Abstract
Selenium is both an essential and a toxic trace element, and the range of concentrations between the two is extremely narrow. Although tellurium is not essential and is only rarely found in the environment, it is considered to be extremely toxic. Several hypotheses have been proposed to account for the toxic effects of selenite and tellurite. However, these potential mechanisms have yet to be fully substantiated. Through screening of an Escherichia coli luxAB transcriptional gene fusion library, we identified a clone whose luminescence increased in the presence of increasing concentrations of sodium selenite or sodium tellurite. Cloning and sequencing of the luxAB junction revealed that the fusion had occurred in a previously uncharacterized open reading frame, termed o393 or yhfC, which we have now designated gutS, for gene up-regulated by tellurite and selenite. Transcription from gutS in the presence of selenite or tellurite was confirmed by RNA dot blot analysis. In vivo expression of the GutS polypeptide, using the pET expression system, revealed a polypeptide of approximately 43 kDa, in good agreement with its predicted molecular mass. Although the function of GutS remains to be elucidated, homology searches as well as protein motif and secondary-structure analyses have provided clues which may implicate GutS in transport in response to selenite and tellurite.
Selenium, as a part of the 21st amino acid, selenocysteine, which is present in numerous enzymes, is essential to all living cells (4, 8, 36). However, at elevated concentrations, selenium is also extremely toxic, and for Escherichia coli the MIC has been observed to be approximately 20 μg of sodium selenite/ml (J. Guzzo, unpublished results). Although tellurium is not an essential nutrient and is relatively rare in the environment, it is also considered to be extremely toxic, and clinical manifestations of toxicity are observed at very low concentrations (39). The MIC for E. coli strains lacking tellurite resistance determinants is approximately 1 μg of sodium tellurite/ml (40). Selenium and tellurium are both semiconductors, and excitation with electromagnetic radiation causes even greater increases in conductivity, making them useful in all photoelectric-based equipment. Furthermore, they are also commonly added to materials to increase their machinability and resistance to heat. Thus, the broad applicability of selenium and tellurium has resulted in a significant increase in production of these elements in the last several decades (22, 40).
Very little is known about the mechanism(s) of toxicity of either of these elements. However, there is mounting evidence that their toxicity in both prokaryotes and eukaryotes relates to their prooxidant capacity. For example, in the reduction of selenite and tellurite by reduced glutathione and other thiols, superoxide (and, in some cases, hydrogen peroxide) can be generated (19, 31, 35, 41). Thus, at least in the case of selenite, toxicity is thought to become manifest when the prooxidant conditions exceed cellular antioxidant defenses (35), and it appears that this may also be the case for toxicity caused by tellurite. Much less information is available regarding the potential mechanism(s) of tellurite toxicity. However, it has been postulated that at elevated concentrations, tellurite may replace sulfur in various proteins, rendering them nonfunctional (43).
In an attempt to understand the potential mechanism(s) of toxicity, our laboratory has utilized luciferase reporter gene fusions to search for and identify a gene(s) whose expression is increased or decreased in the presence of environmental toxins (16). The reporter system we chose to use consists of the promoterless luxAB genes from Vibrio harveyi, which were randomly inserted into the Escherichia coli chromosome in single copies by using a modified Tn5 transposon (16). The resulting library of clones was then screened in the absence of and in the presence of increasing concentrations of sodium selenite, and a clone (strain LF20116) in which luxAB was inserted within a gene whose apparent expression increased in the presence of increasing concentrations of sodium selenite was identified (17). We determined that luminescence from this clone also increased in the presence of tellurite, and we report here on the identification and characterization of the tellurite- and selenite-inducible gene and its putative role in the metabolism of selenite and tellurite.
MATERIALS AND METHODS
Bacterial strains, phages, and media.
Escherichia coli DH1 (F− recA endA1 gyrA96 thi hsdR17 [rk−, mk−] supE44 relA1) (18) was used to prepare the library of luxAB transcription fusion clones (16). Strain NM522 [supE thi Δ(lac-proAB) Δhsd-5 (rk−, mk−) (F′ proAB lac1q ZΔM15)] (15) was used for subcloning the chromosomal DNA from gene fusions originally present in strain DH1. Strain MG1655 (F−) (21) was used to clone the wild-type gutS gene. Strain LF20116 is strain DH1 with a Tn5-luxAB element inserted in the gutS gene (17). Strain LF25005 is strain MG1655 with a Tn5-luxAB element inserted in the gutS gene. Strain BL21(DE3) (hsdS gal [λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1]) (37) was used for expression of the gutS polypeptide from plasmid pJUL8. Phage P1vir was a kind gift from Rick Stewart (University of Maryland, College Park).
Bacterial strains were routinely propagated at 37°C, unless otherwise indicated, in Luria-Bertani (LB) broth or on LB plates containing 1.5% agar, as described by Miller (25), and supplemented with antibiotics as mentioned. Ampicillin was used at a final concentration of 40 μg/ml, and tetracycline was used at final concentrations of 10 μg/ml in broth and 20 μg/ml in plate media. Sodium selenite (21.9-mg/ml stock solution; Anachemia Canada Inc., Montréal, Quebec, Canada), prepared in sterile deionized water, was added to achieve various final concentrations of selenite in the culture media. Sodium tellurite (1.74-mg/ml stock solution; Anachemia Canada Inc.), prepared in sterile deionized water, was added to achieve various final concentrations of tellurite in the media.
DNA manipulations.
Restriction endonuclease hydrolyses were performed in a solution containing 6 mM Tris-HCl (pH 7.5), 6 mM MgCl2, 75 mM NaCl, 6 mM β-mercaptoethanol, and 0.25 mg of bovine serum albumin/ml at 37°C for 2 h, using 3 U of enzyme per μg of DNA. Ligations were performed at 15°C for 18 h in T4 DNA ligase buffer (Gibco BRL, Burlington, Ontario, Canada), using 2 U of T4 DNA ligase (Gibco BRL) per μg of DNA. DNA was subjected to electrophoresis in 0.75% agarose gels, unless otherwise indicated, in TAE buffer (40 mM Tris-acetate [pH 8.0], 1 mM EDTA). Size-selected DNA fragments were purified by using a Geneclean II kit (Bio101, Mississauga, Ontario, Canada) according to the manufacturer's directions.
Small-scale plasmid DNA isolations were performed by the alkaline lysis procedure described by Sambrook et al. (29). Large-scale plasmid DNA isolations were performed from 1-liter LB cultures, and plasmid amplifications were performed at a cell density of 4 × 108/ml by treatment with a 75-μg/ml chloramphenicol solution for 16 h at 32°C. DNA was extracted by using the cleared-lysate technique of Clewell and Helinski (10) followed by ultracentrifugation in cesium chloride-ethidium bromide gradients (24). DNA transformations were performed by the rubidium chloride method as described by Hanahan (18). Two hundred nanograms of plasmid DNA was routinely used for transformation.
Isolation of total cellular DNA.
Ten milliliters of a cell culture grown for 18 h was pelleted, and the cells were resuspended in 1.4 ml of 100 mM Tris-HCl–10 mM EDTA (pH 8.0). Sodium dodecyl sulfate (SDS; 10%, wt/vol) and RNase A (1 mg/ml in 10 mM Tris-HCl, pH 7.6) were added to final concentrations of 0.53% (wt/vol) and 0.21 mg/ml, respectively, and the mixture was incubated at 37°C for 2 h. Pronase (20 mg/ml in 10 mM Tris-HCl, pH 7.6) was then added to a final concentration of 1.9 mg/ml, and the mixture was incubated at 37°C for 2 h. The DNA was purified by two phenol and three ether extractions and precipitated with 70% ethanol. The flocculent DNA fibers were immediately isolated with a micropipette, air dried, and resuspended in a total volume of 50 μl of 1× TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA).
Construction of strains LF25005 and LF25008.
Strain LF25005 was constructed by P1vir transduction of strain LF20116 into E. coli MG1655 and selection on plates containing tetracycline as described by Miller (25). The resultant tetracycline-resistant clones were tested for luminescence induction in the presence of sodium selenite, and the location of the Tn5-luxAB element was confirmed by Southern blot analysis using the luxAB genes as a probe as described in Materials and Methods. Strain LF25008 was constructed by transformation of plasmid pJUL8 into strain BL21(DE3) as previously described.
Amplification of the gutS gene by PCR.
Synthetic oligonucleotide primers used for amplification of the 1,268-bp DNA fragment containing the gutS gene, 5′-CTCGAGCATATGACTAACAGCAATCGCATCAAGC-3′ and 5′-GGGCTCGAGGCACGTAGCGGGGGAAGAGAG-3′, were purchased from the Sheldon Biotechnology Center, McGill University. XhoI and NdeI sites were engineered into the 5′ end of the upstream primer, and a XhoI site was engineered into the 3′ end of the downstream primer. PCR amplification of the 1,268-bp gutS sequence was carried out in a reaction mixture (50 μl) containing 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 1.5 mM MgCl2, 1 mg of bovine serum albumin/ml, 400 μM (each) deoxynucleoside triphosphate, 0.2 mM each primer, 100 ng of E. coli MG1655 DNA, and 2.5 U of Taq polymerase (Gibco BRL). The mixture was overlaid with mineral oil and placed in a model PTC100 temperature cycler (MJ Research Inc., Waltham, Mass.) programmed to carry out the following cycles: first cycle, 94°C for 2 min, 55°C for 5 min, and 72°C for 10 min; subsequent 24 cycles, 94°C for 1 min, 58°C for 1 min, and 72°C for 3 min; and last cycle, 94°C for 1 min, 58°C for 1 min, and 72°C for 15 min. The amplified DNA was phenol and ether extracted, ethanol precipitated, and resuspended in 50 μl of 1× TE buffer.
Construction of plasmids.
To isolate the junction and downstream portion of the selenite-inducible gene, 4.0 μg of BamHI-cleaved total cellular DNA isolated from strain LF20116 (the luxAB gene fusion strain inducible by selenite and tellurite [see Results]) was ligated to 1.0 μg of BamHI-cleaved pUC119 (42) and transformants were selected on tetracycline-medium (16). This plasmid was designated pJUL2. The 1.7-kb BamHI-PstI fragment from pJUL2 (0.3 μg) was then ligated to 0.15 μg of pUC119 (42) also cleaved with BamHI-PstI, transformed into E. coli NM522 (15), and selected on medium containing ampicillin. The resulting plasmid was designated pJUL5.
To clone and express the GutS polypeptide, plasmid pJUL7 was constructed by ligating the 1,268-bp PCR product described above (with XhoI sites engineered at the 5′ end of the upstream primer and the 3′ end of the downstream primer) into XhoI-digested pBluescript SK(−) (33). Plasmid pJUL8 was cloned as a 1,262-bp NdeI-XhoI fragment from pJUL7 into NdeI- and XhoI-hydrolyzed pET29b (Novagen). Thus, the ATG in the NdeI site acts as the gutS translational start site for polypeptide expression.
Luminescence assay.
The assay for light emission was modified from that described by Miyamoto et al. (26). Briefly, 1-cm2 patches of cells were grown overnight on an LB agar plate. Upon addition of 100 μl of dodecyl aldehyde (Aldrich Chemical Co. Inc., Milwaukee, Wis.) to the petri dish covers, the dishes were placed in an inverted position and exposed to Kodak XAR-5 film (Eastman Kodak, Rochester, N.Y.) at 23°C. The X-ray films were developed; the resulting spots on the films were scanned, and their densities were quantified by using the Imagequant software program (Molecular Dynamics, Sunnyvale, Calif.).
DNA sequencing.
To determine the sequence at the junction between IS50R and the chromosomal DNA in pJUL2, an oligonucleotide (5′-AAGGTTCCGTTCAGGAC-3′) corresponding to bp 1497 to 1513 of IS50R (1) was synthesized (Sheldon Biotechnology Center) and used as a primer. To determine the sequence at the BamHI site in the chromosomal DNA, the BamHI-PstI fragment of plasmid pJUL2 (containing the tetracycline resistance gene, IS50R, and adjacent chromosomal DNA sequences) was cloned into BamHI- and PstI-cleaved pUC119, resulting in plasmid pJUL5, and the −40 primer provided in the Sequenase kit was used according to the manufacturer's (U.S. Biochemical Corp., Cleveland, Ohio) directions. Sequencing was performed by isolating single-stranded phagemid DNA from plasmid pJUL5 as described by Vieira and Messing (42). Dideoxy sequencing reactions with [α-35S]dATP (500 Ci/mmol; DuPont Canada Inc., Mississauga, Ontario, Canada) were performed with a Sequenase kit according to the manufacturer's protocol.
Southern blotting.
Total cellular DNA from strain LF20116 was isolated as previously described. Ten micrograms of chromosomal DNA was hydrolyzed with BamHI, EcoRI, HindIII, SacI, or SalI. The hydrolyzed DNA was then subjected to electrophoresis through a 0.75% agarose gel and blotted to Hybond-N nylon membranes (Amersham Ltd., Oakville, Ontario, Canada) saturated in 20× SSC (3 M NaCl, 0.3 M sodium citrate; pH 7.0) as described by Sambrook et al. (29). Blotted DNA was cross-linked to the membrane by using a UV Stratalinker 1800 cross-linker (Stratagene, La Jolla, Calif.) and used in the subsequent hybridization reaction.
The DNA fragment used to probe strain LF20116 was isolated from plasmid pFUSLUX as a 3.25-kb BamHI fragment containing the luxAB genes (16) as previously described. Subsequently, the probe was antigenically labeled by using a DIG Labelling and Detection Kit (Boehringer Mannheim, Laval, Quebec, Canada) as described by the manufacturer. Hybridization, washing, antibody binding, and chemiluminescence reactions were also performed according to the manufacturer's instructions. Finally, the membranes were exposed to Kodak XAR-5 film under DuPont Cronex intensifying screens to visualize the hybridized probe.
Expression of the GutS polypeptide.
Plasmid pJUL8 was used to transform E. coli BL21(DE3) (37) as previously described; this was followed by selection for kanamycin-resistant transformants. The in vivo expression assay was carried out by isopropyl-β-d-thiogalactopyranoside (IPTG) induction of the PT7 promoter as described by Studier et al. (38). Briefly, overnight cultures of strain BL21(DE3) containing plasmid pET29b or plasmid pJUL8 were diluted 50-fold in fresh LB broth. When an A550 of 0.5 was reached, 1 ml of cell culture was centrifuged at 14,000 × g for 1 min and the pelleted cells were resuspended in 100 μl of SDS sample buffer (50 mM Tris-HCl [pH 8.0], 2% [wt/vol] SDS, 0.1% [wt/vol] bromphenol blue, 10% [vol/vol] glycerol) containing 0.1 M dithiothreitol. Samples were subsequently incubated 5 min at 95°C and then centrifuged 5 min at 14,000 × g, and the supernatant fluid was collected. Expression from PT7 was induced by addition of a final concentration of 0.4 mM IPTG. After 2 h of IPTG induction, 1 ml of cell culture was treated as described above. The gene products were visualized by Coomassie blue staining after electrophoresis through an SDS–15% polyacrylamide gel as described by Sambrook et al. (29).
Isolation of total cellular RNA.
Total cellular RNA was isolated from selenite-induced cells by a CsCl purification procedure modified from that of Glisin et al. (14) as described by Deretic et al. (11). Briefly, overnight cultures of E. coli MG1655 were diluted into fresh medium. After an initial growth period of 2 h, sodium selenite was added to cultures to obtain final concentrations of 0, 1.0, 2.5, 5.0, 10.0, and 20.0 μg of selenite/ml. Alternately, sodium tellurite was added to cultures to obtain final concentrations of 0, 0.01, 0.05, 0.1, 0.25, and 0.5 μg of tellurite/ml. Cell cultures were allowed to incubate for an additional 2.5 h. Subsequently, the cultures were rapidly cooled in a dry-ice–ethanol bath and subjected to centrifugation in a Sorvall GSA rotor for 10 min at 6,000 × g and 4°C. The cell pellets were each washed with 5 ml of ice-cold lysis buffer (50 mM Tris-HCl, pH 7.0) and resuspended in another 5-ml volume of lysis buffer at room temperature. One milliliter of a 20% (wt/vol) SDS solution was added to each lysed pellet, and the mixtures were incubated at 65°C for 5 min. Solid CsCl (4 g) was dissolved in, and an additional 5 ml of lysis buffer was added to each sample. After the suspensions were mixed, the cell debris was removed by centrifugation in a Sorvall SM-24 rotor for 10 min at 11,000 × g and room temperature. The clear supernatant liquids were removed, and each was layered over a 2-ml cushion of 5.7 M CsCl. RNA was pelleted by centrifugation at 35,000 rpm and 15°C in a Beckman SW50.1 rotor for 18 h. RNA pellets were each resuspended in 300 μl of water (diethylpyrocarbonate [DEPC] treated), extracted with chloroform-isoamyl alcohol (24:1, vol/vol), and precipitated by adding 0.1 volume of 3 M sodium acetate (DEPC treated) and 2.5 volumes of ethanol. RNA samples were aliquoted and stored at −70°C. The RNA concentration of each sample was spectrophotometrically determined by using a UV-1201 spectrophotometer (Shimadzu Scientific Instruments Inc., Kyoto, Japan) at 260 nm.
RNA dot blotting.
Total cellular RNAs (2.5 and 5.0 μg) from selenite-induced and unexposed E. coli MG1655 cells were loaded onto a Hybond-N nylon membrane (Amersham Ltd.) in a slot blot apparatus (Bio-Rad, Mississauga, Ontario, Canada) as previously described (29). The RNA was fixed to the membrane by using a UV Stratalinker 1800 cross-linker. The gutS probe was α-32P labeled as previously described (12), and hybridization reactions were performed as described by Cai and DuBow (9). The membrane was washed to remove unhybridized probe and exposed to Kodak XAR-5 film as described by Cai and DuBow (9).
RESULTS
Identification of a gene whose luminescence is increased in the presence of selenite or tellurite.
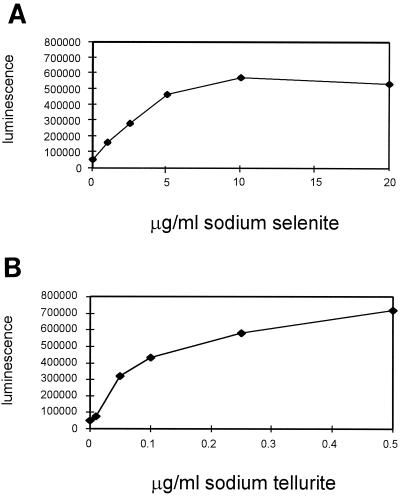
Luminescence from each clone of the luxAB gene fusion library was monitored in the presence of sodium selenite at concentrations of 2.5 and 10.0 μg/ml (17). One clone, designated LF20116, was shown to contain the luxAB insertion within a gene whose luminescence increased in the presence of increasing concentrations of sodium selenite (17). Light emission from this clone was further studied by measurement in the presence of sodium selenite at 0, 1.0, 2.5, 5.0, 10.0, and 20.0 μg/ml; an 11.2-fold increase over basal luminescence (0 μg of additional selenite/ml) was observed at a 10-μg/ml concentration of sodium selenite (Fig. 1A). Because tellurium is just below selenium on the periodic table, we tested the luminescence of this clone after exposure to increasing concentrations of sodium tellurite. This clone was also found to exhibit increased luminescence (approximately 14-fold above its basal level) in the presence of sodium tellurite at 0, 0.01, 0.05, 0.1, 0.25, and 0.5 μg/ml (Fig. 1B). Concentrations of sodium tellurite higher than 0.5 μg/ml could not be assayed because too little growth was observed. Luminescence was also confirmed after P1vir transduction of the Tn5-luxAB element into E. coli MG1655, resulting in strain LF25005 (data not shown), suggesting that the strain's genetic background has little effect on these results. Finally, light emission from LF20116 and LF25005 was also measured in the presence of selenate, sulfate, sulfite, phosphate, zinc, nickel, arsenate, antimonite, seleno-dl-methionine, and 6-selenoguanosine individually. However, no significant increases or decreases in luminescence were observed (data not shown).
FIG. 1.
Light emission from strain LF20116 in the presence of sodium selenite at 0, 0.1, 1.0, 2.5, 5.0, 10.0, or 20.0 μg/ml (A) or sodium tellurite at 0, 0.01, 0.05, 0.1, 0.25, or 0.5 μg/ml (B). Quantification of light emission was performed as described in Materials and Methods. The results shown are the averages of data from three experiments. Standard errors are below 1% and hence are not shown.
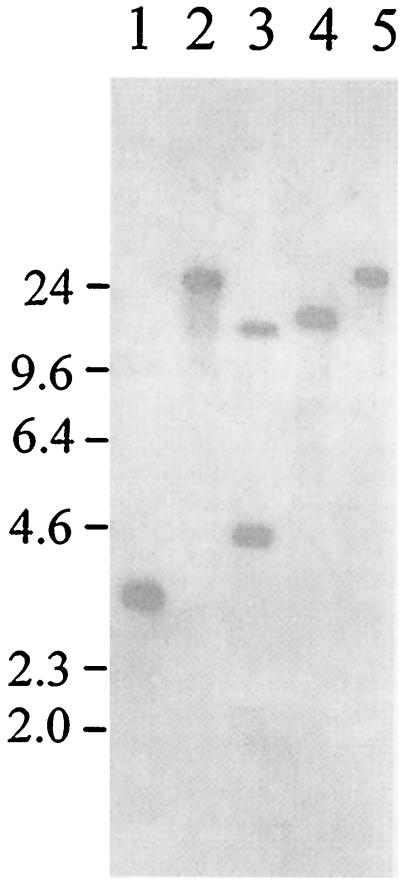
The Tn5-luxAB insertion in strain LF20116 is present in a single copy.
To verify that the Tn5-luxAB element had integrated in a single location in the E. coli chromosome, the number of chromosomal insertions was determined. Upon digesting total chromosomal DNA from strain LF20116 with EcoRI, BamHI, HindIII, SacI, or SalI; blotting; and probing with the luxAB genes, it was shown that the Tn5-luxAB element was integrated at a single chromosomal location (Fig. 2). For example, a single band was observed upon BamHI, HindIII, SacI, or SalI digestion, and two bands were observed following EcoRI digestion, one representing an internal EcoRI fragment and one representing the segment spanning the EcoRI site in luxAB to the adjacent chromosomal EcoRI site (Fig. 2). The maps derived from these results also allowed confirmation of the insertion site of the Tn5-luxAB element in the E. coli chromosome (see below).
FIG. 2.
Southern blot analysis of strain LF20116 hydrolyzed with BamHI (lane 1), HindIII (lane 2), EcoRI (lane 3), SalI (lane 4), or SacI (lane 5). The blots were probed with the luxAB genes as described in Materials and Methods. The migration positions of molecular mass markers are shown on the left (in kilodaltons).
Cloning and mapping of the selenite- and tellurite-inducible gene.
The junction and downstream portion of the selenite- and tellurite-inducible gene were isolated in a single step by hydrolyzing total cellular DNA from strain LF20116 with BamHI, ligating the resultant fragments to BamHI-cleaved pUC119, transforming the plasmids into E. coli NM522, and selecting tetracycline-resistant colonies. Only clones that contain the Tcr cassette, IS50R, and the chromosomal sequence between IS50R and the adjacent BamHI site should survive this selection. Plasmid pJUL2 was cloned in this manner. Subsequently, plasmid pJUL2 was hydrolyzed with BamHI and PstI and subcloned into plasmids pUC119 and pUC120 hydrolyzed with the same enzymes, resulting in plasmids pJUL5 and pJUL6.
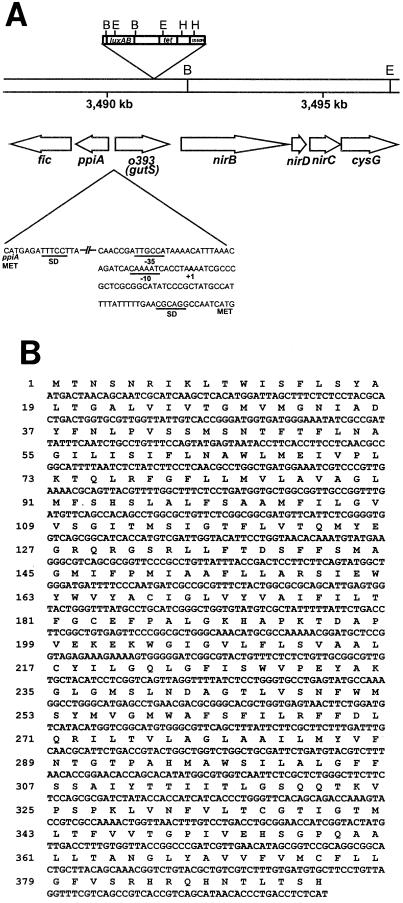
A synthetic oligonucleotide corresponding to bp 1497 to 1513 of IS50R was used as a primer to sequence a portion of IS50R and the adjacent chromosomal DNA from plasmid pJUL2. The resulting sequence was analyzed by a BLAST homology search and was found to correspond to a previously uncharacterized open reading frame, designated o393 or yhfC (Fig. 3). Gene yhfC is located at 75.2 map units and 3,492 kbp on the E. coli chromosome. Because of the tellurite- and selenite-inducible expression of the yhfC “orphan” gene (28), we have named this gene gutS, for gene up-regulated by tellurite and selenite.
FIG. 3.
(A) Location of the gutS gene on the E. coli chromosome. BamHI (B), HindIII (H), and EcoRI (E) restriction endonuclease sites are shown. The promoter region predicted by Genemark is illustrated with corresponding ς70 −35 and −10, Shine-Dalgarno (SD), and translational start site (MET) sequences. (B) Amino acid (one-letter code) and nucleotide sequences of the gutS gene.
Analysis of transcription from the selenite- and tellurite-inducible gene.
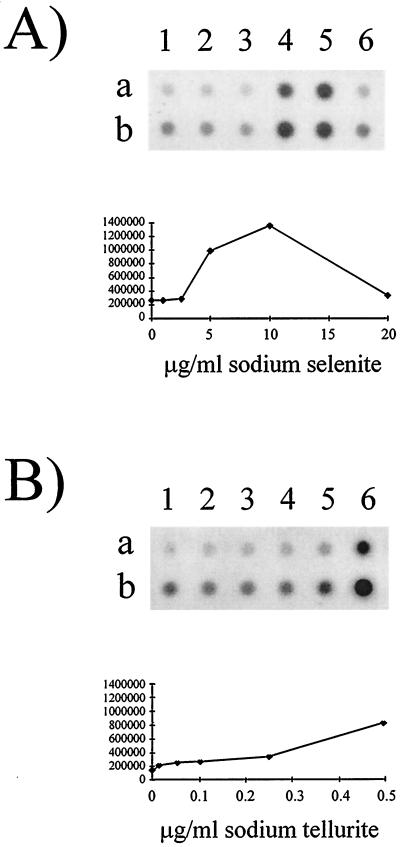
To confirm that the luminescence results correlate with the fact that the level of transcription from the selenite- and tellurite-inducible gene increases in the presence of these elements, RNA dot blot analyses were performed with the gutS gene to probe total RNA from E. coli MG1655 cells grown in the presence of 0, 1.0, 2.5, 5.0, 10.0, or 20.0 μg of sodium selenite/ml (Fig. 4A) or 0, 0.01, 0.05, 0.1, 0.25, or 0.5 μg of sodium tellurite/ml (Fig. 4B). Quantification of hybridized RNA using the Imagequant software program (Molecular Dynamics) confirmed that transcription of gutS does increase (approximately fivefold from its basal level) in the presence of either selenite or tellurite (Fig. 4).
FIG. 4.
RNA dot blot analysis of total cellular RNA extracted from E. coli MG1655 cells grown in the presence of sodium selenite at 0 (lane 1), 1.0 (lane 2), 2.5 (lane 3), 5.0 (lane 4), 10.0 (lane 5), or 20.0 (lane 6) μg/ml (A) or sodium tellurite at 0 (lane 1), 0.01 (lane 2), 0.05 (lane 3), 0.1 (lane 4), 0.25 (lane 5), or 0.5 (lane 6) μg/ml (B). Rows a and b contain 2.5 and 5.0 μg of blotted RNA, respectively. The amount of hybridization was quantified from three experiments using 2.5 μg of blotted RNA as described in Materials and Methods. Standard errors are below 1% and hence are not shown.
Visualization of the GutS polypeptide.
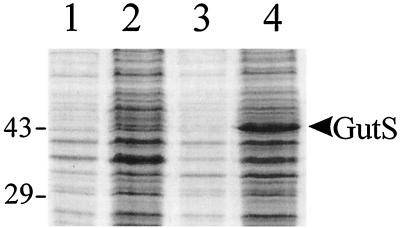
Denaturing SDS-polyacrylamide gel electrophoresis was used to visualize polypeptides expressed from E. coli BL21(DE3) cells containing plasmid pET29b and plasmid pJUL8. A polypeptide with a molecular mass of approximately 43 kDa was observed in protein extracts from E. coli BL21(DE3) cells containing plasmid pJUL8, but not in extracts from BL21(DE3) cells containing pET29b (Fig. 5). This molecular mass is consistent with that of the predicted protein as calculated by the SWISS-PROT program (3). Quantification of the GutS protein was not performed because these levels were not indicative of the presence of endogenous GutS in the cell in response to selenite.
FIG. 5.
In vivo expression of the GutS polypeptide in expression plasmid pET29b. Lanes: 1, strain BL21(DE3) containing plasmid pET29b; 2, plasmid pET29b following IPTG induction for 2 h; 3, plasmid pJUL8; 4, plasmid pJUL8 following IPTG induction for 2 h. The migration positions of molecular mass markers are shown on the left (in kilodaltons).
DISCUSSION
Using a luxAB transcriptional gene fusion library, we have identified a novel gene (designated gutS) whose expression is inducible in the presence of 0, 1.0, 2.5, 5.0, or 10.0 μg of sodium selenite/ml (Fig. 1A). Furthermore, this clone also exhibited increased light emission in the presence of 0, 0.01, 0.05, 0.1, 0.25, or 0.5 μg of sodium tellurite/ml (Fig. 1B). Maximal light emission from these clones was observed at sodium selenite and sodium tellurite concentrations of 10.0 and 0.5 mg/μl, respectively. Furthermore, almost no growth was observed at sodium tellurite concentrations higher than 0.5 μg/ml, illustrating the extremely toxic nature of this element and the different effects of the two elements on the same cells (39). Consequently, cell growth, as well as light emission from these clones, could not be measured at tellurite concentrations higher than those shown here. This transcriptional response appears to be specific to these compounds, since no increases or decreases in luminescence were observed following exposure to selenate, sulfate, sulfite, phosphate, zinc, nickel, arsenate, antimonite, seleno-dl-methionine, or 6-selenoguanosine (data not shown).
To ensure that the Tn5-luxAB element had been integrated at a single location in this clone, the number of chromosomal insertions was determined via Southern blotting and probing with the luxAB genes (Fig. 2). The results show a single Tn5-luxAB insertion within strain LF20116, as demonstrated by a single band obtained following BamHI, HindIII, SalI, or SacI digestion and two bands obtained following EcoRI digestion, owing to an EcoRI restriction site within the luxAB genes (Fig. 2).
To confirm that the gutS gene exhibited increased transcription in the presence of selenite or tellurite, RNA dot blot analyses were performed on total cellular RNA from unexposed cells as well as selenite- or tellurite-exposed cells. An approximate fivefold increase in transcription was demonstrated in the presence of sodium selenite at 10.0 μg/ml (Fig. 4A) or sodium tellurite at 0.5 μg/ml (Fig. 4B). These results correlate with the increases in luminescence observed in the presence of selenite and tellurite. Consistent with the luminescence response, maximal gutS expression was detected at concentrations of 10.0 and 0.5 μg/ml for sodium selenite and sodium tellurite, respectively (Fig. 4), although higher concentrations of tellurite were not tested to determine maximal induction. Hence, although these responses are concordant, a lack of linear correlation between luciferase activity from reporter gene fusions and transcription measured by RNA analyses is frequently observed (9, 13).
Upon sequencing a portion of chromosomal DNA adjacent to the Tn5-luxAB insertion in strain LF20116, the insertion was found to be located at bp 3491247, within a previously uncharacterized open reading frame, called o393 or yhfC, which we have now designated gutS (Fig. 3). This gene is located at 75.2 map units and bp 3490240 on the E. coli chromosome. This open reading frame (Fig. 3B) appears to represent a single-cistron operon, because the open reading frame located upstream of gutS, ppiA, is transcribed in the opposite direction and the promoter of the downstream nirB operon, encoding an NADPH-dependent nitrate reductase, has been well characterized (20). The gutS promoter appears to be a ς70 promoter, as illustrated in Fig. 3. The GutS polypeptide was visualized by SDS-polyacrylamide gel electrophoresis, and a 43-kDa protein, consistent with the size predicted by SWISS-PROT (3), was observed (Fig. 5).
The DNA sequence corresponding to the gutS gene was also used as a probe in Southern blot analysis, and homologous sequences were identified in Citrobacter freundii, Shigella flexneri, Salmonella typhimurium, and Enterobacter cloacae, but not in Proteus mirabilis or members of the genera Pseudomonas, Burkholderia, and Caulobacter (data not shown). To date, only one closely related homolog (exhibiting 79.9% sequence identity and 95.7% sequence similarity), in Yersinia pestis, has been identified by BLAST and FASTA database searches (data not shown). These database searches also revealed that GutS exhibits significant homology to numerous subcellular integral membrane transport proteins in the secondary transporter class of proteins, including drug efflux proteins as well as sugar transporters such as the glucose/galactose transporter (SWISS-PROT accession no. Q44623) of Brucella abortus (27). Furthermore, secondary-structure analysis of the GutS polypeptide using the PROSITE software (2) revealed 12 predicted transmembrane domains along with a sugar transport protein signature (STPS2) motif, highly conserved among the secondary transporter class of proteins (27). Although this consensus sequence is common in glucose transporters, from which it gets its name, not all proteins belonging to this class are involved in sugar transport, and some members of this family have been found to transport inorganic phosphate, citrate, or aromatic amino acids (7, 30, 32, 44).
Little is known about the transport of selenite and tellurite into cells and within the cellular milieu. However, several lines of evidence suggest an as-yet-unidentified mechanism of transport for selenite. First, a mechanism of transport for sulfate has been previously shown to be responsible for transporting selenate and selenite (23, 34). However, the specificity and affinity of this transport system were higher for sulfate than for either of the two selenium compounds, as shown by competitive-uptake inhibition. Furthermore, although very little difference in specificity for either of the two selenium dianions was observed, a higher affinity was shown for selenate (23). A previous study of selenite assimilation by Salmonella typhimurium revealed that selenite was assimilated independently of sulfate and selenate. Selenite uptake could be detected both in wild-type cells repressed for sulfate transport and in mutants that lacked a functional sulfate permease. In contrast, selenate was assimilated by the same process as that used to assimilate sulfate (5). Thus, a specific mechanism for selenite transport is possible, since appreciable amounts of selenium-containing metabolites may be synthesized at sublethal concentrations of selenium, even when relatively high concentrations of sulfur are present (6).
Even less is known regarding the transport of tellurite. Although several tellurite resistance genes have been identified (both chromosomal and plasmid encoded) and one of the chromosomal determinants has been shown to be an integral membrane protein, it has also been shown that resistance (by either chromosomal or plasmid-encoded determinants) is not mediated by either reduced uptake or increased efflux (41). Rather, it has since been shown that tellurite can be detoxified through interactions with cellular thiols, such as glutathione, or by a methyltransferase-catalyzed reaction, although neither process appears to be involved in plasmid-mediated tellurite resistance, the mechanism of which remains to be elucidated (40). Thus, it is possible that several alternate mechanisms of selenite and tellurite transport exist. We have examined the sensitivity of strains LF20116 and LF25005 to increased concentrations of these compounds and have found no differences between their sensitivities and those of the parental strains (data not shown). Furthermore, we expressed GutS in a wild-type strain and found that there was also no change in sensitivity or resistance to selenite or tellurite (data not shown). It is thus apparent that if this protein is implicated in the export or import of selenite and/or tellurite, it is not the major protein involved.
Thus, we have identified in E. coli a previously uncharacterized orphan gene, which we now call gutS, whose expression is induced by sublethal concentrations of selenite or tellurite. The gutS gene product, a 43-kDa protein, has been shown to share homology and consensus patterns with subcellular membrane-located permeases and transport proteins. Future studies of the gutS gene product will focus on dissecting its role in selenite and tellurite metabolism and on the mechanism(s) underlying the selenite- and tellurite-mediated regulation of gutS gene expression. Such studies may provide important insights into the metabolism and toxicity associated with these compounds.
ACKNOWLEDGMENTS
This work was supported by an operating grant (OGP0003222) from the Natural Sciences and Engineering Research Council of Canada (NSERC).
We are grateful to M. A. Costanzo, C. Diorio, K. Salmon, and A. Guzzo for technical advice as well as many useful discussions and to the reviewers for insightful comments and suggestions.
REFERENCES
- 1.Auerswald E A, Ludwig G, Schaller H. Structural analysis of Tn5. Cold Spring Harbor Symp Quant Biol. 1980;45:107–113. doi: 10.1101/sqb.1981.045.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1991;19:2241–2245. doi: 10.1093/nar/19.suppl.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bairoch A, Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1991;19:2247–2249. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böck A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 5.Brown T A, Shrift A. Assimilation of selenate and selenite by Salmonella typhimurium. Can J Microbiol. 1980;26:671–675. doi: 10.1139/m80-117. [DOI] [PubMed] [Google Scholar]
- 6.Brown T A, Shrift A. Selective assimilation of selenite by Escherichia coli. Can J Microbiol. 1982;28:307–310. doi: 10.1139/m82-045. [DOI] [PubMed] [Google Scholar]
- 7.Bun-Ya M, Nishimura M, Harashima S, Oshima Y. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol. 1991;11:3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burk R F, Hill K E. Regulation of selenoproteins. Annu Rev Nutr. 1993;13:65–81. doi: 10.1146/annurev.nu.13.070193.000433. [DOI] [PubMed] [Google Scholar]
- 9.Cai J, DuBow M S. Expression of the Escherichia coli chromosomal ars operon. Can J Microbiol. 1996;42:662–671. doi: 10.1139/m96-091. [DOI] [PubMed] [Google Scholar]
- 10.Clewell D B, Helinski D R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc Natl Acad Sci USA. 1969;62:1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deretic V, Gill J F, Chakrabarty A M. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987;169:351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diorio C, Cai J, Marmor J, Shinder R, DuBow M S. An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J Bacteriol. 1995;177:2050–2056. doi: 10.1128/jb.177.8.2050-2056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsberg Å J, Pavitt G D, Higgins C F. Use of transcriptional fusions to monitor gene expression: a cautionary tale. J Bacteriol. 1994;176:2128–2132. doi: 10.1128/jb.176.7.2128-2132.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glisin V, Crkvenjakov R, Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974;13:2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- 15.Gough J, Murray N. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983;166:1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- 16.Guzzo A, DuBow M S. Construction of stable, single-copy luciferase gene fusions in Escherichia coli. Arch Microbiol. 1991;156:444–448. doi: 10.1007/BF00245390. [DOI] [PubMed] [Google Scholar]
- 17.Guzzo A, DuBow M S. Selenium-induced gene expression to create luminescent biosensors and to elucidate genetically programmed responses to selenium exposure. In: Allan R J, Nriagy J O, editors. 9th International Conference on Heavy Metals in the Environment. Vol. 1. Edinburgh, United Kingdom: CEP Consultants; 1993. pp. 407–410. [Google Scholar]
- 18.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 19.Ip C, Hayes C, Budnick R M, Ganther H E. Chemical form of selenium, critical metabolites, and cancer prevention. Cancer Res. 1991;51:595–600. [PubMed] [Google Scholar]
- 20.Jayaraman P S, Peakman T C, Busby S J W, Quincey R V, Cole J A. Location and sequence of the promoter of the gene for the NADH-dependent nitrite reductase of Escherichia coli and its regulation by oxygen, the Fnr protein and nitrite. J Mol Biol. 1987;196:781–788. doi: 10.1016/0022-2836(87)90404-9. [DOI] [PubMed] [Google Scholar]
- 21.Jensen K F. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabata-Pendias A. Geochemistry of selenium. J Environ Pathol Toxicol Oncol. 1998;17:173–177. [PubMed] [Google Scholar]
- 23.Lindblow-Kull C, Kull F J, Shrift A. Single transporter for sulfate, selenate, and selenite in Escherichia coli K-12. J Bacteriol. 1985;163:1267–1269. doi: 10.1128/jb.163.3.1267-1269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 26.Miyamoto C M, Graham A D, Boylan M, Evans J F, Hasel K W, Meighen E A, Graham A F. Polycistronic mRNAs code for polypeptides of the Vibrio harveyi luminescence system. J Bacteriol. 1985;161:995–1001. doi: 10.1128/jb.161.3.995-1001.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulsen I T, Sliwinski M K, Saier M H., Jr Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J Mol Biol. 1998;277:573–592. doi: 10.1006/jmbi.1998.1609. [DOI] [PubMed] [Google Scholar]
- 28.Rudd K E. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol Mol Biol Rev. 1998;62:985–1019. doi: 10.1128/mmbr.62.3.985-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sasatsu M, Misra T K, Chu L, Laddaga R, Silver S. Cloning and DNA sequence of a plasmid-determined citrate utilization system in Escherichia coli. J Bacteriol. 1985;164:983–993. doi: 10.1128/jb.164.3.983-993.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seko Y, Saito Y, Kitahara J, Imura N. Active oxygen generation by the reaction of selenite with reduced glutathione in vitro. In: Wendel A, editor. Selenium in biology and medicine. Berlin, Germany: Springer-Verlag; 1989. pp. 70–73. [Google Scholar]
- 32.Seol W, Shatkin A J. Escherichia coli kgtP encodes an alpha-ketoglutarate transporter. Proc Natl Acad Sci USA. 1991;88:3802–3806. doi: 10.1073/pnas.88.9.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Short J M, Fernandez J M, Sorge J A, Huse W D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sirko A, Hryniewicz M, Hulanicka D, Böck A. Sulfate and thiosulfate transport in Escherichia coli K-12: nucleotide sequence and expression of the cysTWAM gene cluster. J Bacteriol. 1990;172:3351–3357. doi: 10.1128/jb.172.6.3351-3357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spallholz J E. On the nature of selenium toxicity and carcinostatic activity. Free Radic Biol Med. 1994;17:45–64. doi: 10.1016/0891-5849(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 36.Stadtman T C. Selenocysteine. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 37.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 38.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct the expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 39.Taylor A. Biochemistry of tellurium. Biol Trace Elem Res. 1996;55:231–239. doi: 10.1007/BF02785282. [DOI] [PubMed] [Google Scholar]
- 40.Taylor D E. Bacterial tellurite resistance. Trends Microbiol. 1999;7:111–115. doi: 10.1016/s0966-842x(99)01454-7. [DOI] [PubMed] [Google Scholar]
- 41.Turner R J, Weiner J H, Taylor D E. The tellurite-resistance determinants tehAtehB and klaAklaBtelB have different biochemical requirements. Microbiology. 1995;141:3133–3140. doi: 10.1099/13500872-141-12-3133. [DOI] [PubMed] [Google Scholar]
- 42.Vieira J, Messing C M. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 43.Walter E G, Taylor D E. Plasmid-mediated resistance to tellurite: expressed and cryptic. Plasmid. 1992;27:52–64. doi: 10.1016/0147-619x(92)90006-v. [DOI] [PubMed] [Google Scholar]
- 44.Whipp M J, Camakaris H, Pittard A J. Cloning and analysis of the shiA gene, which encodes the shikimate transport system of Escherichia coli K-12. Gene. 1998;209:185–192. doi: 10.1016/s0378-1119(98)00043-2. [DOI] [PubMed] [Google Scholar]