Abstract
Purpose:
CALGB 80405 compared the combination of first-line chemotherapy with cetuximab or bevacizumab in the treatment of advanced or metastatic colorectal cancer (mCRC). While similar clinical outcomes were observed in the cetuximab-chemotherapy group and the bevacizumab-chemotherapy group, biomarkers could identify patients deriving more benefit from either biologic agent.
Experimental design:
In this exploratory analysis, the Angiome, a panel of 24 soluble protein biomarkers were measured in baseline plasma samples in CALGB 80405. Prognostic biomarkers were determined using univariate Cox proportional hazards models. Predictive biomarkers were identified using multivariable Cox regression models including interaction between biomarker level and treatment.
Results:
In the total population, high plasma levels of Ang-2, CD73, HGF, ICAM-1, IL-6, OPN, TIMP-1, TSP-2, VCAM-1, and VEGF-R3 were identified as prognostic of worse progression-free survival (PFS) and overall survival (OS). PlGF was identified as predictive of lack of PFS benefit from bevacizumab (bevacizumab HR = 1.51, 95% CI 1.10–2.06; cetuximab HR = 0.94, 95% CI 0.71–1.25; Pinteraction = 0.0298) in the combined FOLFIRI/FOLFOX regimens. High levels of VEGF-D were predictive of lack of PFS benefit from bevacizumab in patients receiving FOLFOX regimen only (FOLFOX/bevacizumab HR = 1.70, 95% CI 1.19–2.42; FOLFOX/cetuximab HR = 0.92, 95% CI 0.68–1.24; Pinteraction = 0.0097).
Conclusions:
In this exploratory, hypothesis-generating analysis, the Angiome identified multiple prognostic biomarkers and two potential predictive biomarkers for mCRC patients enrolled in CALGB80405. PlGF and VEGF-D predicted lack of benefit from bevacizumab in a chemo-dependent manner.
Keywords: predictive biomarker, prognostic biomarker, bevacizumab, cetuximab, metastatic colorectal cancer
Introduction
Colorectal cancer (CRC) is a leading cause of cancer mortality in the United States. Significant progress has been made in the past 20 years in both detection and treatment of CRC(1). In 1990s fluorouracil became the only cytotoxic drug in CRC as continuous infusion of fluorouracil improved the overall survival (OS) from 12 to 15 months(2). Since then, combination therapies of fluorouracil with leucovorin and either irinotecan [FOLFIRI regimen(3)] or oxaliplatin [FOLFOX regimen(4)] became the first line chemotherapeutic treatment for CRC. In 2000s, targeted therapies were added to chemotherapy, including monoclonal antibodies targeting epidermal growth factor receptor (EGFR) and vascular endothelial growth factor (VEGF) (5,6).
Two anti-EGFR antibodies, panitumumab and cetuximab, have been approved by Food and Drug Administration (FDA) to treat mCRC patients as monotherapies and combined with chemotherapy(7,8). EGFR binding to its ligands results in the activation of the mitogenic MAPK/ERK signaling cascade via RAS GTPases (9). Patients whose tumors harbor activating RAS mutations do not respond to EGFR-targeting agents (10–12). Current guidelines recommend the use of panitumumab and cetuximab only for CRC patients with wild-type RAS (13).
There are currently four FDA approved biological agents for targeting the VEGF-pathway in CRC patients: first line bevacizumab which binds VEGF-A(5); second line ramucirumab which binds VEGFR-2 (14), and ziv-aflibercept, which binds VEGFs and PlGF (15); and third line regorafenib which blocks activity of all VEGF receptors and other tyrosine kinases(16,17). Extensive effort has been made to identify biomarkers to guide anti-VEGF drugs, including markers at the molecular, cellular, and tissue levels(18).
Circulating protein biomarkers have gained increasing attention given the advantages of ease to obtain, cost-effectiveness, and amenability to repeated sampling. We have developed and optimized the Angiome, a panel of circulating protein biomarkers and analyzed their predictive potential in multiple trials involving thousands of patients in different disease settings(19–22). Ang-2, SDF-1, and VEGF-D plasma levels were identified as predictive biomarkers for bevacizumab in patients with pancreatic cancer treated with gemcitabine ± bevacizumab in a study conducted by Cancer and Leukemia Group B (CALGB), now part of the Alliance for Clinical Trials in Oncology cooperative group, CALGB 80303 (20). Benefit from bevacizumab was associated with high levels of Ang-2 and SDF-1 and low levels of VEGF-D. The VEGF-D effect was strongest in patients in the lowest quartile of plasma VEGF-D (20). Consistently, negative predictive effect of VEGF-D has been reported in metastatic CRC (mCRC) patients treated in the Australian Gastro-Intestinal Trials Group testing Mitomycin, Avastin, and Xeloda (AGITG MAX trial) (23). Despite a number of promising candidates including VEGF-D, currently there are no validated biomarkers to guide the applications of anti-angiogenic agents in any disease settings(24).
CALGB and the Southwest Oncology Group (SWOG), conducted the intergroup study, CALGB/SWOG 80405, to better guide clinical decision making for patients with advanced or metastatic CRC (25). CALGB 80405 was initially designed to compare bevacizumab, cetuximab, and the combination of both agents in addition to FOLFIRI or FOLFOX chemotherapy regimen chosen by the physician and patient. Early in the course of the study, the combination regimen was discontinued due to results from the CAIRO2 and PACCE studies showing increased toxicity and a lack of increased survival benefit in the arms with dual VEGF and EGFR inhibition (26,27). CALGB80405 was negative for overall survival (OS), the primary endpoint for this study. The clinical results of CALGB 80405 showed similar progression-free survival (PFS) and OS benefits from either bevacizumab-chemotherapy or cetuximab-chemotherapy groups (25).
To improve outcomes in future clinical trials of bevacizumab in mCRC, a homogenous selected patient population would be pivotal. Disease parameters and circulating biomarkers would greatly facilitate patient selection to enrich patient deriving the most benefit from anti-angiogenic drugs. As stated earlier, our Angiome analysis has successfully identified prognostic and predictive biomarkers for anti-angiogenic agents, bevacizumab included. While prognostic biomarkers reflect how well a patient perform, independent of treatments; predictive biomarkers have the potential to identify patients benefitting the most from a specific treatment.
In this study, we assessed the Angiome expression in plasma samples collected at baseline in the primary cohort of CALGB 80405. These patients habored KRAS wild-type tumors, received either bevacizumab or cetuximab in addition to FOLFIRI or FOLFOX. Angiome analysis provided a snapshot of tumor angiogenesis, inflammation, and immune status at baseline. Potential prognostic and predictive biomarkers were identified.
Materials and Methods
Study Design and Patients
The study design of CALGB 80405 and clinical results have been reported (25) and detailed information can be found at ClinicalTrials.gov (NCT00265850). Briefly, patients with locally advanced or metastatic CRC were randomized to receive either bevacizumab, cetuximab, or the combination of both in conjunction with chemotherapy (FOLFIRI or FOLFOX) at the discretion of the treating physician. The study was later amended to stop enrollment in the combination arm and to implement KRAS mutation testing. The samples used in this study were collected at baseline from the primary cohort of CALGB 80405. The primary cohort consists of 1137 KRAS wild type patients randomized to the bevacizumab + chemotherapy or cetuximab + chemotherapy treatment groups. This correlative study of CALGB 80405 was approved by the Institutional Review Board at all participating centers, adhered to Guidelines for Good Clinical Practice and guiding principles laid out in the Declaration of Helsinki. Written informed consent was obtained from patients who opted to participate in this analysis.
Sample Collection
Peripheral venous blood was collected at baseline from consenting patients into vacutainers containing EDTA anticoagulant. Samples were centrifuged at 2500g for 15 min within 30 min of collection. The resulting plasma was spun again at 2500g for 15 min. Double-spun, platelet-poor plasma was aliquoted, frozen, and shipped on dry ice to the Alliance Pathology Coordinating Office for centralized storage. Prior to analyses, samples were shipped to Duke Phase I Biomarker Laboratory, thawed once on ice, re-aliquoted, and stored at −80°C until assayed.
Protein Analysis
Levels of 24 soluble protein biomarkers were measured in duplicates with the personnel blinded to the clinical results. Plasma samples from patients in the primary cohort were analyzed using the multiplex enzyme-linked immunosorbent assay (ELISA) techniques on the Quanterix (Billerica, MA) and Meso Scale Discovery (Rockville, MD) platforms as previously described (20,28). The analysis conforms to the guidelines established by the REMARK criteria.
Statistical Considerations
All biomarker levels were log-transformed prior to analysis, to de-emphasize outliers and better approximate a normal distribution. All analyses were performed using baseline data from patients in the primary cohort with continuous values for the protein analytes. Biomarkers prognostic of clinical outcome [overall survival (OS) or progression free survival (PFS)] were determined using univariate Cox proportional hazards models independent of treatment arm, and the resulting hazard ratios (HR), 95% confidence intervals (CI), and asymptotic P-values based on the score test were reported. Multivariable Cox regression models were used to test for interactions between biomarker levels and treatment arms to identify biomarkers predictive of benefit from bevacizumab or cetuximab and asymptotic P-values based on the Wald test for the interaction terms were reported. All analyses were stratified by type of chemotherapy (FOLFIRI or FOLFOX) to control for any differences in baseline survival rate among these two groups. Biomarker levels were also tested for interaction with treatment arm separately within each chemotherapy subpopulation. The reported p-values were not adjusted for multiple testing.
Hierarchical agglomerative clustering, using Euclidean distance and complete linkage, was used to identify groupings among protein biomarkers based on their baseline levels. The results were illustrated using a dendrogram. Forest plots were created as illustrations of prognostic effect sizes (HRs and corresponding 95% CIs). For the purposes of illustrating interaction effects, biomarker levels were dichotomized as “high” or “low” relative to the median, and Kaplan–Meier plots of OS and PFS were created, with separate curves for each combination of treatment group and dichotomized biomarker level. It has previously been shown that low levels of VEGF-D (first quartile) were predictive of benefit from bevacizumab in advanced pancreatic cancer and colon cancer, respectively (20,23), so we also considered VEGF-D levels separated by quartiles. Unless otherwise stated, the results reported pertain to the analysis of biomarkers as continuous measures.
The Alliance Statistics and Data Center conducted data collection and statistical analyses on the basis of clinical data locked as of December 15, 2015. The R software environment for statistical computing and graphics and the survival extension package were used to conduct the statistical analyses and to generate the figures(29,30). The analyses were carried out with adherence to the principles of reproducible analysis using the knitr package(31) for generation of dynamic reports and GitLab for source code management.
Data Availability Statement
The code for replicating the statistical analysis has been made accessible through a public source code repository (https://gitlab.oit.duke.edu/dcibioinformatics/pubs/calgb-80405-plasma). Codes used for all analyses will be made available upon request.
Results
Patient Characteristics
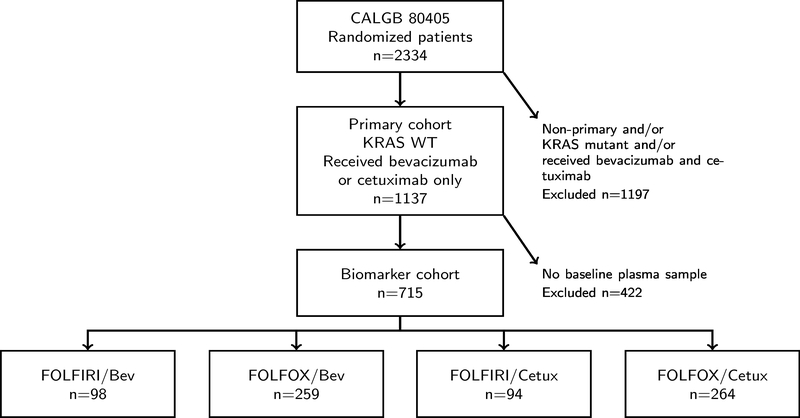
In CALGB 80405 primary cohort, mCRC patients with KRAS wild type were randomized to receive bevacizumab (BEV) or cetuximab (Cetux), in addition to either FOLFIRI or FOLFOX as chemotherapeutic agents per physician choice. The last date of follow up was December 15, 2015 and the median follow up for surviving patients was 47.4 months(25). The demographic and survival outcomes for the total primary cohort population (n=1137) and the biomarker population (n=715) are shown in Figure 1 and Supplementary Table S1. No significant differences were noticed regarding age, race, gender, or clinical outcomes between the biomarker population and the total primary cohort. To better present the four patient cohorts receiving different treatments, a detailed demographic table was generated. As reported in the clinical paper, there is no statistically significant difference in overall survival (OS) and progression free survival (PFS) in the bevacizumab-chemotherapy group and the cetuximab-chemotherapy group(25). All four patient cohorts showed similar baseline characteristics (Table 1).
Figure 1. Patient CONSORT diagram.
Bev: bevacizumab; Cetux: cetuximab.
Table 1.
Distribution of the biomarker subpopulation by treatment.
| Chemotherapy | FOLFIRI/ | FOLFOX/ | FOLFIRI/ | FOLFOX/ | Primary cohort | |
|---|---|---|---|---|---|---|
| Target therapy | Bev | Bev | Cetux | Cetux | ||
| N | 98 | 259 | 94 | 264 | 1137 | |
| Age | Median | 61 | 58 | 60 | 59 | 59 |
| (range) | (34–82) | (22–84) | (36–83) | (23–84) | (20–89) | |
| Race | African American | 9 (9.2%) | 22 (8.5%) | 10 (10.6%) | 31 (11.7%) | 129 (11.3%) |
| Asian | 4 (4.1%) | 4 (1.5%) | 2 (2.1%) | 9 (3.4%) | 35 (3.1%) | |
| Caucasian | 84 (85.7%) | 228 (88.0%) | 81 (86.2%) | 218 (82.6%) | 934 (82.1%) | |
| Multiple | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (0.4%) | 5 (0.4%) | |
| Other | 1 (1.0%) | 5 (1.9%) | 1 (1.1%) | 5 (1.9%) | 34 (3.0%) | |
| Sex | Female | 37 (37.8%) | 95 (36.7%) | 37 (39.4%) | 103 (39.0%) | 440 (38.7%) |
| Male | 61 (62.2%) | 164 (63.3%) | 57 (60.6%) | 161 (61.0%) | 697 (61.3%) | |
| Median Outcome (months) | OS | 35.1 | 30.3 | 31.9 | 28.9 | 29.4 |
| (95% CI) | (29.4–41.3) | (27.1–33.2) | (26.8–40.2) | (25.3–34.6) | (27.6–31.4) | |
| PFS | 12.4 | 10.8 | 10.9 | 10 | 10.6 | |
| (95% CI) | (10.4–14.0) | (9.5–11.7) | (9.2–14.4) | (9.2–11.3) | (9.8–11.1) | |
| ECOG N (%) | 0 | 60 (61.2%) | 153 (59.1%) | 55 (58.5%) | 162 (61.4%) | 657 (57.8%) |
| 1 | 38 (38.8%) | 105 (40.5%) | 39 (41.5%) | 102 (38.6%) | 478 (42.0%) | |
| 2 | 1 (0.4%) | 2 (0.2%) | ||||
| Tumor Biology | Metachronous | 36 (36.7%) | 33 (12.7%) | 34 (36.2%) | 45 (17.1%) | 236 (20.8%) |
| Synchronous | 62 (63.3%) | 225 (86.9%) | 57 (60.6%) | 217 (82.2%) | 892 (78.5%) | |
| unknown | 1 (0.4%) | 3 (3.2%) | 2 (0.8%) | 9 (0.8%) | ||
| CRC Tumor Location N (%) | Left | 63 (64.3%) | 147 (56.8%) | 63 (67.0%) | 164 (62.1%) | 689 (60.6%) |
| Right | 24 (24.5%) | 69 (26.6%) | 16 (17.0%) | 61 (23.1%) | 280 (24.6%) | |
| Transverse | 3 (3.1%) | 18 (7.0%) | 7 (7.5%) | 17 (6.4%) | 62 (5.5%) | |
| Multiple | 1 (0.4%) | 1 (0.1%) | ||||
| unknown | 8 (8.2%) | 25 (9.7%) | 8 (8.5%) | 21 (8.0%) | 105 (9.2%) | |
| Primary tumor unresected at study entry | No | 76 (77.6%) | 192 (74.1%) | 75 (79.8%) | 196 (74.2%) | 845 (74.3%) |
| Yes | 22 (22.4%) | 67 (25.9%) | 19 (20.2%) | 68 (25.8%) | 292 (25.7%) |
Abbreviations: Bev, bevacizumab; Cetux, cetuximab.
Notes: metachronous indicates metastasis subsequent to diagnosis of primary tumor; synchronous indicates metastasis present at diagnosis of primary tumor. Values from the primary cohort are as reported in Venook et al (25).
Biomarker assessment
A total of 24 circulating protein biomarkers were assessed using baseline plasma samples for 715 mCRC patients enrolled in CALGB80405. The median levels and ranges for all biomarkers are shown in Supplementary Table S2. A dendrogram illustrating the associations among these 24 biomarkers is shown in Supplementary Fig. S1. As observed in previous analyses, we noted several consistent relationships among the protein biomarkers. PlGF was tightly associated with general angiogenic biomarkers such as Ang-2 and TSP-2, while VEGF-A, -C, -D were more clustered with factors in TGF-β signaling pathway. CD73 and HER-3, two previously identified biomarkers predictive of benefit from cetuximab(28), were tightly clustered with each other.
Prognostic Analyses
The clinical report showed similar OS and PFS across patients in all treatment groups, therefore, prognostic analysis was conducted with all patients combined. We identified ten biomarkers (Ang-2, CD73, HGF, ICAM-1, IL-6, OPN, TIMP-1, TSP-2, VCAM-1, and VEGFR-3) to be prognostic for both OS and PFS in the overall population, as illustrated by the Forrest plots (Figure 2). All of the biomarkers identified were negative prognostic biomarkers, i.e., higher expression levels were associated with shorter times to event (OS or PFS). The strongest prognostic effects with respect to hazard ratio (HR) for OS and PFS were observed for VCAM-1 (for OS, HR=2.49, 95%CI=2.01–3.09, p<0.0001; for PFS, HR=1.55, 95%CI=1.28–1.88, p<0.0001). A complete list of the prognostic value for all 24 biomarkers for OS and PFS was shown in Supplementary Tables S3 and S4, respectively. All biomarkers tested were independently prognostic from one another. PlGF and VEGF-A were prognostic for OS (PlGF, HR=1.31, 95% CI=1.05–1.63, p=0.0166; VEGF-A, HR=1.10, 95% CI=1.02–1.19, p=0.0143; Supplementary Table S3), but not for PFS (PlGF, HR=1.16, 95% CI=0.95–1.41, p=0.1473; VEGF-A, HR=1.06, 95% CI=0.99–1.14, p=0.0811; Supplementary Table S4).
Figure 2. Prognostic biomarkers.
Forest plots of hazard ratios (HRs) illustrate the prognostic association of biomarkers for both overall survival (A) and progression-free survival (B) in the overall population. All 10 were negative prognostic indicating that higher biomarker levels are associated with higher HR or favors lower expression.
Predictive Analyses
We identified PlGF as the only predictive biomarker of PFS benefit in the overall population. A high baseline level of PlGF was associated with better PFS when treated by cetuximab (BEV arm: HR = 1.51, 95% CI 1.10–2.06; Cetux arm: HR = 0.94, 95% CI 0.71–1.25; Pintx = 0.0298) (Table 2). No other biomarkers were observed to be predictive of OS or PFS in the chemotherapy-pooled populations (Supplemental Tables S5 and S6). A Kaplan-Meier curve illustrating the predictive effect of PlGF is shown in Figure 3. Patients with lower than median PlGF exhibited longer PFS and benefited more from bevacizumab than cetuximab.
Table 2.
Predictive effects of PlGF and VEGF-D.
| OS | PFS | |||||||
|---|---|---|---|---|---|---|---|---|
| Chemo pooled | Chemo pooled | |||||||
| Biomarker | Bev | Cetux | Pintx | Bev | Cetux | Pintx | ||
| PlGF | 1.65 (1.16–2.36) | 1.12 (0.83–1.51) | 0.1157 | PlGF | 1.51 (1.10–2.06) | 0.94 (0.71–1.25) | 0.0298 | |
| VEGF-D | 1.31 (0.98–1.74) | 1.12 (0.84–1.50) | 0.4346 | VEGF-D | 1.34 (1.03–1.73) | 0.97 (0.75–1.27) | 0.1069 | |
| FOLFIRI | FOLFIRI | |||||||
| PlGF | 1.47 (0.74–2.94) | 0.84 (0.45–1.57) | 0.2079 | PlGF | 1.34 (0.71–2.51) | 0.80 (0.46–1.41) | 0.2487 | |
| VEGF-D | 0.84 (0.49–1.45) | 1.10 (0.54–2.26) | 0.5643 | VEGF-D | 0.95 (0.61–1.48) | 1.37 (0.73–2.56) | 0.4336 | |
| FOLFOX | FOLFOX | |||||||
| PlGF | 1.71 (1.13–2.59) | 1.23 (0.88–1.71) | 0.2540 | PlGF | 1.56 (1.08–2.24) | 0.99 (0.72–1.37) | 0.0656 | |
| VEGF-D | 1.65 (1.17–2.33) | 1.12 (0.81–1.54) | 0.1151 | VEGF-D | 1.70 (1.19–2.42) | 0.92 (0.68–1.24) | 0.0097 | |
Note: median overall survival or progression-free survival time are presented in months, with 95% confidence intervals in parenthesis. Abbreviations: Bev, bevacizumab; Cetux, cetuximab; Pintx, interaction p value.
Figure 3. Kaplan-Meier curves of PlGF and treatment for overall survival (Top) and progression-free survival (Bottom).

For illustrative purposes, PlGF levels were dichotomized as “high” or “low” relative to the median. P-values from the test for interaction of the marker as a continuous measure are reported.
To control for the possibility of the interaction effects across chemotherapies used in CALGB 80405, the biomarkers were re-analyzed for predictive associations within chemotherapy subpopulations. In patients receiving FOLFOX, low VEGF-D was associated with better PFS and benefit from bevacizumab (BEV arm: HR = 1.70, 95% CI 1.19–2.42; Cetux arm: HR = 0.92, 95% CI 0.68–1.24; Pintx = 0.0097, Table 2). While a borderline trend was noted for PlGF, the PFS benefit did not reach statistical significance (Pintx =0.0656, Table 2). Kaplan-Meier curves showed the predictive effects of VEGF-D in chemotherapy combined, as well as in FOLFOX and FOLFIRI subpopulation respectively (Figure 4). No other biomarkers were observed to be predictive for OS or PFS when patients were stratified by chemotherapy (Supplementary Table S7 and S8).
Figure 4. Kaplan-Meier curves showing the effects of VEGF-D levels and chemotherapy on overall survival (Top panels) and progression-free survival (Bottom panels).
For illustrative purposes, VEGF-D levels were dichotomized as “high” or “low” relative to the first quartile. P-values from the test for interaction of the maker as a continuous measure are reported. Patients with VEGF-D levels in the lowest quartile receiving FOLFOX as the chemotherapy derived more benefit from bevacizumab (the interaction p-value for the marker as a continuous measure was 0.0097), as shown in the middle bottom figure.
To optimize the cut-point of VEGF-D level, Forest plots were generated to illustrate the effects of VEGF-D by quartile on OS and PFS in patients receiving FOLFOX (Figure 5). Patients with VEGF-D levels in the lowest quartile had better OS (HR =0.62, 95% CI=0.41–0.92) and PFS (HR =0.59, 95% CI=0.41–0.85).
Figure 5. Predictive effect of VEGF-D.
Forest plots of VEGF-D quantiles in the FOLFOX-treated subpopulation for overall survival (A) and progression-free survival (B). Patients with VEGF-D levels in the lowest quantile derived benefit from bevacizumab.
Discussion
CALGB 80405 is the largest phase III trial to directly compare bevacizumab and cetuximab, two of the most commonly used targeted agents, in addition to FOLFIRI or FOLFOX as first-line treatment in advanced or metastatic CRC. Blood samples collected during CALGB 80405 provide a unique and valuable source of materials for biomarker studies that may help guide the use of these therapeutics. Our retrospective biomarker analysis identified PlGF and VEGF-D, two chemo-dependent predictive biomarkers, as well as numerous prognostic biomarkers.
We identified ten biomarkers with highly consistent prognostic impact across both OS and PFS. These biomarkers play important roles in angiogenesis (Ang-2, HGF, TSP-2, VEGF-R3), inflammatory and immune modulating (CD73, IL-6, ICAM-1, VCAM-1), and extracellular matrix remodeling (OPN, TIMP-1). Intrinsic high expression of all these biomarkers was associated with worse outcomes, reflecting the coordinated interplay of angiogenesis, inflammation, and immune modulation. Interestingly, PlGF and VEGF-A were found to be prognostic for OS, but not for PFS. This is likely due to the manifest of stronger prognostic effect at later stage of the disease or over longer time, as the median OS was approximately 20 months longer than median PFS(25).
With respect to EGFR inhibition, we previously identified two predictive biomarkers for cetuximab benefit (HER-3 and CD73) across all patients (KRAS mutant and wild-type) in CALGB 80203(28), a randomized study evaluating the efficacy of cetuximab in combination with FOLFOX or FOLFIRI in mCRC. However, neither biomarker predicted benefit from cetuximab here in CALGB80405 (Supplementary Table S5–S8). HER-3 belongs to the same family of receptor tyrosine kinases as EGFR, with the potential to serve as a resistance mechanism to cetuximab(32). CD73, an immune modulatory AMP phosphatase, was associated with cetuximab benefit in previous studies (28,33). The lack of predictive value in this study may be due to the potential association between CD73 and RAS mutational status(34). In this correlative study, all patients included were KRAS wild type at codons 12 and 13. Exclusion of patients with RAS mutations may confound the analysis. Although RAS wild-type has been widely applied as a prerequisite for patients to receive anti-EGFR inhibitors, not all patients with wild-type RAS respond to EGFR inhibitors. Our Angiome analysis could provide novel insight regarding the patient population most likely to derive benefit from the EGFR-targeting drugs.
With respect to VEGF inhibition, VEGF-A has long been considered a candidate for predictive biomarker for anti-angiogenic therapies, and high levels of VEGF-A may indicate increased biological dependency on this angiogenic factor. Yet a meta-analysis of more than 1800 patients participating in phase III trials in CRC, non-small cell lung cancer, and renal cell carcinoma confirmed that pretreatment VEGF-A levels have only prognostic, but not predictive value (35). In the MERiDiAN trial, pre-selecting metastatic breast cancer patients based on high plasma levels of short isoforms of VEGF-A for bevacizumab treatment revealed no significant association of PFS by VEGF-A interaction, failing to support using baseline VEGF-A to identify patients benefitting most from bevacizumab(36). Similarly, PlGF has prognostic value, but has not been shown to be predictive for anti-angiogenic therapy(37). Our data provide novel insight into the potential predictive role of these VEGF family members.
In this study, we found PlGF was predictive of lack of PFS benefit from bevacizumab independent of the type of chemotherapy used (Table 2 and Figure 3). PlGF belongs to the VEGF family; it binds and activates signaling through VEGF-R1, but does not directly signal through VEGF-R2 (38). PlGF modulates VEGF-A signaling both at the ligand level (by forming heterodimers with VEGF-A) and the receptor level (by engaging VEGF-R1) (39). PlGF has been noted as a pharmacodynamic biomarker with circulating levels consistently increasing in response to anti-VEGF agents (22,40,41), but this is the first report of a potential predictive association between baseline PlGF levels and outcome in CRC. Patients with lower than median levels of PlGF derived more PFS benefit from bevacizumab (Pintx=0.0298, Figure 3), independent of chemotherapy received. It is important to note that the comparative arm used in this study is an active therapy (cetuximab) and not a true placebo. This may explain why we observed PlGF as a predictive biomarker only in this specific study population.
VEGF-D was identified as the strongest predictive biomarker for lack of PFS benefit from bevacizumab; however, this was only observed in the FOLFOX treated patients (Pintx=0.0097, Table 2). VEGF-D is another VEGF family member; it binds VEGF-R2 promoting angiogenesis, as well as VEGF-R3 where it promotes lymphangiogenesis (42). The predictive effect was largest in patients with VEGF-D levels in the lowest quartile, where patients derived the most benefit from bevacizumab (Figure 5). Importantly, far more patients in CALGB 80405 study received FOLFOX (n=523) than FOLFIRI (n=192). It is possible that additional clinical factors related to the use of FOLFOX over FOLFIRI may confound the VEGF-D effect observed here. The exact mechanism for this chemotherapy effect remains unclear. The VEGF-D effect is consistent with results seen previously in patients with pancreatic cancer treated with bevacizumab and gemcitabine in CALGB 80303 (20), and in patients with CRC treated in the Australian Gastro-Intestinal Trials Group testing Mitomycin, Avastin, and Xeloda (AGITG MAX trial) (23). In all cases, low VEGF-D predicts benefit from bevacizumab, while high VEGF-D predicts for lack of benefit from bevacizumab. Excessive VEGF-D can provide compensatory signaling in the VEGF axis through VEGF-R2, serving as a resistance mechanism upon VEGF-A blockage by bevacizumab. Therefore, patients are less likely to benefit from bevacizumab given the presence of a readily available compensatory mechanism. The predictive roles of PlGF and VEGF-D noted here in CALGB 80405 may suggest that alternate VEGF family members may mediate intrinsic resistance mechanisms to bevacizumab, which selectively binds VEGF-A.
While no other biomarkers reach statistical significance, a few interesting trends were noted. First, VEGF-C, an additional VEGF family member with potent angiogenic and lymphangiogenic properties, appears to predict for OS benefit from cetuximab in FOLFIRI subgroup (Pintx=0.0791, Supplementary Table S7). Additionally, in the subgroup of patients receiving FOLFIRI, we identified three TGF-β related biomarkers to be potentially predictive of PFS benefit, including TGF-β1 (Pintx=0.0524), PDGF-AA (Pintx=0.0511), and SDF-1 (Pintx=0.0688; Supplementary Table S8). Higher levels of TGF-β1 and PDGF-AA potentially predict for benefit from cetuximab. These biomarkers may reflect overlapping biologies contributing to cetuximab sensitivity. Interplay between EGFR and TGF-β signaling pathways have been extensively reviewed(43).
Despite extensive effort, no predictive biomarker has been identified that would enable a more personalized use of bevacizumab. Angiogenesis is a complex process involving diverse cell types, cytokines, and growth factors signaling through multiple pathways. The biological redundancies eventually lead to anti-angiogenic drug resistance. As the biology of angiogenesis and its role in tumor development varies across tumor types, suitable biomarkers may be tumor type specific. While angiogenic factors (PlGF and VEGF-D) were noted to be associated with PFS benefit in mCRC patients in CALGB80405, other studies noted the predictive potential of the inflammatory cytokine IL-6 in the context of anti-angiogenic therapy in renal cancer and ovarian cancer(21,44).
This analysis has several limitations. The patients receiving FOLFIRI were much fewer than those receiving FOLFOX, therefore underpowered to explore key subgroups. The unbalanced use of FOLFIRI and FOLFOX may introduce uncontrolled bias into these analyses. While all patients are considered KRAS WT, KRAS testing was only performed at codons 12 and 13. It is possible that some patients with KRAS mutant were included in our analysis. Genetic information such as extended RAS and RAF mutations (45), microsatellites instability and tumor mutational burdens(46), consensus molecular subtypes (CMSs) representing intrinsic heterogeneity of CRC at the genetic expression level (47) will be further investigated in subsequent analysis. Additionally, this retrospective biomarker analysis was intended to be exploratory and hypothesis-generating in nature, hence the p-values presented here were not adjusted for multiple testing. The current findings are promising and biologically plausible but should be considered preliminary and not ready to guide patient care at this time. In order to advance any of these putative biomarkers along the validation path, a predefined cut-point would need to be established and tested. Ideally, any biomarker cut-point would be tested prospectively as a stratification factor or for entry onto the trial. The establishment of a cut-point is a crucial step for clinical decision making when considering the prospective use of any prognostic or predictive biomarker. As such, validation of VEGF-D as a biomarker with a specific cut-point awaits further investigation.
Discovery and validation of predictive biomarkers is challenging and requires a detailed understanding of the target expression and biology in patients, technical optimization of biomarker assays, and clinical validation of any findings. This analysis in CALGB 80405 is the culmination for nearly two decades of work by this group. The findings of predictive biomarkers may be applied to other FDA-approved drugs for mCRC. Notably, ziv-aflibercept, a recombinant receptor containing the ligand-binding domains of VEGF-R1 and -R2, binds both VEGFs and PlGF(15,48); while ramucirumab blocks the binding of both VEGF-A, -C, and -D to VEGF-R2(49,50). It is not known whether these agents may be more active in patients with higher levels of plasma PlGF and VEGF-D. Nevertheless, our data support the rationale for biomarker-driven clinical studies to address this possibility.
In conclusion, our biomarker analysis of CALGB 80405 identified two candidate predictive biomarkers for lack of benefit from bevacizumab: PlGF and VEGF-D. Future studies to validate and refine these findings are warranted.
Supplementary Material
Translational relevance.
CALGB 80405 was a randomized phase III trial in advanced or metastatic colorectal cancer (mCRC) patients testing whether the addition of cetuximab or bevacizumab to the combination of either leucovorin, fluorouracil, and irinotecan (FOLFIRI) or the combination of leucovorin, fluorouracil, and oxaliplatin (FOLFOX) regimen was superior as first-line therapy in this setting. The results of the trial were negative, similar overall survival (OS) and progression-free survival (PFS) were observed for the cetuximab-chemotherapy group and the bevacizumab-chemotherapy group. In this report, we conducted an exploratory, hypothesis-generating, retrospective analysis evaluating a panel of angiogenic and inflammatory biomarkers using baseline plasma samples collected from patients enrolled in CALGB80405 and identified PlGF and VEGF-D as potential predictive biomarkers. High PlGF levels predict lack of PFS benefit from bevacizumab independent of the chemotherapeutic regimen, while low VEGF-D levels predict PFS benefit from bevacizumab in patients receiving FOLFOX. Notably, the predictive potential of VEGF-D for anti-angiogenic agents has been previously reported by several groups. Given the lack of a true control arm, the findings are exploratory and hypothesis-generating in nature.
Acknowledgements
We gratefully acknowledge the invaluable contributions of the patients, their families, and the staff who participated in this study. The authors would also like to thank the Alliance Biorepository at the Ohio State University and administrative staff. More than 680 institutions participated in this study. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of National Cancer Institute.
Support: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821, U10CA180882, and U24CA196171 (to the Alliance for Clinical Trials in Oncology), UG1CA233180, UG1CA233253, UG1CA233290, UG1CA233337, UG1CA233373, U10CA180820 (ECOG-ACRIN), U10CA180888 (SWOG). The following have provided support to Alliance for Clinical Trials in Oncology and Alliance Foundation Trials programs (https://acknowledgments.alliancefound.org). This study was also supported in part by funds from Bristol-Myers, Genentech, Pfizer, and Sanofi. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosure of Potential Conflicts of Interest: A.B. Nixon has received research funding from Genentech, HTG Molecular Diagnostics, MedImmune/AstraZeneca, Medpacto, Promega Corporation, Seattle Genetics; and has received consultant/advisory compensation from AdjuVolt Therapeutics, Eli Lilly, GSK, Leap Therapeutics, Promega Corporation. H.J. Lenz has served as advisory Board and consulting roles in Bayer, BMS, Roche, Genentech, Merck KG, Merck, Fulgent, Oncocyte, G1 Therapeutics, Jazz Therapeutics, Biacara. J.A. Meyerhardt has received institutional research funding from Boston Biomedical, has served as an advisor/ consultant to COTA Healthcare, and served on a grant review panel for the National Comprehensive Cancer Network funded by Taiho Pharmaceutical. E.M. O’Reilly has received funding from Genentech/Roche, Celgene/BMS, BioNTech, BioAtla, AstraZeneca, Arcus, Elicio, Parker Institute, AstraZeneca, Pertzye; and has served consulting role in Cytomx Therapeutics (DSMB), Rafael Therapeutics (DSMB), Sobi, Silenseed, Tyme, Seagen, Molecular Templates, Boehringer Ingelheim, BioNTech, Ipsen, Polaris, Merck, IDEAYA, Cend, AstraZeneca, Noxxon, BioSapien, Cend Therapeutics, Bayer (spouse), Genentech-Roche (spouse), Celgene-BMS (spouse), Eisai (spouse), Research To Practice. H.I. Hurwitz declares ownership of Roche stock. The other authors declare no potential conflicts of interest.
References
- 1.Jemal A, Ward EM, Johnson CJ, Cronin KA, Ma J, Ryerson AB, et al. Annual Report to the Nation on the Status of Cancer, 1975–2014, Featuring Survival. JNCI: Journal of the National Cancer Institute 2017;109(9):djx030–djx doi 10.1093/jnci/djx030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Dwyer PJ, Paul AR, Walczak J, Weiner LM, Litwin S, Comis RL. Phase II study of biochemical modulation of fluorouracil by low-dose PALA in patients with colorectal cancer. J Clin Oncol 1990;8(9):1497–503 doi 10.1200/JCO.1990.8.9.1497. [DOI] [PubMed] [Google Scholar]
- 3.Douillard JY, Cunningham D, Roth AD, Navarro M, James RD, Karasek P, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet 2000;355(9209):1041–7 doi 10.1016/s0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- 4.de Gramont A, Figer A, Seymour M, Homerin M, Hmissi A, Cassidy J, et al. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J Clin Oncol 2000;18(16):2938–47 doi 10.1200/JCO.2000.18.16.2938. [DOI] [PubMed] [Google Scholar]
- 5.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 2004;350(23):2335–42 doi 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004;351(4):337–45 doi 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 7.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26(10):1626–34 doi 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 8.Van Cutsem E, Lenz HJ, Kohne CH, Heinemann V, Tejpar S, Melezinek I, et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol 2015;33(7):692–700 doi 10.1200/JCO.2014.59.4812. [DOI] [PubMed] [Google Scholar]
- 9.Roskoski R Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res 2014;79:34–74 doi 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 2008;26(3):374–9 doi 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 11.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369(11):1023–34 doi 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 12.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359(17):1757–65 doi 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 13.Burtness B, Anadkat M, Basti S, Hughes M, Lacouture ME, McClure JS, et al. NCCN Task Force Report: Management of dermatologic and other toxicities associated with EGFR inhibition in patients with cancer. J Natl Compr Canc Netw 2009;7 Suppl 1:S5–21; quiz S2–4 doi 10.6004/jnccn.2009.0074. [DOI] [PubMed] [Google Scholar]
- 14.Tabernero J, Yoshino T, Cohn AL, Obermannova R, Bodoky G, Garcia-Carbonero R, et al. Ramucirumab versus placebo in combination with second-line FOLFIRI in patients with metastatic colorectal carcinoma that progressed during or after first-line therapy with bevacizumab, oxaliplatin, and a fluoropyrimidine (RAISE): a randomised, double-blind, multicentre, phase 3 study. Lancet Oncol 2015;16(5):499–508 doi 10.1016/S1470-2045(15)70127-0. [DOI] [PubMed] [Google Scholar]
- 15.Van Cutsem E, Tabernero J, Lakomy R, Prenen H, Prausova J, Macarulla T, et al. Addition of aflibercept to fluorouracil, leucovorin, and irinotecan improves survival in a phase III randomized trial in patients with metastatic colorectal cancer previously treated with an oxaliplatin-based regimen. J Clin Oncol 2012;30(28):3499–506 doi 10.1200/JCO.2012.42.8201. [DOI] [PubMed] [Google Scholar]
- 16.Grothey A, Van Cutsem E, Sobrero A, Siena S, Falcone A, Ychou M, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet 2013;381(9863):303–12 doi 10.1016/S0140-6736(12)61900-X. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Qin S, Xu R, Yau TC, Ma B, Pan H, et al. Regorafenib plus best supportive care versus placebo plus best supportive care in Asian patients with previously treated metastatic colorectal cancer (CONCUR): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2015;16(6):619–29 doi 10.1016/S1470-2045(15)70156-7. [DOI] [PubMed] [Google Scholar]
- 18.Murukesh N, Dive C, Jayson GC. Biomarkers of angiogenesis and their role in the development of VEGF inhibitors. Br J Cancer 2010;102(1):8–18 doi 10.1038/sj.bjc.6605483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatch AJ, Clarke JM, Nixon AB, Hurwitz HI. Identifying Blood-Based Protein Biomarkers for Antiangiogenic Agents in the Clinic: A Decade of Progress. Cancer J 2015;21(4):322–6 doi 10.1097/PPO.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 20.Nixon AB, Pang H, Starr MD, Friedman PN, Bertagnolli MM, Kindler HL, et al. Prognostic and predictive blood-based biomarkers in patients with advanced pancreatic cancer: results from CALGB80303 (Alliance). Clin Cancer Res 2013;19(24):6957–66 doi 10.1158/1078-0432.CCR-13-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarez Secord A, Bell Burdett K, Owzar K, Tritchler D, Sibley AB, Liu Y, et al. Predictive Blood-Based Biomarkers in Patients with Epithelial Ovarian Cancer Treated with Carboplatin and Paclitaxel with or without Bevacizumab: Results from GOG-0218. Clin Cancer Res 2020;26(6):1288–96 doi 10.1158/1078-0432.CCR-19-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Starr MD, Bulusu A, Pang H, Wong NS, Honeycutt W, et al. Correlation of angiogenic biomarker signatures with clinical outcomes in metastatic colorectal cancer patients receiving capecitabine, oxaliplatin, and bevacizumab. Cancer Med 2013;2(2):234–42 doi 10.1002/cam4.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weickhardt AJ, Williams DS, Lee CK, Chionh F, Simes J, Murone C, et al. Vascular endothelial growth factor D expression is a potential biomarker of bevacizumab benefit in colorectal cancer. Br J Cancer 2015;113(1):37–45 doi 10.1038/bjc.2015.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia J, Hurwitz HI, Sandler AB, Miles D, Coleman RL, Deurloo R, et al. Bevacizumab (Avastin(R)) in cancer treatment: A review of 15 years of clinical experience and future outlook. Cancer Treat Rev 2020;86:102017 doi 10.1016/j.ctrv.2020.102017. [DOI] [PubMed] [Google Scholar]
- 25.Venook AP, Niedzwiecki D, Lenz HJ, Innocenti F, Fruth B, Meyerhardt JA, et al. Effect of First-Line Chemotherapy Combined With Cetuximab or Bevacizumab on Overall Survival in Patients With KRAS Wild-Type Advanced or Metastatic Colorectal Cancer: A Randomized Clinical Trial. JAMA 2017;317(23):2392–401 doi 10.1001/jama.2017.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hecht JR, Mitchell E, Chidiac T, Scroggin C, Hagenstad C, Spigel D, et al. A randomized phase IIIB trial of chemotherapy, bevacizumab, and panitumumab compared with chemotherapy and bevacizumab alone for metastatic colorectal cancer. J Clin Oncol 2009;27(5):672–80 doi 10.1200/JCO.2008.19.8135. [DOI] [PubMed] [Google Scholar]
- 27.Tol J, Koopman M, Cats A, Rodenburg CJ, Creemers GJ, Schrama JG, et al. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med 2009;360(6):563–72 doi 10.1056/NEJMoa0808268. [DOI] [PubMed] [Google Scholar]
- 28.Hatch AJ, Sibley AB, Starr MD, Brady JC, Jiang C, Jia J, et al. Blood-based markers of efficacy and resistance to cetuximab treatment in metastatic colorectal cancer: results from CALGB 80203 (Alliance). Cancer Med 2016;5(9):2249–60 doi 10.1002/cam4.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria: 2020;URL https://www.R-project.org/. [Google Scholar]
- 30.Therneau T. A package for survival analysis in R. R package version 3.1–11. <URL: https://CRANR-projectorg/package=survival> 2020. [Google Scholar]
- 31.Xie Y. Dynamic Documents with R and knitr. 2nd edition. CRC Press, A Chapman & Hall Book Taylor & Francis Group; 2015;ISBN 13:9781498716970. [Google Scholar]
- 32.Iida M, Brand TM, Starr MM, Huppert EJ, Luthar N, Bahrar H, et al. Overcoming acquired resistance to cetuximab by dual targeting HER family receptors with antibody-based therapy. Mol Cancer 2014;13:242 doi 10.1186/1476-4598-13-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker JB, Dutta D, Watson D, Maddala T, Munneke BM, Shak S, et al. Tumour gene expression predicts response to cetuximab in patients with KRAS wild-type metastatic colorectal cancer. Br J Cancer 2011;104(3):488–95 doi 10.1038/sj.bjc.6606054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol 2007;25(22):3230–7 doi 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 35.Hegde PS, Jubb AM, Chen D, Li NF, Meng YG, Bernaards C, et al. Predictive impact of circulating vascular endothelial growth factor in four phase III trials evaluating bevacizumab. Clin Cancer Res 2013;19(4):929–37 doi 10.1158/1078-0432.CCR-12-2535. [DOI] [PubMed] [Google Scholar]
- 36.Miles D, Cameron D, Bondarenko I, Manzyuk L, Alcedo JC, Lopez RI, et al. Bevacizumab plus paclitaxel versus placebo plus paclitaxel as first-line therapy for HER2-negative metastatic breast cancer (MERiDiAN): A double-blind placebo-controlled randomised phase III trial with prospective biomarker evaluation. Eur J Cancer 2017;70:146–55 doi 10.1016/j.ejca.2016.09.024. [DOI] [PubMed] [Google Scholar]
- 37.Scholch S, Bogner A, Bork U, Rahbari M, Gyorffy B, Schneider M, et al. Serum PlGF and EGF are independent prognostic markers in non-metastatic colorectal cancer. Sci Rep 2019;9(1):10921 doi 10.1038/s41598-019-47429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tjwa M, Luttun A, Autiero M, Carmeliet P. VEGF and PlGF: two pleiotropic growth factors with distinct roles in development and homeostasis. Cell and Tissue Research 2003;314(1):5–14 doi 10.1007/s00441-003-0776-3. [DOI] [PubMed] [Google Scholar]
- 39.Autiero M, Waltenberger J, Communi D, Kranz A, Moons L, Lambrechts D, et al. Role of PlGF in the intra- and intermolecular cross talk between the VEGF receptors Flt1 and Flk1. Nat Med 2003;9(7):936–43 doi 10.1038/nm884. [DOI] [PubMed] [Google Scholar]
- 40.Kopetz S, Hoff PM, Morris JS, Wolff RA, Eng C, Glover KY, et al. Phase II trial of infusional fluorouracil, irinotecan, and bevacizumab for metastatic colorectal cancer: efficacy and circulating angiogenic biomarkers associated with therapeutic resistance. J Clin Oncol 2010;28(3):453–9 doi 10.1200/JCO.2009.24.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieu CH, Tran H, Jiang ZQ, Mao M, Overman MJ, Lin E, et al. The association of alternate VEGF ligands with resistance to anti-VEGF therapy in metastatic colorectal cancer. PLoS One 2013;8(10):e77117 doi 10.1371/journal.pone.0077117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohela M, Bry M, Tammela T, Alitalo K. VEGFs and receptors involved in angiogenesis versus lymphangiogenesis. Curr Opin Cell Biol 2009;21(2):154–65 doi 10.1016/j.ceb.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Koveitypour Z, Panahi F, Vakilian M, Peymani M, Seyed Forootan F, Nasr Esfahani MH, et al. Signaling pathways involved in colorectal cancer progression. Cell Biosci 2019;9:97 doi 10.1186/s13578-019-0361-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nixon AB. Identification of predictive biomarkers of overall survival in patients with advanced renal cell carcinoma treated with interferon alpha +/− bevacizumab: Results from CALGB 90206 (Alliance). CCR, in press 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417(6892):949–54 doi 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 46.Innocenti F, Ou FS, Qu X, Zemla TJ, Niedzwiecki D, Tam R, et al. Mutational Analysis of Patients With Colorectal Cancer in CALGB/SWOG 80405 Identifies New Roles of Microsatellite Instability and Tumor Mutational Burden for Patient Outcome. J Clin Oncol 2019;37(14):1217–27 doi 10.1200/JCO.18.01798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21(11):1350–6 doi 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramlau R, Gorbunova V, Ciuleanu TE, Novello S, Ozguroglu M, Goksel T, et al. Aflibercept and Docetaxel versus Docetaxel alone after platinum failure in patients with advanced or metastatic non-small-cell lung cancer: a randomized, controlled phase III trial. J Clin Oncol 2012;30(29):3640–7 doi 10.1200/JCO.2012.42.6932. [DOI] [PubMed] [Google Scholar]
- 49.Fuchs CS, Tomasek J, Yong CJ, Dumitru F, Passalacqua R, Goswami C, et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 2014;383(9911):31–9 doi 10.1016/S0140-6736(13)61719-5. [DOI] [PubMed] [Google Scholar]
- 50.Garon EB, Ciuleanu TE, Arrieta O, Prabhash K, Syrigos KN, Goksel T, et al. Ramucirumab plus docetaxel versus placebo plus docetaxel for second-line treatment of stage IV non-small-cell lung cancer after disease progression on platinum-based therapy (REVEL): a multicentre, double-blind, randomised phase 3 trial. Lancet 2014;384(9944):665–73 doi 10.1016/S0140-6736(14)60845-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code for replicating the statistical analysis has been made accessible through a public source code repository (https://gitlab.oit.duke.edu/dcibioinformatics/pubs/calgb-80405-plasma). Codes used for all analyses will be made available upon request.