Abstract
Surveillance of bacterial susceptibility to five antimicrobial agents was performed during a 1-year period in and around four freshwater fish farms situated along a stream in western Denmark. Besides assessing the levels of antibiotic resistance among the culturable fraction of microorganisms in fish, water, and sediment samples, two major fish pathogens (88 Flavobacterium psychrophilum isolates and 134 Yersinia ruckeri isolates) and 313 motile Aeromonas isolates, representing a group of ubiquitous aquatic bacteria, were isolated from the same samples. MICs were obtained applying a standardized agar dilution method. A markedly decreased susceptibility of F. psychrophilum isolates to most antimicrobial agents presently available for use in Danish aquaculture was detected, while the collected Y. ruckeri isolates remained largely sensitive to all therapeutic substances. Comparing the inlet and outlet samples, the increase of the antibiotic-resistant proportions observed among the culturable microflora was more pronounced and statistically significant among the motile aeromonads. High levels of individual and multiple antimicrobial resistances were demonstrated within the collected flavobacteria and aeromonads, thus indicating a substantial impact of fish farming on several groups of bacteria associated with aquacultural environments.
The apparent increase of the occurrence of antibiotic resistance among bacteria from various areas of animal production during the past years and its possible implications for public health (2, 29, 43) have in many countries lead to an intensified surveillance of bacterial resistance. In the field of aquaculture, both therapeutic and environmental problems have been addressed, as antimicrobial agents are released into the surrounding water during medical treatment of bacterial fish diseases (2, 6). The impact of these substances on the resident microflora is difficult to assess because of the complexity of the aquatic environment, while the resistance patterns of bacterial fish pathogens often reflect an intensive use of antimicrobial substances (7, 38, 44). Numerous investigators have attempted to elucidate the occurrence and persistence of antibiotic resistance, mostly in marine aquaculture production systems, which predominate in the production of salmonids (19, 25, 32, 40). In contrast, the predominant type of aquacultural production in Denmark is freshwater inland farming, and fewer data are available in this area (12, 14, 30, 41). The aim of the present study was to determine the prevalence and persistence of antimicrobial resistance in a typical Danish freshwater stream with numerous rainbow trout farms. The design of the project allowed the detection of changes of resistance levels in water and sediment sampled at different sites along the river.
In addition to the total resistant bacterial flora, we focused on the three important fish pathogens Yersinia ruckeri, Flavobacterium psychrophilum, and Aeromonas salmonicida, which occur enzootically in Denmark and are associated with enteric redmouth disease (ERM), rainbow trout fry syndrome, and furunculosis, respectively. Furthermore, a group of motile aeromonads was also examined for resistance. The specific resistance patterns of all isolates were studied in order to register local changes and differences between the four fish farms.
Five antimicrobial agents that are currently used in treatment of bacterial fish diseases in Denmark were selected: oxolinic acid (OXA) and sulfadiazine-trimethoprim (S-T) are licensed drugs, while dispensation is required for the use of amoxicillin (AMX), oxytetracycline (OTC), and florfenicol (FLO).
MATERIALS AND METHODS
Sampling and processing of samples.
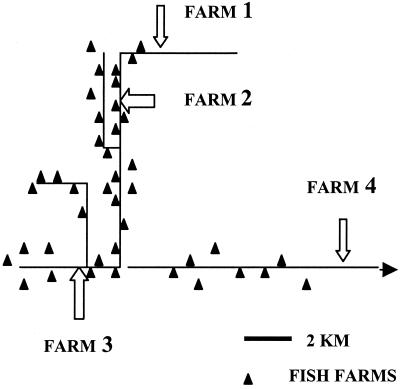
Between October 1997 and February 1999, we sampled four fish farms, situated along the Danish stream Vejle Å, on 11 occasions at approximately monthly intervals (Fig. 1). Farm 1 was located furthest upstream and thus received no effluents from other fish farms. However, the stream had at this point previously received effluents from a sewage treatment plant and some agricultural areas. The farms differed with respect to production size, management, disease incidence, and, consequently, the usage of antibiotic compounds, as listed in Table 1. Figure 1 shows the density and distribution of fish farms in the district and the location of the four test farms.
FIG. 1.
Topography of fish farms (▴) at Vejle Å, showing the relative positions of the test farms (arrows).
TABLE 1.
Summary of antimicrobial use and production of fish on four Danish rainbow trout farms during the sampling period (October 1997 to February 1999)
| Farm | Description | Avg production (tons) in 1996–1998 | Registered consumption of antibiotics (Kg)a
|
|||
|---|---|---|---|---|---|---|
| OXA | S-T | AMX | FLO | |||
| 1 | Earth ponds, fingerling production, high stocking density, high disease incidence | 66 | 0 | 150 | 24 | 0.5 |
| 2 | Earth ponds, fingerlings and adults, ERM vaccination, low disease rate | 55 | 1 | 0.8 | 0.4 | 0 |
| 3 | Concrete ponds, fingerling production, recurrent ERM problems | 132 | 35 | 47.5 | 8.5 | 0 |
| 4 | Earth ponds, rearing only adult fish, few clinical problems | 539 | 31 | 35 | 0 | 0 |
Active compound. Results for OXA, S-T, and AMX are for 1996 to 1998; those for FLO are for 1998. Amounts were annually reported by the fish farmers (40). The 1999 data were not available at the time of submission.
Duplicate water samples from about 10 cm beneath the surface were collected in sterile plastic flasks (100 ml) from inlet water, outlet water, and a pond of each farm. Sediment from the same sites was sampled with a bottom collector (van Veen type), and two portions of the 1-cm top layer were transferred aseptically to plastic flasks. Two to four fry (<5 g) and two larger fish (20 to 100 g) were caught on each farm, preferably from the same pond every time. In the case of clinical disease, two to four affected fish were also collected per farm. All samples were stored on ice and processed in the laboratory within 6 h. Water temperatures were registered on each farm.
Bacteriological examination and culture conditions.
Appropriate 10-fold dilutions of the water and sediment samples in physiological saline (PS) (0.9% [wt/vol] NaCl) were prepared, and 0.1-ml aliquots were plated in duplicate on blood agar (BA) plates (BA base supplemented with 5% citrated calf blood [Difco Laboratories, Detroit, Mich.]), tryptone yeast extract salts (TYES) agar plates (7), and TYES plates containing a fixed amount of one of the selected antimicrobial agents: OTC (10 μg ml−1), OXA (4 μg ml−1), S-T (50-10 μg ml−1), AMX (4 μg ml−1), and FLO (4 μg ml−1). Stock solutions of the respective antibiotics were prepared as previously described (31) (OTC and S-T, European Pharmacopeia, 2nd ed.; OXA and AMX, Sigma Chemical, Poole, United Kingdom; FLO, Schering-Plough Animal Health, Bloomfield, N.J.). All TYES plates were incubated at 15°C for 5 days, while BA plates were incubated at 20°C for 2 days.
BA plates and TYES plates with and without antibiotics were also used to spread out 0.1-ml aliquots of bacterial suspensions and dilutions of samples obtained from fish as follows. From the two large fish, the second gill arch in both sides was excised and vortexed for 1 minute in 10 ml of PS, and one additional 10-fold dilution of the suspension was prepared before plating. From the same fish, mucus was collected by scraping a skin area of 1 by 2 cm with a sterile scalpel. Subsequently the sample was suspended by vortexing in 10 ml of PS and diluted 10 times more. The fry were weighed, cut into small pieces, and homogenized with the amount of PS resulting in a 10−1 dilution, followed by three additional 10-fold dilution steps.
Moreover, all samples from fish and sediment from the pond were routinely plated on selective media designed for the recovery of Y. ruckeri (ribose-ornithin-dextrin agar [ROD] [13]), A. salmonicida (Coomassie brilliant blue [10]), and Aeromonas hydrophila (Pril-ampicillin-dextrin-ethanol [PADE] agar [21]). Coomassie brilliant blue and PADE agar plates were incubated at 20°C for 2 days, and ROD agar plates were incubated at 20°C for 4 days. If diseased fish were found, they were also analyzed for the presence of bacterial pathogens in kidney, spleen, and brain by plating organ samples on BA and plain TYES plates.
Identification.
Final identification of presumptive Y. ruckeri colonies on BA or ROD agar was based on biochemical tests (13) and on a PCR method (3). Oligonucleotide primers were provided by DNA Technology A/S, Aarhus, Denmark, and the thermal cycler was a GeneAmp PCR system 9700 (Perkin-Elmer, Foster City, Calif.).
Similarly, F. psychrophilum-like yellow colonies were picked from TYES plates with or without antibiotics for further biochemical characterization as previously described by Pacha (34) and confirmation by a species-specific PCR method as proposed by Toyama et al. (45). A. salmonicida isolates were not recovered from any sample throughout the whole sample period.
The motile aeromonads were isolated from BA, PADE agar plates, and TYES plates containing AMX and subsequently identified biochemically to the genus level (23). Isolates were considered to be presumptive aeromonads if they were gram-negative, oxidase- and catalase-positive rods which were facultatively anaerobic and fermented carbohydrates, sometimes producing gas. Included isolates were also nitrate reducing, positive for arginine and lysine decarboxylases, negative for ornithin decarboxylase, and resistant to O/129. The PADE agar allowed the growth of a number of motile Aeromonas species besides A. hydrophila, some of which can be extremely difficult to distinguish by conventional biochemical methods, particularly if they originate from environmental sources (33). As the OTC, OXA, S-T, and FLO resistance levels of the species in question did not vary significantly as determined by MIC testing of their respective type strains (data not shown), it was decided to include all motile Aeromonas isolates in the following analysis of specific antibiotic resistance patterns (8).
MIC testing.
Following identification, MICs were determined for all isolates, using an agar dilution method as suggested by the National Committee for Clinical Laboratory Standards (NCCLS) (31). Mueller-Hinton agar (Difco) was the basic medium but was modified for testing of F. psychrophilum as described by Hawke and Thune (16). Doubling dilutions of antibiotic stock solutions (31) were incorporated into the agar plates, with final concentrations ranging from 0.125 to 1,024 μg ml−1. For the combined S-T values, any given concentration refers to the concentration of sulfadiazine, with the ratio between sulfadiazine and trimethoprim being 5:1.
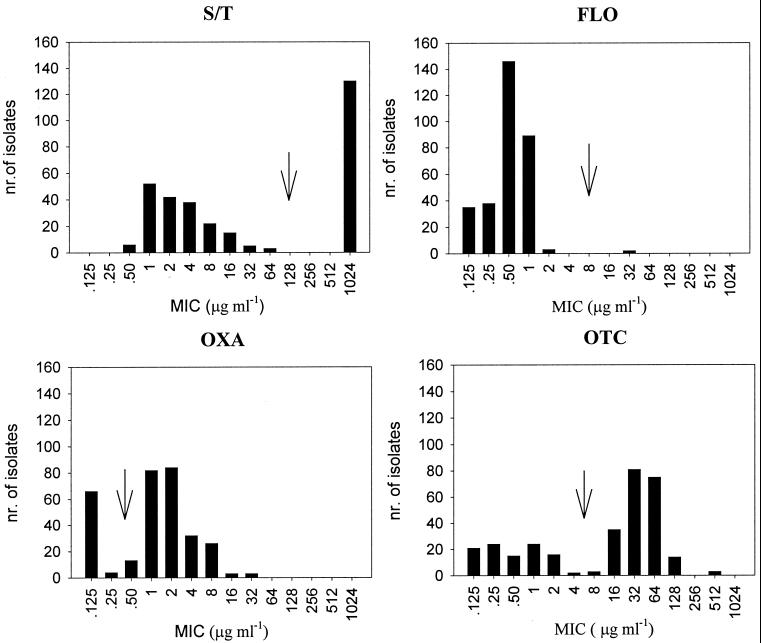
Motile Aeromonas and Yersinia isolates were cultured overnight in Veal infusion broth (Difco) at 20°C, while F. psychrophilum isolates were grown in tryptone yeast extract salts (TYES) broth (7) at 15°C for 2 days. Broth cultures were adjusted to an optical density of a 0.5 McFarland standard (corresponding to 108 CFU ml−1), diluted 1:10 in PS, and applied as 1-μl droplets to the plates, employing a multipoint inoculator (P&R Laboratory Group, St. Helens, United Kingdom). The inoculum was in this way standardized to contain approximately 104 CFU. Every test was run in duplicate on freshly prepared agar plates. The first and the last agar plates did not contain any antibiotics in order to detect possible contamination of the isolates or antibiotic carryover. As recommended in the NCCLS guidelines, the following reference strains were included as internal standards in all tests: Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 29213), Enterococcus faecalis (ATCC 29212), and Pseudomonas aeruginosa (ATCC 27853). Depending on which isolates were tested, the respective type strains were also included (Y. ruckeri ATCC 11476, F. psychrophilum NCIMB 1947, and A. hydrophila ATCC 7699). After 2 days of incubation at 20°C (for F. psychrophilum, 4 days at 15°C), the MIC for each isolate was determined as the lowest concentration of the antimicrobial agent able to inhibit bacterial growth. Subsequently, the isolates were classified as sensitive or resistant to the antibiotic in question, provided that they formed two separate clusters depending on their MIC values (see Fig. 3).
FIG. 3.
Frequencies of MICs for motile Aeromonas isolates (n = 313), determined by a standardized agar dilution method. The arrows indicate the suggested breakpoints for the respective antimicrobial agents. Aeromonads are thought to be intrinsically resistant to AMX.
Statistical methods. (i) Total counts.
CFU were enumerated, and the bacterial numbers per milliliter or per gram of sample were calculated, based on two or three dilution steps. It appeared that duplicate samples from the same site did not differ significantly in their bacterial counts (data not shown), and therefore the mean from both samples was used to calculate the proportion of resistant bacteria compared to the total number of culturable bacteria. It was decided to transform the counts logarithmically in order to stabilize the variance of the data. The assumption of normality of the data was satisfied. An analysis of variance was performed, including the application of a covariance structure for longitudinal data, where measurements closer together in time are considered to be more closely correlated (proc mixed in SAS version 6.12; SAS Institute Inc., Cary, N.C.).
(ii) Resistance counts of F. psychrophilum and Aeromonas isolates.
The proportions of antibiotic-resistant isolates among the F. psychrophilum and Aeromonas isolates were computed for each fish farm and all sampling sites. A logistic regression model was employed (proc genmod in SAS version 6.12) to detect differences between resistance rates from inlets and outlets as well as differences between fish farms.
Resistance proportions among Y. ruckeri isolates were not analyzed in the same manner, because very few of them proved to be antibiotic resistant (see Results).
RESULTS
Total counts.
The overall counts of CFU on TYES plates yielded bacterial counts of between 3.8 × 103 and 7.5 × 104 culturable bacteria per ml of water, while sediment samples ranged between 3.4 × 106 and 7.3 × 108 CFU g−1. Whole homogenized fish contained 3 × 105 to 2.1 × 108 CFU g−1, and the cell suspensions from both gills and mucus were found to have CFU counts of between 2 × 103 and 1.5 × 105 ml−1. The resistance proportions detected within the culturable bacteria did not vary significantly when comparing water, sediment, and fish samples.
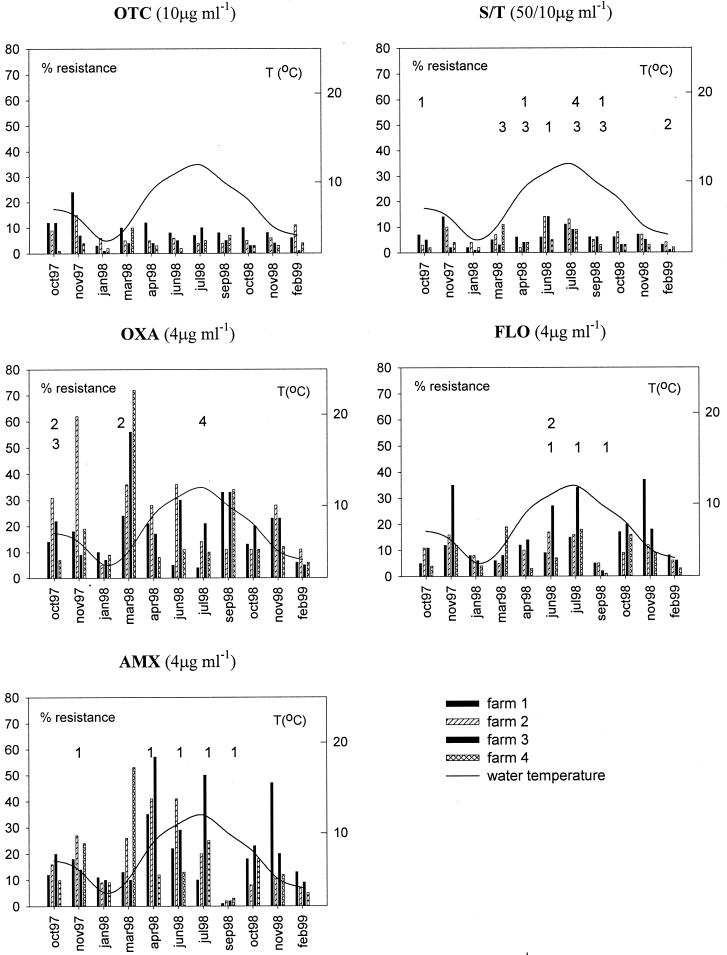
For all five antibiotics tested, it was found that there was significant variation in total resistance levels in regard to different sample times (0.0001 < P < 0.041), as shown in Fig. 2. There appears to be a seasonal variation, where the samples contained lower fractions of resistant bacteria when water temperatures dropped to 3°C in January 1998 and February 1999.
FIG. 2.
Antimicrobial-resistant fractions of culturable bacteria found in water and sediment samples from the four fish farms during the trial period. The numbers above the bars indicate in which farm the relevant agent had been used at the time of sampling or within 3 days before sampling.
On average, 4.8% of the culturable bacteria proved to be OTC resistant, while 4.7% were S-T resistant and 15.8% were OXA resistant. Corresponding proportions for AMX and FLO were 14.5 and 9.5%, respectively (Table 2).
TABLE 2.
Percentages of antimicrobial resistance detected among the total culturable microflora and within Aeromonas and F. psychrophilum isolates in water, sediment, and fish samples from four freshwater fish farms
| Microflora | Farm (no. of samples or isolates) | % Resistance toa:
|
|||||
|---|---|---|---|---|---|---|---|
| OTC | S-T | OXA | AMX | FLO | >2RESb | ||
| Total counts (264 samples) | 1 (66) | 5.7 | 4.7 | 12.8 | 13.7 | 9.9 | |
| 2 (66) | 6.1 | 6.2 | 19.3 | 17.8 | 11.2 | ||
| 3 (66) | 4.8 | 4.2 | 19.5 | 15.8 | 10.7 | ||
| 4 (66) | 2.6c | 3.7 | 11.6c | 10.7c | 6.2c | ||
| Avg | 4.8 | 4.7 | 15.8 | 14.5 | 9.5 | ||
| Aeromonads (313 isolates) | 1 (69) | 65 | 61d | 19 | 100 | 0.3 | 62d |
| 2 (98) | 75 | 42 | 19 | 100 | 0 | 51 | |
| 3 (53) | 68 | 25 | 21 | 100 | 0 | 32 | |
| 4 (93) | 67 | 42 | 22 | 100 | 0 | 46 | |
| Avg | 69 | 43 | 20 | 100 | 0.1 | 48 | |
| F. psychrophilum (89 isolates) | 1 (36) | 81d | 100 | 100 | 81d | 0 | 94d |
| 2 (19) | 84d | 100 | 100 | 53 | 0 | 84d | |
| 3 (18) | 50 | 100 | 98 | 0 | 0 | 44 | |
| 4 (16) | 56 | 100 | 100 | 31 | 0 | 63 | |
| Avg | 71 | 100 | 100 | 50 | 0 | 72 | |
Total-count percentages were derived from differential plating on agar plates with and without the respective antibiotics, while Aeromonas and Flavobacterium isolates were classified as resistant or sensitive according to their relative MICs, which were obtained from an agar dilution assay. Counts from sediment samples were not included, as skewness of the data due to many missing values impaired the statistical analysis.
>2RES, isolates with three or more antibiotic resistance phenotypes.
Statistically significant lower fraction of resistant bacteria compared to water samples from other farms.
Statistically significant higher proportion of resistant strains compared to isolates collected from other farms.
Comparison of inlets, ponds, and outlets of the fish farms showed that water samples from the inlets contained lower fractions of antibiotic-resistant bacteria than the corresponding samples from ponds and outlets (Table 3). However, this difference was statistically significant only for the levels of OTC resistance (3.2% at the inlets, in contrast to 5.5% in ponds [P = 0.037], and 5.4% at outlets [P = 0.038]).
TABLE 3.
Percentages of antimicrobial resistance detected among the total culturable microflora and within Aeromonas and F. psychrophilum isolates in water and sediment sampled at different sites (inlet, pond, and outlet) on four freshwater trout farms
| Microflora | Site (no. of samples or isolates) | % Resistance toa:
|
|||||
|---|---|---|---|---|---|---|---|
| OTC | S-T | OXA | AMX | FLO | >2RESb | ||
| Total counts (528 samples) | Water | ||||||
| Inlet (88) | 3.2c | 3.3 | 13.4 | 14.3 | 9.4 | ||
| Pond (88) | 5.5 | 5.3 | 16.9 | 15.3 | 10.3 | ||
| Outlet (88) | 5.4 | 5.6 | 16.4 | 15.1 | 9.4 | ||
| Sediment | |||||||
| Inlet (88) | 1.7c | 2.6 | 14.8 | 7.4 | 2.8 | ||
| Pond (88) | 4.4 | 3.1 | 8.0 | ||||
| Outlet (88) | 3.9 | 3.1 | 8.7 | 4.8 | 2.2 | ||
| Aeromonads (250 isolates) | Inlet (53) | 49 | 19 | 9 | 100 | 2 | 23 |
| Pond (92) | 71d | 49d | 23 | 100 | 0 | 57d | |
| Outlet (105) | 77d | 47d | 28d | 100 | 0 | 51d | |
| F. psychrophilum (42 isolates) | Inlet (9) | 22 | 100 | 100 | 33 | 0 | 44 |
| Pond (16) | 75d | 100 | 100 | 63 | 0 | 81 | |
| Outlet (17) | 71d | 100 | 100 | 41 | 0 | 71 | |
Total-count percentages were derived from differential plating on agar plates with and without the respective antibiotics, while Aeromonas and Flavobacterium isolates were classified as resistant or sensitive according to their relative MICs, which were obtained from an agar dilution assay. AMX- and FLO-resistant proportions in sediment samples from ponds were not included in the analysis (there were many missing values due to swarming or fungal growth on the agar plates).
>2RES, isolates with three or more antibiotic resistance phenotypes.
Statistically significant lower fraction of resistant bacteria compared to samples from outlets and ponds.
Statistically significant higher proportion of resistant isolates compared to isolates from inlets.
The same trend was found with regard to the proportions of OTC- and S-T-resistant bacteria in the sediment samples. Again, OTC resistance levels were significantly lower at the inlets (1.7%) than in the ponds (4.4%) (P = 0.019) or at the outlets (3.9%) (P = 0.021). In contrast, the percentages of OXA, AMX, and FLO resistance in sediment appeared to be higher in inlet samples than in outlet or pond samples (Table 3). In general, the antibiotic resistance proportions detected in sediment samples were smaller than the corresponding fractions in water samples, with the difference not being statistically significant. Resistance levels in samples from the ponds corresponded closely to those in the outlet counterparts regardless of the sample type.
Regarding the differences between the four fish farms, the results of the statistical analysis are presented in Table 2. Total counts from sediment samples were not included because skewness of the data (there were many missing values from farm 3) impaired a reliable outcome. Interestingly, the comparison of the four farms yielded statistically significant differences for total environmental samples with regard to OTC (P = 0.034), OXA (P = 0.014), AMX (P = 0.038), and FLO (P = 0.026) resistance but not for S-T-resistant bacteria. Farm 4 exhibited the lowest levels of antimicrobial resistance in all samples, while the highest levels were detected in farm 2 (Table 2).
A relationship between periods of antibiotic treatment on the farms and increases in the corresponding resistance levels was not evident (Fig. 2). Thus, high percentages (40 to 55%) of AMX-resistant bacteria were found at various times on farms 2, 3, and 4, although this agent was not used at these sites throughout the trial period. Similarly, only farm 2 had reported use of OXA in March 1998 at the time of sampling, but still levels of OXA resistance were high, up to 70% on farm 4. Conversely, farm 1 experienced several outbreaks of rainbow trout fry syndrome in the summer of 1998, which were treated with FLO and AMX, as well as recurring problems with ERM, which was treated with S-T, yet resistance proportions did not increase significantly at the respective sampling times (Fig. 2).
Inlet samples from the first farm upstream, farm 1, did not contain significantly smaller fractions of resistant bacteria than inlet samples from other farms.
Motile aeromonads.
The 313 Aeromonas isolates were recovered from all types of samples: 110 from water, 140 from sediment, and 63 from gills and mucus from healthy fish. Resistance patterns did not vary significantly between isolates from different sample types. Two hundred sixteen Aeromonas isolates (69%) were resistant to OTC, with MICs of 32 to 256 μg ml−1 (sensitive isolates, 0.125 to 1.0 μg ml−1), while 135 (43%) displayed S-T resistance (MIC values of >1,024-205 μg ml−1 compared to 0.5-0.1 to 8-1.6 μg ml−1 among sensitive isolates). Sixty three isolates (20%) were OXA resistant (MICs of between 4 and 16 μg ml−1; sensitive isolates, 0.125 to 1.0 μg ml−1). Only a single isolate was found to be resistant to FLO (MIC of 32 μg ml−1, versus 0.125 to 1.0 μg ml−1 for sensitive isolates), while all field isolates and type strains were intrinsically AMX resistant (MIC of >256 μg ml−1). The distribution of the isolates according to their MICs and the resulting breakpoints are shown in Fig. 3. Apart from the AMX resistance, 151 isolates (48%) were demonstrated to carry at least two additional antibiotic resistance traits, with the predominant phenotype being AMX, OTC, and S-T resistance (28%).
Results from the logistic regression analysis demonstrated pronounced increases in pond and outlet resistance levels compared to inlet resistance levels (Table 3). The probability of water and sediment samples containing resistant aeromonads was significantly lower at the inlets with regard to resistance to OTC (inlet, 37%; pond, 75% [P = 0.0004]; outlet, 71% [P = 0.0005]) and S-T (inlet, 6%; pond, 57% [P = 0.0001]; outlet, 41% [P = 0.0001]) and multiresistance (inlet, 9%; pond, 63% [P = 0.0001]; outlet, 45% [P = 0.0012]) within fish farms 1, 3, and 4. To prevent a lack of fit of the model, farm 2 resistance rates were removed in the analyses of OTC, S-T, and multiresistance levels. These data were then analyzed separately (by chi-square tests), yielding nonsignificant results when comparing inlet and pond-outlet isolates (for OTC, inlet, 72%; pond, 83%; and outlet, 62%; for S-T, inlet, 44%; pond, 31%; and outlet, 31%; for multiresistance, inlet, 50%; pond, 31%; and outlet, 52%).
OXA resistance levels on all four farms (full model) were significantly higher in isolates sampled at the outlet (outlet, 33%; inlet, 9% [P = 0.014]).
Comparison of Aeromonas isolates from all four farms regardless of sample site (Table 2) rendered significant results for S-T resistance and multiresistance: Farm 1 isolates are more likely to be resistant to S-T (P < 0.005) and at least two additional antibiotics apart from AMX (P < 0.025) than isolates sampled at the other farms.
F. psychrophilum.
Eighty-nine out of 144 presumptive isolates were confirmed to belong to the species F. psychrophilum, and they were all resistant to at least one antibiotic agent in addition to sulfadiazine, to which they are thought to be intrinsically resistant. All isolates proved to be resistant to OXA, with MICs of between 4 and 16 μg ml−1 (the MIC for the sensitive type isolate NCIMB 1947 was 0.25 μg ml−1). Sixty-three isolates (71%) were OTC resistant, with MICs ranging from 1 to 8 μg ml−1, compared to 0.063 to 0.125 μg ml−1 for sensitive isolates. FLO resistance was not found, whereas AMX resistance was detected in 44 (50%) of the isolates, with MICs of between 1 and 2 μg ml−1 (sensitive isolates, 0.016 to 0.125 μg ml−1). Many isolates (72%) expressed three or more resistance phenotypes (Table 2), with S-T, OXA, and OTC resistance being the most prevalent combination.
Only 1 isolate was recovered from sediment, 42 isolates (47%) were isolated from water, and the remaining 46 isolates originated from diseased fish. Resistance levels were not significantly different in water and fish isolates.
Overall comparisons of resistance rates among isolates collected at inlets, ponds, or outlets were possible only for water isolates (Table 3). Among those, the OTC resistance level was significantly higher among isolates from the ponds (75%; P = 0.009) and outlets of farms (71%; P = 0.027) than among those from the inlets (22%). In contrast, AMX resistance rates did not differ significantly among isolates from inlets and outlets (33 versus 41%). As shown in Table 2, OTC-resistant flavobacteria were recovered more frequently from farms 1 and 2 (81 and 84%, respectively) than from farms 3 (50%) and 4 (56%) (P = 0.056). The percentage of AMX-resistant isolates was significantly (P = 0.018) higher among farm 1 isolates (81%) than among isolates from the other fish farms further downstream (farm 2, 53%; farm 3, 0%; farm 4, 31%). The same trend was apparent with regard to the proportion of multiresistant flavobacteria (P = 0.0005); 94% of the isolates from farm 1 and 84% from farm 2 exhibited resistance to at least two antimicrobial agents besides S-T, whereas the proportions in farm 3 and 4 were 44 and 63%, respectively. Curiously, none of the isolates from farm 3 were resistant to AMX. Farm 3 isolates also exhibited the lowest fraction of OTC-resistant and multiresistant isolates (50 and 44%, respectively).
Y. ruckeri.
Of the 134 Y. ruckeri isolates isolated from diseased fish or apparently healthy carriers, none was resistant to OTC or S-T. Forty isolates (30%) exhibited elevated OXA MICs (2 to 8 μg ml−1), but this group of isolates with reduced sensitivity is not very distinct, as MICs are only slightly higher than among the 69 sensitive isolates (0.125 to 0.5 μg ml−1), and 25 isolates were consequently considered to be intermediate sensitive (MIC = 1 μg ml−1). Moreover, the OTC MICs ranged from 2 to 8 μg ml−1 (that for the reference strain ATCC 11476 is 4 μg ml−1), the FLO MICs ranged from 4 to 8 μg ml−1 (ATCC 11476, 8 μg ml−1), the S-T MICs ranged from 0.25-0.05 to 1-0.2 μg ml−1 (ATCC 11476, 2-0.4 μg ml−1), and the AMX MICs ranged from 4 to 16 μg ml−1 (ATCC 11476, 8 μg ml−1). These ranges correspond to MICs reported previously for this species (9, 12). Comparing the four fish farms, the Yersinia isolates did not differ significantly in regard to MICs. No environmental isolates were found.
DISCUSSION
The majority of the fish farms along the river release their effluents after passage of sedimentation ponds and without further treatment. Various amounts of antibiotic residues may still be present in the effluent water following antibiotic therapy on the farms (20, 37, 46) and persist on and around farms (15, 18, 22). Numerous studies suggest a correlation between findings of increased bacterial resistance levels on and around inland fish farms and the antimicrobial agents used at the farms (12, 14, 30, 41). In this investigation, a simple overall correlation between antibiotic usage and emergence of antibiotic resistance was not evident (Fig. 2), perhaps because the sampling strategy was not always correlated with clinical outbreaks of disease and antibiotic therapy at the farms. One of the last fish farms downstream, farm 4, exhibited statistically significant lower levels of resistance to OTC, OXA, FLO, and AMX than the upstream farms investigated (Table 2). This is consistent with the relatively small amounts of antibiotics (OXA and S-T) applied at farm 4 compared to the amount of fish produced (Table 1) and with a lower density of fish farms in this section of the stream. These findings were supported by the resistance patterns found among the flavobacteria and Aeromonas isolates, where isolates from farms 1 and 2 were more likely to carry one or several resistance traits than isolates collected at farms 3 and 4 (Table 2).
Conversely, the relatively high incidences of resistance within the overall culturable microflora at farm 2 did not correspond to its minimal use of antibiotics. Moreover, unusually high fractions of OTC-resistant (72%), S-T-resistant (44%), and multiresistant (50%) aeromonads were found at the inlet of farm 2. As this inlet is situated very close to the outlet of the previous fish farm (Fig. 1), these results could signify that the impact of trout farming may extend beyond the boundaries of the individual farm.
Our study demonstrates the significant impact of trout farming as such on environmental bacteria and fish pathogens (Table 3), as antibiotic resistance levels were higher in pond or outlet samples than in samples from the inlets. Among the culturable bacterial population and the isolated flavobacteria, the effect was statistically significant with regard to OTC resistance, while it was clearly significant among OTC-, S-T-, OXA-, and multiresistant aeromonads (Table 3).
The observed seasonal variations among the total culturable microflora, with lower resistance frequencies during the winter months (Fig. 2), were not detected among the aeromonads and flavobacterial isolates and probably reflect changes in the composition of the sampled microbial population.
The high incidences of OTC-resistant aeromonads (69%) and flavobacteria (72%) were unexpected, considering that this particular compound has been very rarely used in Danish aquaculture during the past 5 years, and the total usage of OTC on all farms along the river had dropped from 16 kg in 1996 to 3 kg in 1997 and 1 kg in 1998 (39). In the absence of residues, decomposing unmedicated fish feed (24, 46) and low-level coresistance to OXA (4, 5) involving outer membrane alterations have been suggested to promote decreased bacterial sensitivity to OTC.
S-T and OXA have been used extensively in Danish aquaculture for many years. With regard to S-T, the resistance proportions among the motile aeromonads (43%) greatly exceeded the average level among the culturable bacteria (4.7%). All Flavobacterium isolates were OXA resistant (aeromonads, 20%; culturable bacteria, 16%). A comparison of the F. psychrophilum isolates collected during this trial to a collection of clinical Danish isolates from 1994 showed an increase of resistance proportions from around 50 to 100% for OXA and from 0 to 36% for AMX (7). OXA has been used in Denmark since 1986, while AMX was introduced in 1993. This rapid emergence of large proportions of antibiotic-resistant isolates in Danish trout farming is alarming, and the underlying mechanisms remain unclear. Chromosomally determined mechanisms of resistance seem to be predominant in F. psychrophilum, as no R plasmids or similar structures have yet been observed.
Interestingly, we did not detect antibiotic resistance among the Y. ruckeri isolates; only a decrease in sensitivity to OXA was found. Similar findings have been reported from other geographical areas where ERM is enzootic (11, 27), although antibiotic-resistant clinical isolates have been described. It remains unclear why antimicrobial resistance within this pathogenic species evolves less frequently, while other bacterial species exposed to the same external influences develop extensive degrees of resistance. The genetic background of the resistant isolates in this study is currently being investigated in order to elucidate the mechanisms of resistance involved. Horizontal spread of resistance genes might have occurred, as several S-T, AMX, FLO, and OTC resistance determinants associated with mobile genetic elements have been described with regard to aquaculture (26, 28, 35, 44). R plasmids have been found in Y. ruckeri (11, 27) and the genus Aeromonas (17, 36, 42).
Our results stress the importance of species- or genus-specific approaches in diversified habitats as well as the advantage of including more than one bacterial group in investigations of antimicrobial resistance. It also is evident that in order to assess the observed impact of freshwater trout farming, further investigation is needed, in particular of the mechanisms of resistance involved. Likewise, preventive measures in freshwater aquaculture should be improved to minimize the usage of antimicrobial agents as well as their release into the effluent water.
ACKNOWLEDGMENTS
This study was supported by the Danish Ministry of Food, Agriculture and Fisheries. We thank Farah S. Bahrani for excellent technical work and Ib Skovgaard and Bo M. Bibby of the Department of Mathematics and Physics, The Royal Veterinary and Agricultural University, for their assistance with the statistical analysis.
REFERENCES
- 1.Alderman D J, Hastings T S. Antibiotic use in aquaculture: development of antibiotic resistance—potential for consumer health risks. Int J Food Sci Technol. 1998;33:139–155. [Google Scholar]
- 2.Aoki T. Present and future problems concerning the development of resistance in aquaculture. In: Michel C, Alderman D, editors. Chemotherapy in aquaculture: from theory to reality. Paris, France: Office International des Epizooties; 1992. pp. 254–262. [Google Scholar]
- 3.Argenton F, De Mas S, Malocco C, Dalla Valle L, Giorgetti G, Colombo L. Use of random DNA amplification to generate specific molecular probes for hybridization tests and PCR-based diagnosis of Yersinia ruckeri. Dis Aquat Org. 1996;24:121–127. [Google Scholar]
- 4.Barnes A C, Lewin C S, Hastings T S, Amyes S G B. Cross resistance between oxytetracycline and oxolinic acid in Aeromonas salmonicida associated with alterations in outer membrane proteins. FEMS Microbiol Lett. 1990;72:337–340. doi: 10.1016/0378-1097(90)90327-m. [DOI] [PubMed] [Google Scholar]
- 5.Barnes A C, Lewin C S, Hastings T S, Amyes S G B. Alterations in outer membrane proteins identified in a clinical isolate of Aeromonas salmonicida subsp. salmonicida. J Fish Dis. 1992;15:279–282. [Google Scholar]
- 6.Bjørklund H. Oxytetracycline and oxolinic acid as antibacterials in aquaculture—analysis, pharmacokinetics and environmental impact. Thesis. Finland: Åbo University; 1991. [Google Scholar]
- 7.Bruun M S, Schmidt A S, Madsen L, Dalsgaard I. Antimicrobial resistance patterns in Danish isolates of Flavobacterium psychrophilum. Aquaculture, 2000;187:201–212. [Google Scholar]
- 8.Burgos A, Qindos G, Martinez R, Rojo P, Cisterna R. In vitro susceptibility of Aeromonas caviae, Aeromonas hydrophila, and Aeromonas sobria to fifteen antimicrobial agents. Eur J Clin Infect Dis. 1990;9:413–417. doi: 10.1007/BF01979472. [DOI] [PubMed] [Google Scholar]
- 9.Burka J F, Hammell K L, Horsberg T E. Drugs in salmonid aquaculture—a review. J Vet Pharmacol Ther. 1997;20:333–349. doi: 10.1046/j.1365-2885.1997.00094.x. [DOI] [PubMed] [Google Scholar]
- 10.Cipriano R C, Ford L A, Teska J D, Hale L E. Detection of Aeromonas salmonicida in the mucus of salmonid fishes. J Aquat Anim Health. 1992;4:114–118. [Google Scholar]
- 11.De Grandis S A, Stevenson R M W. Antimicrobial susceptibility patterns and R plasmid-mediated resistance of the fish pathogen Yersinia ruckeri. Antimicrob Agents Chemother. 1985;27:938–942. doi: 10.1128/aac.27.6.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePaola A, Flynn P A, McPhearson R M, Levy S B. Phenotypic and genotypic characterization of tetracycline- and oxytetracycline-resistant Aeromonas hydrophila from cultured channel fish (Ictalurus punctatus) and their environment. Appl Environ Microbiol. 1988;54:1861–1863. doi: 10.1128/aem.54.7.1861-1863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furones M D, Gilpin M L, Munn C B. Culture media for the differentiation of isolates of Yersinia ruckeri, based on detection of a virulence factor. J Appl Bacteriol. 1993;74:360–366. doi: 10.1111/j.1365-2672.1993.tb05139.x. [DOI] [PubMed] [Google Scholar]
- 14.Guardabassi L, Dalsgaard A, Raffatellu M, Olsen J E. Increase in the prevalence of oxolinic acid resistant Acinetobacter spp. observed in a stream receiving the effluent from a freshwater trout farm following treatment with oxolinic acid-medicated feed. Aquaculture, 2000;188:205–218. [Google Scholar]
- 15.Halling-Sørensen B, Nors Nielsen S, Lanzky P F, Ingerslev F, Holten Luetzhøft H C, Jørgensen S E. Occurrence, fate and effects of pharmacological substances in the environment. Chemosphere. 1998;36:357–393. doi: 10.1016/s0045-6535(97)00354-8. [DOI] [PubMed] [Google Scholar]
- 16.Hawke J P, Thune R L. Systematic isolation and antimicrobial susceptibility of Cytophaga columnaris from commercially reared Channel Catfish. J Aquat Anim Health. 1992;4:109–113. [Google Scholar]
- 17.Hedges R W, Smith P, Brazil G. Resistance plasmids of aeromonads. J Gen Microbiol. 1985;131:2091–2095. [Google Scholar]
- 18.Hektoen H, Berge J A, Hormazabal V, Yndestad M. Persistence of antibacterial agents in marine sediments. Aquaculture. 1995;133:175–184. [Google Scholar]
- 19.Herwig R P, Gray J P, Weston D P. Antibacterial resistant bacteria in surficial sediments near salmon net-cage farms in Puget Sound, Washington. Aquaculture. 1997;149:263–283. [Google Scholar]
- 20.Holten Luetzhøft, H. C., B. Halling-Sørensen, L. Guardabassi, F. Ingerslev, and J. Tjørnelund. Establishing sediment concentrations of oxolinic acid in and around a Danish fish farm. Aquaculture, in press.
- 21.Imziln B, Lafdal O M Y, Barakate M, Hassani L, Ouhdouch Y, Boussaid A, Jana M. Pril-ampicillin-dextrin-ethanol agar for the isolation and quantification of Aeromonas spp. from polluted environmental waters. J Appl Microbiol. 1997;82:557–566. [PubMed] [Google Scholar]
- 22.Jacobsen P, Berglind L. Persistence of oxytetracycline in sediments from fish farms. Aquaculture. 1988;70:365–370. [Google Scholar]
- 23.Joseph S W, Carnahan A. The isolation, identification, and systematics of the motile Aeromonas species. Annu Rev Fish Dis. 1994;4:315–343. [Google Scholar]
- 24.Kapetanaki M, Kerry J, Hiney M, O'Brien C, Coyne R, Smith P. Emergence, in oxytetracycline-free marine mesocosms, of microorganisms capable of colony formation on oxytetracycline-containing media. Aquaculture. 1995;134:227–236. [Google Scholar]
- 25.Kerry J, Coyne R, Gilroy D, Hiney M, Smith P. Spatial distribution of oxytetracycline and elevated frequencies of oxytetracycline resistance in sediments beneath a marine salmon farm following oxytetracycline therapy. Aquaculture. 1996;145:31–39. [Google Scholar]
- 26.Kim E-H, Aoki T. Sequence analysis of the florfenicol resistance gene encoded in the transferable R-plasmid of a fish pathogen, Pasteurella piscida. Microbiol Immunol. 1998;9:665–669. doi: 10.1111/j.1348-0421.1996.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 27.Klein B U, Siesenop U, Boehm K H. Investigations on transferable antibiotic resistance through R-plasmids between obligate and facultative fish pathogenic bacteria. Bull Eur Assoc Fish Pathol. 1996;16:138–142. [Google Scholar]
- 28.Kruse H, Sørum H. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl Environ Microbiol. 1994;60:4015–4021. doi: 10.1128/aem.60.11.4015-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy S B. The challenge of antibiotic resistance. Sci Am. 1998;3:32–39. doi: 10.1038/scientificamerican0398-46. [DOI] [PubMed] [Google Scholar]
- 30.McPhearson R M, DePaola A, Zywno S R, Motes M L, Jr, Guarino A M. Antibiotic resistance in gram-negative bacteria from cultured catfish and aquaculture ponds. Aquaculture. 1991;99:203–211. [Google Scholar]
- 31.National Committe for Clinical Laboratory Standards. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Tentative standard M31-T. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 32.Nygaard K, Lunestad B T, Hektoen H, Berge J A, Hormazabal V. Resistance to oxytetracycline, oxolinic acid and furazolidone in bacteria from marine sediments. Aquaculture. 1992;104:31–36. [Google Scholar]
- 33.Okpowasili G C. Aeromonas hydrophila: variability of biochemical characteristics of environmental isolates. J Basic Microbiol. 1991;31:169–176. doi: 10.1002/jobm.3620310303. [DOI] [PubMed] [Google Scholar]
- 34.Pacha R E. Characteristics of Cytophaga psychrophila isolated during outbreaks of bacterial cold-water disease. Appl Microbiol. 1968;16:97–101. doi: 10.1128/am.16.1.97-101.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosser S J, Young H-K. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J Antimicrob Chemother. 1999;44:11–18. doi: 10.1093/jac/44.1.11. [DOI] [PubMed] [Google Scholar]
- 36.Sandaa R-A, Torsvik V L, Goks¢yr J. Transferable drug resistance in bacteria from fish-farm sediments. Can J Microbiol. 1992;38:1061–1065. [Google Scholar]
- 37.Smith P, Donlon J, Coyne R, Cazabon D. Fate of oxytetracycline in a freshwater fish farm: influence of effluent treatment systems. Aquaculture. 1994;120:319. [Google Scholar]
- 38.Smith P, Hiney M, Samuelsen O B. Bacterial resistance to antimicrobial agents used in fish farming: a critical evaluation of method and meaning. Annu Rev Fish Dis. 1994;4:273–313. [Google Scholar]
- 39.Sørensen A H, Bjerre M. Tilsyn med dambrug 1997 og 1998. Vejle, Denmark: Vejle Amt; 1999. pp. 5–6. [Google Scholar]
- 40.Sørum H, Kvello J H, Håstein T. Occurence and stability of plasmids in Aeromonas salmonicida ss salmonicida isolated from salmonids with furunculosis. Dis Aquat Organisms. 1993;16:199–206. [Google Scholar]
- 41.Spanggaard B, Jørgensen F, Gram L, Huss H H. Antibiotic resistance in bacteria isolated from three freshwater fish farms and an unpolluted stream in Denmark. Aquaculture. 1993;115:195–207. [Google Scholar]
- 42.Starliper C E, Cooper R K. Biochemical and conjugation studies of Romet-resistant strains of Aeromonas salmonicida from salmonid rearing facilities in the Eastern United States. J Aquat Anim Health. 1998;10:221–229. [Google Scholar]
- 43.Tollefson L, Fedorka-Cray P J, Angulo F J. Public health aspects of antibiotic resistance monitoring in the USA. Acta Vet Scand Suppl. 1999;92:67–75. [PubMed] [Google Scholar]
- 44.Toranzo A E, Combarro P, Lemos M L, Barja J L. Plasmid coding for transferable drug resistance in bacteria isolated from cultured rainbow trout. Appl Environ Microbiol. 1984;48:872–877. doi: 10.1128/aem.48.4.872-877.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toyama T, Kika-Tsukamoto K, Wakabayashi H. Identification of Cytophaga psychrophila by PCR targeted 16S ribosomal RNA. Fish Pathol. 1994;29:271–275. [Google Scholar]
- 46.Vaughan S, Coyne R, Smith P. The critical importance of sample site in the determination of the frequency of oxytetracycline resistance in the effluent microflora of a freshwater fish farm. Aquaculture. 1996;139:47–54. [Google Scholar]