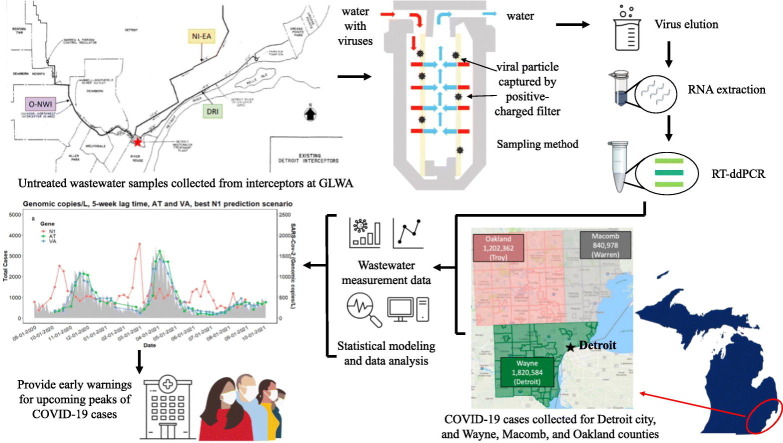
Abstract
Wastewater-based epidemiology (WBE) is useful in predicting temporal fluctuations of COVID-19 incidence in communities and providing early warnings of pending outbreaks. To investigate the relationship between SARS-CoV-2 concentrations in wastewater and COVID-19 incidence in communities, a 12-month study between September 1, 2020, and August 31, 2021, prior to the Omicron surge, was conducted. 407 untreated wastewater samples were collected from the Great Lakes Water Authority (GLWA) in southeastern Michigan. N1 and N2 genes of SARS-CoV-2 were quantified using RT-ddPCR. Daily confirmed COVID-19 cases for the City of Detroit, and Wayne, Macomb, Oakland counties between September 1, 2020, and October 4, 2021, were collected from a public data source. The total concentrations of N1 and N2 genes ranged from 714.85 to 7145.98 gc/L and 820.47 to 6219.05 gc/L, respectively, which were strongly correlated with the 7-day moving average of total daily COVID-19 cases in the associated areas, after 5 weeks of the viral measurement. The results indicate a potential 5-week lag time of wastewater surveillance preceding COVID-19 incidence for the Detroit metropolitan area. Four statistical models were established to analyze the relationship between SARS-CoV-2 concentrations in wastewater and COVID-19 incidence in the study areas. Under a 5-week lag time scenario with both N1 and N2 genes, the autoregression model with seasonal patterns and vector autoregression model were more effective in predicting COVID-19 cases during the study period. To investigate the impact of flow parameters on the correlation, the original N1 and N2 gene concentrations were normalized by wastewater flow parameters. The statistical results indicated the optimum models were consistent for both normalized and non-normalized data. In addition, we discussed parameters that explain the observed lag time. Furthermore, we evaluated the impact of the omicron surge that followed, and the impact of different sampling methods on the estimation of lag time.
Keywords: Wastewater-based-epidemiology (WBE), SARS-CoV-2, COVID-19, Detroit, Early warning, Prediction
Graphical abstract
1. Introduction
Wastewater-based epidemiology (WBE) for the prediction of viral outbreaks was proposed in 2019 and 2020 (Xagoraraki and Brien, 2019; O’Brien and Xagoraraki, 2019; Xagoraraki, 2020), and has been applied for the early detection of COVID-19 (Ahmed et al., 2020a; Ahmed et al., 2021; Brijen et al., 2020). One of the critical utilities of WBE is the possibility to forecast upcoming fluctuations of disease with a lag time. The lag time in our study is defined as the lag between peaks in measured concentrations of SARS-CoV-2 in wastewater and peaks in reported COVID-19 cases based on clinical testing. Lag times observed in published studies since the inception of COVID-19 pandemic (summarized in Table 1 ) vary widely between 2 days and 28 days. The observed lag times may depend on multiple parameters. These parameters include disease characteristics of SARS-CoV-2 such as incubation time and shedding duration summarized in Tables S1 and S2, respectively, that may change with novel variants (Yaniv et al., 2021). The parameters that affect observed lag times may also involve contributing populations and their demographic characteristics, including age (Omori et al., 2021), sex (Syangtan et al., 2021), racial ancestry (Allan-Blitz et al., 2021; Feehan et al., 2021), and traveling history of infected populations (Xiao et al., 2020). Furthermore, the hydraulic influence in the sewage network, including dilution events (Foladori et al., 2020), and sorption and desorption of the virus in wastewater (Ziqiang et al., 2018) as well as the methods of wastewater sampling (that may be include viruses sorbed on particles or supernatant viruses) are critically affecting the observed lag times. Moreover, manners of reporting the clinical data, including the accessibility to the testing (Wiens et al., 2021), and traveling time to the testing sites (Rader et al., 2020) are important.
Table 1.
Lag time in published studies prior to Omicron surge.
| Location | Sample type | Lag time | Test method | References |
|---|---|---|---|---|
| Milan & Rome, Italy | wastewater | within a few days | RT-qPCR | (La Rosa et al., 2020) |
| Ottawa, Canada | wastewater | 2 days | RT-qPCR | (D’Aoust et al., 2021) |
| Montana, USA | wastewater | 2–4 days | RT-qPCR | (Nemudryi et al., 2020) |
| Wisconsin, USA | wastewater | 0–6 days | RT-qPCR | (Feng et al., 2021) |
| New York, USA | wastewater | 3 days | RT-qPCR | (Larsen et al., 2021) |
| New Haven, Connecticut, USA | sludge | 0–2, 1–4, 6–8 days under given scenarios | RT-qPCR | (Peccia et al., 2020) |
| Charlotte, North Carolina | wastewater | 5–12 days | RT-qPCR RT-ddPCR | (Barua et al., 2022) |
| Gandhinagar, Gujarat, India | wastewater | 7–14 days | RT-PCR | (Kumar et al., 2021) |
| Paris, France | wastewater | 8 days | RT-qPCR | (Wurtzer et al., 2020) |
| Minnesota, USA | wastewater | statewide: 15–17 days, regional level: 4–20 days | RT-qPCR | (Melvin et al., 2021) |
| Massachusetts, USA | wastewater | 4–10 days | RT-qPCR | (Wu et al., 2022) |
| Netherlands | wastewater | 4 days | RT-qPCR | (Lodder & de Roda Husman, 2020) |
| Netherlands | sewage samples | 6 days | RT-qPCR | (Medema et al., 2020) |
| Utah, USA | wastewater | 7 days | RT-qPCR | (Weidhaas et al., 2021) |
| Milan Metropolitan Area, Italy, | raw and treated wastewater | 8 days | RT-qPCR | (Rimoldi et al., 2020) |
| Spain | wastewater | 12–16 days | RT-qPCR | (Randazzo et al., 2020) |
| Australia | wastewater | 21 days | RT-qPCR | (Ahmed et al., 2021) |
| Gothenburg, Sweden | wastewater | 19–21 days | RT-qPCR | (Saguti et al., 2021) |
| Australia | wastewater | 28 days | RT-qPCR | (Ahmed et al., 2020, Ahmed et al., 2020, Ahmed et al., 2020) |
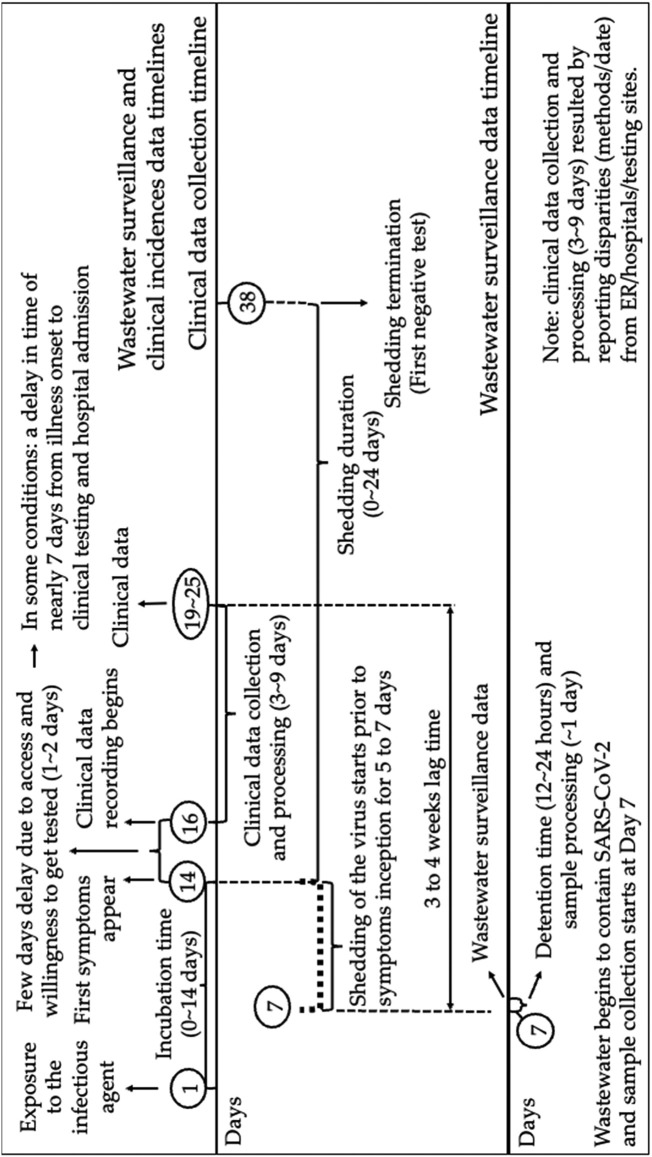
Some of the factors contributing to the lag time between wastewater-based data peaks and clinical data peaks are visualized in the timeline shown in Fig. 1 . The majority of published studies show empirical evidence that the range of the incubation time of SARS-CoV-2 prior to the Omicron surge was 0 to 14 days (shown in Table S1 and summarized in Fig. 1). Clinical testing is often delayed from the manifestation of clinical symptoms by several days, due to limited availability of testing supplies, limited ability to reach testing sites, or resistance of people to seek testing (Rader, 2020; Torres et al., 2021; Wiens et al., 2021). In some cases, the mean delay in the reporting of confirmed COVID-19 cases is 5 days with 15 % of cases reported after day 10 (Harris, 2020). Some clinical studies have demonstrated a delay of approximately 7 days from illness onset to clinical testing (Huang et al., 2020). Henceforth, the delay in gathering clinical data could vary significantly, particularly in demographically and socioeconomically varied populations, like that of Detroit, Michigan. Generally, between day 19 and 25, clinical data will become publicly available, after an estimated delay of 3 to 9 days of clinical data collection and processing time (Garg et al., 2020; Harris, 2020; Rader, 2020). Additionally, Fig. 1 demonstrates the temporal progress of data collection of viral loadings in wastewater. The detention time of wastewater in the collection network is estimated as 12 to 24 h (Table S4). In Fig. 1 it is assumed that in most cases, wastewater laboratory tests are completed within a day upon sample collection. A compilation of all the above-mentioned timelines indicates that the lag time may be estimated to be between 3 and 4 weeks (Fig. 1). This is expected to vary with different variants and different sampling methods.
Fig. 1.
Time scale of clinical data collection and wastewater surveillance (incubation time and shedding duration are summarized in Tables S1 and S2, respectively).
Here we present a twelve-month consecutive study, prior to the omicron surge, using N1 and N2 gene RT-ddPCR to monitor SARS-CoV-2 concentrations in untreated wastewater samples collected from the WRRF of GLWA that serves the city of Detroit, as well as Wayne, Macomb, and Oakland counties in Michigan. We applied a sampling method that captured suspended viruses in the supernatant of wastewater to circumvent the potential input of “old” viruses via desorption of settled viruses during high flows. We investigated lag time through statistical analyses and established four models to predict COVID-19 clinical cases using normalized and non-normalized SARS-CoV-2 concentrations in wastewater. The performance of each model is evaluated using the Root Mean Square Error (RSME) and Pearson correlation between the predicted case number and actual clinical case number. Future incidences of COVID-19 were predicted based on SARS-CoV-2 concentrations in wastewater during the study period, using statistical models. In addition, we examined the influence of the omicron surge to the early waring lag time. We also performed another widely applied sampling method for comparison purposes to demonstrate the benefits of our current method in terms of providing early warning for COVID-19 incidences in Detroit.
2. Materials and methods
2.1. Wastewater treatment plant and sample collection
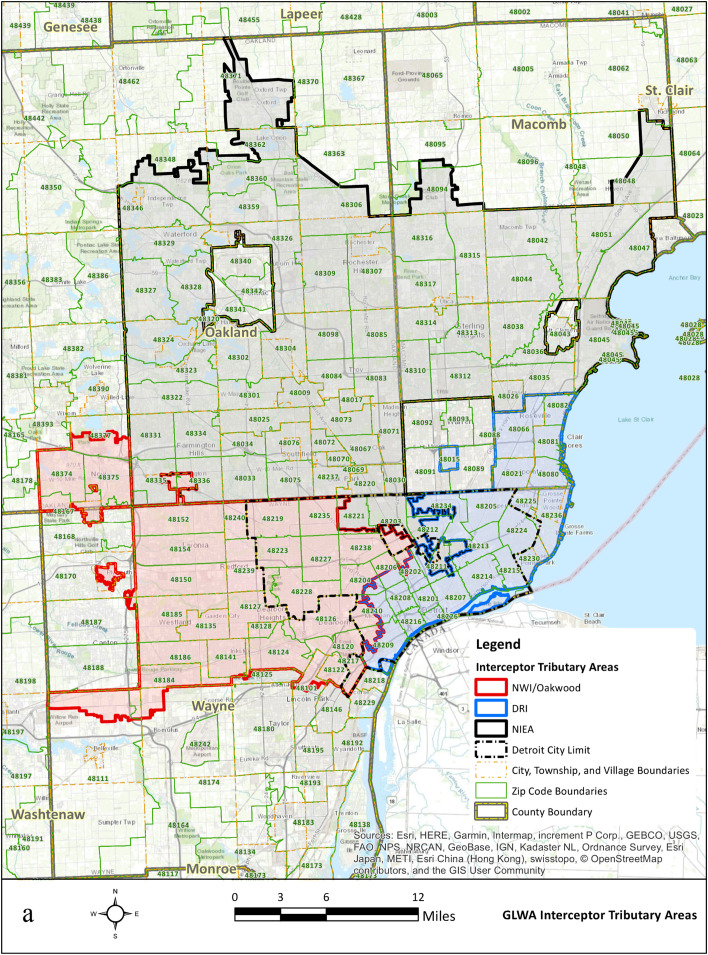
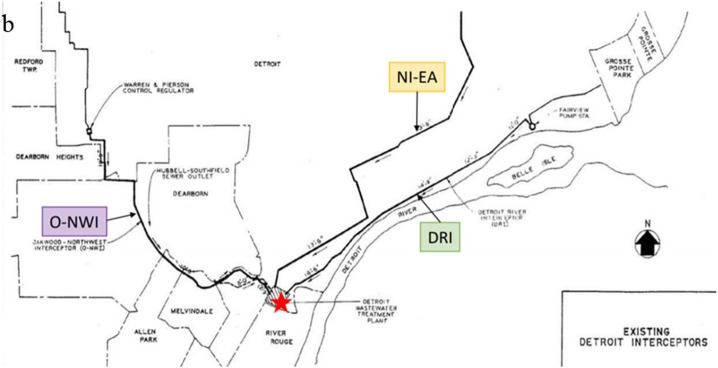
Untreated wastewater samples were collected from the WRRF of GLWA located in southeastern Michigan between September 1, 2020, and August 31, 2021, prior to the Omicron surge. The WRRF in Detroit is the largest single-site wastewater treatment plant in the U.S. with a primary treatment capacity of 1700 million gallons per day (MGD) and a secondary treatment capacity of 930 MGD. GLWA's WRRF has a semi-combined sewer-shed system, which collects and treats stormwater along with residential, industrial, and commercial waste, depending on service areas. The WRRF serves the three most populous Michigan counties: Wayne, Oakland, and Macomb. Fig. 2.a. shows all ZIP codes captured by the three main interceptors that discharge into the WRRF. The WRRF receives wastewater via three main interceptors including the Detroit River Interceptor (DRI), the North Interceptor-East Arm (NIEA), and Oakwood-Northwest-Wayne County Interceptor (ONWI) from its service areas which are shown in Fig. 2.b. The samples were collected from all three interceptors at the point of discharge into the WRRF.
Fig. 2.
a. GLWA WRRF tributary areas; b. Three interceptors and GLWA WRRF locations.
Estimated populations served by each interceptor, daily flows, and other characteristics of the three interceptors, between September 2020 and August 2021, are shown in Table S27. Sampling occurred weekly between September 1, 2020, and August 30, 2021. A total of 407 untreated wastewater samples were collected at the influent of the WRRF, including 146, 117, and 144 samples from ONWI, NIEA, and DRI, respectively.
In addition, to evaluate the effect of omicron surge in the estimated lag time, we performed testing between August 1, 2021, and February 28, 2022, using the same methods, with a total of 249 untreated wastewater samples from the same sites.
2.2. Sampling methods
Viruses were collected and isolated from wastewater using electropositive NanoCeram column filters (Argonide, Sanford, FL) based on the EPA Virus Adsorption-Elution (VIRADEL) method (Xagoraraki et al., 2014; Miyani, Zhao, et al., 2021b). Depending on the suspended solids of wastewater, approximately 10 to 50 L of raw wastewater passed through NanoCeram electropositive cartridge filters at a rate not more than 11 L/min using a previously described method (USEPA, 2001; USEPA, 2014; Miyani et al., 2021b). Flow meter readings were recorded at the inception and termination of each sampling event. After sampling, all NanoCeram column filters were placed in sealed plastic bags, on ice, and transported to the laboratory within 24 h for downstream analysis.
In addition to the VIRADEL method, for method comparison purposes, 1 L of 24-h composite samples were collected weekly between August 1, 2021, and February 28, 2022, to conduct polyethylene glycol precipitation (PEG) for the virus concentration (Ahmed et al., 2020b; Ahmed et al., 2020c; D’Aoust et al., 2021; Kaya et al., 2022).
2.3. Virus elution, RNA extraction, RT-ddPCR, and variants testing
Viruses were eluted within 24 h after sampling based on a previously described method (Miyani et al., 2021b). Bacteriophage Phi6 was used as a proxy virus to estimate losses during virus elution and concentration (Kantor et al., 2021; Ye et al., 2016). The recoveries obtained were from 10.37 % to 58.96 %, with a mean recovery of 24.91 % (±22.89 %). Viral RNA was extracted using Viral RNA QIAGEN kit (QIAGEN, Germantown, Maryland), following the manufacturer's protocol with the modification described previously (Miyani et al., 2021b).
RT-ddPCR was performed on a QX200 AutoDG Droplet Digital PCR system (Bio-Rad, Hercules, CA, USA), using the One-step RT-ddPCR Advanced Kit for Probes (Bio-Rad, Hercules, CA, USA). Per the CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel for SARS-CoV-2 detection (www.cdc.gov), the primers and probe targeting N1 and N2 of SARS-CoV-2 were shown in Table S5, which were proven to be the most sensitive assays to identify SARS-CoV-2 (Ahmed et al., 2022; Bivins et al., 2021) and were chosen in the current study. N1 N2 gene Duplex Assay Reaction Mixture is shown in Table S6. Samples were then run on a C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA, USA) using the conditions shown in Table S7, following a measurement of fluorescence on a QX200 Droplet Reader (Bio-Rad, Hercules, CA, USA).
For each RT-ddPCR run, three positive controls (PTCs) and three negative controls (NTCs), and process negative controls (including virus elution and RNA extraction process controls) were included. 102 gc/μL Twist Bioscience Twist Synthetic SARS-CoV-2 RNA Control 2 (MN908947.3) was used for PTCs. Nuclease-free water was used for NTCs. Nanopure water was used as a substitute for 1.5 % beef extract in virus elution, as process negative controls. Sterile nuclease-free water was used as a substitute for 140 μL of sample for RNA extraction, as process negative controls. All samples were run in triplicate.
Determination of Limit of Blank (LOB) and Limit of Detection (LOD) were based on the methods described in the manufacturer's (Bio-Rad) guidelines for evaluating analytical sensitivity and validation of RT-ddPCR (Bio-Rad, Hercules, CA, USA). The Limit of Blank (LOB) was determined by testing three types of samples using RT-ddPCR, across four consecutive days including the prior-to COVID-19 pandemic samples collected from the same interceptors, nuclease-free water, and negative process control samples from elution and extraction processes. The purpose of testing the LOB across four separate days was to include the unnoticeable impacts when the tests are performed on different days. The prior-to COVID-19 pandemic interceptor samples were collected on February 18, 2018, from the ONWI, NIEA, and DRI interceptors. LOB for N1 gene ddPCR was determined to be 0.09 gc/μL, and the LOB for N2 gene ddPCR was determined to be 0.08 gc/μL. The Limit of Detection (LOD) was determined for each sample using 10−4 and 102 gc/μL of positive SARS-CoV-2 RNA across nine consecutive days using a nonparametric method, as-described in the manufacturer's (Bio-rad) guidelines aforementioned. An LOD of 0.1 gc/μL with 72.92 % confidence for the N1 gene ddPCR and 0.1 gc/μL with 81.25 % confidence for the N2 gene ddPCR were determined.
To elucidate the impact of SARS-CoV-2 variants on the lag time, the mutations of dominant SARS-CoV-2 variants including Alpha, Beta, Gamma, Delta, and Omicron variants were tested in the wastewater samples using the GT Molecular kits (Pulicharla et al., 2021), during the time when N1 and N2 measurements reached three peaks, which were defined in the current study as Peak I (10/6/20–10/28/20) and Peak II (2/17/21–3/8/21). For comparison purposes data were collected during the omicron surge Peak III (12/15/21–1/12/22) and are presented in the supplementary information. Table S8 shows the time of report of the dominant SARS-CoV-2 variants which are corresponded to the peaks' periods that were chosen for variant tests.
2.4. Clinical data of COVID-19
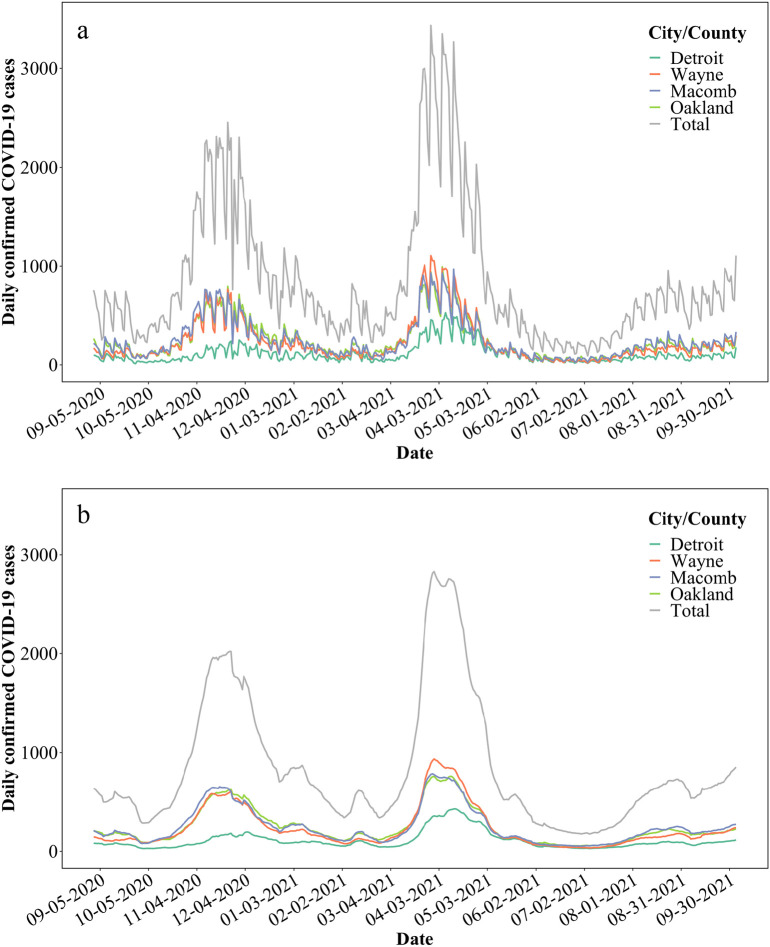
Publicly available data of confirmed COVID-19 incidence in the city of Detroit, as well as Wayne, Macomb, and Oakland counties were used for this study (michigan.gov/coronavirus/). The supplying database was accessed on January 10, 2022 (Fig. 3a). The range of clinical data was between September 1, 2020, and October 4, 2021. The database was accessed again on April 1, 2022, to supply data for the comparative analysis during the omicron surge. Data were reported as follows: “county” is based on the county of residence (or city in the case of Detroit); “cases” are aggregated by the date of onset of COVID-19 symptoms, if known, otherwise by laboratory specimen date, if known, otherwise by case referral date; “confirmed cases” only include individuals who have had a positive diagnostic laboratory test for COVID-19. Clinical data considering a 7-day moving average was used for further statistical analysis (Fig. 3b). COVID-19 data were only available per city/country for the Detroit metropolitan area. Moreover, each interceptor received wastewater from portions of each city/county, thus, only the total SARS-CoV-2 concentrations could be correlated to the total COVID-19 cases of city/counties.
Fig. 3.
a. Total confirmed COVID-19 cases between September 1, 2020, and October 4, 2021, in the city of Detroit, and Wayne, Macomb, and Oakland counties; b. Total confirmed COVID-19 cases between September 1, 2020, and October 4, 2021, with 7-day moving averages in the city of Detroit, and Wayne, Macomb, and Oakland counties.
2.5. Contributing populations, flow rates, and normalization of SARS-CoV-2 concentrations
The contributing population of each interceptor was estimated from 2020 calculations, provided by the Southeast Michigan Council of Governments by traffic analysis zone (TAZ). Geographic information systems (GIS) analysis was adopted to intersect the TAZ boundaries with ZIP Code boundaries and proportionally allocate population from each TAZ to the intersected areas. ZIP Code boundaries were also intersected with the interceptor service areas to allow for a calculation of population by interceptor area.
Daily flow rates for the three interceptors were estimated from the daily influent flow to the WRRF, calculated from GLWA-reported primary influent flow minus WRRF recycle flows, and a calibrated hydrologic and hydraulic model developed for the GLWA collection system. The collection system model was developed with the U.S. Environmental Protection Agency's (EPA's) Stormwater Management Model (SWMM) 5 as part of the GLWA Wastewater Master Plan. The SWMM model represents sanitary wastewater and infiltration/inflow, hydraulics in all physical assets of the collection system and at the WRRF entrances, and stages and flows in the Rouge and Detroit rivers.
To account for the changing flow and dilution of the wastewater where those parameters are highly variable day to day, two approaches of normalizing the N1 and N2 gene concentrations of SARS-CoV-2 in gc/L were adopted using Eqs. (1), (2). The normalized and non-normalized N1 and N2 gene concentrations were used for statistical analysis and modeling.
| (1) |
| (2) |
C(1)= Normalized concentration of SARS-CoV-2 (gc/d)
CN1 or N2 gene= N1 or N2 gene concentrations of SARS-CoV-2 (gc/L)
V=Volume of wastewater flowing into WWTP interceptors during sampling events
f=3.8 × 106, conversion factor between liter and million gallons
C(2)=Normalized concentration of SARS-CoV-2 (gc/L of sanitary flow)
S=Sanitary flow percentage of wastewater flowing into WWTP interceptors during sampling events (%)
2.6. Data analyses and visualization
Data were tracked and organized with Microsoft Excel. MATLAB of a 2019b edition was applied to perform the model regression analyses. All figures were generated using R Statistical Computing Software and MATLAB (2019b), depending essentially on ggplot2 package for visualization, and Forecast and VAR package for autoregression model. The inputs of the models are 7-day moving average of the clinical cases (Barua et al., 2022) and total N1 or N2 gene concentrations for the three interceptors.
Model 1:
Linear regression
| (3) |
where Y is reported clinical cases, x is SARS-CoV-2 concentration from the wastewater samples, b is the slope of the linear regression line, and a is the intercept from the line. The method uses least squares regression.
Model 2:
Linear regression model with autoregressive model (ARIMA model)
Autoregressive Integrated Moving Average (ARIMA) is one of the most widely used forecasting methods for time series data. It applies to time series data which have a trend. In this study, ARIMA model is used to fit the residuals from the linear regression between clinical cases and SARS-CoV-2 concentration. The ARIMA model is characterized by autoregressive (AR) and moving average (MA), and number of differencing required to make the time series stationary. For example, for AR model, Yt depends only on its own lags Y t−1, Y t−2…Y t−p.
| (4) |
In this study, if Y t is clinical case of week t, Y t−1 is the previous one-week (t-1) value of clinical case. β 1 is the coefficient of lag 1-week that the model estimates and α is the intercept term. The criterion to choose the best ARIMA model in this study are: 1) lower Akaike information criterion (AIC) value, a lower AIC score indicates a more predictive model 2) white noise after adjusting residuals 3) low standard error (SE) value.
Model 3:
Regression model with Autoregressive model has a seasonal pattern (SARIMA model)
Seasonal ARIMA or SARIMA model is an extension of ARIMA model that applies to time series data with a seasonal component. In this study, ARIMA has a limitation fitting clinical cases and SARS-CoV-2 concentration time series data because it does not apply to these two time series data which have a seasonal pattern.
Model 4:
Vector autoregressive model (VAR)
VAR model is a forecasting model that can be used when two or more time series influence each other. In this study, clinical cases and SARS-CoV-2 concentration time series data are considered as two time series data, and there is a relationship between these two time series data.
| (5) |
| (6) |
where, Y 1, t is the clinical cases at week t and Y 2, t is the SARS-CoV-2 concentration at week t. Y 1, t−1is the lag of one-week values of clinical cases, Y 2, t−1 is the lag of one-week values of SARS-CoV-2 concentration.
3. Results and discussion
3.1. Observed SARS-CoV-2 RNA in wastewater samples and observed COVID-19 cases
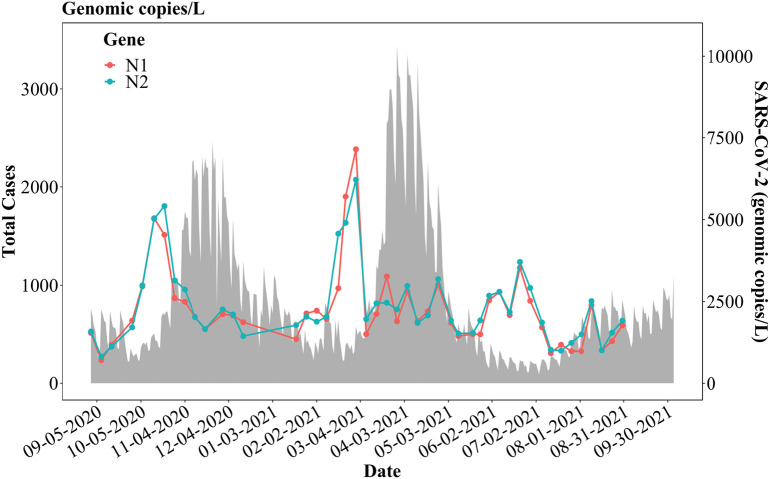
During the 12-month study, N1 and N2 genes of SARS-CoV-2 were detected and quantified in all 407 wastewater samples using RT-ddPCR shown in Table 2 . The overall observed trends of the total N1 and N2 gene concentrations increased steeply from mid-September 2020 and reached a peak in mid-October 2020 (Fig. 4 ), which heralded the first peak of COVID-19 infections, in mid-November 2020. Both N1 and N2 gene concentrations stayed comparatively steady between November 2020 and the end of January 2021, following a sharp increase in February 2021 and reached a peak by the end of the month. This brought about the second peak of COVID-19 infections, from approximately late March to April 2021. Subsequently, the total SARS-CoV-2 concentrations measured by N1 and N2 gene RT-ddPCR in wastewater samples reached comparatively lower peaks in June 2021, which preceded the increase of COVID-19 incidences towards the end of July and August 2021 (Fig. 4). The following decrease of N1 and N2 gene concentrations after each peak was largely due to the termination of shedding events which last a few weeks as demonstrated in Fig. 1 and Table S2.
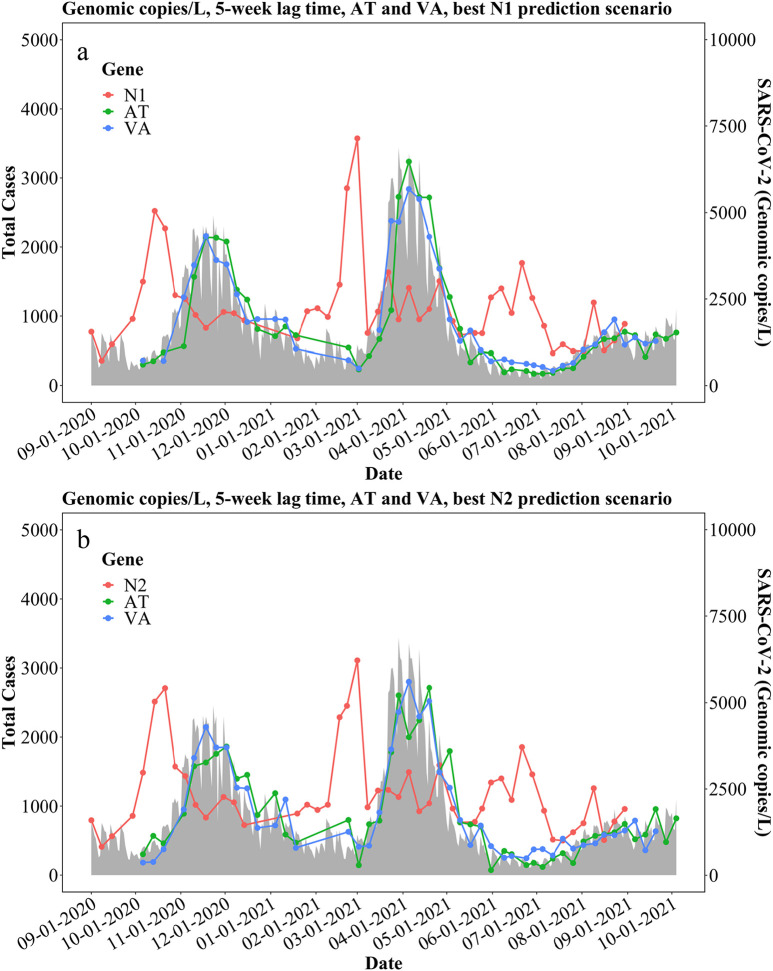
Fig. 5.
Best prediction models based on (a) N1 gene concentrations (gc/L) and (b) N2 gene concentrations (gc/L) with a 5-week lag time.
Table 2.
N1 and N2 gene concentrations measured by RT-ddPCR in wastewater samples collected from GLWA WRRF.
| Unit | Gene | Interceptor |
|||
|---|---|---|---|---|---|
| ONWI | NIEA | DRI | |||
| gc/l | N1 | Maximum | 5773.42 | 1517.10 | 1405.36 |
| Minimum | 256.39 | 204.78 | 185.41 | ||
| Mean | 1069.11 | 659.16 | 550.48 | ||
| N2 | Maximum | 4826.54 | 2583.64 | 1927.34 | |
| Minimum | 281.47 | 202.71 | 210.27 | ||
| Mean | 1048.03 | 729.26 | 600.62 | ||
| gc/d | N1 | Maximum | 5.23E+12 | 1.29E+12 | 1.87E+12 |
| Minimum | 1.66E+11 | 1.57E+11 | 1.61E+11 | ||
| Mean | 7.68E+11 | 4.36E+11 | 4.41E+11 | ||
| N2 | Maximum | 4.38E+12 | 1.75E+12 | 1.87E+12 | |
| Minimum | 1.56E+11 | 1.61E+11 | 1.82E+11 | ||
| Mean | 7.58E+11 | 4.81E+11 | 4.79E+11 | ||
| gc/l of SF | N1 | Maximum | 2.54E+04 | 3.87E+03 | 9.99E+03 |
| Minimum | 8.05E+02 | 4.71E+02 | 8.57E+02 | ||
| Mean | 3.73E+03 | 1.31E+03 | 2.35E+03 | ||
| N2 | Maximum | 2.13E+04 | 5.23E+03 | 9.99E+03 | |
| Minimum | 7.58E+02 | 4.82E+02 | 9.71E+02 | ||
| Mean | 3.68E+03 | 1.44E+03 | 2.55E+03 | ||
Note: SF stands for “Sanitary Flow”.
Fig. 4.
Total N1 and N2 gene concentrations in gc/L for the three interceptors and total confirmed COVID-19 cases in the city of Detroit, as well as Wayne, Macomb, and Oakland counties.
3.2. Correlation between SARS-CoV-2 and confirmed COVID-19 cases
A series of statistical analysis with normalizations using different parameters, including BOD, TSS, wastewater flow volume and sanitary percentage of wastewater were conducted. Wastewater flow volume and sanitary percentage of wastewater were chosen for subsequent analysis in terms of stronger Pearson's correlation. Table 2 presents the N1 and N2 gene concentrations normalized by total flow volume and sanitary percentage, respectively. The fluctuations of the normalized results in gc/d and gc/L of sanitary flow stay relatively comparable to the original results in gc/L of the N1 and N2 genes, which predicted the peaks of COVID-19 incidence by 5 weeks.
SARS-CoV-2 gene concentrations in wastewater were influenced by absolute clinical case numbers of COVID-19 on single consecutive days. Therefore, 7-day moving averages of clinical cases were performed to smooth the data and reduce outliers. Confirmed COVID-19 clinical data were then aligned with SARS-CoV-2 concentrations (N1/N2) on the exact sampling dates. Missing data from samples were filled using linear interpolation (Lepot et al., 2017). After aligning these two datasets, each SARS-CoV-2 concentrations (N1/N2) had a corresponding COVID-19 case data to be analyzed for pairwise correlation and statistical modeling in the next sections.
We proposed the hypothesis that the fluctuations of SARS-CoV-2 concentrations in wastewater correlate to confirmed COVID-19 cases with a prescribed range of lag times. Hence, Pearson's correlations were first conducted to investigate the strength of a linear correlation between the total N1 and N2 gene concentrations in gc/L, gc/d, and gc/L of sanitary flow and total confirmed COVID-19 cases in the study areas with various lag times (Table S28). To estimate the approximate lag time, week-shift of the clinical cases was adopted. Notably, SARS-CoV-2 concentrations in wastewater strongly correlated with the 5-week shifting forward of a 7-day moving average of COVID-19 incidences (Pearson's r = 0.62 for N1 gene in gc/L, and Pearson's r = 0.64 for N2 gene in gc/L).
Our suggested lag time of 5 weeks between peaks in SARS-CoV-2 genes in wastewater and peaks in reported COVID-19 clinical tests is relatively similar to the longest lag times reported in the literature (Table 1). For example, a lag of 21 days was reported in Australia (Ahmed et al., 2021) and Sweden (Saguti et al., 2021), and 28 days in Australia (Ahmed et al., 2020a). The increased lag time may be in partly due to our sampling method (VIRADEL) that focuses on viruses in the supernatant of untreated wastewater rather than in the precipitates and solids. These samples represent the near real-time increase in clinical cases in the community. This is important for samples collected in large interceptors where precipitation and resuspension of solids occurs to a large extent. Factors affecting our observed lag time are discussed in Section 3.4.
Our analysis also demonstrated that the normalization of clinical data using wastewater flow rate and percentage of sanitary flow did not significantly improve the correlation between the SARS-CoV-2 concentrations in wastewater and COVID-19 clinical cases in communities during the study period. Similar observations were also demonstrated in another recent study (Ai et al., 2021).
3.3. Statistical models on the relationship between wastewater surveillance of SARS-CoV-2 and COVID-19 incidence
To better estimate the relationship between the measured SARS-CoV-2 concentrations in wastewater and reported COVID-19 incidence in the communities, four statistical models were established naming: linear model (L), autoregression model (A), autoregression with time effect model (AT), and vector autoregression model (VA). Confirmed COVID-19 cases with lag times of 3, 4, and 5 weeks were chosen to correlate with N1 and N2 gene concentrations in gc/L, gc/d, and gc/L of sanitary flow using the aforementioned models. The Root Mean Square Error (RMSE) and Pearson's coefficient between actual cases and predicted cases were calculated to estimate the performance of each model shown in Supplementary Tables S10 – S15. From the previous result in Section 3.2, a lag time of 5 weeks exhibits a stronger correlation, in agreement with the models in Table 3 and Tables S29 and S30, based on Pearson's r. For 5 weeks, in Table S29, linear regression only considers SARS-CoV-2 concentration as the predictor in the model, the correlation between actual case number and predicted case number is range from 0.4 to 0.62. Seasonal patterns from the residual of linear regression were also observed. Thus, we consider that there is an autocorrelation effect from the case number. The models were therefore improved with autoregressive errors using ARIMA. The correlation then consequently increased to 0.4–0.67. It is important to note that there is a limitation of ARIMA, it does not apply to seasonal data. After a seasonal component was included in the ARIMA model, the correlation further improved to 0.94–0.95. Thus, the analyses suggest that there is a seasonal pattern present in clinical COVID-19 cases. VA is also a better model (r ranges from 0.95 to 0.96) because it considers both SARS-CoV-2 concentration and case number as the predictors in the model and use their past values to predict current case number. Similarly, the modeling results of N2 gene concentrations show agreement with modeling results of N1 gene concentrations. Comparing the non-normalized data with normalized data in Table S29 and Table S30, for the L and A models, using normalized data does not improve the correlation. For AT and VA models, using normalized data shows similar results comparing to using non-normalized data. It may indicate that these two models reduce the effect of normalizing data. It also indicates that normalizations of the original N1 and N2 gene concentrations did not significantly improve the performance of the modeling and vice versa. All other detailed results are shown in Tables S10 – S15. Same models and analysis were also applied to both the measurements by VIRADEL method (results were shown in Tables S16 – S21) and PEG sampling method results (shown in Tables S22 and S23, Fig. S2) of the study period between August 2021 and February 2022 for comparison purposes as explained in Section 3.4.
Table 3.
Statistical modeling results between N1 and N2 gene concentrations and total COVID-19 cases during the 5-week lag time study period in city of Detroit, as well as Wayne, Macomb, and Oakland counties (* is shown in Fig. 5.)
| Lag time |
Model |
N1-based results |
N2-based results |
||||
|---|---|---|---|---|---|---|---|
| RMSE |
RMSE |
||||||
| Unit of N1/N2 gene | gc/L | gc/d | gc/L of sanitary flow | gc/L | gc/d | gc/L of sanitary flow | |
| 3 week | Linear | 7.22 | 135.76 | 11.78 | 12.40 | 926.30 | 5.62 |
| Autoregression | 135.65 | 780.00 | 250.52 | 341.27 | 901.23 | 700.34 | |
| Autoregression+ time effect | 10.18 | 10.18 | 10.97 | 11.34 | 12.34 | 15.33 | |
| Vector Autoregression | 8.32 | 7.85 | 8.97 | 8.89 | 9.90 | 9.90 | |
| 4 week | Linear | 7.26 | 123.56 | 9.18 | 16.37 | 104.45 | 8.33 |
| Autoregression | 182.92 | 234.90 | 635.69 | 132.35 | 730.74 | 500.62 | |
| Autoregression+ time effect | 7.50 | 7.47 | 7.20 | 9.75 | 7.39 | 7.33 | |
| Vector Autoregression | 8.00 | 7.99 | 8.62 | 6.88 | 8.31 | 7.62 | |
| 5 week | Linear | 1.83 | 48.97 | 2.62 | 13.95 | 36.19 | 2.36 |
| Autoregression | 105.81 | 417.57 | 642.83 | 548.14 | 570.56 | 100.95 | |
| Autoregression+ time effect* | 1.47* | 1.60 | 1.60 | 3.21* | 1.60 | 1.42 | |
| Vector Autoregression* | 0.35* | 0.53 | 4.44 | 7.57* | 4.37 | 1.03 | |
3.4. Factors affecting lag time
3.4.1. Variants
All the discussions above are based on the study period between September 2020 and August 2021 prior to the Omicron surge. Variations of lag times could not be explicitly elucidated without addressing the changing epidemiological characteristics of emerging SARS-CoV-2 variants, involving incubation time, shedding durations, shedding dynamics, and so forth. Given the shortened incubation time (median of 3 days, (Baker et al., 2022; Brandal et al., 2021; Jansen et al., 2021)) and shedding duration (less than 10 days, (Lamers et al., 2022)) during the Omicron surge, we identified a 2-week lag time, between August 2021 and February 2022, with the same VIRADEL method and same modeling methods (Fig. S2). The incubation time (Baker et al., 2022) and shedding duration (Lamers et al., 2022) were apparently shorter comparing to the parental and previous variants, inevitably leading to a shorter lag time. Besides, the shedding dynamics changed amid Omicron surge which inevitably affected the lag time (Table S3). The variant test results shown in Table S9 demonstrate the different mutations of dominant variants identified in the samples which in addition correspond to the reported emerging variants shown in Table S8. Changing epidemiological characteristics of SARS-CoV-2 variants play a crucial role in affecting the lag time.
3.4.2. Sampling method
An additional factor that may influence lag time is the sampling method. The VIRADEL electropositive filtration method focuses on supernatant virus in wastewater and avoids the inclusion of large wastewater organic solids where viruses may adsorb onto the surfaces. This method has been recommended by the EPA (USEPA, 2001; USEPA, 2014) and has been extensively applied in the field (Brijen et al., 2020; McCall et al., 2020, McCall et al., 2021; Miyani et al., 2021a). On the other hand, grab or composite sampling followed by PEG precipitation incorporates viruses attached onto larger solid particles, which tend to settle in large interceptors and generally re-suspend during periods of high flow. If these larger particles are included in a grab or composite sample, they may include a portion of the viruses that have been settled for a while, having been excreted earlier into that sewer-shed, thus interfering with the desired prediction which is the objective of this work. This becomes a critical factor in interceptors of large urban centers. For small catchment areas with short hydraulic detention times in neighborhood sewer lines the importance of this factor is expected to decrease.
To compare the two methods, we simultaneously collected samples from the same locations. This comparison took place during the omicron surge and the data are shown in the supplementary information. Between August 2021 and February 2022, 24-h composite samples followed by PEG were collected at the day as the VIRADEL electropositive filtration was performed. This allowed us to investigate on the impact of sampling methods on lag time and demonstrate the potential for providing early warning signals of the VIRADEL method through comparison.
The PEG measurements (Fig. S1) and its Pearson's correlation (Table S26) demonstrated that the N1 and N2 concentrations did not correlate with COVID-19 cases with a lag time. Results from the same models presented above are shown in Table S22 and S23 for PEG measurements. Results demonstrate that N1 and N2 concentrations based on PEG method did not provide an early warning of COVID-19 cases or significant correlations with the COVID-19 cases in the study area in terms of Pearson's r and RMSE.
3.4.3. Clinical data uncertainties
The report time for associated clinical cases tends to be uncertain due to potential reluctance to be tested (Feng et al., 2021), disparities in reporting clinical data and availability of different testing methods (Ai et al., 2021), limited testing supplies and limited testing sites under rapid testing demand and so forth (Hasan and Nasution, 2021). Since the inception of the COVID-19 pandemic, communities in the U.S. as well as many countries across the world had troubles and limited access to COVID-19 testing and real-time reporting, resulting in delay of real-time tracking and monitoring the clinical cases (Bibby et al., 2021; Larsen et al., 2021).
The lag time could not be explicitly elucidated without addressing all the challenges facing clinical testing, especially amid the early stages of the unprecedented pandemic. In the early stage of the pandemic, the health departments were struggling to provide prompt testing across the country, such as in Detroit and its surrounding counties, rendering a notable delay in clinical data collection and reporting, inevitably leading to a longer lag time (Rader, 2020; Torres et al., 2021; Wiens et al., 2021). Throughout the past two years' efforts amid COVID-19 pandemic, governmental agencies and LHDs have been adapting to increasing COVID-19 cases as well as testing demands by gradually building the testing capabilities, improving clinical data processing and organizing, pushing clinical data releasing and so forth (Alexander et al., 2022; Powell et al., 2021). The significant improvement and adaptation of LHD to the rapid changing transmissions and clinical incidences of COVID-19 enable a quicker and prompt clinical data reporting and releasing, inevitably leading to a shorter lag time. Despite these caveats, WBE is widely accepted as an effective tool for forewarning community fluctuations in COVID-19 infections (Bibby et al., 2021). Notwithstanding the uncertainties discussed above, we put forth that WBE is an important tool in predicting future fluctuations of COVID-19 infections.
Overall, this study demonstrates the effectiveness of applying wastewater-based epidemiology (WBE) as an early warning tool for the prediction of fluctuations of COVID-19 cases in communities in the Detroit metropolitan area. To our knowledge, this is one of the first studies to systematically evaluate the lag time between peaks in measured concentrations of SARS-CoV-2 in wastewater and peaks in reported COVID-19 cases based on clinical testing. Also, this is, to our knowledge, one of the first studies to propose the use of an autoregression with seasonal pattern model and a vector autoregression model in predicting clinical COVID-19 incidences based on the N1 and N2 gene measurements in wastewater. Though WBE demonstrates great promise, potential limitations and challenges remain. More research is warranted to establish a standard framework for modeling the latency between early detection of COVID-19 and presentation of clinical cases. Future studies should include establishing predictive models to optimize wastewater surveillance for early warning of clinical manifestation.
4. Conclusions
-
•
During the 12-month study, prior to the omicron surge, 407 wastewater samples were collected and analyzed for SARS-CoV-2 genes using RT-ddPCR. Measured concentrations of SARS-CoV-2 ranged from 714.85 to 7145.98 gc/L by total N1 gene RT-ddPCR and 820.47 to 6219.05 gc/L by total N2 gene RT-ddPCR.
-
•
Lag time, the latency from surge in viral concentration in wastewater and peak in clinical cases was estimated as 5 weeks prior to the Omicron surge.
-
•
As compared to linear regression and autoregression (ARIMA) models, the autoregression model with seasonal patterns and vector autoregression model were more effective in predicting COVID-19 cases during the study period for the 5-week lag scenario.
-
•
Original N1 and N2 gene concentrations were normalized by total flow volumes and sanitary percentage. The statistical results indicated the optimum 5-week prediction models were consistent for both normalized and non-normalized data.
-
•
Surveillance through wastewater sampling and analysis can be employed for predicting infections and monitoring health conditions in large metropolitan areas such as Detroit.
Funding
This study was funded by the Michigan Department of Health and Human Services (MDHHS), Michigan Department of Environment, Great Lakes, and Energy (EGLE), and the Great Lakes Water Authority (GLWA).
CRediT authorship contribution statement
Liang Zhao: Methodology, Investigation, Data curation, Formal analysis, Visualization, Validation, Writing – original draft. Yangyang Zou: Investigation, Software, Formal analysis, Writing – review & editing. Yabing Li: Investigation, Writing – review & editing. Brijen Miyani: Investigation, Writing – review & editing. Maddie Spooner: Investigation, Writing – review & editing. Zachary Gentry: Investigation, Writing – review & editing. Sydney Jacobi: Investigation, Writing – review & editing. Randy E. David: Writing – review & editing. Scott Withington: Writing – review & editing. Stacey McFarlane: Writing – review & editing. Russell Faust: Writing – review & editing. Johnathon Sheets: Investigation, Data curation, Writing – review & editing. Andrew Kaye: Investigation, Data curation, Writing – review & editing. James Broz: Investigation, Data curation, Writing – review & editing. Anil Gosine: Writing – review & editing. Palencia Mobley: Writing – review & editing. Andrea W.U. Busch: Writing – review & editing. John Norton: Funding acquisition, Writing – review & editing. Irene Xagoraraki: Conceptualization, Funding acquisition, Methodology, Investigation, Project administration, Resources, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
We thank the Michigan Department of Health and Human Services (MDHHS), Michigan Department of Environment, Great Lakes and Energy (MI-EGLE), and the Great Lakes Water Authority (GLWA) for funding this work. We furthermore thank Michigan State University AgBioresearch and Michigan State University Institute for Global Health for supporting this work.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.157040.
Appendix A. Supplementary data
Appendix. Supplementary materials
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ,ment. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K.V., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimization and quality control are crucial for generating reliable public health information. Current Opinion in Environmental Science & Health. 2020;17:82–93. doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O’Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. p. Women in Water Quality. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Metcalfe S., Smith W.J.M., Verbyla M.E., Symonds E.M., Simpson S.L. Evaluation of process limit of detection and quantification variation of SARS-CoV-2 RT-qPCR and RT-dPCR assays for wastewater surveillance. Water Res. 2022;213 doi: 10.1016/j.watres.2022.118132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai Y., Davis A., Jones D., Lemeshow S., Tu H., He F., Ru P., Pan X., Bohrerova Z., Lee J. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M., Unruh L., Koval A., Belanger W. United States response to the COVID-19 pandemic, January–November 2020. Health Econ.Policy Law. 2022;17(1):62–75. doi: 10.1017/S1744133121000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan-Blitz L.T., Hertlein F., Klausner J.D. Heterogeneity in SARS-CoV-2 positivity by ethnicity in Los Angeles. J. Racial Ethn. Health Disparities. 2021;2–5 doi: 10.1007/s40615-021-01062-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.A., Rhee C., Tucker R., Badwaik A., Coughlin C., Holtzman M.A., Hsieh C., Maguire A., Mermel Blaeser E., Seetharaman S., Solem O., Vaidya V., Klompas M. Rapid control of hospital-based SARS-CoV-2 Omicron clusters through daily testing and universal use of N95 respirators. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua V.B., Juel M.A.I., Blackwood A.D., Clerkin T., Ciesielski M., Sorinolu A.J., Holcomb D.A., Young I., Kimble G., Sypolt S., Engel L.S., Noble R.T., Munir M. Tracking the temporal variation of COVID-19 surges through wastewater-based epidemiology during the peak of the pandemic: a six-month long study in Charlotte, North Carolina. Sci. Total Environ. 2022;814 doi: 10.1016/j.scitotenv.2021.152503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Bivins A., Wu Z., North D. Making waves: plausible lead time for wastewater based epidemiology as an early warning system for COVID-19. Water Res. 2021;202(July) doi: 10.1016/j.watres.2021.117438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Kaya D., Bibby K., Simpson S.L., Bustin S.A., Shanks O.C., Ahmed W. Variability in RT-qPCR assay parameters indicates unreliable SARS-CoV-2 RNA quantification for wastewater surveillance. Water Res. 2021;203 doi: 10.1016/j.watres.2021.117516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandal L.T., MacDonald E., Veneti L., Ravlo T., Lange H., Naseer U., Feruglio S., Bragstad K., Hungnes O., Ødeskaug L.E., Hagen F., Hanch-Hansen K.E., Lind A., Watle S.V., Taxt A.M., Johansen M., Vold L., Aavitsland P., Nygård K., Madslien E.H. Outbreak caused by the SARS-CoV-2 Omicron variant in Norway, November to December 2021. Eurosurveillance. 2021;26(50) doi: 10.2807/1560-7917.ES.2021.26.50.2101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brijen M., Xavier F., John N., Anna M., Irene X. SARS-CoV-2 in Detroit wastewater. J. Environ. Eng. 2020;146(11) doi: 10.1061/(ASCE)EE.1943-7870.0001830. [DOI] [Google Scholar]
- D’Aoust P.M., Graber T.E., Mercier E., Montpetit D., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Servos M.R., Srikanthan N., MacKenzie M., Figeys D., Manuel D., Jüni P., MacKenzie A.E., Delatolla R. Catching a resurgence: increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feehan A.K., Velasco C., Fort D., Burton J.H., Price-Haywood E.G., Katzmarzyk P.T., Garcia-Diaz J., Seoane L. Racial and workplace disparities in seroprevalence of SARS-CoV-2, Baton Rouge, Louisiana, USA. Emerg. Infect. Dis. 2021;27(1):314–317. doi: 10.3201/eid2701.203808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Roguet A., Mcclary-Gutierrez J.S., Newton R.J., Kloczko N., Meiman J.G., Mclellan S.L., Mclellan S. 1(414) MedRxiv; 2021. Evaluation of sampling frequency and normalization of SARS-CoV-2 wastewater concentrations for capturing COVID-19 burdens in the community. 2021.02.17.21251867. [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S., Kim L., Whitaker M., O’Halloran A., Cummings C., Holstein R., Prill M., Chai S., Kirley P., Alden N., Kawasaki B. Hospitalization Rates and Characteristics of Patients Hospitalized With. Morbidity and Mortality Weekly Report. 69(15) US Department of Health and Human Services/Centers for Disease Control and Prevention; 2020. pp. 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J.E. MedRxiv; 2020. Overcoming reporting delays is critical to timely epidemic monitoring: the case of COVID-19 in New York City. 2020.08.02.20159418. [Google Scholar]
- Hasan A., Nasution Y. A compartmental epidemic model incorporating probable cases to model COVID-19 outbreak in regions with limited testing capacity. ISA Trans. 2021 doi: 10.1016/j.isatra.2021.01.029. xxxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Cao B.… Clinical features of patients infected with 2019 novel coronavirus in Wuhan,China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irene X., Ziqiang Y., Zhassulan S. Fate of viruses in water systems. J. Environ. Eng. 2014;140(7) doi: 10.1061/(ASCE)EE.1943-7870.0000827. [DOI] [Google Scholar]
- Jansen L., Tegomoh B., Lange K., Showalter K., Figliomeni J., Abdalhamid B., Iwen P.C., Fauver J., Buss B., Donahue M. Investigation of a SARS-CoV-2 B.1.1.529 (Omicron) variant cluster - Nebraska, November-December 2021. MMWR Morb. Mortal. Wkly Rep. 2021;70(5152):1782–1784. doi: 10.15585/mmwr.mm705152e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor R.S., Nelson K.L., Greenwald H.D., Kennedy L.C. Challenges in measuring the recovery of SARS-CoV-2 from wastewater. Environ.Sci.Technol. 2021;55(6):3514–3519. doi: 10.1021/acs.est.0c08210. [DOI] [PubMed] [Google Scholar]
- Kaya D., Niemeier D., Ahmed W., Kjellerup B.V. Evaluation of multiple analytical methods for SARS-CoV-2 surveillance in wastewater samples. Sci. Total Environ. 2022;808 doi: 10.1016/j.scitotenv.2021.152033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Joshi M., Patel A.K., Joshi C.G. Unravelling the early warning capability of wastewater surveillance for COVID-19: a temporal study on SARS-CoV-2 RNA detection and need for the escalation. Environ. Res. 2021;196:110946. doi: 10.1016/j.envres.2021.110946. (December 2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Mykytyn A.Z., Breugem T.I., Groen N., Knoops K., Schipper D., van Acker R., van den Doel P.B., Bestebroer T., Koopman C.D., Reusken C., Muraro M.J., GeurtsvanKessel C.H., van Royen M.E., Peters P.J., Zhang J., Haagmans B.L. 2022. SARS-CoV-2 Omicron efficiently infects human airway, but not alveolar epithelium. BioRxiv, 2022.01.19.476898. [DOI] [Google Scholar]
- Larsen D.A., Collins M.B., Du Q., Hill D., Insaf T.Z., Kilaru P., Kmush B.L., Middleton F., Stamm A., Wilder M.L., Zeng T., Green H. 2021. Coupling freedom from disease principles and early warning from wastewater surveillance to improve health security. MedRxiv, 2021.06.11.21258797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepot M., Aubin J.B., Clemens F.H.L.R. Water (Switzerland) vol. 9, issue 10. 2017. Interpolation in time series: an introductive overview of existing methods, their performance criteria and uncertainty assessment. [DOI] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall C., Wu H., Miyani B., Xagoraraki I. Identification of multiple potential viral diseases in a large urban center using wastewater surveillance. Water Res. 2020;184 doi: 10.1016/j.watres.2020.116160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall C., Wu H., O’Brien E., Xagoraraki I. Assessment of enteric viruses during a hepatitis outbreak in Detroit MI using wastewater surveillance and metagenomic analysis. J. Appl. Microbiol. 2021;131(3):1539–1554. doi: 10.1111/jam.15027. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Melvin R.G., Chaudhry N., Georgewill O., Freese R., Simmons G.E., Jr. Predictive power of SARS-CoV-2 wastewater surveillance for diverse populations across a large geographical range. medRxiv. 2021 (Chicago) [Google Scholar]
- Miyani B., McCall C., Xagoraraki I. High abundance of human herpesvirus 8 in wastewater from a large urban area. J. Appl. Microbiol. 2021;130(5):1402–1411. doi: 10.1111/jam.14895. [DOI] [PubMed] [Google Scholar]
- Miyani B., Zhao L., Spooner M., Buch S., Gentry Z., Mehrotra A., Norton J., Xagoraraki I. Early warnings of COVID-19 second wave in Detroit. J. Environ. Eng. 2021;147(8) doi: 10.1061/(asce)ee.1943-7870.0001907. [DOI] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1(6) doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien E., Xagoraraki I. A water-focused one-health approach for early detection and prevention of viral outbreaks. One Health. 2019;7 doi: 10.1016/J.ONEHLT.2019.100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori R., Miura F., Kitajima M. Age-dependent association between SARS-CoV-2 cases reported by passive surveillance and viral load in wastewater. Sci. Total Environ. 2021;792 doi: 10.1016/j.scitotenv.2021.148442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell M.A., Erwin P.C., Bermejo P.M. Comparing the COVID-19 responses in Cuba and the United States. Am. J. Public Health. 2021;111(12):2186–2193. doi: 10.2105/AJPH.2021.306526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulicharla R., Kaur G., Brar S.K. A year into the COVID-19 pandemic: rethinking of wastewater monitoring as a preemptive approach. J.Environ.Chem.Eng. 2021;9(5) doi: 10.1016/j.jece.2021.106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader B. MedRxiv; 2020. Increased travel times to United States SARS-CoV-2 testing sites: a spatial modeling study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader B., Astley C.M., Sy K.T.L., Sewalk K., Hswen Y., Brownstein J.S., Kraemer M.U.G. 2020. Increased travel times to United States SARS-CoV-2 testing sites: a spatial modeling study. 2020.08.02.20159418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguti F., Magnil E., Enache L., Churqui M.P., Johansson A., Lumley D., Davidsson F., Dotevall L., Mattsson A., Trybala E., Lagging M., Lindh M., Gisslén M., Brezicka T., Nyström K., Norder H. Surveillance of wastewater revealed peaks of SARS-CoV-2 preceding those of hospitalized patients with COVID-19. Water Res. 2021;189 doi: 10.1016/j.watres.2020.116620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syangtan G., Bista S., Dawadi P., Rayamajhee B., Shrestha L.B., Tuladhar R., Joshi D.R. Asymptomatic SARS-CoV-2 carriers: a systematic review and meta-analysis. Front. Public Health. 2021;8(January):1–10. doi: 10.3389/fpubh.2020.587374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres T.S., Luz P.M., Coelho L.E., Jalil C., Falco G.G., Sousa L.P., Jalil E., Bezerra D.R.B., Cardoso S.W., Struchiner C.J., Veloso V.G., Grinsztejn B. SARS-CoV-2 testing disparities across geographical regions from a large metropolitan area in Brazil: results from a web-based survey among individuals interested in clinical trials for COVID-19 vaccines. Braz. J. Infect. Dis. 2021;25(4) doi: 10.1016/j.bjid.2021.101600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA . USEPA Manual of Methods of Virology. USEPA; Washington, DC: 2001. “Concentration and processing of waterborne viruses by positive charge 1MDS cartridge filters and organic flocculation.” Chap. 14. [Google Scholar]
- USEPA . 2014. Method 1615 Measurement of Enterovirus and Norovirus Occurrence in Water by Culture and RT-PCR. [Google Scholar]
- Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., Torgersen K., Laan J. Vander, LaCross N. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775 doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens K.E., Mawien P.N., Rumunu J., Slater D., Jones F.K., Moheed S., Caflish A., Bior B.K., Jacob I.A., Lako R.L.L., Guyo A.G., Olu O.O., Maleghemi S., Baguma A., Hassen J.J., Baya S.K., Deng L., Lessler J., Demby M.N., Wamala J.F. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Juba, South Sudan: a population-based study. MedRxiv: The Preprint Server for Health Sciences; 2021. pp. 1–28. [DOI] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M.A., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., Alm E.J. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Eurosurveillance. 2020;25(50) doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xagoraraki I. Can we predict viral outbreaks using wastewater surveillance? (p. J. Environ. Eng., 2020, 146(11): 01820003) J. Environ. Eng. 2020;146(11):01820003-1–01820003-3. [Google Scholar]
- Xagoraraki Irene, Brien Evan O. Wastewater-based Epidemiology for Early Detection of Viral Outbreaks. Springer; 2019. pp. 75–97. p. Women in Water Quality. [Google Scholar]
- Xiao Z., Xie X., Guo W., Luo Z., Liao J., Wen F., Zhou Q., Han L., Zheng T. Examining the incubation period distributions of COVID-19 on Chinese patients with different travel histories. J.Infect.Dev.Countries. 2020;14(4):323–327. doi: 10.3855/JIDC.12718. [DOI] [PubMed] [Google Scholar]
- Yaniv K., Ozer E., Kushmaro A. 2021. SARS-CoV-2 variants of concern, Gamma (P.1) and Delta (B.1.617), sensitive detection and quantification in wastewater employing direct RT-qPCR. MedRxiv, 2021.07.14.21260495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ.Sci.Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Ziqiang Y., C. V.T., V. T.V., Irene X. Sorption of human adenovirus to wastewater solids. J. Environ. Eng. 2018;144(11) doi: 10.1061/(ASCE)EE.1943-7870.0001463. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix. Supplementary materials