and shrimp. J Food Sci. 1984;49:183–187. [Google Scholar]
- 5.Butz P, Ludwig H. Pressure inactivation of microorganisms at moderate temperatures. Physica B. 1986;139/140:875–877. [Google Scholar]
- 6.Carlez A, Cheftel J-C, Rosec J-P, Richard N, Saldana J-L, Balny C. Effects of high pressure and bacteriostatic agents on the destruction of Citrobacter freundii in minced beef muscle. In: Balny C, Hayashi R, Heremans K, Masson P, editors. High pressure and biotechnology. London, United Kingdom: Colloque INSERM/John Libbey Eurotext Ltd.; 1992. pp. 365–368. [Google Scholar]
- 7.Carlez A, Rosec J-P, Richard N, Cheftel J-C. High pressure inactivation of Citrobacter freundii, Pseudomonas fluorescens and Listeria innocua in inoculated minced beef muscle. Lebensm-Wiss Technol. 1993;26:357–363. [Google Scholar]
- 8.Cheftel J-C. Applications des hautes pressions en technologie alimentaire. Actual Ind Aliment Agro-aliment. 1991;108:141–153. [Google Scholar]
- 9.Cheftel J-C. High-pressure, microbial inactivation and food preservation. Food Sci Technol Int. 1995;1:75–90. [Google Scholar]
- 10.Cole R. High pressure processing: a technology of the future. Food Manuf. 1997;72:21–26. [Google Scholar]
- 11.Earnshaw R G. Kinetics of high pressure inactivation of micro-organisms. In: Ledward D A, Johnston D E, Earnshaw R G, Hasting A M P, editors. High pressure processing of foods. Nottingham, United Kingdom: Nottingham University Press; 1995. pp. 37–46. [Google Scholar]
- 12.Farkye N Y, Madkor S A, Atkins H G. Proteolytic abilities of some lactic acid bacteria in a model cheese system. Int Dairy J. 1995;5:715–725. [Google Scholar]
- 13.Fernández de Palencia P, Martín-Hernández C, López-Fandiño R, Peláez C. Proteolytic activity of Lactobacillus casei subsp. casei IFPL 731 in a model cheese system. J Agric Food Chem. 1997;45:3703–3708. [Google Scholar]
- 14.Gervilla R, Sendra E, Ferragut V, Guamis B. Sensitivity of Staphylococcus aureus and Lactobacillus helveticus in ovine milk subjected to high hydrostatic pressure. J Dairy Sci. 1999;82:1099–1107. doi: 10.3168/jds.S0022-0302(99)75332-4. [DOI] [PubMed] [Google Scholar]
- 15.Gilles J, Lawrence R C. The assessment of Cheddar cheese quality by compositional analysis. N Z J Dairy Sci Technol. 1973;8:148–151. [Google Scholar]
- 16.Guinee T P, Pudja P D, Reville W J, Harrington D, Mulholland E O, Cotter M, Cogan T M. Composition, microstructure and maturation of semi-hard cheeses from high protein ultrafiltered milk retentates with different levels of denatured whey protein. Int Dairy J. 1995;5:543–568. [Google Scholar]
- 17.Hauben K J A, Wuytack E Y, Soontjens C C F, Michiels C W. High-pressure transient sensitization of Escherichia coli to lysozyme and nisin by disruption of outer-membrane permeability. J Food Prot. 1996;59:350–355. doi: 10.4315/0362-028X-59.4.350. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi R. Application of high pressure to food processing and preservation: philosophy and development. In: Spiess W E L, Shubert H, editors. Engineering and food. London, United Kingdom: Elsevier Applied Science; 1989. pp. 815–826. [Google Scholar]
- 19.Heinz V, Knorr D. High pressure inactivation kinetics of Bacillus subtilis cells by a three-state model considering distributed resistance mechanisms. Food Biotechnol. 1996;10:149–161. [Google Scholar]
- 20.Hite B H. The effect of pressure in the preservation of milk. Bull WVa Univ Agric Exp Stn. 1899;58:15–35. [Google Scholar]
- 21.Hite B H, Giddings N J, Weakly C E. The effect of pressure on certain microorganisms encountered in the preservation of fruits and vegetables. Bull WVa Univ Agric Exp Stn. 1914;146:1–67. [Google Scholar]
- 22.Hoover D G, Metrick C, Papineau A M, Farkas D F, Knorr D. Biological effects of high hydrostatic pressure on food microorganisms. Food Technol. 1989;43:99–107. [Google Scholar]
- 23.Iandolo J J, Ordal Z J. Repair of thermal injury of Staphylococcus aureus. J Bacteriol. 1966;91:134–142. doi: 10.1128/jb.91.1.134-142.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knorr D. Effects of high-hydrostatic-pressure processes on food safety and quality. Food Technol. 1993;47:156–161. [Google Scholar]
- 25.Ludwig H, Bieler C, Hallbauer K, Scigalla W. Inactivation of microorganisms by hydrostatic pressure. In: Balny C, Hayashi R, Heremans K, Masson P, editors. High pressure and biotechnology. London, United Kingdom: Colloque INSERM/John Libbey Eurotext Ltd.; 1992. pp. 25–32. [Google Scholar]
- 26.Morgan S M, Ross R P, Beresford T, Hill C. Combination of hydrostatic pressure and lacticin 3147 causes increased killing of Staphylococcus and Listeria. J Appl Microbiol. 2000;88:414–420. doi: 10.1046/j.1365-2672.2000.00975.x. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa H, Fukuhisa K, Kubo Y, Fukumoto H. Pressure inactivation of yeasts, moulds, and pectinesterase in satsuma mandarin juice: effects of juice concentration, pH, and organic acids, and comparison with heat sanitation. Agric Biol Chem. 1990;54:1219–1225. [Google Scholar]
- 28.Oxen P, Knorr D. Baroprotective effects of high solute concentrations against inactivation of Rhodotorula rubra. Lebensm-Wiss Technol. 1993;26:220–223. [Google Scholar]
- 29.Patterson M F, Quinn M, Simpson R, Gilmour A. Sensitivity of vegetative pathogens to high hydrostatic pressure treatment in phosphate-buffered saline and foods. J Food Prot. 1995;58:524–529. doi: 10.4315/0362-028X-58.5.524. [DOI] [PubMed] [Google Scholar]
- 30.Ryan M P, Rea M C, Hill C, Ross R P. An application in Cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl Environ Microbiol. 1996;62:612–619. doi: 10.1128/aem.62.2.612-619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sale A J H, Gould G W, Hamilton W A. Inactivation of bacterial spores by hydrostatic pressure. J Gen Microbiol. 1970;60:323–334. doi: 10.1099/00221287-60-3-323. [DOI] [PubMed] [Google Scholar]
- 32.Shakeel-Ur-Rehman P, McSweeney L H, Fox P F. Protocol for the manufacture of miniature cheeses. Lait. 1998;78:607–620. [Google Scholar]
- 33.Smelt J P P M. Recent advances in the microbiology of high pressure processing. Trends Food Sci Technol. 1998;9:152–158. [Google Scholar]
- 34.Smelt J P P M, Rijke G G F. High pressure treatment as a tool for pasteurisation of foods. In: Balny C, Hayashi R, Heremans K, Masson P, editors. High pressure and biotechnology. London, United Kingdom: Colloque INSERM/John Libbey Eurotext Ltd.; 1992. pp. 361–364. [Google Scholar]
- 35.Takahahsi K. Sterilization of microorganisms by hydrostatic pressure at low temperature. In: Balny C, Hayashi R, Heremans K, Masson P, editors. High pressure and biotechnology. London, United Kingdom: Colloque INSERM/John Libbey Eurotext Ltd.; 1992. pp. 303–307. [Google Scholar]
- 36.Weng Y-M, Hotchkiss J H. Headspace gas composition and chitin content as measures of Rhizopus stolonifer growth. J Food Sci. 1991;56:274–275. [Google Scholar]
- 37.Yong R K, Cousin M A. Nonspecific enzyme-linked immunosorbant assay for moulds in foods. J Food Sci. 1995;60:1357–1363. [Google Scholar]