Abstract
The rat model of Pneumocystis carinii pneumonia is frequently used to study human P. carinii infection, but there are many differences between the rat and human infections. We studied naturally acquired P. carinii in wild rats to examine the relevance of the rat model for human infection. P. carinii DNA was detected in 47 of 51 wild rats and in 10 of 12 nonimmunosuppressed laboratory rats. Evidence for three novel formae speciales of rat-derived P. carinii was found, and these were provisionally named Pneumocystis carinii f. sp. rattus-secundi, Pneumocystis carinii f. sp. rattus-tertii, and Pneumocystis carinii f. sp. rattus-quarti. Our data suggest that low-level carriage of P. carinii in wild rats and nonimmunosuppressed laboratory rats is common and that wild rats are frequently coinfected with more than one forma specialis of P. carinii. We also examined the diversity in the internally transcribed spacer (ITS) regions of the nuclear rRNA operon of Pneumocystis carinii f. sp. carinii by using samples from wild rats and laboratory rats and spore trap samples. We report a lack of variation in the ITS1 and ITS2 regions that is consistent with an evolutionary bottleneck in the P. carinii f. sp. carinii population. This study shows that human- and rat-derived P. carinii organisms are very different, not only in genetic composition but also in population structure and natural history.
Animal models have been widely used as a source of Pneumocystis carinii organisms and for studying many aspects of P. carinii infection. This is because sustained in vitro cultivation of P. carinii has not been possible (1, 38), although recently a new method has been reported which is now being evaluated in a number of centers (25). The rat model has been particularly useful in studies of epidemiology (11, 50), drug sensitivity (59), immunology (51), and the biology of the organism (42). Rat- and human-derived P. carinii organisms, however, are known to differ significantly in many respects. Considerable divergence has been shown between the genes of rat- and human-derived P. carinii (36, 43), as well as antigenic (9) and ultrastructural (5) differences.
Two genetically divergent types of P. carinii organisms, known as Pneumocystis carinii f. sp. carinii and Pneumocystis carinii f. sp. ratti, have been found in rat lungs (34). These were originally identified by differences in electrophoretic karyotype (7) and subsequently by DNA sequence variation at a number of genes, including those for the nuclear 26S rRNA (21, 29), the mitochondrial large-subunit (mt LSU) rRNA (14), the mitochondrial small-subunit rRNA (12), the TATA binding factor (45), BiP chaperonin (40), thymidylate synthase (14), and ATPase (24), and the α subunit of the G protein (39). The level of genetic divergence between these two types of rat-derived P. carinii was sufficiently high for them to be classed as different formae speciales (34, 44). Although eight different electrophoretic karyotypes of P. carinii f. sp. carinii and two of P. carinii f. sp. ratti have been identified, very little genetic variation in the form of DNA sequence polymorphisms has been observed among isolates of either P. carinii f. sp. carinii or P. carinii f. sp. ratti (6). In contrast, only one forma specialis has been found in human infections, Pneumocystis carinii f. sp. hominis. However, divergence has been observed within this forma specialis, detected as DNA polymorphisms at a number of loci, including the mt LSU rRNA, the mitochondrial small subunit rRNA, the arom locus and the internal transcribed spacer (ITS) regions of the nuclear rRNA operon (15, 20, 22, 47, 48, 49). The ITS regions have been shown to be more informative in distinguishing between types, and as many as 59 different types have been identified at this locus (19, 32; R. F. Miller and A. E. Wakefield, unpublished results).
One of the major differences between the human and rat infections is that human-derived P. carinii samples are obtained from acutely infected individuals who have been exposed to the environment, whereas the rat model uses laboratory-bred animals housed in close proximity and at some level of isolation from the environment. In this study, we examined P. carinii infection in wild rats. A previous study of P. carinii infection in wild rats, using microscopy for detection of P. carinii cysts, suggested that a significant proportion of rats carried P. carinii, though these studies lacked the sensitivity and discriminatory power of contemporary DNA amplification techniques (35). We have used a PCR technique which can detect P. carinii DNA with great sensitivity and which allowed us to differentiate between the two known formae speciales of rat-derived P. carinii (31). We used amplification at the mt LSU rRNA gene for this study, since all known P. carinii formae speciales have been analyzed at this locus (8, 54, 56) and it is known to be a sensitive and robust target for diagnostic PCR of clinical samples (49, 58), samples of rat lung (50), and environmental samples (52, 53).
In this paper we provide evidence to suggest the presence of five different formae speciales of P. carinii in lung samples from Danish wild rats. We report a high incidence of P. carinii organisms in these samples and a high frequency of mixed infections, and we show that only low levels of sequence heterogeneity were observed in the ITS regions of P. carinii f. sp. carinii in the samples. We compare the results to those from human P. carinii infection, where only one forma specialis has been found, with a large number of polymorphisms in the ITS regions.
MATERIALS AND METHODS
Samples.
Wild brown rats (Rattus norvegicus) were live trapped at various domestic locations throughout Denmark. The 51 nonimmunosuppressed rats were then housed singly or in groups of up to 3 per cage in the laboratory and sacrificed after 0 to 78 days. The animals were all housed initially in a quarantine room and then transferred to a test room, both of which were under negative pressure. There were no immunosuppressed animals housed in the same building. No immunosuppressive drugs were administered, nor were the rats treated with drugs against bacterial infections or worms. Six wild rats were trapped, taken to a different animal facility, and immunosuppressed with corticosteroids. The two rats WR2653 and WR2715 died after 9 and 13 days, but the remaining rats continued on immunosuppressive drugs until they were sacrificed at between 38 and 48 days.
The laboratory rats were male Sprague Dawley rats weighing 200 to 220 g obtained from Harlan, Leicester, United Kingdom. Nonimmunosuppressed laboratory rats were not placed in rooms where immunosuppressed animals were housed, and they were sacrificed immediately on arrival at the animal facility. Immunosuppressed laboratory rats were housed in cages with up to six animals and were treated with dexamethasone (Organon, United Kingdom) and antibiotics as previously described (55). The rat lungs were recovered at sacrifice and stored at −80°C until analysis.
Spore trap samples were collected, and DNA was extracted from them as previously described (52, 53).
DNA extraction.
Approximately one-fifth of each rat lung was used for extraction of total DNA. The tissue was minced using sterile scalpels and digested with 1 mg of proteinase K ml−1 in 10 mM EDTA (pH 8.0) and 0.5% sodium dodecyl sulfate at 50°C overnight, after which a further 1 mg of proteinase K ml−1 was added and the sample was incubated for a further 24 h. The DNA was purified by phenol-chloroform extraction, followed by a DNA binding resin (Wizard DNA Clean-Up system; Promega, Southampton, United Kingdom). All precautions were taken throughout these procedures to eliminate the possibility of cross contamination of samples. All handling of the samples took place in a laminar flow cabinet, and negative controls were included in the extraction procedure to monitor for contamination.
DNA amplification.
PCRs were performed using reagents at the following concentrations: 50 mM KCl, 10 mM Tris (pH 8.0), 0.1% Triton X-100, 3 mM MgCl2, 0.04 mM (each) deoxynucleoside triphosphate, 1 μM oligonucleotide primers, and 0.025 U of Taq polymerase (Promega) ml−1. All PCR experiments were performed on three different dilutions of sample DNA. For samples which tested negative for all types of P. carinii, DNA was reextracted from the tissue and the PCRs were repeated. The utmost care was taken at all times to prevent and monitor for contamination. Negative controls were included for all samples, and all handling of reagents occurred in a laminar flow cabinet, using sterile tubes, pipette tips, and aliquoted reagents.
The primers used to amplify a portion of the mt LSU rRNA were as follows: pAZ102-H and pAZ102-E (57, 58) and pAZ102-X/RI and pAZ102-Z/RI (53); RC1, RC2, RR1, and RR2 were used to specifically detect P. carinii f. sp. carinii and P. carinii f. sp. ratti DNA (31). The ITS1 and ITS2 regions are flanked by rRNA genes that are highly conserved between P. carinii formae speciales and other fungi, whereas the ITS regions themselves are highly divergent. Where the samples were likely to contain DNA from other fungi, it was necessary to use PCR primers designed for the ITS regions which would preferentially amplify P. carinii ITS sequences. The ITS regions from environmental samples were amplified using primers designed specifically for P. carinii f. sp. carinii in both the first (ITS21/HI and ITS22/HI) and second (ITS25/HI and ITS26/HI) rounds of PCR in order to amplify only the desired sequences from this diverse pool of DNA. Primer ITS21/HI (5′-CGGGATCCACCTGCGGAAGGATCATTAAT-3′) was designed to match the 3′ end of the 18S rRNA gene and the first 2 bp of the ITS1 region, and ITS22/HI (5′-CGGGATCCCTGATTTGAGGTCAAAGGTTC-3′) was designed to match the 5′ end of the 26S rRNA gene and the last 6 bp of the ITS2 region. The nested primers ITS25/HI (5′-CGGGATCCGAACTAGTTTATCTGGTTCTTG-3′) and ITS26/HI (5′-CGGGATCCTCTAGGAACAATAGACAAACC-3′) were designed within the ITS regions to confer maximum specificity for P. carinii f. sp. carinii. The ITS region was amplified by nested PCR on all samples from nonimmunosuppressed rats; these specific primers were also used on 10 of the nonimmunosuppressed wild rats. Samples from six wild rats (one sample was amplified using both protocols) were amplified using primers designed for the rRNA genes, ITS30 (5′-TTCCGTAGGTGAACCTGCG-3′) and NITSR (48) in the first round followed by primers ITS21/HI and ITS22/HI in the second round. Primers ITS30 and NITSR were designed for the highly conserved 18S and 26S RNA genes. The ITS regions of heavily infected laboratory rat samples were amplified using a single-round PCR with primers ITS21/HI and ITS22/HI. The ITS regions of the two immunosuppressed wild-rat DNA samples were amplified using a single-round PCR with primers ITS30 and NITSR. In the first round of nested PCRs, the thermal cycling conditions were 94°C for 1.5 min, 55°C for 1.5 min, and 72°C for 2 min. In PCRs with a single round and in the second round of a nested PCR, the thermal cycling conditions were 10 cycles at 94°C for 1.5 min, 55°C for 1.5 min, and 72°C for 2 min, followed by 30 cycles of 94°C for 1.5 min, 63°C for 1.5 min, and 72°C for 2 min.
The PCR products were cloned into pUC18 (Amersham-Pharmacia Biotech) or into pGEM T-Easy (Promega), and the recombinant DNA was sequenced using the Sequenase 2.0 kit (Amersham-Pharmacia Biotech) or the dye terminator kit (Perkin-Elmer) and the ABI Prism 377 DNA sequencer running Data Collection Software version 2.1 (Perkin-Elmer Applied Biosystems). Sequence data analysis was performed using Chromas version 1.44 software (C. McCarthy, Griffith University, Brisbane, Australia). Sequence alignments were performed using the Wisconsin Package version 10 (Genetics Computer Group, Madison, Wis.). Statistical analyses were performed using SPSS 7.0.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the DNA sequence of a portion of the gene encoding the mt LSU rRNA are as follows: P. carinii f. sp. carinii, U20169; P. carinii f. sp. ratti, U20173; Pneumocystis carinii f. sp. rattus-secundi, AF308807; Pneumocystis carinii f. sp. rattus-tertii, AF308808; and Pneumocystis carinii f. sp. rattus-quarti, AF308809.
RESULTS
P. carinii detected in lungs of nonimmunosuppressed Danish wild rats.
PCR was used to search for P. carinii DNA in samples extracted from the lungs of 51 wild rats, live trapped at various locations in Denmark. After a single round of PCR using the primers pAZ102-H and pAZ102-E, 11 of the 51 samples gave a P. carinii-specific PCR product indicative of the presence of P. carinii (Table 1). The first-round PCR products of the 51 samples were used as templates for three different nested PCRs. Primer pair RC1 and RC2 specifically amplifies P. carinii f. sp. carinii DNA (31), and nested PCR with these primers showed that 36 of the 51 samples contained P. carinii f. sp. carinii DNA. Similarly, the primer pair RR1 and RR2 specifically amplifies P. carinii f. sp. ratti DNA (31), and this was found to be present in 32 of the 51 samples (Table 1). The primer pair pAZ102-X/RI and pAZ102-Z/RI amplifies P. carinii DNA from all host species tested to date (A. E. Wakefield, unpublished results). Using nested PCR with primers pAZ102-X/RI and pAZ102-Z/RI, P. carinii DNA could be detected in 47 of the 51 rats. No P. carinii DNA was found in 4 of the 51 samples (Table 1).
TABLE 1.
P. carinii DNA in nonimmunosuppressed wild rats
| Rat sample no. | Date of capture (day-mo-yr) | No. of days in captivity | Sexa | Wt at capture (g) | Presence of DNA fromb:
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Any P. carinii by PCR
|
Formae speciales by specific PCR
|
New formae speciales
|
|||||||||
| Single round (H + E) | Nested (X + Z) | carinii (RC1 + RC2) | ratti (RR1 + RR2) | rattus-secundi (X + Z) | rattus-tertii (X + Z) | rattus-quarti (X + Z) | |||||
| WR2374 | 26-Nov-96 | 74 | M | 173.8 | − | + | + | + | − | − | − |
| WR2377 | 28-Nov-96 | 69 | F | 161.0 | − | + | − | + | − | − | − |
| WR2381 | 4-Dec-96 | 69 | F | 82.5 | − | + | + | + | − | − | − |
| WR2382 | 5-Dec-96 | 61 | M | 302.1 | − | + | + | + | − | − | − |
| WR2385A | 6-Dec-96 | 63 | F | 192.5 | − | + | + | − | − | − | − |
| WR2386 | 12-Dec-96 | 53 | M | 253.4 | − | + | + | + | − | − | − |
| WR2388A | 12-Dec-96 | 35 | F | 114.1 | − | + | + | + | − | − | − |
| WR2395 | 9-Jan-97 | 1 | F | 85.8 | − | + | − | − | − | − | + |
| WR2397A | 14-Jan-97 | 0 | F | 191.2 | − | + | − | + | − | − | − |
| WR2397B | 14-Jan-97 | 0 | M | 158.6 | − | + | − | + | − | − | − |
| WR2398 | 15-Jan-97 | 42 | F | 167.8 | + | + | + | + | − | − | − |
| WR2399 | 15-Jan-97 | 35 | M | 210.4 | + | + | + | + | − | − | − |
| WR2400 | 16-Jan-97 | 6 | F | 246.9 | − | + | + | − | + | − | − |
| WR2401 | 16-Jan-97 | 53 | M | 322.6 | − | + | + | − | − | − | − |
| WR2405 | 17-Jan-97 | 54 | M | 263.1 | − | + | + | + | − | − | − |
| WR2409 | 22-Jan-97 | 49 | F | 97.6 | + | + | + | − | − | − | − |
| WR2414 | 28-Jan-97 | 44 | F | 256.0 | − | + | + | − | − | − | − |
| WR2419 | 30-Jan-97 | 42 | M | 280.4 | − | + | + | + | − | − | − |
| WR2422C | 4-Feb-97 | 37 | M | 433.6 | − | + | − | + | − | − | − |
| WR2422D | 4-Feb-97 | 37 | M | 299.6 | − | − | − | − | − | − | − |
| WR2422E | 4-Feb-97 | 39 | F | 121.6 | − | + | − | + | − | − | − |
| WR2422F | 4-Feb-97 | 39 | F | 201.6 | − | + | + | − | − | − | − |
| WR2424 | 4-Feb-97 | 36 | F | 145.7 | − | + | + | + | − | − | − |
| WR2425 | 5-Feb-97 | 19 | F | 224.8 | − | + | + | − | − | − | − |
| WR2426 | 5-Feb-97 | 8 | F | 67.0 | + | + | + | + | − | − | − |
| WR2428A | 6-Feb-97 | 33 | M | 292.8 | − | + | − | + | − | − | − |
| WR2430A | 7-Feb-97 | 32 | M | 240.2 | − | + | + | + | − | − | − |
| WR2432 | 10-Feb-97 | 7 | M | 118.4 | − | + | + | + | + | − | − |
| WR2433 | 11-Feb-97 | 0 | F | 118.7 | + | + | + | + | − | − | − |
| WR2441D | 19-Feb-09 | 36 | M | 387.8 | − | − | − | − | − | − | − |
| WR2442A | 19-Feb-97 | 37 | F | 123.1 | − | + | + | + | − | − | − |
| WR2442B | 19-Feb-97 | 37 | F | 194.3 | − | + | + | + | − | − | − |
| WR2444D | 19-Feb-97 | 36 | F | 272.5 | + | + | + | + | − | − | − |
| WR2446 | 20-Feb-97 | 78 | F | 127.4 | + | + | + | + | − | − | − |
| WR2447 | 20-Feb-97 | 75 | M | 279.3 | − | + | + | − | − | − | − |
| WR2449A | 21-Feb-97 | 3 | F | 181.3 | − | + | + | − | + | − | − |
| WR2449B | 21-Feb-97 | 4 | F | 225.3 | − | + | + | − | − | − | − |
| WR2455 | 27-Feb-97 | 0 | M | 172.8 | − | − | − | − | − | − | − |
| WR2466 | 5-Mar-97 | 61 | M | 298.9 | − | + | + | − | − | − | − |
| WR2470 | 7-Mar-97 | 59 | F | 219.0 | + | + | + | + | − | − | − |
| WR2471 | 13-Mar-97 | 4 | F | 133.1 | − | + | − | + | − | − | − |
| WR2473 | 18-Mar-97 | 48 | F | 142.7 | − | + | + | + | − | − | − |
| WR2484A | 26-Mar-97 | 1 | F | 145.3 | + | + | + | + | − | − | − |
| WR2486 | 4-Apr-97 | 6 | M | 271.7 | − | + | + | + | − | − | − |
| WR2494 | 15-Apr-97 | 3 | F | 147.2 | + | + | + | + | − | − | − |
| WR2498 | 16-Apr-97 | 3 | F | 144.8 | − | + | − | − | − | + | − |
| WR2502 | 22-Apr-97 | 0 | F | 111.0 | − | + | − | − | − | − | + |
| WR2511 | 29-Apr-97 | 0 | M | 206.7 | + | + | + | + | − | − | − |
| WR2512 | 29-Apr-97 | 6 | F | 188.0 | − | + | − | + | − | − | − |
| WR2515 | 1-May-97 | 0 | M | 216.0 | − | + | + | − | − | − | − |
| WR2519A | 6-May-97 | 0 | M | 44.5 | − | − | − | − | − | − | − |
| Total positive | 11 | 47 | 36 | 32 | 3 | 1 | 2 | ||||
| % Positive | 21.6 | 92.2 | 70.6 | 62.7 | 5.9 | 2.0 | 3.9 | ||||
F, female; M, male.
+, present; −, absent; H, primer pAZ102-H; E, primer pAZ102-E; X, primer pAZ102-X/RI; Z, primer pAZ102-Z/RI.
As a control, a group of 12 nonimmunosuppressed laboratory rats were examined in the same way. The animals were sacrificed immediately on arrival at the animal facility and before they came into contact with any immunosuppressed rats. Single-round PCR was carried out with primer pair pAZ102-H and pAZ102-E, and 2 of the 12 samples were positive for P. carinii. Using nested PCR with the P. carinii special form-specific primers, P. carinii f. sp. carinii DNA alone was detected in 9 of the 12 rats, both P. carinii f. sp. carinii and P. carinii f. sp. ratti DNAs were detected in 1 of the 12, and no P. carinii DNA was found in 2 of the 12 samples (Table 2).
TABLE 2.
P. carinii DNA in nonimmunosuppressed laboratory rats
| Rat sample no. | Presence of DNAa
|
|||||
|---|---|---|---|---|---|---|
| Single-round PCR
|
Nested PCR
|
|||||
| Any P. carinii (H + E)a | f. sp. carinii (RC1 + RC2) | f. sp. ratti (RR1 + RR2) | Any P. carinii (X + Z)b | f. sp. carinii (RC1 + RC2) | f. sp. ratti (RR1 + RR2) | |
| R1632 | + | + | − | + | + | − |
| R1633 | − | − | − | + | + | + |
| R1634 | − | − | − | − | − | − |
| R1635 | + | − | − | + | + | − |
| R1636 | − | − | − | + | + | − |
| R1637 | − | − | − | + | + | − |
| R1682 | − | NDb | ND | ND | − | − |
| R1683 | − | ND | ND | ND | + | − |
| R1684 | − | ND | ND | ND | + | − |
| R1685 | − | ND | ND | ND | + | − |
| R1686 | − | ND | ND | ND | + | − |
| R1687 | − | ND | ND | ND | + | − |
| No. positive/total | 2/12 | 1/6 | 0/6 | 5/6 | 10/12 | 1/12 |
H + E, primers pAZ102-H and pAZ102-E; +, present; −, absent; ND, not determined.
X + Z, primers pAZ102-X and pAZ102-Z.
Evidence for five different formae speciales of P. carinii in wild rats.
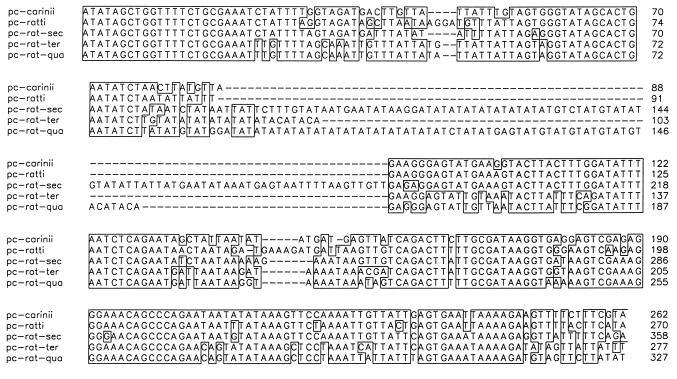
A number of the nested PCRs with primer pair pAZ102-X/RI and pAZ102-Z/RI gave products of unexpected size, which were cloned and sequenced (Table 3). Five distinct sequence types were identified (Fig. 1). One of the sequences corresponded to P. carinii f. sp. carinii, and another corresponded to P. carinii f. sp. ratti. The three other sequences, which were isolated from six rats and sequenced several times in more than one PCR experiment, have not been previously reported.
TABLE 3.
Summary of sequencing performed on P. carinii from nonimmunosuppressed wild rats
| Rat sample no. | No. of PCRs performed | No. of clones sequenced from f. sp.:
|
|||||
|---|---|---|---|---|---|---|---|
| carinii | ratti | rattus-secundi | rattus-tertii | rattus-quarti | Total | ||
| WR2395 | 3 | 0 | 0 | 0 | 0 | 3 | 3 |
| WR2400 | 2 | 2 | 0 | 2 | 0 | 0 | 4 |
| WR2426 | 1 | 1 | 1 | 0 | 0 | 0 | 2 |
| WR2432 | 5 | 6 | 3 | 6 | 0 | 0 | 15 |
| WR2449A | 1 | 0 | 0 | 2 | 0 | 0 | 2 |
| WR2498 | 2 | 0 | 0 | 0 | 6 | 0 | 6 |
| WR2502 | 2 | 0 | 0 | 0 | 0 | 2 | 2 |
| Total | 16 | 9 | 4 | 10 | 6 | 5 | 34 |
FIG. 1.
Alignment of the DNA sequence of a portion of the P. carinii gene encoding the mt LSU rRNA isolated from wild rats showing the sequences from P. carinii f. sp. carinii (pc-carinii) and P. carinii f. sp. ratti (pc-ratti) and the new DNA sequences from P. carinii f. sp. rattus-secundi (pc-rat-sec), P. carinii f. sp. rattus-tertii (pc-rat-ter), and P. carinii f. sp. rattus-quarti (pc-rat-qua). Nucleotides are boxed to highlight regions of sequence identity. Gaps (–) were introduced to improve the alignment.
The three novel sequences were aligned with sequences from all other known P. carinii formae speciales and with other fungi. Each of the three new types had a higher level of sequence identity to the corresponding sequence from P. carinii formae speciales than to those from the other fungi (data not shown). We propose that these sequences were amplified from three new formae speciales, which we have provisionally named P. carinii f. sp. rattus-secundi, P. carinii f. sp. rattus-tertii, and P. carinii f. sp. rattus-quarti. These names are in keeping with the previously published guidelines and were devised in consultation with the Pneumocystis Taxonomy and Nomenclature Committee (34, 44). The P. carinii f. sp. rattus-secundi sequence was found to have the highest identity to P. carinii f. sp. carinii and P. carinii f. sp. ratti sequences, but it contained a 100-bp insertion compared with the sequences from known rat-derived P. carinii special forms. The P. carinii f. sp. rattus-tertii sequence had the highest identity to the P. carinii f. sp. rattus-quarti sequence. The P. carinii f. sp. rattus-quarti sequence contained a 56-bp insertion at the same position as the P. carinii f. sp. rattus-secundi sequence (Fig. 1). Some minor variations were noted in the repetitive regions of the sequences; these could have been due to PCR-induced error and the instability of such repeats in Escherichia coli plasmids (41). At positions 116 to 117 in the P. carinii f. sp. rattus-secundi sequence, TG, GA, and a 2-bp deletion were also observed, and at positions 82 to 83 in P. carinii f. sp. rattus-tertii, TG and a 2-bp deletion were seen. From positions 91 to 120 in P. carinii f. sp. rattus-quarti, a variable number (from 10 to 15) of AT repeats were observed (15 AT repeats are shown in Fig. 1).
Distribution of P. carinii in nonimmunosuppressed wild rats.
The amount of rat-derived P. carinii DNA detected was stratified into three levels: (i) presence of P. carinii-specific amplification product, visualized on an agarose gel after single-round PCR; (ii) presence of P. carinii-specific amplification product, visualized on an agarose gel after nested PCR; (iii) undetectable P. carinii-specific amplification product. When the P. carinii-specific amplification product could be visualized on an agarose gel after a single-round PCR, it was taken to indicate the presence of more P. carinii than if it could only be detected using nested PCR. It was found that all female rats harbored detectable levels of P. carinii DNA, and in 9 of 30, P. carinii DNA could be detected by single-round PCR. In contrast, 4 of 21 male rats contained no detectable P. carinii DNA, and in only 2 of 21 was P. carinii DNA found at the higher level. This distribution suggests a relationship between the amount of P. carinii DNA detected and the sex of the rat (G test, P = 0.011; Williams' correction). There were no associations between the types of P. carinii DNA found and the site of capture, the weight of the rat at capture, or the month of capture.
P. carinii f. sp. rattus-secundi was detected in three samples, P. carinii f. sp. rattus-tertii was found in one sample, and P. carinii f. sp. rattus-quarti was found in two samples (Table 3). Considering all five types of rat-derived P. carinii, there was evidence for more than one type of P. carinii in 26 of the 51 rats (Table 1). After capture, 8 of 51 rats were killed immediately, and the remainder were housed in the animal facility for up to 78 days before sacrifice. We examined the proportions of the five different formae speciales as a function of the time the animals spent in captivity after capture. P. carinii f. sp. rattus-secundi, P. carinii f. sp. rattus-tertii, and P. carinii f. sp. rattus-quarti were detected in 6 of 20 rats housed in captivity for less than 10 days but not in any of the 31 rats housed for 11 days or more. A significant relationship was detected between the presence of the three new formae speciales sequences and the amount of time between capture and sacrifice (P = 0.0027; χ2 test; Yates' correction). In contrast, the proportion of rats in which P. carinii f. sp. carinii was detected increased from 11 of 20 (55%) in captivity for less than 10 days to 25 of 31 (81%) in captivity for 11 days or more (P = 0.05; χ2 test).
P. carinii in immunosuppressed wild rats.
In order to obtain larger amounts of P. carinii DNA, six wild rats were live trapped and immunosuppressed at an animal facility different from that used in the study of the nonimmunosuppressed wild rats, whereupon they developed symptoms of P. carinii pneumonia. DNA was extracted and tested using PCR in the same way as for the nonimmunosuppressed wild rats. After a single round of PCR, four of the six rats had detectable P. carinii DNA. When nested PCR was performed, all six rats were positive for both P. carinii f. sp. carinii and P. carinii f. sp. ratti DNA (Table 4). None of the samples showed evidence of the presence of P. carinii f. sp. rattus-secundi, P. carinii f. sp. rattus-tertii, or P. carinii f. sp. rattus-quarti DNA.
TABLE 4.
P. carinii DNA in immunosuppressed wild rats
| Rat sample no. | Date of capture (day-mo-yr) | No. of days in captivity | Sexa | Wt at capture (g) | Presence of DNA fromb:
|
|||
|---|---|---|---|---|---|---|---|---|
| Any P. carinii by PCR
|
Known types by specific nested PCR
|
|||||||
| Single round (H + E) | Nested (X + Z) | carinii (RC1 + RC2) | ratti (RR1 + RR2) | |||||
| WR2653 | 30-Oct-97 | 9 | F | 83.0 | + | + | + | + |
| WR2661 | 6-Nov-97 | 46 | M | 277.8 | + | + | + | + |
| WR2665 | 11-Nov-97 | 41 | F | 117.0 | − | + | + | + |
| WR2715 | 7-Jan-98 | 13 | F | 308.7 | − | + | + | + |
| WR2722 | 13-Jan-98 | 48 | F | 199.0 | + | + | + | +c |
| WR2724 | 15-Jan-98 | 38 | F | 230.6 | + | + | + | + |
F, female; M, male.
+, present; −, absent; H, primer pAZ102-H; E, primer pAZ102-E; X, primer pAZ102-X/RI; Z, primer pAZ102-Z/RI.
A very low level of P. carinii f. sp. ratti DNA was detected.
Low level of diversity in the ITS regions from P. carinii f. sp. carinii.
In order to further examine diversity within P. carinii f. sp. carinii, we studied DNA polymorphisms in the ITS regions of the nuclear rRNA operon. DNA was isolated from a total of 32 samples: (i) the lungs of 10 immunosuppressed laboratory rats spanning a 10-year period, (ii) 15 nonimmunosuppressed wild rats collected throughout Denmark over a period of 6 months, (iii) 2 wild rats trapped on the Danish island of Bornholm and then immunosuppressed, and (iv) 5 environmental samples collected using spore traps at a rural location in the United Kingdom. Single-round PCR was used to amplify the ITS regions from the immunosuppressed rats, whereas nested PCR was required to amplify the same regions from the environmental samples and the nonimmunosuppressed wild rats. The ITS PCR products were cloned, and the DNA sequences were compared. In all, 13 clones from 10 immunosuppressed laboratory rats, 35 clones from 15 nonimmunosuppressed wild rats, 10 clones from 2 immunosuppressed wild rats, and 12 clones from five spore trap samples were analyzed. A very low level of heterogeneity was detected. A number of single-base polymorphisms were found, but most of these were seen in only 1 of the 70 clones examined. There were 10 positions within the sequences of ITS1 and ITS2 at which a single-base polymorphism was observed in two different samples, and one of these polymorphisms in the ITS1 region was seen in three samples, an immunosuppressed laboratory rat, a nonimmunosuppressed wild rat, and an environmental sample.
DISCUSSION
High incidence of P. carinii infection in rats.
We found P. carinii DNA in 92% of wild rats, which is higher than most previously reported estimates of prevalence. Using a single round of PCR, the incidence of P. carinii was very similar to that found in a previous Danish survey which used the same collection methods (35) and to the incidence of P. carinii in the field vole, Microtus agrestis, and the common shrew, Sorex araneus, in Finland (16). The nested-PCR assay detected low numbers of organisms which would not be seen by histochemical staining or immunocytochemistry (23, 28, 33, 46, 50) and which would not cause pathological changes in the lung (26, 60). Our data show an association between the amount of P. carinii DNA detected by PCR and the sex of the rat, with female rats carrying more P. carinii DNA than males. This is in agreement with data from laboratory rats in which corticosteroid-treated female rats acquired severe P. carinii pneumonitis faster than male rats (30).
P. carinii f. sp. carinii and P. carinii f. sp. ratti can coexist in the same laboratory rat and may compete for resources (6). We found both formae speciales in nonimmunosuppressed wild rats, in solitary infections and also in coinfections with other rat-derived P. carinii types. However, this study cannot conclude that either forma specialis can exist alone in wild rats. Multiple PCR tests on each sample would be needed to totally exclude the presence of other types. With over half of the wild rats carrying at least two formae speciales of P. carinii, we suggest that infection in the wild rat is a complex interaction of several distinct types of organisms. Our findings are consistent with those of another study of the interaction of populations of P. carinii f. sp. carinii and P. carinii f. sp. ratti, which suggested that these two formae speciales may be competing for resources within the lung (13). It contrasts with human infection, where, although large numbers of samples have been analyzed, only one special form, P. carinii f. sp. hominis, has been found (10, 14, 17, 18, 19, 20, 47, 48, 49).
Our data on prevalence suggest that low-level carriage of P. carinii in wild rats and nonimmunosuppressed laboratory rats is common. Immunocompetent rats have been shown to be able to clear all P. carinii organisms if housed under isolator conditions (50), and a model of continual de novo infection via an airborne route and slow elimination is suggested rather than stable latency (2, 11, 18, 52, 53). This may also be the case in human infection, where low levels of P. carinii f. sp. hominis have recently been reported in some patients who are not profoundly immunosuppressed, suggestive of short-term, low-level carriage of P. carinii f. sp. hominis in these patient groups. These include individuals with other pulmonary disease, such as chronic lung disease, and also patients with malignant disease (3, 4, 27, 37).
Evidence for three new formae speciales of P. carinii.
We present data in support of three novel formae speciales of rat-derived P. carinii. We consider that they are likely to be as-yet-undescribed P. carinii types based on DNA sequence homology to known P. carinii and fungal mt LSU rRNA sequences. The majority of other different Pneumocystis formae speciales were first identified by analysis of the mt LSU rRNA sequence, and divergence at this sequence has provided a useful indicator for assigning new formae speciales (8, 32, 36, 56). We consider it highly unlikely that the three new sequences could have been generated purely by a PCR artifact or the cloning procedure; none of the sequences is a mosaic of known types, and all have portions of the sequence which are unique to that sequence. It is unlikely that the new sequences resulted from some form of “contamination,” since it is difficult to postulate a source of such contamination. These sequences had never previously been found in our laboratory; indeed, they have not been described prior to this study, yet they appear to be of Pneumocystis origin, since they form a monophyletic group with the other P. carinii special forms. All of the new sequences were isolated from more than one PCR, and both P. carinii f. sp. rattus-secundi and P. carinii f. sp. rattus-quarti sequences were amplified independently from more than one rat sample. The sequences were isolated using primers which are capable of amplifying many formae speciales of P. carinii. However, we did not find any evidence for the presence of non-rat-derived P. carinii in the samples, although they are present in samples of air spora (52, 53). We therefore consider it likely that these novel sequences were amplified from viable P. carinii organisms within the lungs of the feral rats. We suggest that the sequences detected were amplified from previously unknown formae speciales of rat-derived P. carinii. While we suggest that these new types make up a part of the lung fauna of wild rats, there are significant practical problems in obtaining enough material for further studies. Immunosuppression of a number of wild rats did not result in the isolation of the new formae speciales.
Low level of diversity in the ITS regions of P. carinii f. sp. carinii.
Our data on the analysis of the ITS1 and ITS2 sequences of P. carinii f. sp. carinii suggested that the level of variation was very low. The lack of diversity in the ITS regions of P. carinii f. sp. carinii contrasts strikingly with the extensive diversity found in the ITS regions of P. carinii f. sp. hominis. To date, we have found 34 types from 55 episodes of P. carinii pneumonia (A. G. Tsolaki, R. F. Miller, and A. E. Wakefield, unpublished observations), and Lee et al. have reported 59 combinations of ITS1 and ITS2 types from 207 clinical samples (19). The techniques used in the present study were capable of detecting such heterogeneity if the P. carinii f. sp. carinii locus had a level of variation similar to that of P. carinii f. sp. hominis. All our samples came from northern Europe, however, so we cannot exclude the possibility that other genotypes of P. carinii f. sp. carinii could be found elsewhere. The primers that were used to amplify the ITS regions from the spore trap samples and from 18 of the 45 wild-rat samples were designed for the sequence of the ITS regions of P. carinii f. sp. carinii. Since the ITS regions of the special forms of P. carinii are so divergent, it is conceivable that the primers selectively amplified sequences identical to the prototype sequence. However, the results gained from using the primers designed for the sequence of the conserved rRNA genes were similar. The ITS regions of P. carinii f. sp. carinii are unlikely to form the basis of a useful typing system like that used in P. carinii f. sp. hominis.
There are a number of explanations to account for the low level of diversity seen in the ITS regions. One possibility is that the population of P. carinii f. sp. carinii has passed through a recent genetic bottleneck; alternatively, the ITS regions of P. carinii f. sp. carinii may be under high selective pressure. The genetic-bottleneck hypothesis is consistent with the natural history of the laboratory animals, since many rat colonies were derived from the same small populations of laboratory animals established relatively recently. The lack of diversity seen in the wild-rat P. carinii population may be explained by the recent and rapid expansion of the brown rat, R. norvegicus, through Europe in the 18th century from central or east Asia, displacing the black rat, Rattus rattus, the carrier of plague (61). P. carinii shows strict host specificity, and so one would expect the natural history of the parasite to be greatly influenced by that of the host. This study of P. carinii infection in wild rats may lead to a better understanding of the links between the population structures of other formae speciales of P. carinii and their hosts.
In this study we have shown that P. carinii is frequently found in the lungs of nonimmunosuppressed rats. Our data support the notion of short-term carriage of P. carinii f. sp. hominis in patient groups other than the severely immunocompromised. However, our results also highlight the differences between P. carinii infections in rats and humans and underline the caution needed when using the rat model of infection for investigating the human disease.
ACKNOWLEDGMENTS
This research was supported by the Medical Research Council (R.J.P.) and the Royal Society (A.E.W.) and formed a part of the European Concerted Action Biomed 1 “Pneumocystis and pneumocystosis: impact of the biodiversity of Pneumocystis carinii on epidemiology, pathology, diagnosis, monitoring and prevention of pneumocystosis—new therapeutic approaches” PL941118.
REFERENCES
- 1.Aliouat E M, Dei Cas E, Dujardin L, Tissier J P, Billaut P, Camus D. High infectivity of Pneumocystis carinii cultivated on L2 rat alveolar epithelial cells. J Eukaryot Microbiol. 1996;43:22S. doi: 10.1111/j.1550-7408.1996.tb04960.x. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett M S, Lu J J, Lee C H, Durant P J, Queener S F, Smith J W. Types of Pneumocystis carinii detected in air samples. J Eukaryot Microbiol. 1996;43:44S. doi: 10.1111/j.1550-7408.1996.tb04980.x. [DOI] [PubMed] [Google Scholar]
- 3.Calderon E J, Regordan C, Medrano F J, Ollero M, Varela J M. Pneumocystis carinii infection in patients with chronic bronchial disease. Lancet. 1996;347:977. doi: 10.1016/s0140-6736(96)91468-3. [DOI] [PubMed] [Google Scholar]
- 4.Contini C, Villa M P, Romani R, Merolla R, Delia S, Ronchetti R. Detection of Pneumocystis carinii among children with chronic respiratory disorders in the absence of HIV infection and immunodeficiency. J Med Microbiol. 1998;47:329–333. doi: 10.1099/00222615-47-4-329. [DOI] [PubMed] [Google Scholar]
- 5.Creusy C, Bahon le Capon J, Fleurisse L, Mullet C, Dridba M, Cailliez J-C, Antoine M, Camus D, Dei Cas E. Pneumocystis carinii pneumonia in four mammal species: histopathology and ultrastructure. J Eukaryot Microbiol. 1996;43:47S–48S. doi: 10.1111/j.1550-7408.1996.tb04983.x. [DOI] [PubMed] [Google Scholar]
- 6.Cushion M T. Genetic heterogeneity of rat-derived Pneumocystis. FEMS Immunol Med Microbiol. 1998;22:51–58. doi: 10.1111/j.1574-695X.1998.tb01186.x. [DOI] [PubMed] [Google Scholar]
- 7.Cushion M T, Zhang J, Kaselis M, Giuntoli D, Stringer S L, Stringer J R. Evidence for two genetic variants of Pneumocystis carinii coinfecting laboratory rats. J Clin Microbiol. 1993;31:1217–1223. doi: 10.1128/jcm.31.5.1217-1223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand-Joly I, Wakefield A E, Palmer R J, Denis C M, Creusy C, Fleurisse L, Ricard I, Gut J P, Dei-Cas E. Ultrastructural and molecular characterization of Pneumocystis carinii isolated from a rhesus monkey (Macaca mulatta) Med Mycol. 2000;38:61–72. doi: 10.1080/mmy.38.1.61.72. [DOI] [PubMed] [Google Scholar]
- 9.Gigliotti F. Host species-specific antigenic variation of a mannosylated surface glycoprotein of Pneumocystis carinii. J Infect Dis. 1992;165:329–336. doi: 10.1093/infdis/165.2.329. [DOI] [PubMed] [Google Scholar]
- 10.Helweg-Larsen J, Tsolaki A G, Miller R F, Lundgren B, Wakefield A E. Clusters of Pneumocystis carinii pneumonia: analysis of person-to-person transmission by genotyping. Q J Med. 1999;91:813–820. doi: 10.1093/qjmed/91.12.813. [DOI] [PubMed] [Google Scholar]
- 11.Hughes W T. Natural mode of acquisition for de novo infection with Pneumocystis carinii. J Infect Dis. 1982;145:842–848. doi: 10.1093/infdis/145.6.842. [DOI] [PubMed] [Google Scholar]
- 12.Hunter J A, Wakefield A E. Genetic divergence at the mitochondrial small subunit ribosomal RNA gene among isolates of Pneumocystis carinii from five mammalian host species. J Eukaryot Microbiol. 1996;43:24S–25S. doi: 10.1111/j.1550-7408.1996.tb04962.x. [DOI] [PubMed] [Google Scholar]
- 13.Icenhour C R, Arnold J, Cushion M T. Interactions of two Pneumocystis carinii populations within rat lungs. J Eukaryot Microbiol. 1999;46:107S–108S. [PubMed] [Google Scholar]
- 14.Keely S, Pai H J, Baughman R, Sidman C, Sunkin S M, Stringer J R, Stringer S L. Pneumocystis species inferred from analysis of multiple genes. J Eukaryot Microbiol. 1994;41:94S. [PubMed] [Google Scholar]
- 15.Keely S P, Stringer J R, Baughman R P, Linke M J, Walzer P D, Smulian A G. Genetic variation among Pneumocystis carinii hominis isolates in recurrent pneumocystosis. J Infect Dis. 1995;172:595–598. doi: 10.1093/infdis/172.2.595. [DOI] [PubMed] [Google Scholar]
- 16.Laakkonen J, Henttonen H, Niemimaa J, Soveri T. Seasonal dynamics of Pneumocystis carinii in the field vole, Microtus agrestis, and in the common shrew, Sorex araneus, in Finland. Parasitology. 1999;118:1–5. doi: 10.1017/s0031182098003497. [DOI] [PubMed] [Google Scholar]
- 17.Latouche S, Ortona E, Mazars E, Margutti P, Tamburrini E, Siracusano A, Guyot K, Nigou M, Roux P. Biodiversity of Pneumocystis carinii hominis: typing with different DNA regions. J Clin Microbiol. 1997;35:383–387. doi: 10.1128/jcm.35.2.383-387.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latouche S, Poirot J L, Bertrand V, Roux P. Pneumocystis carinii hominis sequencing for reactivation or de novo contamination and for hypothetic transmission from person to person. APMIS Suppl. 1997;77:11–13. doi: 10.1111/j.1600-0463.1997.tb05374.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee C H, Helweg-Larsen J, Tang X, Jin S, Li B, Bartlett M S, Lu J J, Lundgren B, Lundgren J D, Olsson M, Lucas S B, Roux P, Cargnel A, Atzori C, Matos O, Smith J W. Update on Pneumocystis carinii f. sp. hominis typing based on nucleotide sequence variations in internal transcribed spacer regions of rRNA genes. J Clin Microbiol. 1998;36:734–741. doi: 10.1128/jcm.36.3.734-741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee C H, Lu J J, Bartlett M S, Durkin M M, Liu T H, Wang J, Jiang B, Smith J W. Nucleotide sequence variation in Pneumocystis carinii strains that infect humans. J Clin Microbiol. 1993;31:754–757. doi: 10.1128/jcm.31.3.754-757.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Rocourt M, Pan S, Liu C, Leibowitz M J. Sequence and variability of the 5.8S and 26S rRNA genes of Pneumocystis carinii. Nucleic Acids Res. 1992;20:3763–3772. doi: 10.1093/nar/20.14.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J J, Bartlett M S, Shaw M M, Queener S F, Smith J W, Ortiz Rivera M, Leibowitz M J, Lee C H. Typing of Pneumocystis carinii strains that infect humans based on nucleotide sequence variations of internal transcribed spacers of rRNA genes. J Clin Microbiol. 1994;32:2904–2912. doi: 10.1128/jcm.32.12.2904-2912.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J J, Chen C H, Bartlett M S, Smith J W, Lee C H. Comparison of six different PCR methods for detection of Pneumocystis carinii. J Clin Microbiol. 1995;33:2785–2788. doi: 10.1128/jcm.33.10.2785-2788.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meade J C, Stringer J R. Cloning and characterization of an ATPase gene from Pneumocystis carinii which closely resembles fungal H+ ATPases. J Eukaryot Microbiol. 1995;42:298–307. doi: 10.1111/j.1550-7408.1995.tb01584.x. [DOI] [PubMed] [Google Scholar]
- 25.Merali S, Frevert U, Williams J H, Chin K, Bryan R, Clarkson A B. Continuous axenic cultivation of Pneumocystis carinii. Proc Natl Acad Sci USA. 1999;96:2402–2407. doi: 10.1073/pnas.96.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millard P R, Wakefield A E, Hopkin J M. A sequential ultrastructural study of rat lungs infected with Pneumocystis carinii to investigate the appearances of the organism, its relationships and its effects on pneumocytes. Int J Exp Pathol. 1990;71:895–904. [PMC free article] [PubMed] [Google Scholar]
- 27.Nevez G, Raccurt C, Jounieaux V, Dei-Cas E, Mazars E. Pneumocystosis versus pulmonary Pneumocystis carinii colonization in HIV-negative and HIV-positive patients. AIDS. 1999;13:535–536. doi: 10.1097/00002030-199903110-00020. [DOI] [PubMed] [Google Scholar]
- 28.O'Leary T J, Tsai M M, Wright C F, Cushion M T. Use of semiquantitative PCR to assess onset and treatment of Pneumocystis carinii infection in rat model. J Clin Microbiol. 1995;33:718–724. doi: 10.1128/jcm.33.3.718-724.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortiz-Rivera M, Liu Y, Felder R, Leibowitz M J. Comparison of coding and spacer region sequences of chromosomal rRNA-coding genes of two sequevars of Pneumocystis carinii. J Eukaryot Microbiol. 1995;42:44–49. doi: 10.1111/j.1550-7408.1995.tb01538.x. [DOI] [PubMed] [Google Scholar]
- 30.Oz H S, Hughes W T. Effect of sex and dexamethasone dose on the experimental host for Pneumocystis carinii. Lab Anim Sci. 1996;46:109–110. [PubMed] [Google Scholar]
- 31.Palmer R J, Cushion M T, Wakefield A E. Discrimination of rat-derived Pneumocystis carinii f. sp. carinii and Pneumocystis carinii f. sp. ratti using the polymerase chain reaction. Mol Cell Probes. 1999;13:147–155. doi: 10.1006/mcpr.1999.0229. [DOI] [PubMed] [Google Scholar]
- 32.Peters S E, English K, Laakkonen J, Gurnell J. DNA analysis of Pneumocystis carinii infecting Finnish and English shrews. J Eukaryot Microbiol. 1994;41:108S. [PubMed] [Google Scholar]
- 33.Peters S E, Wakefield A E, Banerji S, Hopkin J M. Quantification of the detection of Pneumocystis carinii by DNA amplification. Mol Cell Probes. 1992;6:115–117. doi: 10.1016/0890-8508(92)90055-3. [DOI] [PubMed] [Google Scholar]
- 34.Pneumocystis Workshop. Revised nomenclature for Pneumocystis carinii. J Eukaryot Microbiol. 1994;41:121S–122S. [PubMed] [Google Scholar]
- 35.Settnes O P, Lodal J. Prevalence of Pneumocystis carinii Delanoë & Delanoë, 1912 in rodents in Denmark. Nord Vet Med. 1980;32:17–27. [PubMed] [Google Scholar]
- 36.Sinclair K, Wakefield A E, Banerji S, Hopkin J M. Pneumocystis carinii organisms derived from rat and human hosts are genetically distinct. Mol Biochem Parasitol. 1991;45:183–184. doi: 10.1016/0166-6851(91)90042-5. [DOI] [PubMed] [Google Scholar]
- 37.Sing A, Roggenkamp A, Autenrieth I B, Heesemann J. Pneumocystis carinii carriage in immunocompetent patients with primary pulmonary disorders as detected by single or nested PCR. J Clin Microbiol. 1999;37:3409–3410. doi: 10.1128/jcm.37.10.3409-3410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sloand E, Laughon B, Armstrong M, Bartlett M S, Blumenfeld W, Cushion M, Kalica A, Kovacs J A, Martin W, Pitt E, Pesanti E L, Richards F, Rose R, Walzer P. The challenge of Pneumocystis carinii culture. J Eukaryot Microbiol. 1993;40:188–195. doi: 10.1111/j.1550-7408.1993.tb04902.x. [DOI] [PubMed] [Google Scholar]
- 39.Smulian A G, Ryan M, Staben C, Cushion M. Signal transduction in Pneumocystis carinii: characterization of the genes (pcg1) encoding the alpha subunit of the G protein (PCG1) of Pneumocystis carinii carinii and Pneumocystis carinii ratti. Infect Immun. 1996;64:691–701. doi: 10.1128/iai.64.3.691-701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stedman T T, Buck G A. Identification, characterization, and expression of the BiP endoplasmic reticulum resident chaperonins in Pneumocystis carinii. Infect Immun. 1996;64:4463–4471. doi: 10.1128/iai.64.11.4463-4471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strauss B S, Sagher D, Acharya S. Role of proofreading and mismatch repair in maintaining the stability of nucleotide repeats in DNA. Nucleic Acids Res. 1997;25:806–813. doi: 10.1093/nar/25.4.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stringer J R. Pneumocystis carinii: what is it, exactly? Clin Microbiol Rev. 1996;9:489–498. doi: 10.1128/cmr.9.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stringer J R, Stringer S L, Zhang J, Baughman R, Smulian A G, Cushion M T. Molecular genetic distinction of Pneumocystis carinii from rats and humans. J Eukaryot Microbiol. 1993;40:733–741. doi: 10.1111/j.1550-7408.1993.tb04468.x. [DOI] [PubMed] [Google Scholar]
- 44.Stringer J R, Wakefield A E, Cushion M T, Dei Cas E. Pneumocystis taxonomy and nomenclature: an update. J Eukaryot Microbiol. 1997;44:5S–6S. doi: 10.1111/j.1550-7408.1997.tb05736.x. [DOI] [PubMed] [Google Scholar]
- 45.Sunkin S M, Stringer J R. Transcription factor genes from rat Pneumocystis carinii. J Eukaryot Microbiol. 1995;42:12–19. doi: 10.1111/j.1550-7408.1995.tb01534.x. [DOI] [PubMed] [Google Scholar]
- 46.Tamburrini E, Mencarini P, De Luca A, Maiuro G, Ventura G, Antinori A, Ammassari A, Visconti E, Ortona L, Siracusano A, Ortona E, Vicari G. Diagnosis of Pneumocystis carinii pneumonia: specificity and sensitivity of polymerase chain reaction in comparison with immunofluorescence in bronchoalveolar lavage specimens. J Med Microbiol. 1993;38:449–453. doi: 10.1099/00222615-38-6-449. [DOI] [PubMed] [Google Scholar]
- 47.Tsolaki A G, Beckers P, Wakefield A E. Pre-AIDS era isolates of Pneumocystis carinii f. sp. hominis: high genotypic similarity with contemporary isolates. J Clin Microbiol. 1998;36:90–93. doi: 10.1128/jcm.36.1.90-93.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsolaki A G, Miller R F, Underwood A P, Banerji S, Wakefield A E. Genetic diversity at the internal transcribed spacer regions of the rRNA operon among isolates of Pneumocystis carinii from AIDS patients with recurrent pneumonia. J Infect Dis. 1996;174:141–156. doi: 10.1093/infdis/174.1.141. [DOI] [PubMed] [Google Scholar]
- 49.Tsolaki A G, Miller R F, Wakefield A E. Oropharyngeal samples for genotyping and monitoring response to treatment in AIDS patients with Pneumocystis carinii pneumonia. J Med Microbiol. 1999;48:897–905. doi: 10.1099/00222615-48-10-897. [DOI] [PubMed] [Google Scholar]
- 50.Vargas S L, Hughes W T, Wakefield A E, Oz H S. Limited persistence in and subsequent elimination of Pneumocystis carinii from the lungs after P. carinii pneumonia. J Infect Dis. 1995;172:506–510. doi: 10.1093/infdis/172.2.506. [DOI] [PubMed] [Google Scholar]
- 51.Vasquez J, Smulian A G, Linke M J, Cushion M T. Antigenic differences associated with genetically distinct Pneumocystis carinii from rats. Infect Immun. 1996;64:290–297. doi: 10.1128/iai.64.1.290-297.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wakefield A E. Detection of DNA sequences identical to Pneumocystis carinii in samples of ambient air. J Eukaryot Microbiol. 1994;41:116S. [PubMed] [Google Scholar]
- 53.Wakefield A E. DNA sequences identical to Pneumocystis carinii f. sp. carinii and Pneumocystis carinii f. sp. hominis in samples of air spora. J Clin Microbiol. 1996;34:1754–1759. doi: 10.1128/jcm.34.7.1754-1759.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wakefield A E. Genetic heterogeneity in Pneumocystis carinii: an introduction. FEMS Immunol Med Microbiol. 1998;22:5–13. doi: 10.1111/j.1574-695X.1998.tb01182.x. [DOI] [PubMed] [Google Scholar]
- 55.Wakefield A E, Hopkin J M, Burns J, Hipkiss J B, Stewart T J, Moxon E R. Cloning of DNA from Pneumocystis carinii. J Infect Dis. 1988;158:859–862. [PubMed] [Google Scholar]
- 56.Wakefield A E, Keely S P, Stringer J R, Christensen C B V, Ahrens P, Peters S E, Bille-Hansen V, Henriksen S A, Jorsal S E, Settnes O P. Identification of porcine Pneumocystis carinii as a genetically distinct organism by DNA amplification. APMIS. 1997;105:317–321. [PubMed] [Google Scholar]
- 57.Wakefield A E, Pixley F J, Banerji S, Sinclair K, Miller R F, Moxon E R, Hopkin J M. Amplification of mitochondrial ribosomal RNA sequences from Pneumocystis carinii DNA of rat and human origin. Mol Biochem Parasitol. 1990;43:69–76. doi: 10.1016/0166-6851(90)90131-5. [DOI] [PubMed] [Google Scholar]
- 58.Wakefield A E, Pixley F J, Banerji S, Sinclair K, Miller R F, Moxon E R, Hopkin J M. Detection of Pneumocystis carinii with DNA amplification. Lancet. 1990;336:451–453. doi: 10.1016/0140-6736(90)92008-6. [DOI] [PubMed] [Google Scholar]
- 59.Walzer P D, Foy J, Steele P, Kim C K, White M, Klein R S, Otter B A, Allegra C. Activities of antifolate, antiviral, and other drugs in an immunosuppressed rat model of Pneumocystis carinii pneumonia. Antimicrob Agents Chemother. 1992;36:1935–1942. doi: 10.1128/aac.36.9.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weisbroth S H, Geistfeld J, Weisbroth S P, Williams B, Feldman S H, Linke M J, Orr S, Cushion M T. Latent Pneumocystis carinii infection in commercial rat colonies: comparison of inductive immunosuppressants plus histopathology, PCR, and serology as detection methods. J Clin Microbiol. 1999;37:1441–1446. doi: 10.1128/jcm.37.5.1441-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zissner H. Rats, lice and history. London, United Kingdom: George Routledge and Sons; 1935. [Google Scholar]