To the Editor —
Open access to software in computational and systems biology, including data, code and models, is widely acknowledged as essential for ensuring reproducibility of research results and reuse of methods1. Although there are software tools that allow sharing of computational pipelines, these systems generally do not allow the integration of software annotation and documentation at each step in the process — elements that are required to understand and run complex and rapidly evolving software, including methods developed in systems biology for inferring biological pathways.
Jupyter notebooks2 allow developers to combine text, code and code output elements in an integrated executable document so that complex analyses can be reproduced, thus also allowing production of tutorials and educational vignettes. Jupyter notebooks are increasingly used in computational and systems biology, including by gene expression and visualization pipeline tools like Biojupies3 and the CoLoMoTo Interactive Notebook4 for containerized Boolean Network modeling; Binder5, Appyters6 and the GenePattern notebook7 extend Jupyter notebooks to web-based applications for a large array of genomic analyses. However, not all notebook tools are seamlessly enabled for end users, and there are few resources that provide executable, exemplar workflows in network biology.
The modeling and inference of gene regulatory networks (GRNs) connecting transcription factors to their target genes presents challenges in methods development and deployment. Genome-wide GRN inference requires inferring tens of millions of regulatory interactions between transcription factors (or other regulators such as miRNAs) and genes and relies on complex calculations involving large matrix operations. Network models are often inferred for different phenotypes and compared to identify edge weights that differ between states, genes or transcription factors that have condition-specific patterns of targeting, or communities of genes and regulators that are unique to, and functionally associated with, each state.
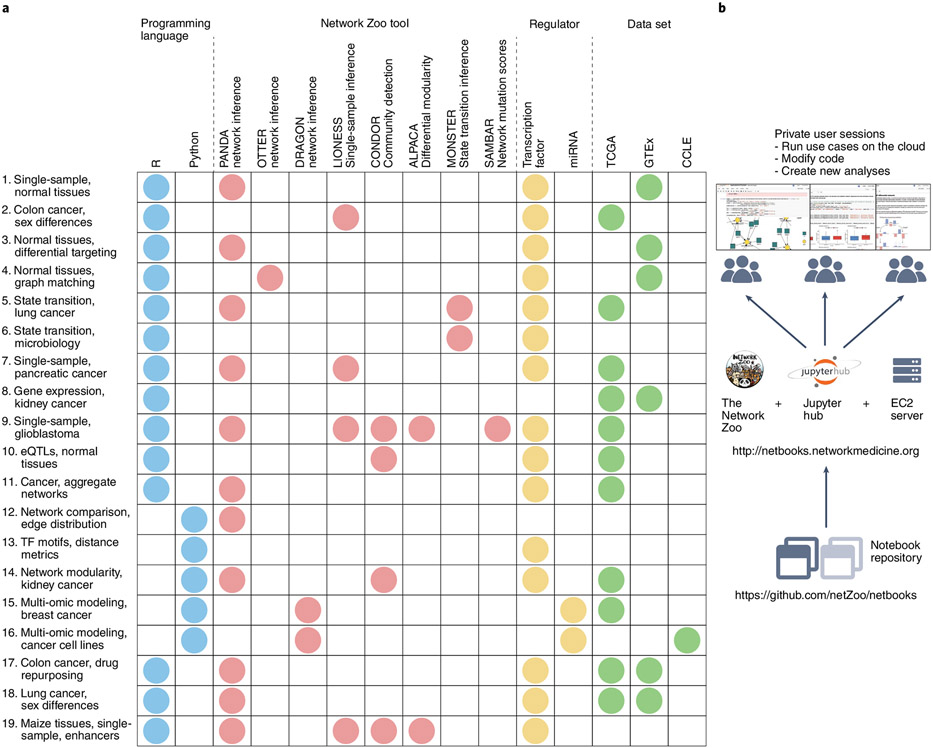
Our research team has been developing network inference and analysis methods, collected into the Network Zoo (http://netzoo.github.io), with implementations in R, C, MATLAB and Python. The growing community of users of these network resources, the increasing interest in learning how to apply network inference methods, and the need to ensure that published analyses are fully reproducible led us to develop Netbooks (http://netbooks.networkmedicine.org), a hosted collection of Jupyter notebooks that provide detailed and annotated step-by-step case studies of GRN analysis. These case studies (Fig. 1a and Supplementary Table 1) include recreation of a published comparison of inferred GRNs between cell lines and their tissues of origin, a comparison of regulatory networks between two pancreatic cancer subtypes, a study of regulatory changes in glioblastoma that implicated PD1 signaling in outcomes, and the inference of trans-regulatory effects in breast cancer.
Fig. 1 ∣. Netbooks contains a catalog of 19 case studies in regulatory genomics using the Network Zoo tools.
a, Netbooks collection includes case studies in cancer genomics and network medicine using a variety of network reconstruction and analysis tools. Row number corresponds to case study number in Supplementary Table 1. TF, transcription factor. b, Web server design enables users to run and modify existing case studies and create new notebooks.
Netbooks allows users on any device to access the resource using a web browser and without login (Supplementary Note 1). Each access creates a new instance on the server with a unique user token and provides read-and-write disk space, memory and dedicated CPU resources (Fig. 1b). The welcome page contains a set of simple notebooks called ‘vignettes’ that detail basic input, calling and output for GRN inference methods installed on the server, together with their usage and program parameters. The case studies are grouped by the programming language (either R or Python) chosen for the example, and users can run and modify the notebook for each case study to understand how changes in input or parameters affect the results. Users can also create their own notebook using R or Python kernels and a preinstalled set of packages and tools.
A primary motivation behind the development of Netbooks has been our group’s longstanding interest in promoting reproducible research. While we have long made our primary code and data available, we recognize that potential users seeking to replicate our results can struggle with not only the installation of the software, but the correct version of the programming language, software environment and associated software dependencies; these issues have been recognized as a barrier to reproducibility in other fields, including machine learning8. Netbooks solves these software environment problems by creating a containerized version of the operating system configuration that allows analyses to be reproducibly rerun over time.
We have used Netbooks to test new network inference pipelines and methods, and to provide reproducible versions of results (including recreation of figures) in both published manuscripts9 and those submitted for review and posted on the arXiv preprint server10. Posting analyses to Netbooks has the advantage of providing anonymous access to new methods or analyses during the peer review process and allows reviewers to investigate questions they might have otherwise raised in their reviews.
We are continuing to expand the catalog of case studies and welcome submissions from the community of Network Zoo users. We will also add examples in Netbooks as we develop new methods that take into account an ever-growing quantity of published multi-omic data, thus ensuring that these methods are also reproducible.
Supplementary Material
Acknowledgements
M.L.K. and R.T.P. are supported by grants from the Norwegian Research Council (313932) and the Norwegian Cancer Society (214871). M.L.K. is also supported by the Norwegian Research Council, Helse Sør-Øst and University of Oslo through the Centre for Molecular Medicine Norway (187615). K.G. is supported by a grant from the US National Heart, Lung, and Blood Institute, National Institutes of Health, K25HL133599. J.P. is supported by US National Heart, Lung, and Blood Institute grant K25HL140186. C.M.L.-R., R.B., D.W., M.B.G. and J.Q. are supported by a grant from the National Cancer Institute, National Institutes of Health, R35CA220523; J.Q. and M.B.G. are also supported by U24CA231846; J.Q. is supported by additional grants from the US National Institutes of Health, 2P50CA127003, 5R01CA205406 and 5R01HL135142. C.M.L.-R. is also supported by a grant from the American Lung Association, LCD-821824 and by the National Heart, Lung, and Blood Institute, 5T32HL007427-41. M.F. is supported by a Marie Sklodowska-Curie grant (PATTERNS 845083). M.A. is supported by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (grant no. 01ZX1912C).
Footnotes
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41592-022-01479-2.
Competing interests
The authors declare no competing interests.
Code availability
Netbooks can be accessed through http://netbooks.networkmedicine.org and the individual Jupyter notebooks are available at https://github.com/netZoo/netbooks.
References
- 1.Tiwari K et al. Mol. Syst. Biol 17, e9982 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kluyver T et al. In Positioning and Power in Academic Publishing: Players, Agents and Agendas (eds Loizides F and Scmidt B) 87–90 (IOS Press, 2016); 10.3233/978-1-61499-649-1-87 [DOI] [Google Scholar]
- 3.Torre D, Lachmann A & Ma’ayan A Biojupies: automated generation of interactive notebooks for RNA-Seq data analysis in the cloud. Cell Syst. 7, 556–561.e553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naldi A et al. Front. Physiol 9, 680 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ragan-Kelley B & Willing C in Proc. 17th Python in Science Conf. (Akici F et al. , eds) 113–120 (2018). [Google Scholar]
- 6.Clarke DJB et al. Patterns (N Y) 2, 100213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reich M et al. Cell Syst. 5,149–151.e141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heil BJ et al. Nat. Methods 18,1132–1135 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopes-Ramos CM et al. Cancer Res. 81, 5401–5412 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weighill D et al. Preprint at arXiv 10.48550/arXiv.2104.01690 (2021). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.