Abstract
Extensive studies and analyses into the molecular features of severe acute respiratory syndrome related coronavirus 2 (SARS-CoV-2) have enhanced the surveillance and investigation of its clusters and transmission worldwide. The whole genome sequencing (WGS) approach is crucial in identifying the source of infection and transmission routes by monitoring the emergence of variants over time and through communities. Varying SARS-CoV-2 genomics capacity and capability levels have been established in public health laboratories across different Australian states and territories. Therefore, laboratories performing SARS-CoV-2 WGS for public health purposes are recommended to participate in an external proficiency testing program (PTP). This study describes the development of a SARS-CoV-2 WGS PTP. The PTP assessed the performance of laboratories while providing valuable insight into the current state of SARS-CoV-2 genomics in public health across Australia. Part 1 of the PTP contained eight simulated SARS-CoV-2 positive and negative specimens to assess laboratories' wet and dry laboratory capacity. Part 2 involved the analysis of a genomic dataset that consisted of a multi-FASTA file of 70 consensus genomes of SARS-CoV-2. Participating laboratories were required to (1) submit raw data for independent bioinformatics analysis, (2) analyse the data with their processes, and (3) answer relevant questions about the data. The performance of the laboratories was commendable, despite some variation in the reported results due to the different sequencing and bioinformatics approaches used by laboratories. The overall outcome is positive and demonstrates the critical role of the PTP in supporting the implementation and validation of SARS-CoV-2 WGS processes. The data derived from this PTP will contribute to the development of SARS-CoV-2 bioinformatic quality control (QC) and performance benchmarking for accreditation.
Key words: SARS-CoV-2, whole genome sequencing, proficiency testing, bioinformatics analysis
Introduction
The early release of the whole genome sequence of SARS-CoV-2 in January 20201 enabled the design and rapid development of one of the first reverse transcriptase polymerase chain reaction (RT-PCR) assays, as reported by Corman et al. 2 It also facilitated our understanding of the dynamics of subtype evolution through its application in understanding the clinical outcome of the disease,3 developing whole genome sequencing (WGS)-based coronavirus disease (COVID-19) diagnostics4 and vaccines,5 evolution tracking6 and investigation of virus transmission using phylogenetic analysis.7
Different WGS protocols and approaches have been developed by many research groups and can be broadly categorised as targeted and non-targeted. This includes an amplicon-based method of SARS-CoV-2 WGS on the Oxford Nanopore Technologies (ONT) platform developed by the ARTIC network (https://artic.network/ncov-2019), which has been adapted for other sequencing platforms, allowing the genome of the virus to be studied by more laboratories. Other protocols utilised by laboratories include shotgun metagenomic approaches8 , 9 and target capture sequencing using Twist custom target enrichment.10 Many of these protocols are available on GitHub (https://github.com/CDCgov/SARS-CoV-2_Sequencing), which contains a comprehensive list of protocols, tools and resources for SARS-CoV-2 WGS on various platforms, including Illumina, ONT, PacBio and Ion Torrent. The ARTIC protocol using the ARTIC primer set is the most widely adopted targeted amplicon approach for WGS SARS-CoV-2. This approach requires ongoing modifications and optimisation to achieve high genome coverage while addressing mutations in the primer binding sites resulting in amplicon drop-offs in the variant of concern (VoC).
The sharing and analysis of genomics data during outbreaks within a country and on a global level is now a fundamental part of the outbreak response.11, 12, 13 The Global Initiative on Sharing All Influenza Data (GISAID)14 , 15 is an international data repository that applies a phylogenetic analysis tool that classifies sequences (at the time of writing) into four major clades (S, L, V and G) with several subclades. As of 31 March 2022, GISAID has made sequence data sharing possible by compiling approximately nine million SARS-CoV-2 complete genomes contributed by researchers globally. Alternative assignment of similar but not identical lineages has also been added to the GISAID clades by an additional tool, Pangolin (Phylogenetic Assignment of Named Global Outbreak LINeages).16 Other data sharing and analysis platforms include the AusTrakka platform,17 designed and developed by the Australian Communicable Disease Genomics Network (CDGN). AusTrakka is a secure platform that allows sharing and analysis of pathogen genomics data across Australian jurisdictions and New Zealand, with SARS-CoV-2 being the first pathogen to be implemented. The development of the system for combined data analysis based on a model for collaborative exploration of WGS and metadata in a protected sharing environment18 will fully capitalise on the potential added value of WGS for public health decision making. This approach is a plausible way to facilitate cross jurisdictional sequence data analyses.
The use of SARS-CoV-2 genomics is continually evolving, creating the need for WGS data to meet defined quality metrics in identifying and characterising the virus and the surveillance of emerging strains using this data. Defined quality metrics are essential due to the significant variations in genome coverage and single nucleotide polymorphisms (SNPs) detection found across these protocols. While studies comparing different SARS-CoV-2 WGS protocols may address these issues,19 , 20 an external proficiency testing program (PTP) for SARS-CoV-2 WGS is critical to ensure that the performance of laboratories in applying these relatively new WGS protocols is assessed using standard quality metrics. In line with this, the Royal College of Pathologists of Australasia Quality Assurance Programs (RCPAQAP) Biosecurity developed a SARS-CoV-2 WGS PTP to assess Australian laboratories' capacity for SARS-CoV-2 genomics. The RCPAQAP is an Australian government funded program since 2009. It has provided PTP to assess laboratories' diagnostic and technical proficiency in Australia and internationally for Security Sensitive Biological Agents (SSBA) and emerging and re-emerging pathogens. This study aimed to present the process of developing a SARS-CoV-2 WGS PTP. The two-part PTP assessed the pathogen genomics capacity and capability levels of laboratories while providing valuable insight into the current state of SARS-CoV-2 genomics in public health across Australia.
Materials and methods
Organisation and planning
The SARS-CoV-2 WGS PTP was initiated by the RCPAQAP in collaboration with the CDGN, an expert reference panel under the Australian Public Health Laboratory Network (PHLN). The planning of this PTP was based on the relevant information collected from a workshop facilitated by the RCPAQAP in October 2020. The workshop focused on the determination of key metrics and evaluation criteria, including: (1) criteria used in post-processing practice (trimming of low quality reads, assemblies of the genome data); (2) the quality of reads; (3) the accuracy of lineage designation, identification and characterisation; (4) the percentage genome coverage for samples with varying amounts of the virus; (5) the phylogenetic tree building capacity, and (6) the de novo sequencing and genome assembly capacity. Following the workshop, 11 Australian research, public health and private pathology laboratories experienced in analysing WGS data sets for infectious agents, were invited to participate in the SARS-CoV-2 WGS PTP. By agreeing to participate in this PTP, laboratories agreed that the data submitted would be treated confidentially, and all data to be shared would be de-identified.
Survey specimens and survey instructions
In Part 1 of the PTP, participating laboratories were supplied with a specimen panel consisting of eight samples, including one SARS-CoV-2 negative and seven SARS-CoV-2 positive specimens. Details of the specimen panel are listed in Table 1, Table 2 . Participants were only provided with the RCPAQAP sample ID, and no other information about the content of each sample was supplied. All samples were confirmed stable and homogenous using an in-house process, as described previously,21 while samples characteristics were validated by an external laboratory. The in-house preparation of all samples was as follows. All SARS-CoV-2 positive samples contained 0.5 mL diluted viral total RNA (isolated from live SARS-CoV-2) in nuclease-free water, simulating respiratory samples. RNA extractions were performed using the Qiagen QIAamp 96 DNA QIAcube HT kit, which co-purified DNA and RNA. This process involved using lysis buffer (VXL buffer), which has been shown to inactivate SARS-CoV-2.22 Inactivation was confirmed by subsequent negative virus isolation attempts with sub-culturing (blind passage) of negative cultures verified as non-replicative by RT-PCR confirmation. All samples were safe to use without risk of infection using standard Physical Containment 2 (PC2) practices and handling protocols. The survey specimens were dispatched on dry ice to preserve the integrity of the RNA, and in-house testing had confirmed the stability of the specimens under these conditions. Participating laboratories received the specimen panel within 24 h. They were advised to store the survey specimens at –80°C at all times, perform RNA extraction and WGS, and analyse the sequence data using their choice of bioinformatics software. When determining lineages for the specimens, they were required to use the following for their analysis: pangolin v2.1.7 (https://github.com/cov-lineages/pangolin) with pangoLEARN version 2021-01-11 (https://github.com/cov-lineages/pangoLEARN).
Table 1.
Content of each specimen of Part 1 of the SARS-CoV-2 whole genome sequencing proficiency testing program, with its associated GISAID ID and the average CT
| RCPAQAP Sample ID |
Contenta (GISAID ID) | In-house NAT (E-gene) Average CT |
External NAT (E-gene) Average CT |
|---|---|---|---|
| BS01 | SARS-CoV-2 RNA (EPI_ISL_406844) | 16.8 | 16.2 |
| BS02 | SARS-CoV-2 RNA (EPI_ISL_419750) | 17.0 | 16.5 |
| BS03 | SARS-CoV-2 RNA (EPI_ISL_480701) | 17.3 | 16.7 |
| BS04 | SARS-CoV-2 RNA (EPI_ISL_519314) | 17.3 | 16.9 |
| BS05 | SARS-CoV-2 RNA (EPI_ISL_563416) | 17.3 | 17.6 |
| BS06 | Negative specimen | Not detected | Not detected |
| BS07 | SARS-CoV-2 RNA 1:102 dilution (EPI_ISL_419750) | 24.1 | 22.4 |
| BS08 | SARS-CoV-2 RNA 1:103 dilution (EPI_ISL_419750) | 27.5 | 26.3 |
CT, cycle threshold; GISAID, Global Initiative on Sharing All Influenza Data; NAT, nucleic acid testing.
BS02, BS07 and BS08 were prepared from the same RNA preparation, with BS07 and BS08 serially diluted from BS02 at 1:102 and 1:103, respectively.
Table 2.
In-house sequence analysis for each specimen of Part 1 of the SARS-CoV-2 whole genome sequencing proficiency testing program
| RCPAQAP Sample ID |
Genome recovered (%) | Pango lineagea | Single-nucleotide polymorphism (SNP)b | Amino acid replacementc |
|---|---|---|---|---|
| BS01 | 99.6 | B | T19065C, T22303G, G26144T | S:S247R; ORF3a:G251V |
| BS02 | 99.6 | B.1 | C241T, C1059T, C3037T, C14408T, A23403G, G25563T | nsp2:T85I; nsp12:P323L; S:D614G; ORF3a:Q57H |
| BS03 | 98.9 | B.1.1.136 | C241T, C3037T, C14408T, A23063T, A23403G, C26984T, T28196C, G28881A, G28882A, G28883C | nsp12:P323L; S:N501Y; S:D614G; N:R203K; N:G204R |
| BS04 | 99.6 | D.2 | C241T, A1163T, C3037T, T7540C, C14408T, G16647T, C18555T, G22992A, G23401A, A23403G, C28647T, G28881A, G28882A, G28883C | nsp2:I120F; nsp12:P323L; S:S477N; S:D614G; N:A125V; N:R203K; N:G204R |
| BS05 | 99.2 | D.2 | C241T, A1163T, C3037T, T7540C, C9996T, C10279T, C14408T, C14599T, G16647T, C18555T, G22205C, G22927T, G22992A, G23401A, A23403G, A28416T, G28881A, G28882A, G28883C, C29585T | nsp2:I120F; nsp4:S481L; nsp12:P323L; S:D215H; S:L455F; S:S477N; S:D614G; N:N48I; N:R203K; N:G204R; ORF10:P10S |
| BS06 | 1.00 | N/A | N/A | N/A |
| BS07 | 98.8 | B.1 | C241T, C1059T, C3037T, C14408T, A23403G, G25563T | nsp2:T85I; nsp12:P323L; S:D614G; ORF3a:Q57H |
| BS08 | 98.3 | B.1 | C241T, C1059T, C3037T, C14408T, A23403G, G25563T | nsp2:T85I; nsp12:P323L; S:D614G; ORF3a:Q57H |
Pangolin v2.1.7 (https://github.com/cov-lineages/pangolin) with pangoLEARN version 2021-01-11 (https://github.com/cov-lineages/pangoLEARN) were used in the analysis.
Sequence base pairs deletion and N(s) are excluded. C241T is a common SNP introduced to the sequence likely due to degradation of material and therefore absence of C241T, as reported by participants is considered concordant.
nsp2:T85I is equivalent to ORF1a:T265I, ORF1ab:T265I and ORF1ab:T85I; nsp12:P323L is equivalent to nsp12b:P314L, ORF1b:P314L, ORF1ab:P4715L, and ORF1ab:P323L; nsp2:I120F is equivalent to ORF1a:I300F, ORF1ab:I300F and ORF1ab:I120F; N:R203K and N:G204R are equivalent to N:RG608-609KR; N:R203K is equivalent to N:R50K; N:G204R is equivalent to N:G50R and ORF14:G50R; nsp4:S481L is equivalent to ORF1a:S3244L, ORF1ab:S3244L and ORF1ab:S481L.
Part 2 of the PTP involved analysing a genomic dataset file obtained from the GMI benchmarking repository (https://github.com/globalmicrobialidentifier-WG3/datasets), available as the multi-FASTA file of 70 consensus genomes of SARS-CoV-2. Details of the dataset are shown in Table 3 . The phylogenetic tree for this dataset can be found online (https://itol.embl.de/tree/15811123610035991611254254) and is displayed in Supplementary Fig. 1 (Appendix A). Using their preferred pipeline, participating laboratories were required to classify the downloaded sequences into one or more genomic clusters of interest to epidemiological investigations. All survey instructions are available in the Supplementary Data (Appendix A).
Table 3.
Cluster ID of the 70 SARS-CoV-2 consensus genomes included in Part 2 of the SARS-CoV-2 whole genome sequencing proficiency testing program
| Accession | RCPAQAP Sample ID |
Cluster ID | Accession | RCPAQAP Sample ID |
Cluster ID | Accession | RCPAQAP Sample ID |
Cluster ID |
|---|---|---|---|---|---|---|---|---|
| MT520173.1 | RCPA-PTP-2021-S001 | A | MT520273.1 | RCPA-PTP-2021-S025 | A | MT520445.1 | RCPA-PTP-2021-S049 | A |
| MT520175.1 | RCPA-PTP-2021-S002 | A | MT520285.1 | RCPA-PTP-2021-S026 | A | MT520448.1 | RCPA-PTP-2021-S050 | A |
| MT520196.1 | RCPA-PTP-2021-S003 | A | MT520288.1 | RCPA-PTP-2021-S027 | A | MT520453.1 | RCPA-PTP-2021-S051 | A |
| MT520202.1 | RCPA-PTP-2021-S004 | B | MT520295.1 | RCPA-PTP-2021-S028 | A | MT520454.1 | RCPA-PTP-2021-S052 | A |
| MT520206.1 | RCPA-PTP-2021-S005 | A | MT520300.1 | RCPA-PTP-2021-S029 | A | MT520460.1 | RCPA-PTP-2021-S053 | A |
| MT520208.1 | RCPA-PTP-2021-S006 | A | MT520309.1 | RCPA-PTP-2021-S030 | A | MT520464.1 | RCPA-PTP-2021-S054 | A |
| MT520216.1 | RCPA-PTP-2021-S007 | A | MT520312.1 | RCPA-PTP-2021-S031 | A | MT520479.1 | RCPA-PTP-2021-S055 | C |
| MT520219.1 | RCPA-PTP-2021-S008 | A | MT520318.1 | RCPA-PTP-2021-S032 | A | MT520480.1 | RCPA-PTP-2021-S056 | A |
| MT520222.1 | RCPA-PTP-2021-S009 | A | MT520324.1 | RCPA-PTP-2021-S033 | A | MT520482.1 | RCPA-PTP-2021-S057 | A |
| MT520226.1 | RCPA-PTP-2021-S010 | A | MT520330.1 | RCPA-PTP-2021-S034 | A | MT520483.1 | RCPA-PTP-2021-S058 | A |
| MT520230.1 | RCPA-PTP-2021-S011 | A | MT520334.1 | RCPA-PTP-2021-S035 | A | MT520495.1 | RCPA-PTP-2021-S059 | A |
| MT520232.1 | RCPA-PTP-2021-S012 | A | MT520340.1 | RCPA-PTP-2021-S036 | C | MT520504.1 | RCPA-PTP-2021-S060 | A |
| MT520234.1 | RCPA-PTP-2021-S013 | A | MT520349.1 | RCPA-PTP-2021-S037 | A | MT520505.1 | RCPA-PTP-2021-S061 | A |
| MT520235.1 | RCPA-PTP-2021-S014 | A | MT520369.1 | RCPA-PTP-2021-S038 | A | MT520507.1 | RCPA-PTP-2021-S062 | A |
| MT520239.1 | RCPA-PTP-2021-S015 | A | MT520374.1 | RCPA-PTP-2021-S039 | A | MT520510.1 | RCPA-PTP-2021-S063 | A |
| MT520244.1 | RCPA-PTP-2021-S016 | A | MT520380.1 | RCPA-PTP-2021-S040 | A | MT520514.1 | RCPA-PTP-2021-S064 | A |
| MT520246.1 | RCPA-PTP-2021-S017 | A | MT520400.1 | RCPA-PTP-2021-S041 | C | MT520525.1 | RCPA-PTP-2021-S065 | A |
| MT520247.1 | RCPA-PTP-2021-S018 | A | MT520407.1 | RCPA-PTP-2021-S042 | A | MT520529.1 | RCPA-PTP-2021-S066 | A |
| MT520248.1 | RCPA-PTP-2021-S019 | A | MT520417.1 | RCPA-PTP-2021-S043 | A | MT520530.1 | RCPA-PTP-2021-S067 | B |
| MT520256.1 | RCPA-PTP-2021-S020 | A | MT520420.1 | RCPA-PTP-2021-S044 | A | MT520536.1 | RCPA-PTP-2021-S068 | A |
| MT520263.1 | RCPA-PTP-2021-S021 | B | MT520421.1 | RCPA-PTP-2021-S045 | A | MT520538.1 | RCPA-PTP-2021-S069 | A |
| MT520268.1 | RCPA-PTP-2021-S022 | A | MT520426.1 | RCPA-PTP-2021-S046 | A | MT520539.1 | RCPA-PTP-2021-S070 | A |
| MT520270.1 | RCPA-PTP-2021-S023 | A | MT520435.1 | RCPA-PTP-2021-S047 | A | |||
| MT520271.1 | RCPA-PTP-2021-S024 | A | MT520440.1 | RCPA-PTP-2021-S048 | B |
Reporting of proficiency testing (PT) results
All laboratories were required to submit results within a 4-week timeframe, from 18 February to 17 March 2021. Participants were to report results from Part 1 in an online questionnaire and upload the raw reads of the sequence data without pre-processing or trimming and the derived consensus sequence data to a server provided by the RCPAQAP. In Part 2, laboratories were required to upload the clustering results and the phylogenetic tree file and report all analysis results in the questionnaire. The questionnaire for Parts 1 and 2 is available in the Supplementary Data (Appendix A). The questionnaire for Part 1 contained four sections: (1) the snapshot of sequence information; (2) the protocol used to generate the sequence data; (3) the criteria used to analyse the sequence data; (4) the quantitative or qualitative criteria used for QC checks. Part 2 consisted of the questionnaire on the analysis of the genomic dataset. Laboratories were also invited to provide feedback on the survey. The responses were collected as single or multiple options with free text for remarks and comments.
Assessment of PT results
The quality of the sequence data uploaded in Part 1 was determined according to the suitability of a sequence for downstream analyses, based on the consensus sequence submitted. Assessment of results for all SARS-CoV-2 positive samples was based on three quality metrics decided with input from CDGN Genomics Implementation and Bioinformatics working groups: (1) the proportion of the SARS-CoV-2 genome recovered (a minimum of 50% is considered adequate); (2) pango lineage (correct lineage); and (3) sequencing accuracy (accuracy >95%, based on the number of variant sites that were identified and not identified in the consensus sequence). The sequencing accuracy recorded the following four criteria: (1) true positive (TP), i.e., the number of variant sites or single nucleotide polymorphisms (SNPs) that were identified in the consensus sequence; (2) true negative (TN), i.e., the number of non-missing sites correctly identified as non-variant; (3) false positive (FP), i.e., the number of variant sites identified (SNPs) where no variant site was present; (4) and false negative (FN), i.e., the number of sites that were variant (SNPs) in the specimen but not identified by the participant. The sequencing accuracy (%) was defined as:
The CDGN Bioinformatics working group analysed the FASTQ data for Part 1. FASTQ quality metrics were generated using minimap2 (version 2.18; https://github.com/lh3/minimap2) and samtools (version 1.12; https://github.com/samtools/samtools) to assess the depth of sequencing coverage, base quality and mapping quality. Participants were assessed as ‘pass’ if there was a combination of the following for the consensus sequences submitted for all eight survey specimens (BS01–BS08): a minimum of 50% genome recovered (for SARS-CoV-2 positive samples), a correct pango lineage, and >95% sequencing accuracy.
For Part 2, the assessment of clustering performed by participants was compared to the results provided on the GMI link (https://itol.embl.de/tree/15811123610035991611254254), which was used as the benchmarked cluster designation. The clustering performed by participants was considered concordant if a participant's interpretation of the genomic relationships did not contradict the benchmarked cluster designation. Results with individual isolates clustered into the same cluster as in the benchmarked dataset and identified subclusters within a cluster were considered concordant. A result was considered not clustered if the participants did not identify a sequence as part of a cluster where it was identified as part of the outbreak in the benchmarked dataset.
Participant report
The report issued to the participating laboratories was an individual report divided into two sections. Section 1 consisted of the bioinformatics QC report, including the assessment of the participants' performance (either ‘pass’ or ‘fail’), based on the results submitted for all specimens. It also included the summary of responses from all participants for the questionnaire of Part 1 of the PTP, including the percentage genome recovered, the average read depth, the pango lineage, the SNPs, the amino acid replacement and the sequencing site. Section 1 also included a summary of sequencing and analysis protocols used by participating laboratories, and the details of this are available in the Supplementary Data (Appendix A). Section 2 of the report consisted of: (1) the phylogenetic tree generated by participants for the data in Part 2 of the PTP; and (2) the summary of the cluster data, as submitted by participants and the cluster data summary. The cluster data summary section, which outlined the concordance group and the number of sequences assessed based on each benchmark cluster ID (A, B and C), allowed participants to perform a self assessment. Section 2 also contained the summary of responses submitted for the questionnaire of Part 2 of the PTP, and the details of this are available in the Supplementary Data (Appendix A).
Results
Performance assessment
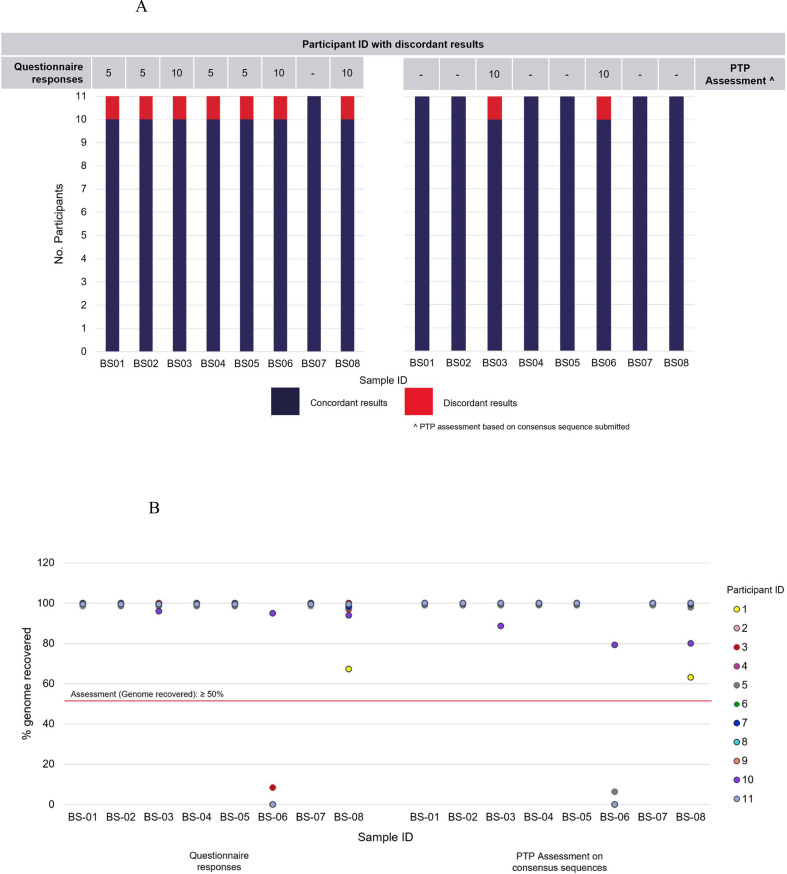
The majority of participants (except participants 1 and 10) achieved high genome coverage (over 90%) for all samples. Additionally, all participants submitted an over 99% sequencing accuracy for all samples. All participants except two were assessed as ‘pass’ for all specimens (Table 4 ). One of the participants (participant 3) did not submit any consensus sequence, and therefore their results were not assessed. Based on the sequence submitted, one participant (participant 10) was assessed as ‘fail’ (genome recovered: 79.3%; pango lineage: B.1, sequencing accuracy: not calculated) for sample BS06, which was a SARS-CoV-2 negative specimen and did not contain any viral genetic material. We also noted some discrepancies in the questionnaire responses from participants on the reported pango lineage compared to the analysis performed on the consensus sequences submitted (Fig. 1 A). Participant 5 reported discordant results for 4/8 samples in the questionnaire; however, upon analysis of their submitted consensus sequences, pango lineages for all samples were identified as concordant. On the other hand, participant 10 reported discordant results in their questionnaire responses for 3/8 samples; however, upon analysis of their submitted consensus sequences, only pango lineage assignments for samples BS03 and BS06 were identified as discordant (Fig. 1A). The results on the pango lineage and the genome recovered based on the analysis of the consensus sequences submitted by participants were compared with their questionnaire responses. The comparisons of these are available in Fig. 1A,B.
Table 4.
Performance assessment of participants based on the consensus sequence submitted for Part 1 of the SARS-CoV-2 whole genome sequencing proficiency testing program
| Sample ID | Performance assessment based on consensus sequence |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| BS01 | Pass | Pass | NA | Pass | Pass | Pass | Pass | Pass | Pass | Pass | Pass |
| BS02 | Pass | Pass | NA | Pass | Pass | Pass | Pass | Pass | Pass | Pass | Pass |
| BS03 | Pass | Pass | NA | Pass | Pass | Pass | Pass | Pass | Pass | Pass | Pass |
| BS04 | Pass | Pass | NA | Pass | Pass | Pass | Pass | Pass | Pass | Pass | Pass |
| BS05 | Pass | Pass | NA | Pass | Pass | Pass | Pass | Pass | Pass | Pass | Pass |
| BS06 | Pass | Pass | NA | Pass | Pass | Pass | Pass | Pass | Pass | Fail | Pass |
| BS07 | Pass | Pass | NA | Pass | Pass | Pass | Pass | Pass | Pass | Pass | Pass |
| BS08 | Pass | Pass | NA | Pass | Pass | Pass | Pass | Pass | Pass | Pass | Pass |
NA, not assessed.
Fig. 1.
Comparisons of results on the (A) pango lineage and the (B) genome recovered for samples BS01–BS08, based on the analysis of the consensus sequences submitted by participants and their questionnaire responses in Part 1 of the SARS-CoV-2 whole genome sequencing proficiency testing program.
Average read depth
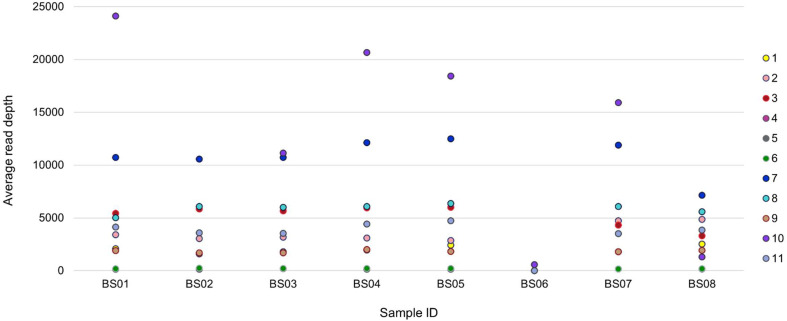
An overview of the average read depth based on the questionnaire responses is available in Fig. 2 . Participant 5 was the only participant that did not submit any results for the average read depth of all samples; the participant indicated that read depth was subsampled to 200× coverage maximum as per Artic 1.2.1 and as described.23 Participant 10 submitted unusually high reads at 83509× for sample BS02, which has been excluded from the figure. Participants who returned results for the negative sample BS06 either indicated the average read depth as 0 (n=3) or close to 0 (n=2), except participant 10, who returned 570× average read depth.
Fig. 2.
Overview of the average read depth for samples BS01–BS08, as reported by participants in Part 1 questionnaire responses.
Single-nucleotide polymorphisms and amino acid replacement
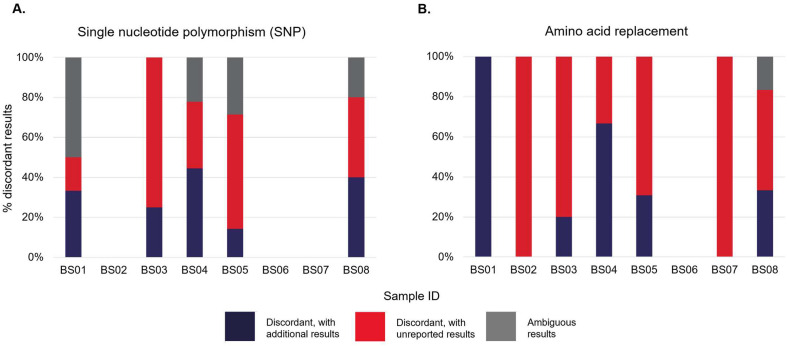
As reported by participants, snapshots of the discordant results for the SNPs and amino acid replacement are available in Fig. 3 A,B. Ambiguous SNP results included any nomenclature indication of M, S, R and Y, which did not match the sequence analysis results performed in-house during the pre-issue testing. Ambiguous amino acid replacement results were any results that did not match with the sequence analysis results performed in-house at the pre-issue testing. The details of these results are available in Supplementary Tables 1 and 2 and (Appendix A). No discordant or ambiguous SNP results were recorded for samples BS02, BS06 and BS07, while concordant amino acid replacement results were only recorded for sample BS06.
Fig. 3.
Discordant results for the single nucleotide polymorphism (SNP) and amino acid replacement for samples BS01–BS08, as reported by participants in Part 1 questionnaire response.
The SNP results reported for sample BS08 (1:103 dilution) were inconsistent, as reported by three participants (Supplementary Table 1, Appendix A), compared to samples BS02 (undiluted) and BS07 (1:102 dilution), which contained the same samples and were serially diluted from the same stock. Similar findings were also observed for amino acid replacement results for sample BS08, as submitted by the same three participants (Supplementary Table 2, Appendix A). Only two participants had 100% concordance for the SNP results, while none scored 100% concordance for the amino acid replacement results submitted for all samples.
Participants' protocols and approaches
Most participants (n=10) performed the WGS within their laboratory, while only one laboratory sequenced the survey specimens at an external sequencing facility. The choice of protocols and approaches used in generating the WGS data are available in the Supplementary Data (Appendix A). All participants indicated that the percentage genome coverage/recovery is one of the QC criteria used to evaluate the sequence data quality. A total of 8/11 participants nominated sequencing depth as another QC criterion that they had used. The majority (7/11) did not consider SNP distance from the reference genome, and 8/11 participants nominated ambiguous bases/sites as one of their QC criteria. A total of 7/11 participants considered contamination as a QC criterion. Thresholds or conditions used for all these criteria are summarised in the Supplementary Data (Appendix A). Only one participant indicated that heterozygous sites would be included in the QC process, with a threshold of 40.
Phylogenetic analysis
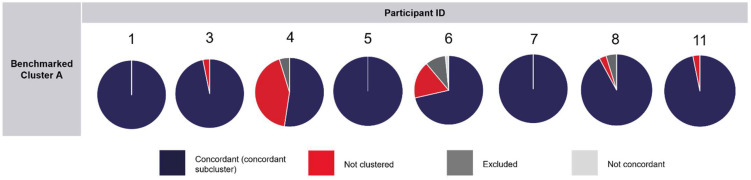
A total of 8/11 participants submitted their phylogenetic analysis results for Part 2 of the PTP. After the closing date for this PTP, the report provided to participants included the phylogenetic tree generated using the genomic clusters of interest to epidemiological investigations, as classified by each participant in their responses. This tree was compared to the phylogenetic tree in Supplementary Fig. 1 (Appendix A), which consisted of the 70 sequences and their respective benchmarked clusters. There was 100% concordance with benchmarked cluster C in the results submitted by participants. A 100% concordance (including concordant sub-clustering) with benchmarked cluster B was also observed in the results submitted by most of the participants (n=7), with only one participant recording 75% concordance. Three participants recorded results of 100% concordance with benchmarked cluster A, as designated in-house at the pre-issue testing. The assessment of results in concordance with benchmarked cluster A is available in Fig. 4 . Only one participant (participant 6) interpreted the cluster of an isolate that contradicted the benchmarked cluster A (not concordant). The summary of responses submitted for the questionnaire of Part 2 of the PTP is available in the Supplementary Data (Appendix A).
Fig. 4.
Assessment of result concordance with benchmarked cluster A, as reported by participants in the phylogenetic analysis of 70 SARS-CoV-2 consensus genomes in Part 2 of the SARS-CoV-2 whole genome sequencing proficiency testing program.
Discussion
WGS data can reveal the genetic makeup of the virus and can be used to discriminate between mutation patterns of the SARS-CoV-2 virus from different clinical samples. 24, 25, 26 This approach is crucial in identifying the source of infection and transmission routes by monitoring the emergence of variants over time and through communities.27 , 28 It can also determine whether the virus was acquired overseas or locally from a known or unknown contact. At the beginning of the pandemic or with the emergence of new VoC, limited viral diversity results in identical SARS-CoV-2 genomes in epidemiologically unrelated cases and makes the interpretation with epidemiological context vital. All this information is critical in planning and delivering public health measures to minimise the impact of the diseases. With the rapid introduction and broader application of new SARS-CoV-2 WGS protocols across laboratories for public health purposes, little is known about whether a laboratory has effectively and accurately implemented these protocols in its SARS-CoV-2 WGS processes. Therefore, the development of a SARS-CoV-2 WGS PTP is required to assess SARS-CoV-2 WGS protocols and the performance of individual laboratories, which includes the associated bioinformatics capacity in the clinical setting.
To our knowledge, the PTP described in this study that opened in February 2021 was the first SARS-CoV-2 WGS PTP offered worldwide, approximately a year after the pandemic began. The results submitted by participants of the RCPAQAP SARS-CoV-2 WGS PTP demonstrated the capability and proficiency of laboratories across Australia in performing SARS-CoV-2 WGS, both the laboratory-based component (wet lab) and the bioinformatic analysis of the sequence data (dry lab). Overall, the participants' performance was commendable. Based on the percentage genome recovered, pango lineage, and sequencing accuracy for SARS-CoV-2 samples, most participants who submitted the consensus sequences passed the assessment of Part 1 of the PTP. However, these criteria should be set more stringently for future SARS-CoV-2 WGS PTP, for instance, a minimum of 90% genome coverage and over 99% sequencing accuracy. The negative specimen was added to the specimen panel to assess the laboratory-based component of the WGS process. The participant who failed the SARS-CoV-2 negative specimen assessment potentially had a contamination issue while processing the survey specimens. By comparing the results (questionnaire responses versus PTP assessment), it became apparent that WGS protocols used in the whole genome sequence data analysis ultimately determine the outcome of the SARS-CoV-2 WGS. The use of the most suitable SARS-CoV-2 WGS protocols and bioinformatics methods, as reported in a comprehensive benchmark study19 which ranked the performance of protocols based on six different metrics, is not only crucial for the research community but also for diagnostics.
We found that the correct identification of SNPs and amino acid replacement was particularly challenging to participating laboratories, with low concordance recorded for results submitted by most participants. Interestingly, there were inconsistent findings in the SNP and amino acid replacement results reported for sample BS08 compared to samples BS02 and BS07. These samples contained the same viral RNA and were serially diluted from the one stock solution to represent patient specimens presented to the laboratory as positive cases with low viral load in a clinical setting. The cause of inconsistencies observed in the SNP and amino acid replacement results reported for sample BS08 (1:103 dilution) is unknown. However, it may be associated with the lower viral load in this sample compared to its counterparts with higher viral load, samples BS02 (undiluted) and BS07 (1:102 dilution). According to Liu et al.,19 WGS performed on samples with low viral loads may return lower genome coverage and impact the WGS quality and confidence in SNP or insertion or deletion (indel) detection calls. However, this pattern was not observed across all participants' WGS data, which returned high genome coverage of above 80% for all specimens (except sample BS08 sequenced by participant 1).
In conclusion, our study highlights the importance of PTPs for WGS. The PTP offered by RCPAQAP provides insight into the current state of SARS-CoV-2 genomics in public health. The results were positive and demonstrated the critical role of the PTP in supporting the implementation and validation of WGS processes and the potential to provide ongoing performance benchmarking for accreditation of test processes employing this technology. The ongoing participation of clinical and public health laboratories in a WGS PTP will improve the quality of the SARS-CoV-2 WGS processes, and the RCPAQAP will continue to offer this annually, depending on the demand. Further study on the data derived from this PTP will contribute to the development of SARS-CoV-2 bioinformatic QC for test processes and standards or the performance benchmarking for accreditation.
Acknowledgement
We thank the contribution of the scientists and pathologists of Melbourne Health in supplying the biospecimens in this project.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pathol.2022.04.002.
Conflicts of interest and sources of funding
The RCPAQAP Biosecurity is funded by the Australian Government Department of Health (Grant no. 4-4GCFA17). The authors state that there are no conflicts of interest to disclose.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., et al. A novel Coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corman V.M., Landt O., Kaiser M., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X., Tan Y., Ling Y., et al. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583:437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 4.First NGS-based COVID-19 diagnostic. Nat Biotechnol. 2020;38:777. doi: 10.1038/s41587-020-0608-y. [DOI] [PubMed] [Google Scholar]
- 5.Thanh Le T., Andreadakis Z., Kumar A., et al. The COVID-19 vaccine development landscape. Nat Rev Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 6.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci USA. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedford T., Greninger A.L., Roychoudhury P., et al. Cryptic transmission of SARS-CoV-2 in Washington state. Science. 2020;370:571–575. doi: 10.1126/science.abc0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler D.J., Mozsary C., Meydan C., et al. Shotgun transcriptome and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. bioRxiv. 2020 doi: 10.1038/s41467-021-21361-7. May 1: 2020.04.20.048066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurano M.T., Ramaswami S., Zappile P., et al. Sequencing identifies multiple early introductions of SARS-CoV-2 to the New York City region. Genome Res. 2020;30:1781–1788. doi: 10.1101/gr.266676.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polonsky J.A., Baidjoe A., Kamvar Z.N., et al. Outbreak analytics: a developing data science for informing the response to emerging pathogens. Philos Trans R Soc Lond B Biol Sci. 2019;374 doi: 10.1098/rstb.2018.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modjarrad K., Moorthy V.S., Millett P., Gsell P.-S., Roth C., Kieny M.-P. Developing global norms for sharing data and results during public health emergencies. PLOS Med. 2016;13 doi: 10.1371/journal.pmed.1001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gire S.K., Goba A., Andersen K.G., et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Eurosurveillance. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Toole Á, Scher E., Underwood A., et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7:veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoang T., da Silva A.G., Jennison A.V., Williamson D.A., Howden B.P., Seemann T. AusTrakka: fast-tracking nationalized genomics surveillance in response to the COVID-19 pandemic. Nat Commun. 2022;13:865. doi: 10.1038/s41467-022-28529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amid C., Pakseresht N., Silvester N., et al. The COMPARE Data Hubs. Database (Oxford) 2019 doi: 10.1093/database/baz136. 2019: baz136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T., Chen Z., Chen W., et al. A benchmarking study of SARS-CoV-2 whole-genome sequencing protocols using COVID-19 patient samples. iScience. 2021;24 doi: 10.1016/j.isci.2021.102892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hourdel V., Kwasiborski A., Balière C., et al. Rapid genomic characterization of SARS-CoV-2 by direct amplicon-based sequencing through comparison of MinION and Illumina iSeq100(TM) System. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.571328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau K.A., Theis T., Gray J., Rawlinson W.D. Ebola preparedness: diagnosis improvement using rapid approaches for proficiency testing. J Clin Microbiol. 2017;55:783–790. doi: 10.1128/JCM.02173-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pastorino B., Touret F., Gilles M., Luciani L., de Lamballerie X., Charrel R.N. Evaluation of chemical protocols for inactivating SARS-CoV-2 infectious samples. Viruses. 2020;12:624. doi: 10.3390/v12060624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loman N., Rowe W., Rambaut A. 23 Jan 2020. nCoV-2019 novel coronavirus bioinformatics protocol.https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html [Google Scholar]
- 24.Oude Munnink B.B., Nieuwenhuijse D.F., Stein M., et al. Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in The Netherlands. Nat Med. 2020;26:1405–1410. doi: 10.1038/s41591-020-0997-y. [DOI] [PubMed] [Google Scholar]
- 25.Umair M., Ikram A., Salman M., et al. Whole-genome sequencing of SARS-CoV-2 reveals the detection of G614 variant in Pakistan. PLoS One. 2021;16 doi: 10.1371/journal.pone.0248371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muttineni R., Kammili N., Bingi T.C., et al. Clinical and whole genome characterization of SARS-CoV-2 in India. PLoS One. 2021;16 doi: 10.1371/journal.pone.0246173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersson P., Sherry N.L., Howden B.P. Surveillance for SARS-CoV-2 variants of concern in the Australian context. Med J Aust. 2021;214:500–502.e1. doi: 10.5694/mja2.51105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seemann T., Lane C.R., Sherry N.L., et al. Tracking the COVID-19 pandemic in Australia using genomics. Nat Commun. 2020;11:4376. doi: 10.1038/s41467-020-18314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.