Abstract
Objective
We aimed to develop and test a deep learning algorithm (DLA) for fully automated measurement of the volume and signal intensity (SI) of the liver and spleen using gadoxetic acid-enhanced hepatobiliary phase (HBP)-magnetic resonance imaging (MRI) and to evaluate the clinical utility of DLA-assisted assessment of functional liver capacity.
Materials and Methods
The DLA was developed using HBP-MRI data from 1014 patients. Using an independent test dataset (110 internal and 90 external MRI data), the segmentation performance of the DLA was measured using the Dice similarity score (DSS), and the agreement between the DLA and the ground truth for the volume and SI measurements was assessed with a Bland-Altman 95% limit of agreement (LOA). In 276 separate patients (male:female, 191:85; mean age ± standard deviation, 40 ± 15 years) who underwent hepatic resection, we evaluated the correlations between various DLA-based MRI indices, including liver volume normalized by body surface area (LVBSA), liver-to-spleen SI ratio (LSSR), MRI parameter-adjusted LSSR (aLSSR), LSSR × LVBSA, and aLSSR × LVBSA, and the indocyanine green retention rate at 15 minutes (ICG-R15), and determined the diagnostic performance of the DLA-based MRI indices to detect ICG-R15 ≥ 20%.
Results
In the test dataset, the mean DSS was 0.977 for liver segmentation and 0.946 for spleen segmentation. The Bland-Altman 95% LOAs were 0.08% ± 3.70% for the liver volume, 0.20% ± 7.89% for the spleen volume, -0.02% ± 1.28% for the liver SI, and -0.01% ± 1.70% for the spleen SI. Among DLA-based MRI indices, aLSSR × LVBSA showed the strongest correlation with ICG-R15 (r = -0.54, p < 0.001), with area under receiver operating characteristic curve of 0.932 (95% confidence interval, 0.895–0.959) to diagnose ICG-R15 ≥ 20%.
Conclusion
Our DLA can accurately measure the volume and SI of the liver and spleen and may be useful for assessing functional liver capacity using gadoxetic acid-enhanced HBP-MRI.
Keywords: Deep learning, Gadoxetic acid, Magnetic resonance imaging, Volumetry, Liver
INTRODUCTION
Liver volume assessment is an important component of a clinical workup before hepatic resection or living donor liver transplantation because remnant liver volume is a major predictor of post-hepatectomy liver failure and morbidity [1,2,3,4]. Volumes of the liver and spleen are usually assessed using cross-sectional imaging, such as CT or MRI. Gadoxetic acid-enhanced MRI has potential advantages when used for liver volumetry because its signal intensity (SI) measured on hepatobiliary phase (HBP) images reflects liver function [5,6]. Therefore, gadoxetic acid-enhanced HBP images allow for simultaneous assessment of liver function and volume. Previous studies have demonstrated that liver SI normalized using spleen SI on HBP images, alone or in combination with liver volume measurement, positively correlates with liver function [7] and predicts the risk of post-hepatectomy liver failure [8,9].
Despite the clinical value of volumetric and functional liver assessment using gadoxetic acid-enhanced MRI, the time-consuming nature of the segmentation process limits its routine use. Although some image processing-based algorithms have been developed to facilitate the segmentation process [10,11,12,13,14,15], they are not fully automated and require user intervention. Additionally, deep learning has recently gained attention by showing promising results in liver segmentation using CT or MRI [16,17,18,19,20,21], but most previous studies in this regard were preliminary and focused mainly on its technical feasibility [18,19,20,21]. Furthermore, no deep learning algorithm (DLA) has yet been validated for its utility in assessing functional liver volume using gadoxetic acid-enhanced HBP-MRI. To accomplish this, an accurate measurement of not only liver volume but also SI of both the liver and spleen is required, with the spleen data used as the internal signal reference. This study aimed to develop and test a DLA for fully automated measurement of the volume and SI of the liver and spleen using gadoxetic acid-enhanced HBP-MRI and evaluate the clinical utility of DLA-assisted assessment of functional liver capacity.
MATERIALS AND METHODS
This study was approved by the Institutional Review Boards of the three participating institutions (Asan Medical Center, Pusan National University Hospital, and Pusan National University Yangsan Hospital) (IRB No. 2018-0507). The requirement for informed consent from the patient was waived due to the retrospective nature of the analysis.
Datasets for the Development and Testing of the DLA
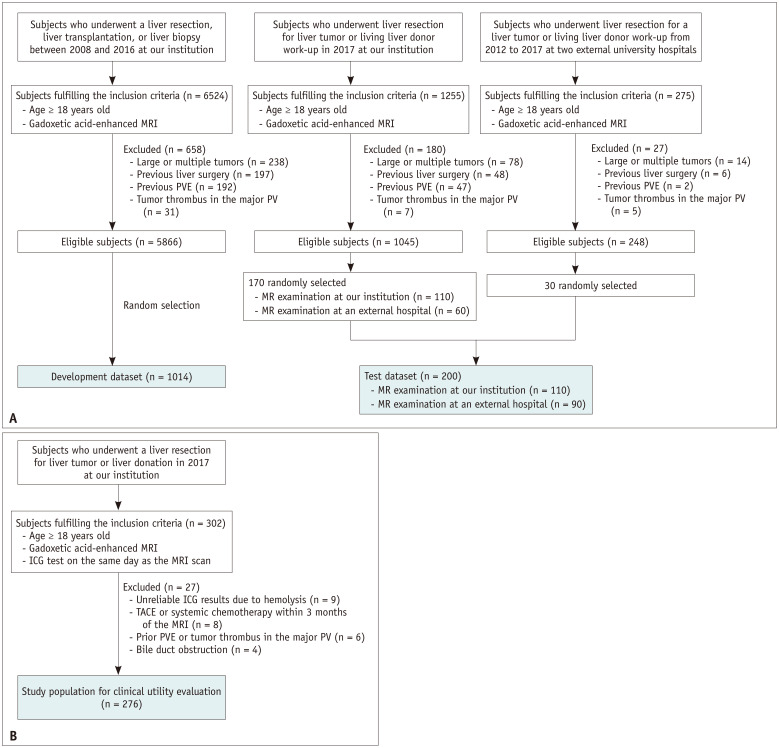
The development dataset consisted of gadoxetic acid-enhanced HBP-MRI data from 1014 subjects. The eligibility criteria for the development dataset included an age of ≥ 18 years, liver biopsy, hepatic resection, or liver transplantation conducted between January 2008 and December 2016 at our institution (Asan Medical Center), and the availability of gadoxetic acid-enhanced liver MRI results. Subjects were excluded if they had large (≥ 10 cm) or multiple (≥ 10) hepatic tumors on imaging, a history of liver or spleen surgery, a history of portal vein embolization, or a tumor thrombus in the major portal vein on imaging. Among the 5866 eligible subjects, 1014 were randomly selected and constituted the development dataset.
DLA performance was tested using data from 200 randomly selected patients who underwent a liver donor workup or resection for hepatic neoplasm in our institution in 2017 (n = 170) or two other tertiary institutions (Pusan National University Hospital and Pusan National University Yangsan Hospital) (n = 30) between 2012 and 2017. The exclusion criteria for this test dataset were the same as those used for the development dataset. To test the DLA using MRI data acquired with various imaging techniques, we included MRI data obtained from outside institutions for 60 patients who had been referred to our hospital. Therefore, the test dataset included 110 internal and 90 external MRI data points (Fig. 1). MRI data with imaging artifacts were not excluded from the development and test datasets to address the performance of DLA in real-world clinical practice.
Fig. 1. Study materials.
A. Flow diagram of the development and test datasets for the development and technical testing of the deep learning algorithm. B. Flow diagram for the study population for evaluating the clinical utility of functional liver capacity assessment. ICG = indocyanine green, PV = portal vein, PVE = portal vein embolization, TACE = transarterial chemoembolization
MRI examinations were performed at 1.5T or 3T using various types of scanners. HBP-MRI was performed using the breath-hold T1-weighted fat-suppressed three-dimensional spoiled gradient echo sequence acquired 15–30 minutes after a bolus injection of 0.025 mmoL/kg gadoxetic acid (Bayer Health Care). Supplementary Table 1 presents details of the MRI techniques used in the included study subjects.
Ground Truth
We generated ground truth segmentation maps of the liver and spleen by outlining organ margins while excluding any focal hepatic lesions and vessels on HBP-MRI using in-house software plugged into ImageJ software (https://imagej.nih.gov/ij/). To obtain a sufficient amount of labeled training data in a time-efficient manner, we first trained the prototype DLA using the 100 HBP-MRI data selected from the development dataset and the ground truth segmentation maps that were manually drawn by an abdominal radiologist. The remaining MRI data in the development dataset were first processed by this prototype DLA, and then a radiology technician manually edited the liver and spleen masks generated by the algorithm. For the test dataset, the ground truth segmentation maps were drawn manually by a technician without using the prototype DLA. An abdominal radiologist reviewed all ground truth segmentation maps in the development and test datasets and corrected any inaccuracies.
Development and Technical Testing of the DLA
The DLA for automated segmentation of the liver and spleen on HBP-MRI was designed by modifying a previously reported algorithm for CT-based liver and spleen segmentation [17]. The development dataset was divided into training and tuning sets (8:2 ratio). The tuning set was used to select the architecture and hyperparameters of the DLA. The final DLA was based on the DeepLabV3+ [22]. After processing the input images, the DLA generated segmentation masks with the same resolution as that of the input images. The source code is available at https://github.com/wltjr1007/MRSeg. Further details of this DLA development process are provided in Supplementary Materials and Supplementary Figure 1.
Technical testing of the DLA was performed using the test dataset. The segmentation performance of the DLA was analyzed using the Dice similarity score (DSS), which was calculated as 2 × true positive pixels/([2 × true positive pixels] + false negative pixels + false positive pixels). Volumes and SIs of the whole liver and spleen were measured using the DLA-generated segmentation results and then compared with those measured using the ground truth segmentation results. Volumes were calculated by summing consecutive areas of the organ, multiplied by section intervals, and expressed in cm3.
Adjusted Liver-to-Spleen SI Ratio Corrected for MRI Parameter Effects
We adopted liver-to-spleen SI ratio (LSSR), that is, liver SI divided by spleen SI, as the liver functional metric on HBP-MRI, as used in previous studies [7,8,9]. However, the LSSR is affected by the MRI parameters, especially the repetition time (TR) and flip angle (FA), which determine the T1 contrast. Therefore, we attempted to correct for the effects of the MR parameter on the LSSR through computer simulation. Details of this simulation and the clinical feasibility study are provided in the Supplementary Materials. Briefly, the equation for the LSSR on T1-weighted spoiled gradient echo MRI can be simplified as follows:
where α is FA, T1Liver is liver T1, and T1Spleen is spleen T1. Through a computer simulation, we found combinations of T1Liver, T1Spleen, and resultant spleen-to-liver T1 ratios (i.e., the reciprocals of liver-to-spleen T1 ratio) that corresponded to a given LSSR, TR, and FA. An adjusted LSSR (aLSSR) was then calculated as the median value of the spleen-to-liver T1 ratio, as shown in Supplementary Figure 2.
The feasibility of aLSSR in correcting for the effects of the MRI parameter on LSSR was evaluated in a group of 221 patients who underwent two sets of HBP-MRI using FAs of 10° and 19° (see Supplementary Materials). The LSSRs were measured on two sets of HBP images, and the corresponding aLSSRs were calculated. The correlation between the values obtained from the FA 10° images and those from the FA 19° images was evaluated.
Clinical Utility of DLA-Assisted Assessment of Functional Liver Capacity
The clinical utility of DLA-assisted assessment of functional liver capacity using HBP-MRI in assessing liver function was evaluated in 276 patients who underwent an indocyanine green (ICG) clearance test and gadoxetic acid-enhanced liver MRI on the same day as part of the living liver donor workup or preoperative evaluation before liver resection in 2017 at our institution (Fig. 1). The MRI techniques used are summarized in Supplementary Table 1.
The DLA was implemented using commercial software (GoCDSS; SmartCareworks Inc.). When the MRI data are uploaded, the software automatically performs liver and spleen segmentation. An abdominal radiologist reviewed the DLA-generated segmentation results and corrected any errors, and the time required to correct the segmentation error was recorded. The volume and SI of the whole liver and spleen were measured, and the following MRI-derived indices were calculated: liver volume normalized by body surface area (LVBSA) according to the Du Bois formula [23], LSSR, aLSSR, and combined volume-SI indices including LSSR × LVBSA and aLSSR × LVBSA. An ICG excretion test was performed as described previously [24], and the ICG plasma disappearance rate (ICG-PDR) and retention rate at 15 minutes (ICG-R15) were recorded. We then analyzed the correlation between the MRI-derived indices and the results of the ICG test (ICG-PDR and ICG-R15). For the selected MRI-derived indices, the diagnostic performance in predicting an ICG-R15 ≥ 20%, which is considered a contraindication for a major hepatectomy [25], was assessed.
Statistical Analysis
In the test dataset, the DSSs were compared among the subgroups of MRI and patient characteristics using the independent sample t test or the analysis of variance test. In the subset of 145 patients who underwent ICG test within 1 month of liver MRI, DSS was compared according to ICG-R15 categories (i.e., ICG-R15 ≤ 10%, 10% < ICG-R15 < 20%, and ICG-R15 ≥ 20%). The agreement between DLA and ground truth for volume and SI measurements was evaluated using the Bland-Altman 95% limit of agreement (LOA) presented as the mean difference ± 1.96 × standard deviation (SD) of the difference on a percentage scale, in which the mean difference represents systematic bias, and 1.96 × SD of the difference represents the magnitude of the measurement error [26]. The correlation of LSSR and aLSSR between the two sets of HBP images was evaluated using linear regression analysis, and differences in regression slope from 1 and intercept from 0 were assessed.
In the clinical utility study, the correlations of MRI-derived indices with ICG-PDR and ICG-R15 were evaluated using the Pearson’s correlation coefficient [27]. The diagnostic performance of the MRI-derived index to predict ICG-R15 ≥ 20% was evaluated using receiver operating characteristic (ROC) analysis. Statistical analyses were performed with SPSS version 21 (IBM Corp.) and MedCalc 12.7.0 (MedCalc Software). Statistical significance was set at p < 0.05.
RESULTS
Technical Testing of the DLA
The characteristics of the development and test datasets are summarized in Table 1 and Supplementary Table 2. In the test dataset, the mean ± SD of DSS was 0.977 ± 0.009 (range, 0.916–0.988) for the liver and 0.946 ± 0.025 (range, 0.872–0.989) for the spleen. The segmentation performance of the DLA was not significantly different between the internal (n = 110) and external (n = 90) test data and between the MRI field strengths for the liver and spleen (p ≥ 0.083). The DSS for liver segmentation did not differ according to the fibrosis stage (p = 0.766), while the DSS for spleen segmentation was higher in F4 than in F0 (p = 0.005) and F2 (p = 0.021). The DSSs for the liver (p = 0.207) and spleen (p = 0.998) did not differ according to the liver function categories as assessed by ICG-R15 (Table 2). Representative segmentation results are shown in Figure 2.
Table 1. Characteristics of the Development and Test Datasets and the Study Population Used for Clinical Utility Evaluation of the DLA.
| DLA Development and Test | Study Population for Clinical Utility Evaluation | ||||
|---|---|---|---|---|---|
| Development Dataset | Test Dataset | ||||
| Number of patients | 1014 | 200 | 276 | ||
| Male:female | 738:276 | 153:47 | 191:85 | ||
| Age, years* | 54 ± 13 | 55 ± 13 | 40 ± 15 | ||
| Underlying liver disease, % | |||||
| None | 227 (22.4) | 32 (16.0) | 103 (37.3) | ||
| Hepatitis B | 572 (56.4) | 142 (71.0) | 76 (27.5) | ||
| Hepatitis C | 35 (3.5) | 8 (4.0) | 4 (1.5) | ||
| Alcohol-induced | 27 (2.7) | 4 (2.0) | 3 (1.1) | ||
| NAFLD | 100 (9.9) | 4 (2.0) | 82 (29.7) | ||
| Other† | 53 (5.2) | 10 (5.0) | 8 (2.9) | ||
| Hepatic mass | |||||
| Absent, % | 178 (17.6) | 28 (14.0) | 179 (64.9) | ||
| Present, % | 836 (82.4) | 172 (86.0) | 97 (35.1) | ||
| Malignant | 791 (78.0) | 169 (84.5) | 94 (34.1) | ||
| Benign‡ | 45 (4.4) | 3 (1.5) | 3 (1.1) | ||
| Largest mass size, cm* | 4.2 ± 4.1 | 3.1 ± 1.7 | 4.4 ± 3.7 | ||
| Pathologic liver fibrosis stage, % | |||||
| F0 | 354 (34.9) | 60 (30.0) | 179 (64.9) | ||
| F1 | 42 (4.1) | 21 (10.5) | 8 (2.9) | ||
| F2 | 113 (11.1) | 29 (14.5) | 21 (7.6) | ||
| F3 | 156 (15.4) | 40 (20.0) | 20 (7.2) | ||
| F4 | 349 (34.4) | 50 (25.0) | 32 (11.6) | ||
| Laboratory results* | |||||
| AST, IU/mL | 35.9 ± 46.6 | 30.7 ± 18.5 | 26.9 ± 25.6 | ||
| ALT, IU/mL | 31.9 ± 39.6 | 29.0 ± 17.9 | 26.0 ± 25.1 | ||
| Total bilirubin, ng/mL | 1.4 ± 4.3 | 0.7 ± 0.4 | 0.7 ± 0.3 | ||
| Platelet count, 109/L | 182.9 ± 77.9 | 179.7 ± 62.8 | 243.2 ± 69.6 | ||
| PT (INR) | 1.1 ± 0.3 | 1.1 ± 0.1 | 1.0 ± 0.1 | ||
Unless otherwise indicated, data are number of subjects, and data in parentheses are percentages. *Data are mean values ± standard deviation, †Including toxic hepatitis, hepatitis A, autoimmune hepatitis, Wilson’s disease, and hepatolithiasis, ‡Including hemangioma, angiomyolipoma, focal nodular hyperplasia, reactive lymphoid hyperplasia, adenoma, and cyst. ALT = alanine aminotransferase, AST = aspartate aminotransferase, DLA = deep learning algorithm, INR = international normalized ratio, NAFLD = non-alcoholic fatty liver disease, PT = prothrombin time
Table 2. Performance of Deep Learning Algorithm in Liver and Spleen Segmentation in the Test Dataset.
| Patient Number (%) | Liver Segmentation | Spleen Segmentation | ||||
|---|---|---|---|---|---|---|
| Mean DSS ± SD | P | Mean DSS ± SD | P | |||
| Total | 200 (100.0) | 0.977 ± 0.009 | NA | 0.946 ± 0.025 | NA | |
| MRI data source | 0.289 | 0.083 | ||||
| Internal | 110 (55.0) | 0.976 ± 0.007 | 0.943 ± 0.024 | |||
| External | 90 (45.0) | 0.978 ± 0.103 | 0.949 ± 0.259 | |||
| MRI field strength | 0.874 | 0.508 | ||||
| 1.5T | 60 (30.0) | 0.978 ± 0.006 | 0.946 ± 0.02 | |||
| 3T | 140 (70.0) | 0.976 ± 0.009 | 0.945 ± 0.027 | |||
| Fibrosis stage | 0.766 | 0.003* | ||||
| F0 | 60 (30.0) | 0.978 ± 0.007 | 0.942 ± 0.027 | |||
| F1 | 21 (10.5) | 0.976 ± 0.01 | 0.943 ± 0.026 | |||
| F2 | 29 (14.5) | 0.976 ± 0.007 | 0.938 ± 0.029 | |||
| F3 | 40 (20.0) | 0.976 ± 0.009 | 0.948 ± 0.021 | |||
| F4 | 50 (25.0) | 0.977 ± 0.011 | 0.954 ± 0.023 | |||
| Liver function† | 0.207 | 0.998 | ||||
| ICG-R15 ≤ 10% | 35 (24.1) | 0.976 ± 0.013 | 0.946 ± 0.024 | |||
| 10% < ICG-R15 < 20% | 103 (71.0) | 0.978 ± 0.007 | 0.946 ± 0.025 | |||
| ICG-R15 ≥ 20% | 7 (4.8) | 0.971 ± 0.012 | 0.946 ± 0.026 | |||
*Post-hoc Bonferroni test revealed statistically significant difference in DSS for F4 compared with those for F0 (p = 0.005) and F2 (p = 0.021), †Data were obtained from the 145 patients who had indocyanine green test within 1 month of liver MRI. DSS = Dice similarity score, ICG-R15 = indocyanine green retention rate at 15 minutes, SD = standard deviation
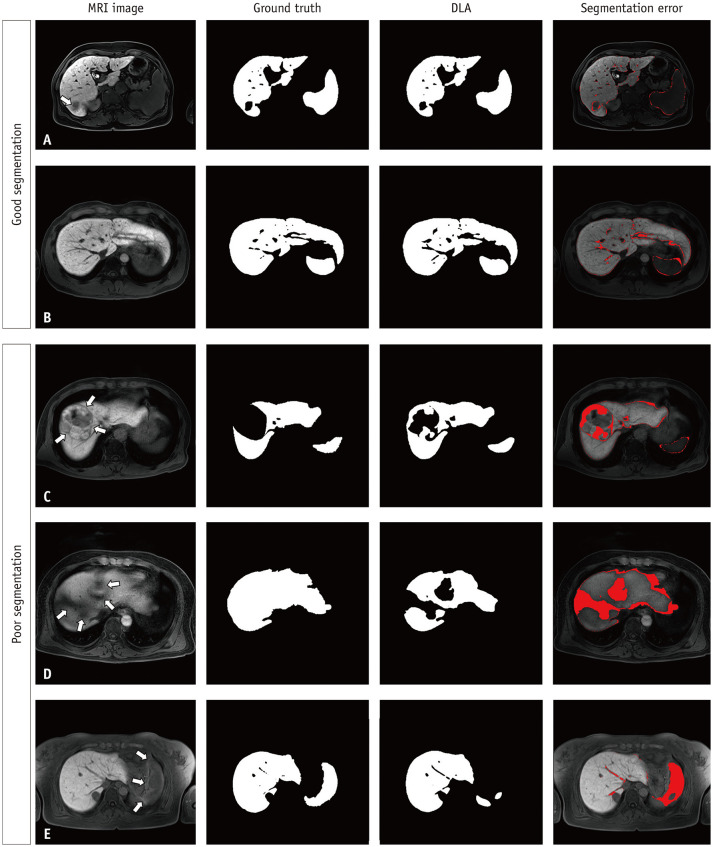
Fig. 2. Example of good (A-B) and poor (C-E) segmentation results from the DLA.
A-E. The segmentation maps produced by the DLA and the ground truth segmentation maps exhibit good agreement with a small segmentation error in a patient with a 2.7-cm hepatocellular carcinoma (arrow, A) and in another with liver cirrhosis (B). The use of the DLA resulted in large segmentation errors in a hepatocellular carcinoma (arrows) showing heterogeneous isointense-to-hypointense signals on a hepatobiliary phase image (C), in a liver dome area (arrows) due to artifact-induced image degradation (D), and in a spleen (arrows) due to uneven fat suppression in the upper abdomen (E). DLA = deep learning algorithm
The Bland-Altman 95% LOAs between the DLA and ground truth were 0.08% ± 3.70% and 0.20% ± 7.89% for liver and spleen volumes, respectively (Supplementary Fig. 3). In both internal and external test data, automated volume measurements using DLA did not result in a significant bias (range of mean difference, -0.06%–0.37%, p ≥ 0.200), and the measurement error was less than 4.28% and 8.01% for liver and spleen volumes, respectively (Table 3). Regarding the SI, the Bland-Altman 95% LOAs between the DLA and ground truth were -0.02% ± 1.28% for the liver SI and-0.01% ± 1.70% for the spleen SI; no significant bias was found for any of the SI indices measured by the DLA (p ≥ 0.115).
Table 3. Agreement between the DLA and Ground Truth Measurements of the Volume and SI in the Test Dataset.
| Parameter | All MRI Data (n = 200) | Internal MRI Data (n = 110) | External MRI Data (n = 90) | |||
|---|---|---|---|---|---|---|
| 95% LOA (%)* | P † | 95% LOA (%)* | P † | 95% LOA (%)* | P † | |
| Liver volume | 0.08 ± 3.70 | 0.542 | 0.20 ± 3.15 | 0.200 | −0.06 ± 4.28 | 0.793 |
| Spleen volume | 0.20 ± 7.89 | 0.480 | 0.37 ± 8.01 | 0.351 | 0.00 ± 7.76 | 0.997 |
| Liver SI | −0.02 ± 1.28 | 0.645 | −0.07 ± 0.94 | 0.115 | 0.04 ±1.61 | 0.638 |
| Spleen SI | −0.01 ± 1.70 | 0.822 | −0.09 ± 1.45 | 0.195 | 0.08 ± 1.94 | 0.433 |
*Data represent Bland-Altman 95% LOA values expressed as a percentage mean difference ± 1.96 × standard deviation of the difference, †p values for statistically significant differences in the mean difference from zero. DLA = deep learning algorithm, LOA = limit of agreement, SI = signal intensity
Adjusted LSSR Corrected for Effects of MRI Parameters
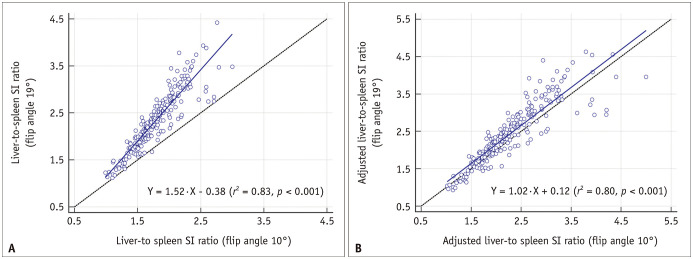
A web-based algorithm to calculate an aLSSR by inputting a measured LSSR, TR, and FA is provided at https://i-pacs.com/adjustedSIR. In patients who underwent two sets of HBP-MRI using different MRI parameters, as shown in the scatter plots (Fig. 3), the LSSRs measured on FA 19° images were overestimated compared with those measured on FA 10° images, with a regression slope of 1.52 (95% confidence interval [CI], 1.43–1.61). However, the use of aLSSR resulted in nearly equivalent estimates for the two HBP-MRI sets, as indicated by a regression slope close to 1 (slope, 1.02; 95% CI, 0.95–1.08) and an intercept close to 0 (intercept, 0.12; 95% CI, -0.05–0.28).
Fig. 3. Scatterplots of the liver-to-spleen SI ratio (A) and adjusted liver-to-spleen SI ratio (B) measured using the two different hepatobiliary magnetic resonance image sets.
A. The liver-to-spleen SI ratio measured on the hepatobiliary phase images using a 19° flip angle (echo time, 1.39 ms; repetition time, 3.83 ms) were overestimated compared with those measured on the images using a 10° flip angle (echo time, 1.33 ms; repetition time, 3.39 ms), with a regression slope of 1.52 (95% CI, 1.43–1.61). B. The adjusted liver-to-spleen SI ratios obtained using the two hepatobiliary phase image sets were equivalent, with a regression slope of 1.02 (95% CI, 0.95–1.08) and an intercept of 0.12 (95% CI, -0.05–0.28). CI = confidence interval, SI = signal intensity
Clinical Utility of DLA-Assisted Assessment of Functional Liver Capacity
In the 276 patients included in the clinical utility evaluation (191 males and 85 females; mean age, 40 years) (Table 1), a review of the DLA-generated segmentation results revealed a minor segmentation error in 130 (47.1%) patients, which was associated with a short correction time (mean, 59.9 seconds; range, 18–124 seconds) and a small change in the measured values (Bland-Altman 95% LOAs of 0.35% ± 2.17%, -0.01% ± 0.23%, -0.45% ± 3.33%, and 0.05% ± 1.14% for liver volume, liver SI, spleen volume, and spleen SI, respectively).
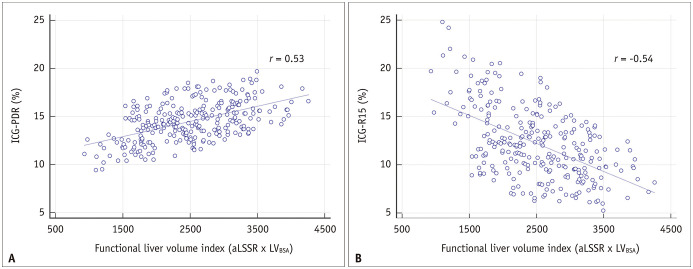
The correlations between the MRI-derived indices and the results of the ICG test are summarized in Table 4. The aLSSR showed a stronger correlation with the ICG test results than the LSSR (r = 0.49 vs. 0.42 for ICG-PDR; r = -0.50 vs. -0.43 for ICG-R15). The combined volume-SI indices (i.e., LSSR × LVBSA and aLSSR × LVBSA) showed a stronger correlation with the ICG test results than LSSR or aLSSR alone, and the aLSSR × LVBSA showed the strongest correlation with ICG-PDR (r = 0.53) and ICG-R15 (r = -0.54) among all MRI-derived indices (Fig. 4).
Table 4. Correlation between MRI-Derived Indices and ICG Excretion Results.
| MRI Indices | ICG-PDR | ICG-R15 | ||
|---|---|---|---|---|
| Correlation Coefficient | P | Correlation Coefficient | P | |
| aLSSR x LVBSA | 0.53 (0.44, 0.61) | < 0.001 | −0.54 (−0.61, −0.45) | < 0.001 |
| LSSR x LVBSA | 0.46 (0.36, 0.55) | < 0.001 | −0.47 (−0.56, −0.37) | < 0.001 |
| aLSSR | 0.49 (0.40, 0.58) | < 0.001 | −0.50 (−0.58, −0.40) | < 0.001 |
| LSSR | 0.42 (0.31, 0.51) | < 0.001 | −0.43 (−0.52, −0.32) | < 0.001 |
| LVBSA | 0.16 (0.04, 0.27) | 0.008 | −0.17 (−0.28, −0.05) | 0.005 |
Data in parentheses are 95% confidence intervals. aLSSR = adjusted LSSR, BSA = body surface area, ICG = indocyanine green, ICG-PDR = ICG plasma disappearance rate, ICG-R15 = ICG retention rate at 15 minutes, LSSR = liver-to-spleen signal intensity ratio, LVBSA = liver volume normalized by BSA
Fig. 4. Scatterplots showing the correlation between the functional liver volume index (aLSSR × LVBSA) and ICG excretion test results.
A, B. As the functional liver volume index, the aLSSR multiplied by the LVBSA was used and was compared with ICG-PDR (A) and ICG-R15 (B). aLSSR = adjusted liver-to-spleen signal intensity ratio, ICG = indocyanine green, ICG-PDR = ICG plasma disappearance rate, ICG-R15 = ICG retention rate at 15 minutes, LVBSA = liver volume normalized by BSA
As a functional liver volume index, the aLSSR × LVBSA achieved area under the ROC curve of 0.932 (95% CI, 0.895–0.959) in predicting an ICG-R15 ≥ 20%. Using the cut-off value of 1957.0 cm3/m2, this index showed a sensitivity of 100.0% (8/8) and a specificity of 76.1% (204/268) (Fig. 5). Examples of DLA-assisted functional liver volume assessment are shown in Supplementary Figure 4.
Fig. 5. Receiver operating characteristics curve of the functional liver volume index (aLSSR × LVBSA) for predicting the ICG-R15 ≥ 20%.
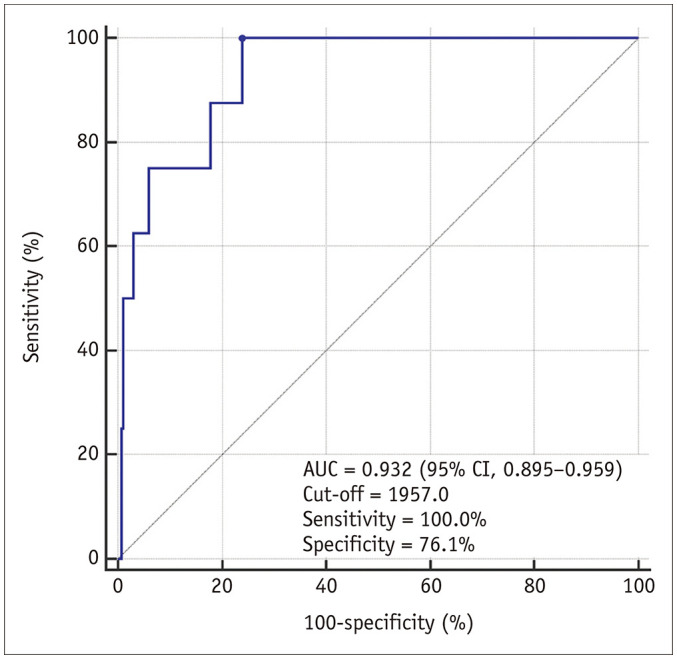
aLSSR = adjusted liver-to-spleen signal intensity ratio, AUC = area under the curve, CI = confidence interval, ICG-R15 = indocyanine green retention rate at 15 minutes, LVBSA = liver volume normalized by body surface area
DISCUSSION
We have developed a DLA for fully automated measurement of the volume and SI of the liver and spleen on gadoxetic acid-enhanced HBP-MRI. We then tested this algorithm using an independent test dataset obtained using various imaging techniques. We observed that our DLA performed well in liver segmentation (mean DSS, 0.977), which is superior to previously reported CT- or MRI-based DLAs (mean DSS, 0.920–0.973) [17,18,19,20,21] and thus enables accurate liver volume measurement with a small measurement error (3.70%). The performance of DLA in spleen segmentation was slightly poorer (mean DSS, 0.946) than that found for liver segmentation, which may be attributed to the fact that the spleen has a significantly lower organ-to-background contrast than the liver on HBP-MRI. However, the segmentation performance of our DLA produced a very accurate automated measurement of the liver and spleen SI, as indicated by their nearly perfect agreement with the ground truth values (measurement error of 1.28% and 1.70% for the liver and spleen, respectively).
For the assessment of liver function on gadoxetic acid-enhanced HBP-MRI, spleen SI has been commonly used as an internal reference for normalizing liver SI [7,8,9,28]. However, LSSR is influenced by MRI parameters, which can hinder the use of this index in clinical practice when HBP-MRI is performed using various scanning parameters. In this regard, we devised a method to adjust the effects of the MRI parameters on LSSR and developed a web calculator for the instant calculation of the aLSSR (https://i-pacs.com/adjustedSIR). In the study, including patients who underwent HBP-MRI using two different parameter settings, the aLSSR obtained from the two sets of HBP-MRI data yielded equivalent estimates, in contrast to the considerable bias observed for the LSSR. Furthermore, in subjects who underwent an ICG excretion test and gadoxetic acid-enhanced MRI, the aLSSR showed stronger correlations with the ICG excretion index than the LSSR, further supporting the usefulness of the aLSSR index.
We also conducted a preliminary study to evaluate the clinical utility of MRI-based, DLA-assisted assessment of functional liver capacity. Although DLA produced segmentation errors in approximately half of the patients (47.1%; 130/276), these errors were minor and could be quickly corrected by the radiologist and were associated with errors of less than 3% of the measured volume. We also found that among the MRI-derived indices, the functional liver capacity index, aLSSR × LVBSA, showed the strongest correlation with ICG-PDR and ICG-R15. Although the correlation coefficients between the aLSSR × LVBSA and the ICG indices were only moderate (r = 0.53 and -0.54 for ICG-PDR and ICG-R15, respectively), these were stronger than the correlation between MRI index and ICG-R15 reported in a previous study [29] that assessed the volumetric hepatic extraction fraction based on signal modeling of multiphasic dynamic and HBP-MRI. Furthermore, aLSSR × LVBSA also allowed an accurate prediction of an ICG-R15 ≥ 20% (AUC of 0.932), a measure that is considered a contraindication for major hepatectomy [25]. Taken together, our findings highlight the usefulness of DLA-assisted assessment of functional liver capacity using gadoxetic acid-enhanced HBP-MRI in assessing liver function.
Different approaches have been used to assess liver function using gadoxetic acid-enhanced HBP-MRI. Compared with the SI-based approach used in our present study [7,8,9], the methods based on liver T1 mapping [30,31] may have a theoretical advantage as T1 measurements more directly reflect the amount of contrast material in the liver than SI. However, liver T1 mapping has limitations in terms of clinical application as it requires dedicated pulse sequences that may not be available with some MRI scanners; due to their longer scan time, T1-mapping sequences are not suitable for high-resolution whole liver imaging, which is crucial for accurate assessment of liver volume. In contrast, our present approach based on routine HBP-MRI would be advantageous for clinical application, as it allows for simultaneous assessment of liver function and volume. Furthermore, the use of DLA can facilitate the clinical use of MRI-based assessments of functional liver capacity. MRI data analysis with this DLA can be performed as a background process before radiologist image review. Therefore, if our DLA is successfully implemented in daily clinical practice, radiologists may be able to review MR images along with DLA-generated organ segmentation results, and thus measure organ volume and SI with little additional time or effort required. Indeed, there may be multiple potential applications of our DLA in clinical practice, including the prediction of remnant liver volume and function prior to liver resection and the functional liver reservoir in patients with liver disease. The real-life utility and clinical implications of our DLA should be evaluated in future studies.
Our study has several limitations. First, since our DLA provides segmentation of the whole liver, additional user intervention is required to assess the volume and function of the liver in accordance with different regions. Second, although our development and test datasets include MRI data obtained using various techniques, MRI techniques are continually progressing. The applicability of our DLA when using MRI data obtained with newer techniques must be evaluated. Third, the feasibility of aLSSR was evaluated using MR scanners from a single vendor in our study. Its validity across different platforms and vendors should be evaluated in future studies. Fourth, technical tests and clinical utility evaluation of DLA were performed in subjects with relatively preserved liver function (i.e., living liver donors and patients who underwent liver resection). Therefore, our results may not be directly generalizable to patients with severe liver dysfunction. Larger-scale validation of our DLA is warranted in patients with diverse liver functions. Lastly, future studies should evaluate the actual clinical impact of our DLA on patient care as a comprehensive and versatile method of assessing functional liver capacity.
In conclusion, we have developed and tested a DLA for fully automated segmentation of the liver and spleen using gadoxetic acid-enhanced HBP-MRI. Our DLA enables the accurate measurement of the volume and SI of the liver and spleen and may be useful for assessing functional liver capacity using gadoxetic acid-enhanced HBP-MRI.
Acknowledgments
Seung Soo Lee was responsible for the clinical study, and Heung-Il Suk was responsible for the development of the deep learning algorithm. Both authors contributed equally to this study and assume the role of corresponding authors.
Footnotes
Conflicts of Interest: Seung Soo Lee who is on the editorial board of the Korean Journal of Radiology was not involved in the editorial evaluation or decision to publish this article. All remaining authors have declared no conflicts of interest.
- Conceptualization: Seung Soo Lee, Heung-Il Suk.
- Data curation: Hyo Jung Park, Jee Seok Yoon, Seung Soo Lee, Heung-Il Suk, Seung Baek Hong, Hwaseong Ryu.
- Formal analysis: Hyo Jung Park, Seung Soo Lee.
- Funding acquisition: Seung Soo Lee.
- Investigation: Hyo Jung Park, Jee Seok Yoon, Seung Soo Lee, Heung-Il Suk, Bumwoo Park.
- Methodology: Hyo Jung Park, Seung Soo Lee.
- Project administration: Seung Soo Lee, Heung-Il Suk.
- Resources: Seung Soo Lee, Heung-Il Suk, Seung Baek Hong, Hwaseong Ryu.
- Software: Jee Seok Yoon, Heung-Il Suk, Bumwoo Park, Yu Sub Sung.
- Supervision: Seung Soo Lee, Heung-Il Suk.
- Validation: Hyo Jung Park, Seung Soo Lee.
- Visualization: Hyo Jung Park, Jee Seok Yoon, Seung Soo Lee, Heung-Il Suk, Yu Sub Sung.
- Writing—original draft: Hyo Jung Park, Seung Soo Lee.
- Writing—review & editing: Seung Soo Lee, Heung-Il Suk.
Funding Statement: This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2020R1F1A1048826).
Availability of Data and Material
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.
Supplement
The Supplement is available with this article at https://doi.org/10.3348/kjr.2021.0892.
References
- 1.Schindl MJ, Redhead DN, Fearon KC, Garden OJ, Wigmore SJ Edinburgh Liver Surgery and Transplantation Experimental Research Group (eLISTER) The value of residual liver volume as a predictor of hepatic dysfunction and infection after major liver resection. Gut. 2005;54:289–296. doi: 10.1136/gut.2004.046524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim MC, Tan CH, Cai J, Zheng J, Kow AW. CT volumetry of the liver: where does it stand in clinical practice? Clin Radiol. 2014;69:887–895. doi: 10.1016/j.crad.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 3.Ogasawara K, Une Y, Nakajima Y, Uchino J. The significance of measuring liver volume using computed tomographic images before and after hepatectomy. Surg Today. 1995;25:43–48. doi: 10.1007/BF00309384. [DOI] [PubMed] [Google Scholar]
- 4.Prodeau M, Drumez E, Duhamel A, Vibert E, Farges O, Lassailly G, et al. An ordinal model to predict the risk of symptomatic liver failure in patients with cirrhosis undergoing hepatectomy. J Hepatol. 2019;71:920–929. doi: 10.1016/j.jhep.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Motosugi U, Ichikawa T, Sou H, Sano K, Tominaga L, Kitamura T, et al. Liver parenchymal enhancement of hepatocyte-phase images in Gd-EOB-DTPA-enhanced MR imaging: which biological markers of the liver function affect the enhancement? J Magn Reson Imaging. 2009;30:1042–1046. doi: 10.1002/jmri.21956. [DOI] [PubMed] [Google Scholar]
- 6.Kim JY, Lee SS, Byun JH, Kim SY, Park SH, Shin YM, et al. Biologic factors affecting HCC conspicuity in hepatobiliary phase imaging with liver-specific contrast agents. AJR Am J Roentgenol. 2013;201:322–331. doi: 10.2214/AJR.12.9478. [DOI] [PubMed] [Google Scholar]
- 7.Yamada A, Hara T, Li F, Fujinaga Y, Ueda K, Kadoya M, et al. Quantitative evaluation of liver function with use of gadoxetate disodium-enhanced MR imaging. Radiology. 2011;260:727–733. doi: 10.1148/radiol.11100586. [DOI] [PubMed] [Google Scholar]
- 8.Araki K, Harimoto N, Kubo N, Watanabe A, Igarashi T, Tsukagoshi M, et al. Functional remnant liver volumetry using Gd-EOB-DTPA-enhanced magnetic resonance imaging (MRI) predicts post-hepatectomy liver failure in resection of more than one segment. HPB (Oxford) 2020;22:318–327. doi: 10.1016/j.hpb.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Kim DK, Choi JI, Choi MH, Park MY, Lee YJ, Rha SE, et al. Prediction of posthepatectomy liver failure: MRI with hepatocyte-specific contrast agent versus indocyanine green clearance test. AJR Am J Roentgenol. 2018;211:580–587. doi: 10.2214/AJR.17.19206. [DOI] [PubMed] [Google Scholar]
- 10.Gotra A, Sivakumaran L, Chartrand G, Vu KN, Vandenbroucke-Menu F, Kauffmann C, et al. Liver segmentation: indications, techniques and future directions. Insights Imaging. 2017;8:377–392. doi: 10.1007/s13244-017-0558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakayama Y, Li Q, Katsuragawa S, Ikeda R, Hiai Y, Awai K, et al. Automated hepatic volumetry for living related liver transplantation at multisection CT. Radiology. 2006;240:743–748. doi: 10.1148/radiol.2403050850. [DOI] [PubMed] [Google Scholar]
- 12.Fananapazir G, Bashir MR, Marin D, Boll DT. Computer-aided liver volumetry: performance of a fully-automated, prototype post-processing solution for whole-organ and lobar segmentation based on MDCT imaging. Abdom Imaging. 2015;40:1203–1212. doi: 10.1007/s00261-014-0276-9. [DOI] [PubMed] [Google Scholar]
- 13.Huynh HT, Karademir I, Oto A, Suzuki K. Computerized liver volumetry on MRI by using 3D geodesic active contour segmentation. AJR Am J Roentgenol. 2014;202:152–159. doi: 10.2214/AJR.13.10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, Kim KW, Kim SY, Kim B, Lee SJ, Kim HJ, et al. Feasibility of semiautomated MR volumetry using gadoxetic acid-enhanced MRI at hepatobiliary phase for living liver donors. Magn Reson Med. 2014;72:640–645. doi: 10.1002/mrm.24964. [DOI] [PubMed] [Google Scholar]
- 15.Grieser C, Denecke T, Rothe JH, Geisel D, Stelter L, Cannon Walter T, et al. Gd-EOB enhanced MRI T1-weighted 3D-GRE with and without elevated flip angle modulation for threshold-based liver volume segmentation. Acta Radiol. 2015;56:1419–1427. doi: 10.1177/0284185114558975. [DOI] [PubMed] [Google Scholar]
- 16.Park HJ, Park B, Lee SS. Radiomics and deep learning: hepatic applications. Korean J Radiol. 2020;21:387–401. doi: 10.3348/kjr.2019.0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn Y, Yoon JS, Lee SS, Suk HI, Son JH, Sung YS, et al. Deep learning algorithm for automated segmentation and volume measurement of the liver and spleen using portal venous phase computed tomography images. Korean J Radiol. 2020;21:987–997. doi: 10.3348/kjr.2020.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Mamidipalli A, Retson T, Bahrami N, Hasenstab K, Blansit K, et al. Automated CT and MRI liver segmentation and biometry using a generalized convolutional neural network. Radiol Artif Intell. 2019;1:180022. doi: 10.1148/ryai.2019180022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu P, Wu F, Peng J, Liang P, Kong D. Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol. 2016;61:8676–8698. doi: 10.1088/1361-6560/61/24/8676. [DOI] [PubMed] [Google Scholar]
- 20.Huo Y, Terry JG, Wang J, Nair S, Lasko TA, Freedman BI, et al. Fully automatic liver attenuation estimation combing CNN segmentation and morphological operations. Med Phys. 2019;46:3508–3519. doi: 10.1002/mp.13675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Gastel MDA, Edwards ME, Torres VE, Erickson BJ, Gansevoort RT, Kline TL. Automatic measurement of kidney and liver volumes from MR images of patients affected by autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2019;30:1514–1522. doi: 10.1681/ASN.2018090902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder-decoder with atrous separable convolution for semantic image segmentation. [cited 2020 May 11];arXiv [Preprint] 2018 Available at: https://arxiv.org/abs/1802.02611v3 . [Google Scholar]
- 23.Du Bois D, Du Bois EF. A formula to estimate the approximate surface area if height and weight be known. 1916. Nutrition. 1989;5:303–311. discussion 312-313. [PubMed] [Google Scholar]
- 24.Lee SG, Hwang S. How I do it: assessment of hepatic functional reserve for indication of hepatic resection. J Hepatobiliary Pancreat Surg. 2005;12:38–43. doi: 10.1007/s00534-004-0949-9. [DOI] [PubMed] [Google Scholar]
- 25.Imamura H, Sano K, Sugawara Y, Kokudo N, Makuuchi M. Assessment of hepatic reserve for indication of hepatic resection: decision tree incorporating indocyanine green test. J Hepatobiliary Pancreat Surg. 2005;12:16–22. doi: 10.1007/s00534-004-0965-9. [DOI] [PubMed] [Google Scholar]
- 26.Hernaez R. Reliability and agreement studies: a guide for clinical investigators. Gut. 2015;64:1018–1027. doi: 10.1136/gutjnl-2014-308619. [DOI] [PubMed] [Google Scholar]
- 27.Zou KH, Tuncali K, Silverman SG. Correlation and simple linear regression. Radiology. 2003;227:617–622. doi: 10.1148/radiol.2273011499. [DOI] [PubMed] [Google Scholar]
- 28.Kudo M, Gotohda N, Sugimoto M, Kobayashi T, Kojima M, Takahashi S, et al. Evaluation of liver function using gadolinium-ethoxybenzyl-diethylenetriamine pentaacetic acid enhanced magnetic resonance imaging based on a three-dimensional volumetric analysis system. Hepatol Int. 2018;12:368–376. doi: 10.1007/s12072-018-9874-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon JH, Choi JI, Jeong YY, Schenk A, Chen L, Laue H, et al. Pre-treatment estimation of future remnant liver function using gadoxetic acid MRI in patients with HCC. J Hepatol. 2016;65:1155–1162. doi: 10.1016/j.jhep.2016.07.024. [DOI] [PubMed] [Google Scholar]
- 30.Yoon JH, Lee JM, Kang HJ, Ahn SJ, Yang H, Kim E, et al. Quantitative assessment of liver function by using gadoxetic acid–enhanced MRI: hepatocyte uptake ratio. Radiology. 2019;290:125–133. doi: 10.1148/radiol.2018180753. [DOI] [PubMed] [Google Scholar]
- 31.Haimerl M, Schlabeck M, Verloh N, Zeman F, Fellner C, Nickel D, et al. Volume-assisted estimation of liver function based on Gd-EOB-DTPA-enhanced MR relaxometry. Eur Radiol. 2016;26:1125–1133. doi: 10.1007/s00330-015-3919-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.