Summary
The human gastrointestinal tract harbours an abundance of viruses, collectively known as the gut virome. The gut virome is highly heterogeneous across populations and is linked to geography, ethnicity, diet, lifestyle, and urbanisation. The currently known function of the gut virome varies greatly across human populations, and much remains unknown. We review current literature on the human gut virome, and the intricate trans-kingdom interplay among gut viruses, bacteria, and the mammalian host underlying health and diseases. We summarise evidence on the use of the gut virome as diagnostic markers and a therapeutic target. We shed light on novel avenues of microbiome-inspired diagnosis and therapies. We also review pre-clinical and clinical studies on gut virome-rectification-based therapies, including faecal microbiota transplantation, faecal virome transplantation, and refined phage therapy. Our review suggests that future research effort should focus on unravelling the mechanisms exerted by gut viruses/phages in human pathophysiology, and on developing phage-prompted precision therapies.
Keywords: Gut virome, Microbiome, Virus, Phage, Faecal virome transplantation, Phage therapy
Introduction
The human gastrointestinal (GI) tract extends from the oral cavity to the anus and harbours an immense number of microorganisms. The GI microorganism communities and their constituent genomes/genes range from the small intestine, large intestine, rectum, to faeces. These communities collectively known as the gut microbiome and the majority of them are bacteria and bacterial genes.1 The gut microbiome plays a crucial role in human immune development and homeostasis by interacting with epithelial and immune cells, and regulating metabolism (such as bile acid and short-chain fatty acids metabolism).2, 3, 4, 5 In addition to bacteria, a substantial quantity of viruses (that outnumber bacteria at a ratio of 1:1-10:1) co-colonise the human gut (grow or live in the GI tract), and are collectively referred to as the gut virome.6 The human virome is composed of eukaryotic and prokaryotic viruses, including viruses that infect human cells, viruses that infect microbes (such as bacteria, fungi, and archaea), and plant viruses that are primarily derived from environment and diet.7 Akin to the bacterial microbiome (bacteriome), the gut virome also plays an important role in the pathogenesis of disease, including inflammatory bowel disease (IBD),8 Clostridioides difficile infection (CDI),9 obesity,10 diabetes, SARS-CoV-2 infection,11,12 liver diseases,13 colorectal cancer (CRC),14 as well as malnutrition.15 Gut virome rectification has shown great potential as disease therapeutics through faecal microbiota transplantation (FMT), faecal virome transplantation (FVT), and phage therapy.9,16,17 All these therapies were reported to be highly efficacious in treating antibiotics-induced gut dysbiosis,18 recurrent CDI (rCDI),16 obesity,19 and bacterial infectious diseases.20,21 Taken together, these studies demonstrated that the gut virome is critical for disease pathogenesis and therapeutics, and may inspire novel therapeutic strategies based on the gut virome. FVT and phage therapy could provide benefits in clinical practice beyond contemporary microbiome therapeutics (including FMT, bacterial probiotics consumption, or antibiotics usage), which may introduce opportunistic pathogens and further distort the bacterial microbiome assembly linking to an unwanted dysbiosis and hence health concerns.
Owing to prior technological restraints in sequencing technology, screening, and culturomics, our recognition into the landscape of the human gut microbiome was deficient. General understanding of the human gut bacteriome has improved over the past decade, due to the rapid development of metagenomic sequencing (a next-generation sequencing approach that simultaneously sequences all genetic materials in a sample in a high-throughput manner), bioinformatic analyses, and molecular biology techniques. However, specific studies of the gut virome as significant component of the gut microbiome are lagging. Since the first glimpse of the diversity and richness of the human gut virome in 2003,22 there has been a rapid growth in peer-reviewed studies on the community composition and genetic composition of the human gut virome. As of January 2022, over 11,551 viral genome sequences and 4507 viral species have been deposited into the National Center of Biotechnology Information (NCBI) GenBank and Refseq databases. A recent study reported an updated collection of gut viral lineages titled the Metagenomic Gut Virus (MGV), which catalogued 189,680 viral draft genomes and estimated 54,118 viral species.23 In this review, we will summarise the current knowledge regarding the composition and function of the human gut virome and we will discuss its clinical implications and therapeutic applications.
The composition of the gut virome
The human virome has a site-specific composition across the different anatomical compartments of the human body, including blood, GI tract, respiratory tract, urogenital system, and skin.7 The quantity of gut viruses in an adult human has been estimated at a similar order of magnitude to gut bacteria, with over 1012 virus-like particles (VLPs) per person, proportional to individual body size.6 The GI tract harbours the highest number of viruses across the human body, and is estimated to have ∼109-1010 VLPs per gram of faeces.7 A healthy adult gut virome is composed of DNA viruses (both single-stranded DNA [ssDNA] and double-stranded DNA [dsDNA] viruses) and RNA viruses (both single-stranded RNA [ssRNA] and double-stranded RNA [dsRNA] viruses). Studies by us and those by others have consistently shown that humans have highly individualised gut virome configurations, that vary across geography, lifestyle, diet, and age.24,25
The human gut virome contains eukaryotic viruses (viruses that infect eukaryotic cells, primarily human cells in the gut) and prokaryotic viruses (viruses that infect prokaryotic cells, mostly bacteria), of which the eukaryotic viruses account for <10%.6 Eukaryotic DNA viruses (such as herpesviruses, anelloviruses and adenoviruses) are mostly latent and dormant in steady-state, calibrating host immunity.26 Meanwhile, eukaryotic RNA viruses are rare in the healthy state, and most intestinal eukaryotic RNA viruses are reported to be plant viruses.7,27 Pathogenic RNA viruses may appear in the gut when the human host is under infection-induced stress,12,28,29 and is a cause of significant health concern due to transmission via the faecal-oral route, contaminated food/water, and person-to-person contact. Generally, the RNA virome in the human gut is significantly less studied than the DNA virome, due to that RNA viruses appear to be less stable in samples compared to DNA viruses, causing difficulty in their identification by metagenomic sequencing.30 Compared to eukaryotic viruses, a substantially greater abundance of phages (also known as bacteriophages and bacterial viruses) are observed in the human gut, accounting for >90% of the gut virome (the phageome), whereas RNA phages are significantly less abundant (Figure 1).6 In recent studies, a higher diversity of RNA phages have been reported to be more widely distributed than previously recognised in several environmental niches, including animal faeces.30,31 Most of the known intestinal phages in the human gut are DNA encoded. The most abundant taxa in the gut virome are Caudovirales (dsDNA viruses) at the order level, followed by Microviridae (ssDNA viruses) at the family level.6,23 Only ∼56·6% of the gut virome could be annotated at the family and lower taxonomic levels, and the major unannotated viruses were predicted to belong to Caudovirales.23 The Caudovirales in the phylum Caudoviricetes was recently disbanded by the International Committee on Taxonomy of Viruses (ICTV) in 2021, and six new families (Ackermannviridae, Chaseviridae, Herelleviridae, Demerecviridae, Drexlerviridae and Autographiviridae) have been officially ratified to replace the previous Caudovirales order.32 Of note, the taxonomic entities mentioned in the present review are as reported in the original publication. CrAssphage is a prevalent phage family (Intestiviridae family) within the Crassvirales order (a new order created by ICTV in 2021). Diverse variants of crAssphage have been found in human faeces globally, accounting for ∼22%-90% of relative abundance (percentage of a microbial taxon in the whole gut microbial community) within the human gut virome.32,33 Although crAssphages are hypothesised to be stable colonisers in the human gut, their phenotypic linkages to human health and disease are still unclear.34
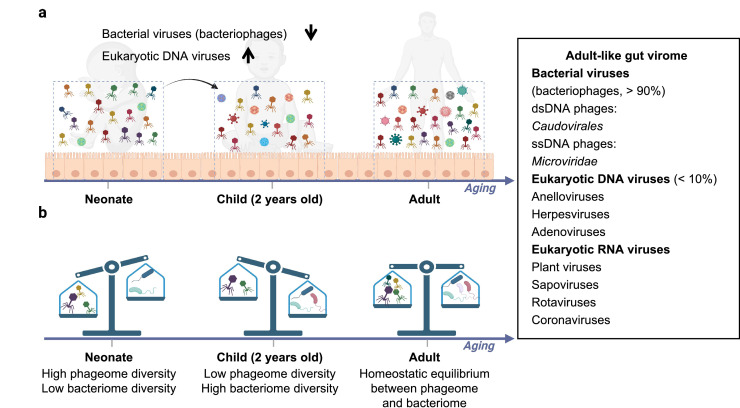
Figure 1.
The composition of the human gut virome. a) The composition, richness, and diversity of the gut virome change as a function of age. b) The ratio of bacteriophage to bacteria abundance changes as a function of age.
The composition of the human gut virome develops as a function of age (Figure 1). A longitudinal study of the composition of the infant GI virome revealed that eukaryotic, bacterial, and archaeal viruses were established during early life.35 Eukaryotic viruses are present in the earliest time window after birth and expand during the first two years of life.35 In contrast, the richness and diversity of phages are greatest at birth, gradually decreasing from birth to two years old.35 An inverse relationship in the gut microbiome between the phagome diversity and the bacteriome diversity is present in human infants, shifting from a high phageome, low bacteriome diversity community at birth (1-4 days) to a low phageome, high bacteriome diversity community at the age of two years.35 A recent study showed that persistent immature gut microbiota in children, which is characterised by low α diversity (the ecological diversity of a given taxonomical entity in a sample or environment) of the gut microbiota and disproportionately higher levels of Proteobacteria [recently renamed Pseudomonadota in 202136], is closely related to stunting, suggesting an important role of gut microbiota in child development.37 A 2·5-year follow-up analysis of an adult gut virome showed that almost 80% of the identified viral contigs persisted throughout the study.38 Consistently, another year-long longitudinal study of faecal VLPs (n=10 healthy individuals) showed stable and individual-specific gut virome constitution, dominated by crAss-like and Microviridae bacteriophages.25 In another longitudinal study on the human gut virome (n=12 healthy adults), only a small proportion (∼5%) of virotypes fluctuated over one year.39 In conclusion, after establishment at birth, the human gut virome constantly evolves during babyhood/childhood, then stabilised during adulthood in a healthy state.
The function of the gut virome
Cross-kingdom interactions between phages and bacteria and between viruses/phages and the host immune system underly the function of the human gut virome in health and disease.7 As natural parasites of bacteria, phages can shape the composition, to regulate metabolism, evolution, and adaptability of the bacteriome in the gut.40 Beyond that, gut viruses are crucial players in the establishment, development, and function of the human immune system. In return, the composition and functionality of the gut virome are reciprocally shaped and regulated by both the gut bacteriome and host immunity.40,41
Interactions between Gut Virome and Bacteriome
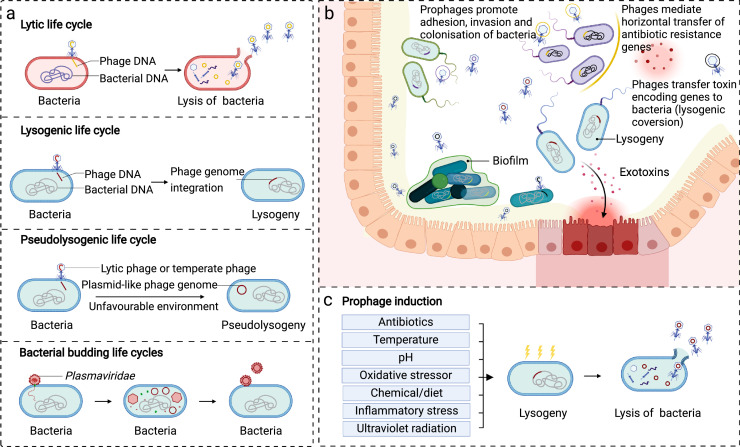
Phages are widely distributed in the intestinal lumen and faeces, and on the intestinal mucosal surfaces,7 interacting profoundly with the co-residing bacteria in the human gut. The ability of phages to shape the composition of gut bacterial communities largely depends on their replication cycles (also known as life cycles) (Figure 2a). Four types of life cycles (including lytic, temperate/lysogenic, pseudolysogenic, and bacterial budding life cycles) are reported for phages, and the lytic and lysogenic life cycles are the two classical forms.42 Lytic phages (also known as virulent phages) inject their genome into the host bacterium, followed by the production of viral macromolecules and particles, which are critical biological event potently regulating the composition of the gut bacteriome.42 The lytic phage phenotype is pervasive under environmental stress and gut inflammation. The lysogenic life cycle is another typical replication cycle, carried out by lysogenic phages (also known as temperate phages).42 In most cases, temperate phages integrate their genomes into the chromosome of their host bacteria and maintain a quiescent “prophage” state unless encountering unfavourable environmental factors.42 Antibiotics, ultraviolet radiation, alterations in temperature or pH, chemical/diet inducers, and oxidative/inflammatory stressors are all effective factors that promote excision of the phage genome, leading to a switch to a lytic life cycle (Figure 2c).43, 44, 45, 46 In general, prophage induction is phage-dependent and inducer-dependent, whereas many phages do not have such a life cycle.47 In addition, some phages can lead a pseudolysogenic life cycle under unfavourable environmental conditions, where the phage genome exists in the bacterial cell as a plasmid-like (episomal) construct without integration or replication.48 Occasionally, some phages, such as Plasmaviridae, lead a special replication mode, that can bud out of the host cells, protecting the bacterial host from lysis or death.49 Although the underlying mechanisms remain unclear, evidence has confirmed the gut virome can undergo an age-dependent lytic-to-lysogenic phage shift50 and a disease-dependent lysogenic-to-lytic switch (Figure 2c).15,51 However, whether this transformation is a cause or a consequence of disease, and how phages change during the disease course remains elusive and warrants extensive investigation.
Figure 2.
Interactions between gut viruses and bacteria. a) Four primary life cycles of phages. b) Infection of phages facilitates phenotypic alterations of the bacterial host. Phages are a natural predator of bacteria, and how their predatory behaviour shapes the bacterial community composition largely depends on their replication cycles. In the lytic replication cycle, phages inject their genome into the host bacterium and end with the release of viral offspring and lysis of the host bacteria cell. The lysogenic replication cycle is a life cycle led by temperate phages under favourable conditions in the human gut. The integration of prophages facilitates alterations in bacterial phenotypes via transfer of phage-coding factors and horizontal transfer of genes encoding for antibiotic-resistance or toxins between bacterial communities, and thus enhance metabolism capacity, environmental adaptation, and pathogenicity of gut bacteria. c) The inducing factors of prophages. Temperate phages remain at a prophage state unless encountering stimulating factors, such as antibiotics, ultraviolet radiation, alterations in temperature or pH, chemical/diet inducers as well as oxidative/inflammatory stress.
In addition to a direct regulation through “predator-prey” relationships between phages and bacteria, intestinal phages can also influence gut bacteriome functions via regulating bacterial metabolism and viability (Figure 2b). For instance, Escherichia coli infected with the bacteriophage Φ24B was found to be more resistant to acid.52 Some phages, such as filamentous phages, can improve the environmental adaptability and virulence of the host by modulating gene expression of the bacterial host to change the bacterial metabolism and biofilm structure and/or up-regulating the exogenous toxins production of bacteria.53 Furthermore, phages can promote the horizontal transfer of antibiotic-resistant genes (ARGs) across bacterial communities during the lysogenic process.54,55 The acquisition of ARG mediated by phage transduction has been observed in clinical bacterial pathogens, such as Enterococcus faecalis,56 Staphylococcus aureus,57 and C. difficile,58 which partly accounts for the antibiotic resistance issues frequently observed in the clinical settings of bacterial infections. Of note, some recent studies showed contrasting results that human-associated and animal-associated phageomes rarely carried ARGs,59,60 suggesting that bona fide ARG attributed to phages in viromes might be overestimated.
Factors encoded by phages can also influence the pathogenicity of intestinal bacteria via promoting their adhesion, invasion, colonisation and production and delivery of toxins (Figure 2b).40 An elegant example is ankyrin protein (ANKp), a protein encoded by phages that was found to attenuate endothelial innate immune defence against E. coli, leading to the expansion of pathogenic E. coli.61 Adenosine-diphosphate-ribosyltransferases (ADPRTs), a class of enzymes encoded by certain phages, were demonstrated to increase adherence and colonisation of the bacterial pathogen (C. difficile) on the host mucosa.62 A recent study revealed that ADPRTs could enhance the overall energy utilisation and mucosal colonisation of gut microbes.63 From an evolutionary biology perspective, phages function as a reservoir or intermediary pool of certain genes, facilitating phenotypical alterations during chromosome exchange between bacteria, due to repeated cycles of phage infection or lysogenic conversion.64 The combined mechanisms of action exerted by phages contribute to the functionality, adaptability, stability, and consequently evolution of the bacterial communities within the human gut.
Interactions between the gut virome and host immunity
In addition to the intestinal lumen and faeces, ample amounts of viruses/phages are present in the GI mucosa, which regulate both the innate and adaptive immunity of the host.40 A set of phages that have variable Ig-like domains on viral capsids (such as T4 phage with Hoc proteins) were reported to bind to mucin glycoproteins on the intestinal mucous layer of the host via the interaction between Ig-like proteins and mucin-displayed glycans.65 These abundant, adherent phages on mucosal surfaces can contribute to the establishment of an innate immune barrier by providing an antimicrobial upfront defence against luminal bacterial pathogens (Figure 3a).40 Weakly adherent T4 phages with Hoc proteins (Ig-like domain) at the mucosal surface exhibit abnormal sub-diffusive motion behaviour, effectively reducing bacterial infection of human mucosal/epithelial cells.66
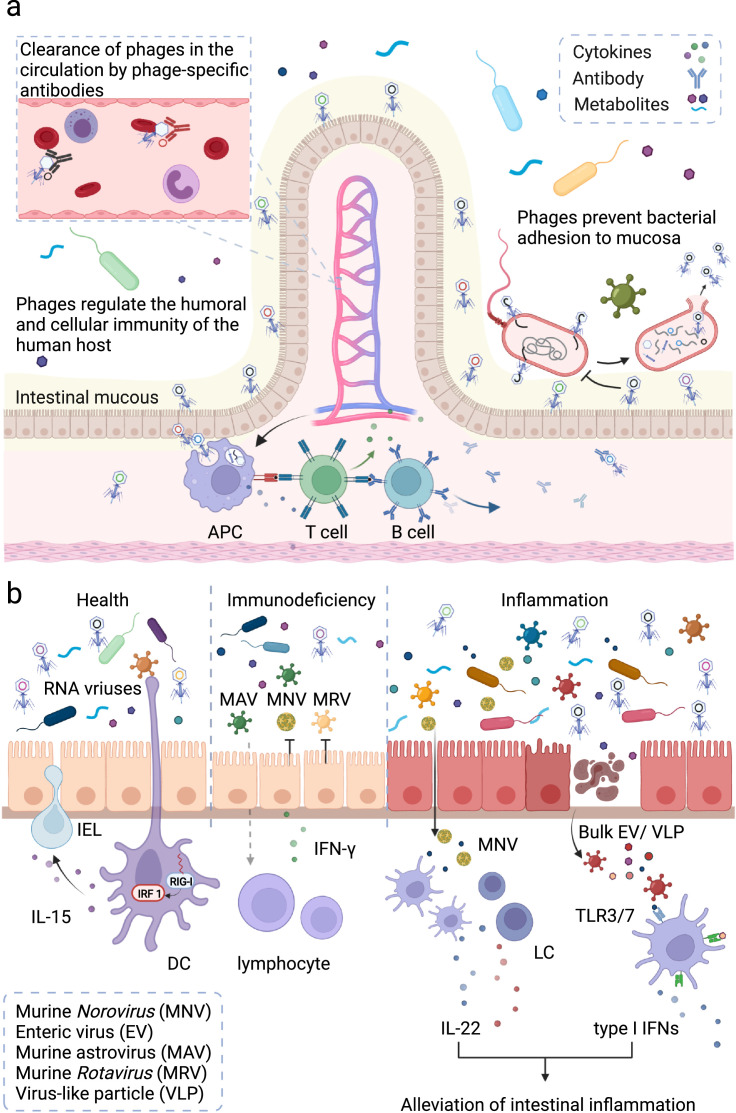
Figure 3.
Gut virome and host immunity. a) The interaction between phages and the human immune system. b) Eukaryotic viruses play a key role in maintaining the homeostasis of the human gut and host immunity. In the human GI tract, phages adhere to mucosal surfaces to prevent luminal bacterial pathogens from adhesion. Numerous phages can be transported across the intestinal epithelial cells towards the systemic circulation and directly influence the human immune system. Innate immune cells recognise these viruses and regulate release of cytokines (such as IFN-γ) and enhance opsonization and recognition of bacteria. Antigen presenting cells (APC) cells present phage-derived peptides to naive T cells and activate the humoral and cellular immunity. Activated B cells differentiate into plasmacytes, to release phage-specific antibodies to clear phages from the body circulation. Colonisation of eukaryotic viruses is also crucial for maintaining the homeostasis of gut and host immunity. In the steady state, RNA virus can be recognised by the RIG-I receptor of dendritic cells (DCs) to maintain the homeostasis of IELs. In the inflammation state, murine Rotavirus (MRVs) activates IFN-I/IL-22-dependent pathways to prevent mice from intestinal injury. The bulk eukaryotic viruses trigger surface TLR3/TLR7-dependent production of IFN-β to protect the host from inflammation. In an immunodeficient state, the supplementation of astrovirus can protect mice from viral (MNVs/MRVs) infection via production of IFN-γ. Abbreviations: LC, lymphoid cell; IEL, intestinal intraepithelial lymphocyte; TLR, Toll-like receptor; IL, interleukin; IFN, interferon.
Beyond direct defence against bacterial invasion at the mucin layer, phages can also interact with the human immune system to maintain immune homeostasis and affect the disease process.67 It is estimated that approximately 31 billion phage particles are daily transported across human GI epithelial cells and into the human circulation.68 Most phages in circulation are captured and filtered by the mononuclear phagocyte system in the liver and spleen and are neutralised by phage-specific antibodies within 24 hours (Figure 3a).69 Several studies have revealed that phages influence the human immune system by regulating the release of cytokines, enhancing opsonization and recognition of bacteria, and tuning the activities of T and B cell activity (Figure 3a).70 A recent study showed that feeding germ-free (GF) mice with E. coli phages or T4 phages can increase infiltration of interferon gamma (IFN-γ)-producing CD4+ and CD8+ T cells in the intestinal mucosa. In dextran sodium sulfate (DSS)-induced IBD mice model, E. coli phages were shown to exacerbate colitis by activating Toll-like receptors 9 (TLR9) and IFN-γ dependent pathways.71 These studies suggest that phages play an important role in the pathogenesis of IBD and are very likely to be potential therapeutic targets.
Colonisation of the gut with eukaryotic viruses is also crucial for the maintenance of gut homeostasis and host immunity (Figure 3b). Recognition of bulk enteric viruses via the surface receptors TLR3 or TLR7 induces IFN-β production, which protects the host from inflammation.72 Receptor retinoic acid inducible gene-1 (RIG-I) is the viral RNA recogniser in the cytosol, which can trigger interleukin-15 (IL-15) production to maintain homeostasis of intraepithelial lymphocytes (IELs).73 Colonising murine Norovirus (MNV) in GF mice elicited a host-protective immune response, which provided critical protection against Citrobacter rodentium infection and promoted proliferation of intestinal epithelial cells.74 Murine astrovirus (MAV) supplementation in immunocompromised mice protected the animals from enteric pathogens via synthesis of IFN-γ, and this protection was transferable by cohousing and faecal transplantation.41 These lines of evidence suggest that eukaryotic viruses in the gut regulate host homeostasis by orchestrating both the bacteria and human immunity. Overall, tri-kingdom interactions between viruses, bacteria and the human host are a burgeoning area of research.
The gut virome in human diseases
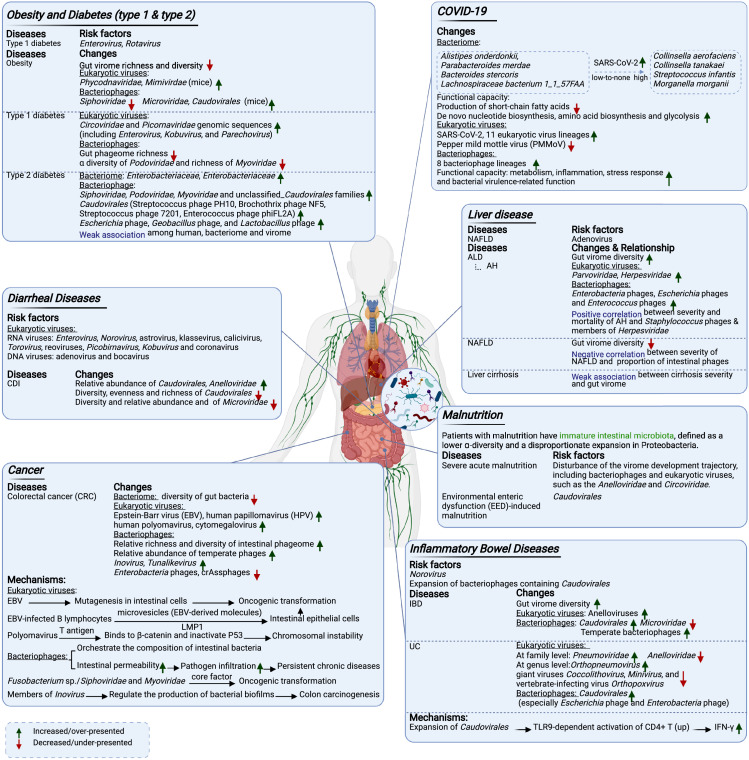
Mounting evidence has confirmed that the gut microbiome affects the onset and the development of both intestinal and extra-intestinal diseases (Figure 4). The gut virome is increasingly reported to play an important role in the pathogenesis of many diseases, and thus represents one of the frontiers in gut microbiome research.
Figure 4.
The gut virome in human diseases.
Diarrheal diseases
Diarrheal diseases can arise from multiple aetiologies,75 and some eukaryotic viruses (particularly RNA viruses) are potent inducers of foodborne diarrheal diseases.76 Rotavirus was reported as the leading cause of death among diarrheal patients aged <5 years (27%), followed by adenovirus and Norovirus.28 Some of these viruses are opportunistic pathogens that be detected occasionally in healthy individuals, and only becoming pathogenic under certain conditions (such as hypoimmunity, gut microbiota dysbiosis, and intestinal injuries).77,78
Studies have also shown significant alterations in the gut phageome in diarrheal diseases, including C. difficile and SARS-CoV-2 infection-induced diarrhoea.9,11,12 Our prior study in CDI showed that CDI-diarrhoea-specific gut virome dysbiosis was characterised by a high relative abundance of Caudovirales and Anelloviridae, along with a low relative abundance of Microviridae.9 In a murine diarrhoea model induced by Salmonella Typhimurium infection, intestinal inflammation boosted free phage production and subsequent lysogenic conversion, and the increase in lysogenic conversion of the P2-like phage (SopEΦ) could in turn aggravate intestinal inflammation.79 However, caution should be exercised when interpreting the virome and bacteriome changes in diarrhoeal diseases, as prolonged symptoms of diarrhoea will significantly decrease the total mass of intestinal microbes. Furthermore, diarrhoea causes a high dilution rate and altered transit time, providing an environment that benefits fast-growing microorganisms and is to the detriment of slow-growing microorganisms. Therefore, the changes in the relative abundance of viruses or bacteria in diarrheal diseases might reflect or is a result of such pathophysiological changes in the bowel.
Inflammatory bowel disease
IBD is a chronic inflammatory disease of the GI tract.80 It is primarily subdivided into two categories depending on the anatomical sites affected by GI inflammation: ulcerative colitis (UC, clinical inflammation primarily manifests in the large bowel) and Crohn's disease (CD, clinical inflammation primarily manifests in the small bowel, the large bowel, and can inflict any part of the GI tract).81 Although the exact cause of IBD remains unknown, a disturbed gut microbiome is hypothesised to underpin the pathogenesis of IBD,82 which is corroborated by a large number of animal studies that demonstrate the gut microbiome to have a causal role in IBD.83
The richness and diversity of the faecal virome were reported to increase in both UC and CD patients, characterised by an expansion of Caudovirales and a reduction in Microviridae abundance.82 However, a subsequent study re-analysed the published dataset based on a whole-virome analysis approach and revealed no significant differences between gut virome richness in IBD patients and healthy controls.51 This study also observed a predominance of temperate virions (mostly Caudovirales taxa) in the gut virome of patients with IBD, suggesting an increase in the lysogenic conversion of phages.51 Our recent study on the intestinal mucosal virome also found an expansion of Caudovirales, especially Escherichia and Enterobacteria phage in patients with UC. The expansion was particularly pronounced in inflamed mucosa compared to non-inflamed mucosa in UC patients.84 A recent preprint study observed that transplantation of VLPs from UC patient faeces to human-microbiota-associated (HMA) mice significantly aggravated colitis in mice and changed the bacterial taxa associated with IBD pathogenesis.8 In another murine study, several phage taxa belonging to Caudovirales were shown to mediate TLR9-dependent activation of CD4+ T cells to exacerbate intestinal inflammation, by enhancing the TLR-dependent production of IFN-γ.71 Together, these results emphasised the potential central role of phages in activating the gut mucosal immune responses and exacerbating colitis.
Our study also showed that eukaryotic viruses were altered in the intestinal mucosa of IBD patients.84 At the family level, a higher relative abundance of Pneumoviridae was observed in UC patients, in contrast to the higher relative abundance of Anelloviridae in healthy controls. At the Genus level, Orthopneumovirus was more prevalent in UC, whereas a reduction in the giant viruses Coccolithovirus, Minivirus, and the vertebrate-infecting virus Orthopoxvirus was observed in UC patients.84 In accordance, other studies reported that anelloviruses are more prevalent in IBD mucosal specimens compared to healthy controls.82,85 In IBD mice model, Norovirus was reported to trigger intestinal inflammation through microbial interaction with IBD susceptibility genes.85 These studies together implicate a role for eukaryotic gut viruses in IBD.
Obesity and diabetes
Obesity and diabetes are two forms of metabolic disorders that are prevalent worldwide. Our recent study in humans showed dysbiosis of the gut virome in both obesity and type 2 diabetes (T2D).10 Obese subjects, particularly those with T2D, had decreased viral richness and diversity in the gut virome compared to lean controls.10 Intensive trans-kingdom correlations between gut viruses (including prokaryotic and eukaryotic viruses) and bacteria were observed in lean controls, the ecological network of which became sparse in obese subjects with T2D.10 However, it is worth to mention that the observed correlations between certain eukaryotic viruses and bacteria in obesity and T2D might be an artefact based on pure bioinformatics analysis, or a mere reflection of the mediating effect of host immunity calibrating the homeostasis between eukaryotic viruses and bacteria in the host. A human study found an increased relative abundance of Siphoviridae, Podoviridae, Myoviridae, and unclassified Caudovirales families in T2D patients.86 In addition, synergistic alterations of intestinal bacteria and phages in the composition of T2D patients have been discovered, including increases in the bacterial families Enterobacteriaceae and their predatory phages such as members from Caudovirales.87 Considering natural “predator-prey” relationship and the complicated interactions between bacteria and phages, it is not surprising to see concurrent changes in both the bacteriome and phageome.
In a prospective cohort with longitudinal sampling of children with genetic susceptibility to type 1 diabetes (T1D), enteroviruses infection was found to be a risk factor for T1D.88 Later, researchers further validated the association between the indicted viral exposures [rotaviruses89 and enteroviruses90] and T1D. Consistently, a recent study revealed that change in the gut virome predated the onset of autoimmunity in paediatric T1D cases, and reported a higher prevalence of Circoviridae and Picornaviridae genomic sequences in faecal samples of T1D children compared to healthy controls.91 These studies have presented different alteration patterns of the gut virome between T1D and T2D, which may underlie the different pathogenesis mechanisms of diabetes.
A study employing western diet-induced obese mice showed a loss of spatial compartmentalization between the mucosal and luminal viromes, and revealed a high richness of lysogenic phages from the Caudovirales order in both mucosal and luminal viromes.92 High-fat diet (HFD)-induced obese mice showed a significant reduction in the Siphoviridae family and an increase in the virus families Microviridae, Phycodnaviridae, and Mimivirdae in the faecal virome.93 The study showed a reduction in lysogenic phages in the HFD group, especially in the family Siphoviridae which is known to have a lysogenic lifestyle.93 Recently, scientists transplanted faecal VLPs from slim mice to obese mice induced by HFD, which significantly decreased weight gain and symptoms of T2D in recipient obese mice.19 These studies together highlight the importance of the gut virome in obesity.
Liver diseases
The liver is a pivotal organ for host metabolism, which maintains bi-directional communications with the gut via the gut-liver axis.94 A recent study on patients with alcohol-associated liver disease (ALD) reported a disease-specific alteration in the gut virome and identified gut viruses as potent drivers of ALD.13 High gut viral diversity was observed in patients with ALD, especially in those with alcoholic hepatitis (AH). Meanwhile, the authors found an increase in eukaryotic viruses such as Parvoviridae and Herpesviridae, along with increases in intestinal phages such as Enterobacteria phages, Escherichia phages, and Enterococcus phages in the AH group.13 Multiple lineages of viruses such as Staphylococcus phages and members of Herpesviridae were demonstrated to be associated with increased severity and mortality of AH.13 Studies on non-alcoholic fatty liver disease (NAFLD) reported a decrease in gut viral diversity in patients, an association between the severity of NAFLD and a low proportion of gut phages in NAFLD patients,95 and identified human adenovirus as a significant risk factor for NAFLD progression.96 In liver cirrhosis, the alterations in the gut virome were reported to correlate with the progression of cirrhosis.97
Cancer
Recently, accumulating evidence suggests that the gut virome contributes to the onset and development of cancers, particularly GI cancers. Metagenomic analysis of stool samples from CRC patients revealed a distinctive faecal virome which showed an increase in the richness and diversity of the intestinal virome.26 The enteric virus markers that differentiated CRC from non-CRC controls comprised Orthobunyavirus, Tunalikevirus, Phikzlikevirus, and 19 other viral genera.26 Phage enrichment from Inovirus and Tunalikevirus were detected,26 wherein some species of Inovirus were reported to regulate the production of bacterial biofilms contributing to the carcinogenesis of the colon.14 A study of the gut virome in CRC patients reported decreased diversity of bacteria (especially butyrate-producing and anti-inflammatory microbes) and significant reductions of Enterobacteria phages and crAssphages compared to healthy controls.98
Regarding eukaryotic viruses, especially carcinogenic viruses in humans, a higher prevalence of Epstein-Barr virus (EBV), human papillomavirus (HPV), human polyomaviruses, and Cytomegalovirus (CMV) was seen in CRC versus non-CRC tissues.99 Preliminary evidence also showed that EBV infection could contribute to CRC development by inducing mutagenesis in intestinal cells.100 In addition, EBV-infected B lymphocytes were reported to produce microvesicles, transmitting EBV-derived molecules to intestinal epithelial cells and subsequently triggering oncogenic transformation.101 Polyomavirus was reported to produce T antigen and inactivate key regulatory tumour suppressor proteins, thus inducing chromosomal instability and malignant transformation of colonic cells.102
Significant alterations of the intestinal phageome were observed in CRC,26,100 but whether the alterations are pivotal in malignant transformation still needs further investigation. The mainstream view is that phages exert a dual-effect on the initiation and progression of CRC by orchestrating the composition of intestinal bacteria.71 Another hypothesis is that phages increase intestinal permeability, known as “leaky gut”, which facilitates the infiltration of pathogens and triggers chronic inflammation.103 A pilot study identified phages, especially those from Siphoviridae and Myoviridae, as a vital driving factors of gut microbiome dysbiosis during the transformation from the healthy intestine to intestinal adenocarcinoma and to CRC.104 Collectively, these findings highlight the significant role of gut viruses in CRC.
Malnutrition
Children with malnutrition were reported to have an intestinal microbiota composition that was immature when compared to healthy controls, defined by lower ɑ-diversity and a disproportionate expansion of Proteobacteria.37 Similar to the gut bacteriome, the gut virome undergoes a malleable developmental process in children after birth.35 Disturbance of the development trajectory of the virome, including that for intestinal phages and members of eukaryotic viruses, can increase the risk of severe acute malnutrition.105 Another study found that phage species from the order Caudovirales differentially contributed to stunted growth in malnutrition induced by environmental enteric dysfunction (EED).106 Consistent with this finding, a pilot study found that the gut phageome/bacteriome of in stunted children were substantially distinct from those of non-stunted children. The researchers detected a co-blooming of Caudovirales phages (especially Siphoviridae) and Proteobacteria (one of the major bacterial hosts of Caudovirales phages),107 suggesting a co-occurring phage-bacteria dynamic in the gut of stunted children gut.15 In vitro bacteria-phage cross-infection experiment showed that phages isolated from stunted children up-regulated the abundance of Proteobacteria (known pathogenic bacteria), in the faeces of non-stunted children.15 These results suggest that both viruses/phages may contribute to the severity of malnutrition.
COVID-19
Coronavirus disease 2019 (COVID-19) is caused by the RNA virus SARS-CoV-2, a virus that induced respiratory disease.11 Among COVID-19 patients, 5%-33% had GI symptoms (including diarrhoea, nausea, and vomiting), indicating enteric involvement in COVID-19.11 Building on that, we conducted the study to investigate microbiome alterations in COVID-19 infected individuals and its association with disease severity and presentations.12,108,109 In these studies, we found transcriptionally active SARS-CoV-2 in faeces and such transcriptional activity persisted even after the respiratory clearance of SARS-CoV-2.29 These results add to the that live SARS-CoV-2 may remain in the gut, and suggesting faecal-oral transmission risk for SARS-CoV-2. Patients with high SARS-CoV-2 infectivity in the gut displayed a high relative abundance of the bacterial species Collinsella aerofaciens, Collinsella tanakaei, Streptococcus infantis, Morganella morganii, and a high functional capacity for de novo nucleotide biosynthesis, amino acid biosynthesis, and glycolysis, which is completely different from patients with low-to-no SARS-CoV-2 infectivity in the gut.29 Moreover, we identified an increased richness of 11 eukaryotic virus lineages and eight bacteriophage lineages in the gut virome of COVID-19 patients.29 Meanwhile, pepper mild mottle virus (a plant-derived RNA virus) and 16 prokaryotic virus lineages were significantly underrepresented in COVID-19 cases.29 Surprisingly, the dysbiosis of both the gut bacteriome and the gut virome persisted for a long time, even after discharge from the hospital.12,29 These data indicate that SARS-CoV-2 infection has detrimental effects on the overall configuration of the gut microbiome and the host physiology.
Implications of the gut virome in disease therapeutics
Faecal virome transplantation (FVT)
With the gradual establishment of the relationship between gut dysbiosis and human health/diseases, strategies to effectively manipulate the gut microbiome have become the focus of intense research, such as the use of prebiotics, probiotics, synbiotics,110 FMT,9 and phage-based therapies [including FVT111 and refined phage therapy112]. Among these, FMT is recognised as one of the most effective and accepted approaches for modulating the gut microbiota, by restoring homeostasis of the gut microbiome through reintroduction of beneficial microbes from a healthy donor. It has been widely applied in the clinical treatment of various diseases, such as CDI,9 IBD,113 sepsis,114 metabolic syndrome,115 blood disorders,116 mental disorders,117 and malignant tumours.118
Our previous FMT study in patients with CDI first demonstrated that FMT can restore the human gut virome, where decreases in Caudovirales and similarities to the donor virome post FMT were observed.9 Consistently, another study also found a high similarity in the phage population after FMT between donors and CDI patients who benefited from FMT treatment (referred to as “responders”) compared to those who did not respond to FMT (referred to as “non-responders”), and such similarities persisted up to 12 months after FMT.119 In addition, our study showed that FMT responders had a higher proportion of donor-derived faecal viral taxa (especially Caudovirales) transferred from donor to recipient after FMT than non-responders.9 Intriguingly, among the CDI patients, when the richness of donor Caudovirales was higher than that of the recipients, the recipients were prone to be cured after FMT.9 Similarly, a recent study revealed that the relative abundance change in Caudovirales was closely related to the response to FMT in IBD patients, and those patients who had no response to FMT had an increased relative abundance of Caudovirales.71
Upon initial proof of the significance of the gut virome during FMT, several studies successfully investigated the utility and feasibility of FVT in treating diseases (Table 1). FVT (also called faecal filtrate transplantation, FFT) is a refined method of FMT that removes faecal bacteria, and therefore decreases the risk of bacterial infection associated with FMT.111 A pilot study in a murine model revealed that autochthonous FVT can reshape the dysbiotic gut microbiome caused by antibiotic treatment.18 Moreover, FVT was also demonstrated to successfully eliminate the symptoms of patients with rCDI in a the clinical setting.16 Concordantly, the intestinal phageome of these patients changed in response to FVT and tended to be like that of the donor six weeks after FVT. The similar treatment efficacy between FMT and FVT in rCDI patients suggests that live microorganisms in faeces maybe not be necessary for FMT to treat rCDI.120 Another study also evaluated the effect of FMT versus FVT in preventing piglets from necrotising enterocolitis and found that FVT was superior to FMT without any recognised side effects.121 A significant alteration in the gut microbiome was observed after FVT, including an increase in the diversity of the gut virome and a decrease in the relative abundance of Proteobacteria in the ileal mucosa.121 Recently, transfer of the caecal virome from lean mice to obese recipient mice was also shown to decrease weight gain and blood glucose parameters, and decrease the expression of genes related to T2D.19 In this HFD-induced obese mice study19 and another targeted phage transfer study,122 phage transfer was shown to mediate the shift of the metabolic profile in plasma or faeces, suggesting that FVT and phage transfer may change the metabolic repertoire of the bacteria communities/mammalian host and/or regulate the expression profile of metabolic genes in the bacteriome. Although the efficacy of FVT has been established, we should be cautious of FVT implementation as eukaryotic viruses are co-transferred along with phages during FVT. The transfer of unwanted eukaryotic viruses might be pathogenic and potentially pose health concerns, particularly to immuno-compromised hosts. Overall, FVT presents as an effective therapy for treating microbiome dysbiosis-related diseases, but the mechanisms and functional components of FVT require further investigation.
Table 1.
Exemplary faecal virome transplantation studies.
| Disease | Model | Donor | Route | Effect | Year | Refs. |
|---|---|---|---|---|---|---|
| Antibiotics-induced dysbiosis | Murine model | Autochthonous stool | Orogastric administration | The gut bacteriome was reshaped towards pre-antibiotic treated mice | 2020 | 18 |
| Recurrent Clostridioides difficile infection (rCDI) | Patients with rCDI (n=5) | Stool from healthy donors | Nasojejunal infusion | Restored normal stool habits; eliminated symptoms of CDI for a minimum period of six months | 2016 | 16 |
| Recurrent Clostridioides difficile infection (rCDI) | Patients with rCDI (n=4) | Uncertain | Oral take capsules of lyophilized sterile faecal filtrate (LSFF) | 75% (3/4) patients achieved no CDI recurrence at the end of eighth weeks | 2019 | 120 |
| Necrotizing Enterocolitis | Caesarean-delivered preterm piglets | Breastfed, term pigs | Rectal or orogastric administration | Orogastric FVT increased gut virome diversity, reduced Proteobacteria relative abundance and completely resolved NEC | 2021 | 121 |
| Obesity Type 2 Diabetes (T2D) | Murine model | Low-fat (LF) diet-fed mice (lean mice) | Orogastric administration | Alleviated symptoms of T2D and obesity; mediate the shift of the metabolic profile among obesity mice. | 2020 | 19 |
Phage therapy
Apart from FMT/FVT which targets broadly on the gut bacteriome, phage therapy is a more targeted therapeutic approach by attacking specific pathogenic bacteria to treat diseases associated with overgrowth of invasive bacteria.17,123 Phage therapy has great advantages in treating pathogenic and/or drug-resistant bacteria infections.
Adherent invasive E. coli (AIEC) is a pathovar E. coli, that causes acute and persistent diarrhoea, including IBD.20 In clinical practice, the treatment of AIEC infection/colonisation by antibiotics is confronted with the problem of multi-drug-resistance, making phage therapy an appealing alternative treatment.124 Research shows that a lytic phage cocktail targeting AIEC significantly decreased the abundance of LF82 and ameliorated intestinal inflammation in mice.20 PDX, a strictly lytic Myoviridae phage isolated from sewage, was found to lyse enteroaggregative E. coli (EAEC, a diarrhoeal pathogen), both in vitro and in vivo in mice model.21 Therefore, PDX might be an effective therapy for EAEC-induced paediatric and travel-related diarrhoea.21 Another study in AIEC-aggravated intestinal tumours in mice showed that AIEC-specific phage treatment could significantly decrease the tumour burden and prolong the survival of mice by reducing the AIEC colonisation and activating the TLR9-dependent immune response.71 Exploratory studies have demonstrated a curative role of phage therapy in the treatment of extra-intestinal diseases. For example, phages of the cytolytic bacterium Enterococcus faecalis were detected to target cytolysin-positive E. faecalis in the intestine, to relieve the translocation of cytolytic E. faecalis to the liver, thereby attenuating AH.17 In conclusion, phage therapy appears to be an effective method to eliminate culprit bacteria in diseases, particularly drug-resistant pathogens.
Phages targeting C. difficile are an alternative to antibiotics and FMT for the treatment of CDI.125 Nearly all identified naturally occurring C. difficile phages display a lysogenic or pseudolysogenic life cycle with low lytic activity once entering the GI tract, resulting in the limited efficacy of phage therapy.125 Owing to recent advances in gene editing and synthetic biology techniques, scientists have directionally engineered the functions and structure of phages, and promoted the refinement of phage therapy.126,127 A recent study found that CRISPR-Cas3 system-induced removal of the lysogeny-related genes in C. difficile phages successfully converted them to an obligately virulent (lytic) type.126 The following experiments demonstrated that it could efficiently target and lyse C. difficile both in vitro and in vivo in mice model. In another study, scientists loaded phages targeting Fusobacterium nucleatum (a tumour-causing bacterium) and irinotecan (an anti-tumour drug) together into one nanosystem, and deployed these nanoparticles to CRC mice.127 The engineered phage-guided nanoparticles precisely targeted tumour, and thus inhibited the growth of F. nucleatum, promoted the proliferation of Clostridium butyricum (a bacterium which has anti-tumour effect by secreting butyrate).127 Though lack of clinical trial validation, these refined phage therapies have achieved great success both in vitro and in vivo.
Research has demonstrated the safety of phage therapy, for human immune system could well tolerate lytic phage therapy via the orogastric path.123 Compared with FMT/FVT, phage therapy can target specific bacteria, which comes at the cost of losing a broad target and sometimes efficacy when implemented in complex diseases involving the GI dysbiosis of a large bacteria set. There are many barriers to the wide application of phage therapy. Firstly, it remains largely unclear whether lytic phages would turn temperate and hence lose efficiency after entering the human body; and if so, how to develop strategies to counteract this issue. Secondly, a majority of current phage therapy studies are still exploratory and conducted in mice which need to be translated to humans. Thirdly, regulatory challenges hamper commercial interests and obstruct the clinical application of phage therapy (including protecting patents regarding phage isolation/purification/combination). Further research efforts should directed toward streamlining and standardizing phage therapy. Overall, phage therapy has been a long-standing, conventional therapy against bacterial infections and was extensively reviewed in other literatures.128,129
Conclusion
Owing to the development and application of state-of-the-art metagenomic sequencing and bioinformatic technologies, the gut virome “dark matter” is being unveiled. In this review, we systematically discussed the features of the human gut virome in health and disease, and dissected the intricate tri-kingdom interactions among gut bacteria, viruses/phages, and human host. Appropriate and equilibrated gut virome configurations are crucial for an individual to maintain a healthy gut microbiome, whereas gut virome dysbiosis may predispose an individual to disease. Hence, precision targeting of the gut virome holds a promising prospect for clinical diagnostics and therapeutic interventions.
Outstanding questions
Existing studies on alterations to the human gut virome in different diseases imply that there is a close association between gut virome dysbiosis and disease pathophysiology. There are multiple outstanding questions that should be addressed in future research. The correlation or causation relationship between gut virome dysbiosis and disease is largely unclear in different diseases, and the virome studies are limited by computational tools and incomplete reference databases. The totality of the gut virome is significantly under-explored, and more than 50% of viral sequences generated by metagenomic sequencing remain unclassified. This hinders the accuracy of the generalisations and conclusions gleaned from the current studies. The RNA virome in the human gut is generally less studied than the DNA virome. The mechanistic underpinning of various intestinal viruses/phages linking to a host phenotype merits an in-depth investigation to guide next-generation microbe-based products (such as FVT and refined phage therapy) to enhance human health and treat a specific disease. The faecal virome/phageome is worthy of exploration for non-invasive disease diagnosis.
Search strategy and selection criteria
References of this review were collected using PubMed for relevant articles published in 2003-2022, using the terms “human virome”, “virus”, “gut microbiome”, “gut virus”, “intestinal virus”, “phage”, “bacteriophage”, “faecal microbiota transplantation”, “faecal virome transplantation”, and “phage therapy”.
Contributors
T.Z. conceived the manuscript. Z.R.C. and T.Z. wrote the manuscript. P.L., N.S., E.B., and M.P.E. provided significant intellectual contribution and edited the manuscript. T.Z. and P.L. supervised this study and drafting of this manuscript. All authors read and approved the final version of the manuscript.
Declaration of interests
The authors declare no conflict of interest.
Acknowledgements
TZ was supported by the National Natural Science Foundation of China (NSFC grant Nos. 82172323 and 32100134), the Key Research and Development Program of Guangzhou (grant No. 202206010014), and a seed fund from the sixth affiliated hospital of Sun Yat-sen University and Sun Yat-sen University. PL was supported by National Natural Science Foundation of China (NSFC grant Nos. U21A20344 and 81970452). Figures were adapted and created via BioRender.com. None of the funders had any role in paper design, data collection, data analysis, interpretation, and writing of the paper.
Contributor Information
Tao Zuo, Email: zuot@mail.sysu.edu.cn.
Ping Lan, Email: lanping@mail.sysu.edu.cn.
References
- 1.Berg G, Rybakova D, Fischer D, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. 2020;8:103. doi: 10.1186/s40168-020-00875-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3:4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivanov II, Littman DR. Modulation of immune homeostasis by commensal bacteria. Curr Opin Microbiol. 2011;14:106–114. doi: 10.1016/j.mib.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddiqui MT, Cresci GAM. The Immunomodulatory Functions of Butyrate. J Inflamm Res. 2021;14:6025–6041. doi: 10.2147/JIR.S300989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai J, Sun L, Gonzalez FJ. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe. 2022;30:289–300. doi: 10.1016/j.chom.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shkoporov AN, Hill C. Bacteriophages of the human gut: the “Known Unknown” of the microbiome. Cell Host Microbe. 2019;25:195–209. doi: 10.1016/j.chom.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Liang G, Bushman FD. The human virome: assembly, composition and host interactions. Nat Rev Microbiol. 2021 doi: 10.1038/s41579-021-00536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha A LY, Mirzaei MK, Shamash M, Samadfam R, King IL, Maurice CF. Transplantation of bacteriophages from ulcerative colitis patients shifts the gut bacteriome and exacerbates severity of DSS-colitis. bioRxiv. 2021 doi: 10.1101/2021.09.10.459444. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuo T, Wong SH, Lam K, et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 2018;67:634–643. doi: 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang K, Niu J, Zuo T, et al. Alterations in the gut virome in obesity and type 2 diabetes mellitus. Gastroenterology. 2021;161:1257–1269.e13. doi: 10.1053/j.gastro.2021.06.056. [DOI] [PubMed] [Google Scholar]
- 11.Liang W, Feng Z, Rao S, et al. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141–1143. doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- 12.Zuo T, Zhan H, Zhang F, et al. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology. 2020;159:1302–1310.e5. doi: 10.1053/j.gastro.2020.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang L, Lang S, Duan Y, et al. Intestinal virome in patients with alcoholic hepatitis. Hepatology. 2020;72:2182–2196. doi: 10.1002/hep.31459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson CH, Dejea CM, Edler D, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21:891–897. doi: 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan Mirzaei M, Khan MAA, Ghosh P, et al. Bacteriophages isolated from stunted children can regulate gut bacterial communities in an age-specific manner. Cell Host Microbe. 2020;27:199–212.e5. doi: 10.1016/j.chom.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ott SJ, Waetzig GH, Rehman A, et al. Efficacy of sterile fecal filtrate transfer for treating patients with clostridium difficile infection. Gastroenterology. 2017;152:799–811.e7. doi: 10.1053/j.gastro.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Duan Y, Llorente C, Lang S, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. 2019;575:505–511. doi: 10.1038/s41586-019-1742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Draper LA, Ryan FJ, Dalmasso M, et al. Autochthonous faecal viral transfer (FVT) impacts the murine microbiome after antibiotic perturbation. BMC Biol. 2020;18:173. doi: 10.1186/s12915-020-00906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen TS, Mentzel CMJ, Kot W, et al. Faecal virome transplantation decreases symptoms of type 2 diabetes and obesity in a murine model. Gut. 2020;69:2122–2130. doi: 10.1136/gutjnl-2019-320005. [DOI] [PubMed] [Google Scholar]
- 20.Galtier M, De Sordi L, Sivignon A, et al. Bacteriophages targeting adherent invasive escherichia coli strains as a promising new treatment for Crohn's disease. J Crohns Colitis. 2017;11:840–847. doi: 10.1093/ecco-jcc/jjw224. [DOI] [PubMed] [Google Scholar]
- 21.Cepko LCS, Garling EE, Dinsdale MJ, et al. Myoviridae phage PDX kills enteroaggregative Escherichia coli without human microbiome dysbiosis. J Med Microbiol. 2020;69:309–323. doi: 10.1099/jmm.0.001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breitbart M, Hewson I, Felts B, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nayfach S, Paez-Espino D, Call L, et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat Microbiol. 2021;6:960–970. doi: 10.1038/s41564-021-00928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuo T, Sun Y, Wan Y, et al. Human-Gut-DNA virome variations across geography, ethnicity, and urbanization. Cell Host Microbe. 2020;28 doi: 10.1016/j.chom.2020.08.005. 741–51 e4. [DOI] [PubMed] [Google Scholar]
- 25.Shkoporov AN, Clooney AG, Sutton TDS, et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe. 2019;26 doi: 10.1016/j.chom.2019.09.009. 527–541.e5. [DOI] [PubMed] [Google Scholar]
- 26.Nakatsu G, Zhou H, Wu WKK, et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology. 2018;155 doi: 10.1053/j.gastro.2018.04.018. 529–541.e5. [DOI] [PubMed] [Google Scholar]
- 27.Zhang T, Breitbart M, Lee WH, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fulci V, Stronati L, Cucchiara S, Laudadio I, Carissimi C. Emerging roles of gut virome in pediatric diseases. Int J Mol Sci. 2021;22:4127–4139. doi: 10.3390/ijms22084127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuo T, Liu Q, Zhang F, et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. 2021;70:276–284. doi: 10.1136/gutjnl-2020-322294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishnamurthy SR, Janowski AB, Zhao G, Barouch D, Wang D. Hyperexpansion of RNA bacteriophage diversity. PLoS Biol. 2016;14 doi: 10.1371/journal.pbio.1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callanan J, Stockdale SR, Shkoporov A, Draper LA, Ross RP, Hill C. Expansion of known ssRNA phage genomes: from tens to over a thousand. Sci Adv. 2020;6:eaay5981. doi: 10.1126/sciadv.aay5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner D, Kropinski AM, Adriaenssens EM. A roadmap for genome-based phage taxonomy. Viruses. 2021;13:506–516. doi: 10.3390/v13030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutilh BE, Cassman N, McNair K, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun. 2014;5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koonin EV, Yutin N. The crAss-like phage group: how metagenomics reshaped the human virome. Trends Microbiol. 2020;28:349–359. doi: 10.1016/j.tim.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Lim ES, Zhou Y, Zhao G, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oren A, Garrity GM. Valid publication of the names of forty-two phyla of prokaryotes. Int J Syst Evol Microbiol. 2021;71 doi: 10.1099/ijsem.0.005056. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian S, Huq S, Yatsunenko T, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proc Natl Acad Sci USA. 2013;110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reyes A, Haynes M, Hanson N, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popescu M, Van Belleghem JD, Khosravi A, Bollyky PL. Bacteriophages and the immune system. Annu Rev Virol. 2021;8:415–435. doi: 10.1146/annurev-virology-091919-074551. [DOI] [PubMed] [Google Scholar]
- 41.Ingle H, Lee S, Ai T, et al. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-lambda. Nat Microbiol. 2019;4:1120–1128. doi: 10.1038/s41564-019-0416-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Federici S, Nobs SP, Elinav E. Phages and their potential to modulate the microbiome and immunity. Cell Mol Immunol. 2021;18:889–904. doi: 10.1038/s41423-020-00532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yue WF, Du M, Zhu MJ. High temperature in combination with UV irradiation enhances horizontal transfer of stx2 gene from E. coli O157:H7 to non-pathogenic E. coli. PLoS One. 2012;7:e31308. doi: 10.1371/journal.pone.0031308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Lin S, Liu T, et al. Xenogeneic silencing relies on temperature-dependent phosphorylation of the host H-NS protein in Shewanella. Nucleic Acids Res. 2021;49:3427–3440. doi: 10.1093/nar/gkab137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boling L, Cuevas DA, Grasis JA, et al. Dietary prophage inducers and antimicrobials: toward landscaping the human gut microbiome. Gut Microbes. 2020;11:721–734. doi: 10.1080/19490976.2019.1701353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korkina L, Ozben T, Saso L. Modulation of oxidative stress: pharmaceutical and pharmacological aspects. Oxid Med Cell Longev. 2016;2016 doi: 10.1155/2016/6023417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutcliffe SG, Shamash M, Hynes AP, Maurice CF. Common oral medications lead to prophage induction in bacterial isolates from the human gut. Viruses. 2021;13:455–469. doi: 10.3390/v13030455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Los M, Wegrzyn G. Pseudolysogeny. Adv Virus Res. 2012;82:339–349. doi: 10.1016/B978-0-12-394621-8.00019-4. [DOI] [PubMed] [Google Scholar]
- 49.Buchmann JP, Holmes EC. Cell walls and the convergent evolution of the viral envelope. Microbiol Mol Biol Rev. 2015;79:403–418. doi: 10.1128/MMBR.00017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manrique P, Dills M, Young MJ. The human gut phage community and its implications for health and disease. Viruses. 2017;9:141–160. doi: 10.3390/v9060141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clooney AG, Sutton TDS, Shkoporov AN, et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe. 2019;26:764–778.e5. doi: 10.1016/j.chom.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 52.Veses-Garcia M, Liu X, Rigden DJ, Kenny JG, McCarthy AJ, Allison HE. Transcriptomic analysis of Shiga-toxigenic bacteriophage carriage reveals a profound regulatory effect on acid resistance in Escherichia coli. Appl Environ Microbiol. 2015;81:8118–8125. doi: 10.1128/AEM.02034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mai-Prochnow A, Hui JG, Kjelleberg S, Rakonjac J, McDougald D, Rice SA. Big things in small packages: the genetics of filamentous phage and effects on fitness of their host'. FEMS Microbiol Rev. 2015;39:465–487. doi: 10.1093/femsre/fuu007. [DOI] [PubMed] [Google Scholar]
- 54.Modi SR, Lee HH, Spina CS, Collins JJ. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499:219–222. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown-Jaque M, Calero-Caceres W, Muniesa M. Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid. 2015;79:1–7. doi: 10.1016/j.plasmid.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 56.Mazaheri Nezhad Fard R, Barton MD, Heuzenroeder MW. Bacteriophage-mediated transduction of antibiotic resistance in enterococci. Lett Appl Microbiol. 2011;52:559–564. doi: 10.1111/j.1472-765X.2011.03043.x. [DOI] [PubMed] [Google Scholar]
- 57.Varga M, Kuntova L, Pantucek R, Maslanova I, Ruzickova V, Doskar J. Efficient transfer of antibiotic resistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiol Lett. 2012;332:146–152. doi: 10.1111/j.1574-6968.2012.02589.x. [DOI] [PubMed] [Google Scholar]
- 58.Goh S, Hussain H, Chang BJ, Emmett W, Riley TV, Mullany P. Phage varphiC2 mediates transduction of Tn6215, encoding erythromycin resistance, between Clostridium difficile strains. mBio. 2013;4 doi: 10.1128/mBio.00840-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Enault F, Briet A, Bouteille L, Roux S, Sullivan MB, Petit MA. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 2017;11:237–247. doi: 10.1038/ismej.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Billaud M, Lamy-Besnier Q, Lossouarn J, et al. Analysis of viromes and microbiomes from pig fecal samples reveals that phages and prophages rarely carry antibiotic resistance genes. ISME Commun. 2021;1:1–10. doi: 10.1038/s43705-021-00054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jahn MT, Arkhipova K, Markert SM, et al. A phage protein aids bacterial symbionts in eukaryote immune evasion. Cell Host Microbe. 2019;26 doi: 10.1016/j.chom.2019.08.019. 542-50 e5. [DOI] [PubMed] [Google Scholar]
- 62.Aktories K, Schwan C, Jank T. Clostridium difficile toxin biology. Annu Rev Microbiol. 2017;71:281–307. doi: 10.1146/annurev-micro-090816-093458. [DOI] [PubMed] [Google Scholar]
- 63.Brown EM, Arellano-Santoyo H, Temple ER, et al. Gut microbiome ADP-ribosyltransferases are widespread phage-encoded fitness factors. Cell Host Microbe. 2021;29:1351–1365.e11. doi: 10.1016/j.chom.2021.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ogilvie LA, Jones BV. The human gut virome: a multifaceted majority. Front Microbiol. 2015;6:918. doi: 10.3389/fmicb.2015.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barr JJ, Auro R, Furlan M, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci USA. 2013;110:10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barr JJ, Auro R, Sam-Soon N, et al. Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc Natl Acad Sci USA. 2015;112:13675–13680. doi: 10.1073/pnas.1508355112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huh H, Wong S, St Jean J, Slavcev R. Bacteriophage interactions with mammalian tissue: therapeutic applications. Adv Drug Deliv Rev. 2019;145:4–17. doi: 10.1016/j.addr.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 68.Bichet MC, Chin WH, Richards W, et al. Bacteriophage uptake by mammalian cell layers represents a potential sink that may impact phage therapy. iScience. 2021;24 doi: 10.1016/j.isci.2021.102287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dabrowska K. Phage therapy: What factors shape phage pharmacokinetics and bioavailability? Systematic and critical review. Med Res Rev. 2019;39:2000–2025. doi: 10.1002/med.21572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dery KJ, Gorski A, Miedzybrodzki R, Farmer DG, Kupiec-Weglinski JW. Therapeutic perspectives and mechanistic insights of phage therapy in allotransplantation. Transplantation. 2021;105:1449–1458. doi: 10.1097/TP.0000000000003565. [DOI] [PubMed] [Google Scholar]
- 71.Gogokhia L, Buhrke K, Bell R, et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe. 2019;25:285–299.e8. doi: 10.1016/j.chom.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang JY, Kim MS, Kim E, et al. Enteric viruses ameliorate gut inflammation via toll-like receptor 3 and toll-like receptor 7-mediated interferon-beta production. Immunity. 2016;44:889–900. doi: 10.1016/j.immuni.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 73.Liu L, Gong T, Tao W, et al. Commensal viruses maintain intestinal intraepithelial lymphocytes via noncanonical RIG-I signaling. Nat Immunol. 2019;20:1681–1691. doi: 10.1038/s41590-019-0513-z. [DOI] [PubMed] [Google Scholar]
- 74.Neil JA, Matsuzawa-Ishimoto Y, Kernbauer-Holzl E, et al. IFN-I and IL-22 mediate protective effects of intestinal viral infection. Nat Microbiol. 2019;4:1737–1749. doi: 10.1038/s41564-019-0470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Younis M, Rastogi R, Chugh A, Rastogi S, Aly H. Congenital diarrheal diseases. Clin Perinatol. 2020;47:301–321. doi: 10.1016/j.clp.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 76.Julio-Pieper M, Lopez-Aguilera A, Eyzaguirre-Velasquez J, et al. Gut susceptibility to viral invasion: contributing roles of diet, microbiota and enteric nervous system to mucosal barrier preservation. Int J Mol Sci. 2021;22:4734–4747. doi: 10.3390/ijms22094734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koonin EV, Dolja VV, Krupovic M. The healthy human virome: from virus-host symbiosis to disease. Curr Opin Virol. 2021;47:86–94. doi: 10.1016/j.coviro.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 78.Kapusinszky B, Minor P, Delwart E. Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol. 2012;50:3427–3434. doi: 10.1128/JCM.01589-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Diard M, Bakkeren E, Cornuault JK, et al. Inflammation boosts bacteriophage transfer between Salmonella spp. Science. 2017;355:1211–1215. doi: 10.1126/science.aaf8451. [DOI] [PubMed] [Google Scholar]
- 80.Zuo T, Kamm MA, Colombel JF, Ng SC. Urbanization and the gut microbiota in health and inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2018;15:440–452. doi: 10.1038/s41575-018-0003-z. [DOI] [PubMed] [Google Scholar]
- 81.Flynn S, Eisenstein S. Inflammatory bowel disease presentation and diagnosis. Surg Clin North Am. 2019;99:1051–1062. doi: 10.1016/j.suc.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 82.Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caruso R, Lo BC, Nunez G. Host-microbiota interactions in inflammatory bowel disease. Nat Rev Immunol. 2020;20:411–426. doi: 10.1038/s41577-019-0268-7. [DOI] [PubMed] [Google Scholar]
- 84.Zuo T, Lu XJ, Zhang Y, et al. Gut mucosal virome alterations in ulcerative colitis. Gut. 2019;68:1169–1179. doi: 10.1136/gutjnl-2018-318131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Basic M, Keubler LM, Buettner M, et al. Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm Bowel Dis. 2014;20:431–443. doi: 10.1097/01.MIB.0000441346.86827.ed. [DOI] [PubMed] [Google Scholar]
- 86.Ma Y, You X, Mai G, Tokuyasu T, Liu C. A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome. 2018;6:24. doi: 10.1186/s40168-018-0410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Q, Ma X, Li C, et al. Enteric phageome alterations in patients with type 2 diabetes. Front Cell Infect Microbiol. 2020;10 doi: 10.3389/fcimb.2020.575084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sadeharju K, Hamalainen AM, Knip M, et al. Enterovirus infections as a risk factor for type I diabetes: virus analyses in a dietary intervention trial. Clin Exp Immunol. 2003;132:271–277. doi: 10.1046/j.1365-2249.2003.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rogers MAM, Basu T, Kim C. Lower incidence rate of type 1 diabetes after receipt of the rotavirus vaccine in the United States, 2001-2017. Sci Rep. 2019;9:7727. doi: 10.1038/s41598-019-44193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oikarinen M, Tauriainen S, Oikarinen S, et al. Type 1 diabetes is associated with enterovirus infection in gut mucosa. Diabetes. 2012;61:687–691. doi: 10.2337/db11-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao G, Vatanen T, Droit L, et al. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc Natl Acad Sci USA. 2017;114:E6166–E6175. doi: 10.1073/pnas.1706359114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim MS, Bae JW. Spatial disturbances in altered mucosal and luminal gut viromes of diet-induced obese mice. Environ Microbiol. 2016;18:1498–1510. doi: 10.1111/1462-2920.13182. [DOI] [PubMed] [Google Scholar]
- 93.Schulfer A, Santiago-Rodriguez TM, Ly M, et al. Fecal viral community responses to high-fat diet in mice. mSphere. 2020;5:e00833–19. doi: 10.1128/mSphere.00833-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J Hepatol. 2020;72:558–577. doi: 10.1016/j.jhep.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 95.Lang S, Demir M, Martin A, et al. Intestinal virome signature associated with severity of nonalcoholic fatty liver disease. Gastroenterology. 2020;159:1839–1852. doi: 10.1053/j.gastro.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tarantino G, Citro V, Cataldi M. Findings from studies are congruent with obesity having a viral origin, but what about obesity-related NAFLD? Viruses. 2021;13:1285–1302. doi: 10.3390/v13071285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bajaj JS, Sikaroodi M, Shamsaddini A, et al. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut. 2021;70:1162–1173. doi: 10.1136/gutjnl-2020-322470. [DOI] [PubMed] [Google Scholar]
- 98.Gao R, Zhu Y, Kong C, et al. Alterations, interactions, and diagnostic potential of gut bacteria and viruses in colorectal cancer. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.657867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Massimino L, Lovisa S, Antonio Lamparelli L, Danese S, Ungaro F. Gut eukaryotic virome in colorectal carcinogenesis: is that a trigger? Comput Struct Biotechnol J. 2021;19:16–28. doi: 10.1016/j.csbj.2020.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marongiu L, Landry JJM, Rausch T, et al. Metagenomic analysis of primary colorectal carcinomas and their metastases identifies potential microbial risk factors. Mol Oncol. 2021;15:3363–3384. doi: 10.1002/1878-0261.13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Delecluse S, Tsai MH, Shumilov A, et al. Epstein-barr virus induces expression of the LPAM-1 integrin in B cells in vitro and in vivo. J Virol. 2019;93:e01618–18. doi: 10.1128/JVI.01618-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ricciardiello L, Baglioni M, Giovannini C, et al. Induction of chromosomal instability in colonic cells by the human polyomavirus JC virus. Cancer Res. 2003;63:7256–7262. [PubMed] [Google Scholar]
- 103.Tetz G, Tetz V. Bacteriophage infections of microbiota can lead to leaky gut in an experimental rodent model. Gut Pathog. 2016;8:33. doi: 10.1186/s13099-016-0109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hannigan GD, Duhaime MB, Ruffin Mtt, Koumpouras CC, Schloss PD. Diagnostic potential and interactive dynamics of the colorectal cancer virome. mBio. 2018;9:e02248–18. doi: 10.1128/mBio.02248-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reyes A, Blanton LV, Cao S, et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci USA. 2015;112:11941–11946. doi: 10.1073/pnas.1514285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Desai C, Handley SA, Rodgers R, et al. Growth velocity in children with Environmental Enteric Dysfunction is associated with specific bacterial and viral taxa of the gastrointestinal tract in Malawian children. PLoS Negl Trop Dis. 2020;14 doi: 10.1371/journal.pntd.0008387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Low SJ, Dzunkova M, Chaumeil PA, Parks DH, Hugenholtz P. Evaluation of a concatenated protein phylogeny for classification of tailed double-stranded DNA viruses belonging to the order Caudovirales. Nat Microbiol. 2019;4:1306–1315. doi: 10.1038/s41564-019-0448-z. [DOI] [PubMed] [Google Scholar]
- 108.Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–955.e8. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yeoh YK, Zuo T, Lui GC, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ke A, Parreira VR, Goodridge L, Farber JM. Current and future perspectives on the role of probiotics, prebiotics, and synbiotics in controlling pathogenic cronobacter spp. in infants. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.755083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rasmussen TS, Koefoed AK, Jakobsen RR, et al. Bacteriophage-mediated manipulation of the gut microbiome - promises and presents limitations. FEMS Microbiol Rev. 2020;44:507–521. doi: 10.1093/femsre/fuaa020. [DOI] [PubMed] [Google Scholar]
- 112.Tsonos J, Vandenheuvel D, Briers Y, De Greve H, Hernalsteens JP, Lavigne R. Hurdles in bacteriophage therapy: deconstructing the parameters. Vet Microbiol. 2014;171:460–469. doi: 10.1016/j.vetmic.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 113.Lee M, Chang EB. Inflammatory bowel diseases (IBD) and the microbiome-searching the crime scene for clues. Gastroenterology. 2021;160:524–537. doi: 10.1053/j.gastro.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Keskey R, Cone JT, DeFazio JR, Alverdy JC. The use of fecal microbiota transplant in sepsis. Transl Res. 2020;226:12–25. doi: 10.1016/j.trsl.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Leong KSW, Jayasinghe TN, Wilson BC, et al. Effects of fecal microbiome transfer in adolescents with obesity: the gut bugs randomized controlled trial. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.30415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bilinski J, Grzesiowski P, Sorensen N, et al. Fecal microbiota transplantation in patients with blood disorders inhibits gut colonization with antibiotic-resistant bacteria: results of a prospective, single-center study. Clin Infect Dis. 2017;65:364–370. doi: 10.1093/cid/cix252. [DOI] [PubMed] [Google Scholar]
- 117.Kelly JR, Borre Y, OB C, et al. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res. 2016;82:109–118. doi: 10.1016/j.jpsychires.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 118.Baruch EN, Youngster I, Ben-Betzalel G, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602–609. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 119.Draper LA, Ryan FJ, Smith MK, et al. Long-term colonisation with donor bacteriophages following successful faecal microbial transplantation. Microbiome. 2018;6:220. doi: 10.1186/s40168-018-0598-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kao DH, Roach B, Walter J, Lobenberg R, Wong K. Effect of lyophilized sterile fecal filtrate vs lyophilized donor stool on recurrent clostridium diffcile infection (RCDI): preliminary results from a randomized, double-blind pilot study. J Canad Assoc Gastroenterol. 2019;2:101–102. doi: 10.1093/jcag/gwz006.050. [DOI] [Google Scholar]
- 121.Brunse A, Deng L, Pan X, et al. Fecal filtrate transplantation protects against necrotizing enterocolitis. ISME J. 2021;16:686–694. doi: 10.1038/s41396-021-01107-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hsu BB, Gibson TE, Yeliseyev V, et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe. 2019;25:803–814.e5. doi: 10.1016/j.chom.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Maronek M, Link R, Ambro L, Gardlik R. Phages and their role in gastrointestinal disease: focus on inflammatory bowel disease. Cells. 2020;9:1013–1029. doi: 10.3390/cells9041013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gutierrez B, Domingo-Calap P. Phage therapy in gastrointestinal diseases. Microorganisms. 2020;8:1420–1432. doi: 10.3390/microorganisms8091420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Heuler J, Fortier LC, Sun X. Clostridioides difficile phage biology and application. FEMS Microbiol Rev. 2021;45:1–23. doi: 10.1093/femsre/fuab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Selle K, Fletcher JR, Tuson H, et al. In vivo targeting of clostridioides difficile using phage-delivered CRISPR-Cas3 antimicrobials. mBio. 2020;11:e00019–e00020. doi: 10.1128/mBio.00019-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zheng DW, Dong X, Pan P, et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat Biomed Eng. 2019;3:717–728. doi: 10.1038/s41551-019-0423-2. [DOI] [PubMed] [Google Scholar]
- 128.Kortright KE, Chan BK, Koff JL, Turner PE. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe. 2019;25:219–232. doi: 10.1016/j.chom.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 129.Hesse S, Adhya S. Phage therapy in the twenty-first century: facing the decline of the antibiotic era; is it finally time for the age of the phage? Annu Rev Microbiol. 2019;73:155–174. doi: 10.1146/annurev-micro-090817-062535. [DOI] [PubMed] [Google Scholar]