Summary
Background
Immunoglobulin G (IgG) antibodies serve a crucial immuno-protective function mediated by IgG Fc receptors (FcγR). Absence of fucose on the highly conserved N-linked glycan in the IgG Fc domain strongly enhances IgG binding and activation of myeloid and natural killer (NK) cell FcγRs. Although afucosylated IgG can provide increased protection (malaria and HIV), it also boosts immunopathologies in alloimmune diseases, COVID-19 and dengue fever. Quantifying IgG fucosylation currently requires sophisticated methods such as liquid chromatography-mass spectrometry (LC-MS) and extensive analytical skills reserved to highly specialized laboratories.
Methods
Here, we introduce the Fucose-sensitive Enzyme-linked immunosorbent assay (ELISA) for Antigen-Specific IgG (FEASI), an immunoassay capable of simultaneously quantitating and qualitatively determining IgG responses. FEASI is a two-tier immunoassay; the first assay is used to quantify antigen-specific IgG (IgG ELISA), while the second gives FcγRIIIa binding-dependent readout which is highly sensitive to both the IgG quantity and the IgG Fc fucosylation (FcγR-IgG ELISA).
Findings
IgG Fc fucosylation levels, independently determined by LC-MS and FEASI, in COVID-19 responses to the spike (S) antigen, correlated very strongly by simple linear regression (R2=0.93, p < 0.0001). The FEASI method was then used to quantify IgG levels and fucosylation in COVID-19 convalescent plasma which was independently validated by LC-MS.
Interpretation
FEASI can be reliably implemented to measure relative and absolute IgG Fc fucosylation and quantify binding of antigen-specific IgG to FcγR in a high-throughput manner accessible to all diagnostic and research laboratories.
Funding
This work was funded by the Stichting Sanquin Bloedvoorziening (PPOC 19-08 and SQI00041) and ZonMW 10430 01 201 0021.
Keywords: COVID-19, Anti-spike IgG, IgG glycosylation, Fucosylation
Research in context.
Evidence before this study
The activity of immunoglobulin (IgG) antibodies depends on the subclass and the Fc glycan composition. The absence of core fucose on the conserved N297-linked Fc glycan considerably elevates the affinity of IgG to Fc-gamma receptor III family (FcγRIII), and consequently enhances the IgG effector function. While afucosylated IgG seem to be protective in HIV infections as well as in natural immune responses to erythroid stages of Plasmodium falciparum, it can also induce inflammation, e.g. in dengue fever, and has been identified as a hallmark of COVID-19 severity. Investigation of the IgG Fc glycan structure requires a combination of technologies, sophisticated equipment and complex analytical and computational processing. This is particularly true when investigating minute amounts of antigen-specific responses. Investigation of IgG glycans has therefore never been implemented as a standard research or diagnostic tool. Previous reports on systems serology platforms making use of FcγRs as probes did not make a direct, quantifiable link to IgG Fc fucosylation.
Added value of this study
Here, we present a two-tier ELISA based method for simultaneous determination of antigen-specific IgG quantity and quality of binding to its cognate IgG receptors, which is reflected in IgG Fc fucosylation. The quantifiable nature of this assay will allow investigation of IgG fucosylation in virtually any immune response measurable by ELISA, which was previously impossible. Although absolute quantification of IgG Fc fucosylation requires a calibrated standard, relative quantification is possible without.
Implications of all the available evidence
This method, named FEASI, can be easily applied in all research and diagnostic laboratories worldwide and used for high-throughput screening of plasma with lowered IgG Fc fucosylation, and thus higher potential for antibody dependent cellular cytotoxicity (ADCC). This is important as such plasma can be used for treatment of COVID-19. Moreover, FEASI can be adapted to rapidly examine binding profiles of existing antibodies to novel viral variants of SARS-CoV-2 as they continue to emerge. It can also be adapted to profile other antigen-specific IgG suitable for ELISA, or to retrospectively analyse convalescent plasma used in clinical trials in order to discover antibody functions associated with passive antibody efficacy.
Alt-text: Unlabelled box
Introduction
Immunoglobulin G (IgG) is a crucial mediator of human immune responses and the most abundant antibody isotype in human serum where it accounts for 10-20% of the total protein content.1 IgG antibodies are glycoproteins and carbohydrate units, also known as glycans, are found on both antigen-binding variable regions and on highly conserved constant regions (Fc). IgG Fc domains are responsible for mediating IgG effector functions by binding and activating the Fc-gamma receptors (FcγR) on myeloid and natural killer (NK) cells, or by activating the complement cascade. IgG antibodies are classified according to the Fc domain structure, into four subclasses (IgG1-4), with different affinities for different FcγR family members (FcγRI-III) and therefore different capacities to activate FcγR-expressing white blood cells.1,2 Protein antigens, abundantly presented during viral infections, predominantly stimulate IgG1 and IgG3 responses that show efficient interaction with all FcγR.3 IgG2 are uniquely formed as a response to T-cell independent antigens such as bacterial polysaccharides, and IgG4, found after repeated antigen exposure, are often elevated in allergies and tolerance responses.4
Aside from subclass, the activity of IgG antibodies depends on Fc glycan composition.2,5 IgG glycans influence folding and function of the IgG, and changes in IgG glycosylation patterns were first reported in autoimmune inflammatory diseases such as rheumatoid arthritis,6,7 and later associated with aging.8 All IgG subclasses have a single conserved N-linked glycosylation site at position 297. The core of the N297-linked glycan is composed of N-acetylglucosamine (GlcNAc) and mannose residues organized in a biantennary structure. This core structure can be modified by a bisecting GlcNAc, a core fucose and one or two galactoses, which can be further elongated by one or two sialic acids.6,9 Of these monosaccharide residues, core fucose has greatest effect on FcγR binding and activity, as its absence alone elevates the affinity of IgG to FcγRIIIa/b by approximately 20-fold.10, 11, 12, 13, 14 This increase in affinity to target receptors can transform quiescent antibodies into potent mediators of phagocytosis and antibody dependent cellular cytotoxicity (ADCC).15, 16, 17, 18, 19 Next to fucosylation, galactosylation has a secondary effect on FcγR binding as it further elevates the affinity of afucosylated IgG to FcγRIIIa and FcγRIIIb by a factor of two, while sialylation was shown to have a minor negative effect.18,20 In addition, increased galactosylation elevates the hexamerization potential of IgG1, promoting both binding and activation of C1q, the first component of the complement cascade.21,22
The majority (∼94%) of adult human IgG contains core fucose in the Fc domain,23 suggesting that the absence of fucose, found only in specific types of immune responses, is a highly regulated process. The clinical importance of low IgG Fc fucosylation was first identified in pregnancy-associated alloimmunizations, where afucosylated IgG is one of the contributing factors to disease severity and clinical outcome.24, 25, 26, 27, 28 More recently, low IgG Fc fucosylation was correlated with severe disease in dengue fever29,30 and COVID-19.31, 32, 33 By contrast, in human immunodeficiency virus (HIV) infections and malaria, afucosylated IgG responses seem to have a protective role.34,35
Among FcγR, only the FcγRIII family is sensitive to the presence of core fucose on IgG, due to a unique N162-linked glycan on the FcγRIIIa itself that extends directly into the IgG Fc.12 The presence of the IgG core fucose sterically impedes the positioning of the N162-linked glycan of the FcγRIII, resulting in increased affinity of afucosylated IgG to FcγRIIIa/b.13 IgG without core fucose is most commonly found among IgG1 and IgG3 subclasses,36,37 produced as a response to alloimmunization in pregnancy or by blood transfusion24, 25, 26, 27, 28 and in infections by enveloped viruses.30,31,34 According to our current understanding, the display of target antigen on the cell surface is a common denominator for all immune responses known to give rise to afucosylated IgG.31
The study of the IgG Fc glycan structure requires a combination of technologies, sophisticated equipment and complex analytical and computational processing. Moreover, the analysis of antigen-specific IgG Fc glycosylation starts with stepwise purification, followed by glycan release methods or digestion of the IgG fraction by proteolytic enzymes (for subclass specific analyses).36 This can be followed by a variety of methods, commonly involving capillary-gel electrophoresis (CGE) with fluorescence detection of labelled glycans or liquid-chromatography and mass spectrometry (LC-MS). Since the concentration of antigen-specific IgG in human serum is typically in the range of micrograms per millilitre,38 achieving high sensitivity and selectivity is challenging. Glycoprofiling of antigen-specific IgG has provided crucial information for clinical research of alloimmune cytopenia,24 dengue fever30 and COVID-19.31, 32, 33 However, the lack of a widely accessible and high-throughput approach to analyse IgG glycan structures has limited extensive testing and thus the development of our understanding of antibody-mediated human immune responses.
Here, we present fucose-sensitive enzyme-linked immunosorbent assay (ELISA)-based method for quantification of antigen-specific IgG Fc fucosylation (FEASI). Since binding of the FcγRIIIa to IgG is highly sensitive to the absence of core fucose in IgG Fc,18,39 we generated a site-specifically c-terminally biotinylated FcγRIIIa-based reagent that can be used to measure IgG Fc fucosylation. FEASI requires two ELISA assays; one to quantify antigen-specific IgG, and the second to provide a signal sensitive to both antigen-specific IgG quantity and the absence of core fucose by measuring soluble FcγRIIIa binding. This approach circumvents the requirement for sophisticated equipment and computational analysis, requiring only an ELISA platform accessible with standard laboratory infrastructure. Moreover, it eliminates the need for stepwise purification of antigen-specific IgG, as FEASI is done directly on plasma samples, with antigen-coated microtiter plates ensuring antigen specificity.
Methods
mAb production and glycoengineering
Expression vector (pcDNA3.1) encoding human monoclonal IgG1 specific for spike protein of SARS-CoV-2 (COVA1–18, described by Brouwer and co-workers40) was transfected in HEK 293 Freestyle cells using PEI MAX (Polysciences) as previously reported.41 To enrich for afucosylated IgG1, 0.2 mM 2-deoxy-2-fluoro-l-fucose (Carbosynth) was added to the culture 1 h prior to transfection. After 5 days, the supernatant was harvested, filtered, and antibodies were purified on an ÄKTAprime plus system (GE Healthcare Life Sciences) by affinity chromatography using a protein A HiTrap HP column (GE Healthcare Life Sciences), as previously described.18 Similar procedure was applied for production of low and high fucosylated anti-Rhesus D (anti-RhD) IgG1. Briefly, variable regions for anti-RhD IgG1 were cloned into a pEE6.4 expression vector containing coding sequence for an anti-RhD heavy chain and produced as recently reported.42
FcγRIIIa-V158 production
C-terminally biotinylated human FcγRIIIa-V158 was produced using the genetic sequence encoding for the extracellular domain of the receptor with the addition of a C-terminal tail containing polyhistidine (10xHis)-tag and an Avi-Tag (GLNDIFEAQKIEWHE). The DNA sequence was codon-optimized using GeneArt Tools (Invitrogen) as described by Temming et al.43 and cloned in the pcDNA3.1 mammalian expression vector. The receptor was produced in HEK293 Freestyle cells as described previously.44 Five days after transfection, cell supernatant was harvested, filtered through a 0.2 µm filter and isolated with affinity chromatography on an ÄKTAprime plus system (GE Life Sciences) using a His-trap column (GE Life Sciences) according to the manufacturer's protocol. Site-specific C-terminal BirA-mediated biotinylation was performed as described previously.39 Briefly, for biotinylation of 1 µM FcγRIIIa, 3.3 nM BirA ligase was added and Amicon Ultra centrifugal filter units (MWCO 10 kDa) (Merck, Millipore) were used to concentrate the sample and to remove unbound biotin. Final FcγRIIIa-biotinylated products were validated by confirming reactivity to IgG subclasses as previously described.18
Patient samples
All blood samples were collected in the presence of heparin to prevent coagulation and diluted in equal amount of PBS. Plasma fractions were collected after Ficoll-PlaqueTM-Plus (GE Healthcare) gradient centrifugation, heat inactivated for 30 min at 56°C and stored at -20°C. SARS-CoV-2 patient samples from ICU patients from the Amsterdam UMC COVID study group were included, as well as SARS-CoV-2 seropositive blood donors from Sanquin.
Ethics
The Amsterdam cohort study was conducted in accordance with the ethical principles set out in the declaration of Helsinki and all participants provided written informed consent. Study on COVID-19 biobanked patient samples was approved by the Academic Medical Center institutional Medical Ethics Committee of the Amsterdam UMC (reference number 2020.182) previously.31
Mass spectrometric IgG Fc glycosylation analysis
SARS-CoV-2-specific antibodies were purified and analysed by nano liquid chromatography (LC) reverse phase (RP) electrospray (ESI) mass spectrometry as described previously.24,31,37 Briefly, the Nunc MaxiSorp™ flat-bottom microtiter plates (Thermo Scientific) were coated overnight with 5 μg/ml of recombinant trimerized spike protein40 in PBS at 4°C without shaking. Next, the plates were washed three times with PBS supplemented with 0.05% TWEEN 20® (PBS-T) and incubated with plasma samples diluted in PBS (1:10; 1 h; shaking). Following the incubation, the plates were washed three times with PBS-T, twice with PBS, and twice with ammonium bicarbonate (50 mM). Spike-specific antibodies were eluted into low-bind PCR plates (Eppendorf) using formic acid (100 mM; 5 min; shaking).
The plates containing eluted antibodies were dried by vacuum centrifugation and reconstituted in 20 µl of 50 mM ammonium bicarbonate by shaking for 5 min at room temperature. Neutral pH was confirmed and the samples were digested with 20 µl of 15 ng/µl TPCK-treated trypsin (Sigma-Aldrich) in ice cold water. After 5 min of shaking, the digests were incubated at 37°C overnight without shaking and stored at -20°C. Prior to the LC-MS analysis the digests were thawed and centrifuged at 780× g for 5 min to pellet any precipitate.
Tryptic digests of the eluted proteins (5 µl aliquots) were separated by liquid chromatography (LC) using the UltiMate™ 3000 RSLCnano System (Thermo Scientific) equipped with a PepMap 100 trap column and a nanoEase M/Z peptide column. The glycopeptides were separated with a solvent-A (0.1% TFA in water) and solvent-B (95% ACN) gradient (Table S6). The LC was coupled by electrospray ionization to an Impact HD quadrupole time-of-flight mass spectrometer (q-TOF-MS; Bruker Daltonics) equipped with a nanoBooster (Bruker Daltonics). Ionization was enhanced by applying acetonitrile-doped nebulizing nitrogen gas at 0.2 bar. Profile spectra were recorded in an m/z range from 550 to 1800 with a frequency of 1 Hz. The collision energy was 5 eV, the transfer time 110 μs and the pre-pulse storage 21 μs.
Mass spectrometry data were extracted and evaluated using Skyline software (version 4.2.19107).45 IgG glycopeptides were detected by creating extracted ion chromatograms (XIC) using the monoisotopic signals of the 3+ and 2+ charge states of previously reported major glycoforms, with a tolerance of +/- 0.055 Th. IgG1 glycopeptides were identified using standard reference samples and rentention time windows were defined for each glycopeptide. The first 3 isotopic peaks of the identified glycopeptides were quantified using an extraction window of +/- 0.055 Th and the total signals of individual glycopeptides, within the defined retention time windows, were used to calculate the abundance of glycosylation traits. Data were judged reliable when the sum of the signal intensities of all accepted glycopeptide species (listed in Tables S7 and S8) were higher than negative samples plus 10 times its standard deviation. The list of analysed glycopeptides differed slightly between cohorts, as a reflection of higher signal intensities in the ICU cohort that enabled us to reliably quantify a broader selection of analytes. Each analyte signal was assessed individually to ensure that no overlapping signals were present in the integrated area. Signals under the threshold and signals with overlapping peaks were excluded from the analysis. Glycosylation traits were calculated as the relative abundance of fucose and bisecting N-acetylglucosamine, and the relative antenna occupancy of galactose and sialic acids as shown in Table S9.
FEASI
Nunc MaxiSorp 96-well flat-bottom plates (Thermo Fisher Scientific) were coated overnight with 1 µg/ml recombinant spike protein in PBS. The following day, plates were washed five times with PBS supplemented with 0.02% polysorbate-20 (PBS-T-02) and incubated for 1 h with the dilution range of plasma from COVID-19 ICU patients and convalescent donors. Dilutions were done in PBS supplemented with 0.02% polysorbate-20 and 0.3% gelatine (PTG). In both the IgG and the FcγR-IgG ELISA, a serially diluted plasma pool, obtained by combining plasma from a collection of COVID-19 blood donors,46 was used as a calibrator. This plasma pool was assigned the value of 100 arbitrary units (AU), which corresponds to approximately 21 µg/ml,47 determined using the recombinant anti-spike monoclonal antibody (clone COVA1–18) as a standard. After incubation, plates were washed five times with PBS-T-02. Biotinylated FcγRIIIa (1 µg/ml) was added to the FcγR-IgG ELISA, incubated for 1 h, washed again five times in PBS-T-02 and incubated with 0.1 µg/ml streptavidin polymerized horse-radish peroxidase (poly-HRP, M2032, Sanquin). For the IgG ELISA, anti-human IgG-HRP (catalogue number M1268, clone: MH16.1, Sanquin) was used at 1 µg/ml. Reactivity of this clone to human IgG was confirmed by testing binding to all known human IgG allotypes (Figure S1) and by verifying lack of reactivity to mouse sera. Both assays were developed with 50% water diluted tetramethylbenzidine substrate (1-step ultra TMB, #34029, Thermo Scientific). Absorbance was measured at 450 and 540 nm and reported as optical density (OD).
Serial dilutions of the calibrator sample (plasma pool) were used to obtain a standard curve and AU values of individual samples were calculated from the standard curve which was formed using a logistic-log function embedded in a software program developed by Ed Nieuwenhuys and publicly available at https://github.com/ednieuw/ELISA-logit-regression. The two-fold dilution range of the calibrator for the IgG ELISA was between 1:1000 and 1:64000, and between 1:100 and 1:6400 for the FcγR-IgG ELISA. Each sample was probed with at least four two-fold dilutions, and AU were determined for dilutions whose OD at 450/540 nm falls within the range of the calibrator. If all four dilutions were outside the range, sample was measured again with the adjusted dilution range. Geometric mean of all AU values for all available dilutions of each sample was taken as the AU value of the given sample. For comparison, the same calibrator was used with the same dilution range in all assays.
For total IgG assays, plates were coated with 1 µg/ml Goat F(ab’)2 Anti-Human IgG-UNLB (Southern Biotech) and the same procedure was followed as described above except that serial dilutions of glycoengineered anti-RhD IgG1 were used instead of plasma.
Statistics
Statistical analyses were performed using GraphPad Prism (version 9.1.1) for Windows (GraphPad Software Inc., La Jolla, CA, www.graphpad.com) and MATLAB® version R2021b (MathWorks, MA, matlab.mathworks.com). To analyse contribution of galactosylation to FEASI outcome, multiple linear regression was used, accounting for two-way interaction between galactosylation and sialylation. Contribution of individual sugar residue was deemed significant if p < 0.05. To examine the difference between FEASI and LC-MS verified fucosylation percentages, Bland-Altman analysis was used and 95% limits of agreement were calculated.
Role of Funders
The funders had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Anti-Spike IgG Fc glycoprofiling by LC-MS
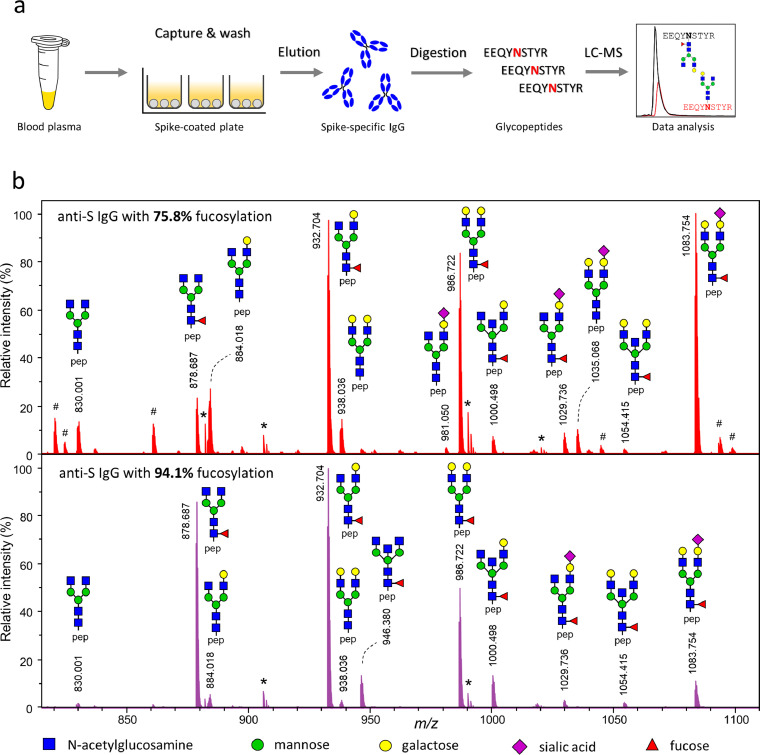
To quantify anti-SARS-CoV-2 spike (S) IgG1 Fc fucosylation by mass spectrometry, immunoglobulins were affinity-captured from serum and/or plasma samples of COVID-19 convalescent blood donors and intensive care unit (ICU) COVID-19 patients. Subsequently, samples were digested with trypsin and measured by LC-MS (Figure 1a) and IgG1 Fc glycopeptides were identified based on accurate mass and characteristic retention times (Figure 1b). IgG1 Fc glycopeptide signals were quantified and normalized to the sum intensity of all IgG1 Fc glycopeptides, resulting in relative abundances (percentages) of all IgG1 Fc glycopeptide species. For downstream analysis, the data were simplified by summing the relative abundance of aggregated types of glycan modifications, including IgG1 Fc fucosylation, as described previously.31,37
Figure 1.
IgG-glycosylation analysis workflow and results by LC-MS. a, Schematic representation of the workflow for antigen(spike)-specific IgG1 glycosylation analysis by liquid chromatography-mass spectrometry (LC-MS); b, Representative mass spectra of N297 IgG1-Fc glycopeptides. Shown are anti-spike IgG1 of two individuals with low (top) and high (bottom) anti-spike IgG fucosylation. The IgG1 Fc glycopeptides elute as a cluster under selected LC conditions, and we display a sum spectrum of their elution range. Many non-glycosylated peptides are outside this elution and m/z window and therefore not displayed. All assigned signals are triple protonated tryptic IgG1 Fc glycopeptides. Pep, tryptic IgG1 Fc peptide portion; #, missed cleavage IgG1 glycopeptide signals; *, single charged, non-glycopeptide species. Spectra are normalized to the highest glycopeptide signal.
FEASI experimental design
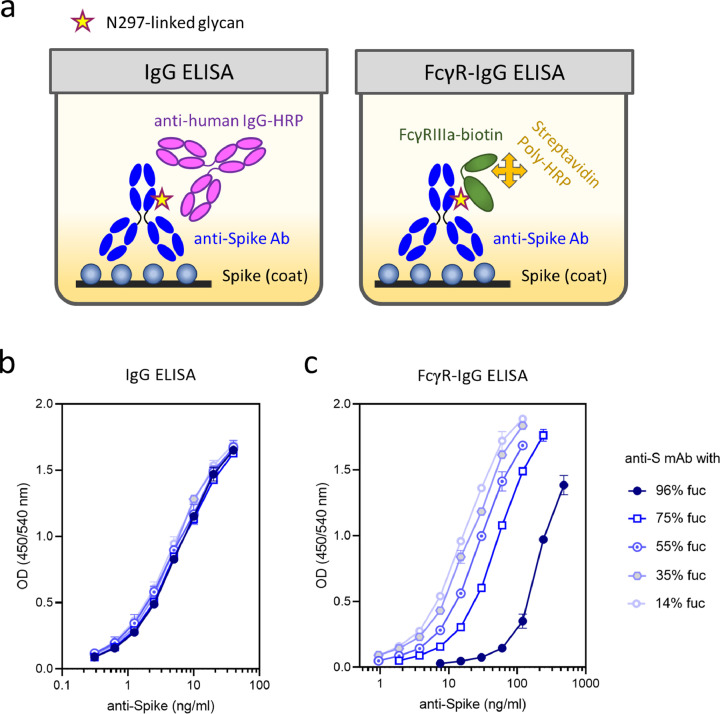
To quantify the presence of afucosylated anti-S IgG formed in response to infection with SARS-CoV-2, we coated the microtiter plates with spike protein as previously described.31,46 First, we quantified the anti-S IgG in plasma by conventional ELISA (IgG ELISA). Second, we used biotinylated FcγRIIIa-based reagent to detect the same antigen-specific IgG in a fucose-dependent assay (FcγR-IgG ELISA), (Figure 2a). The IgG ELISA was sensitive to the quantity of anti-S IgG in the plasma, irrespective of the glycan profile (Figure 2b) and the IgG subclass (Figure S1), while the FcγR-IgG ELISA was sensitive to both quantity as well as the binding affinity of the FcγRIIIa to anti-S IgG (Figure 2c), reflecting the proportion of IgG Fc fucosylation. The results of the FcγR-IgG ELISA were then normalized using the IgG ELISA to determine the degree of fucosylation as detailed below.
Figure 2.
Setup and basic principle of FEASI. a, Schematic overview of the anti-IgG (left) and the FcγR-IgG ELISA (right); b, Recombinant anti-spike monoclonal antibody COVA1-1840 glycoengineered to contain high and low levels of fucose probed with the anti-IgG, and c, the FcγR-IgG ELISA. Error bars represent standard deviation of three measurements.
To validate this approach, we produced human monoclonal anti-S IgG1 antibodies, glycoengineered to contain high or low levels of core fucose. Analysis by LC-MS identified the fucosylation percentages of 96% and 14%, respectively, with comparable galactosylation levels (Table S1). We mixed these variants in different proportions (3:1, 1:1, and 1:3), resulting in preparations containing varying degrees of IgG Fc fucosylation (75%, 55%, and 35%, respectively). Upon testing, identical dose-response relations were observed in the IgG ELISA for all samples irrespective of the degree of fucosylation (Figure 2b). On the other hand, the FcγR-IgG ELISA showed a progressively weaker signal with increasing degrees of IgG Fc fucosylation (Figure 2c).
Developing the FEASI model
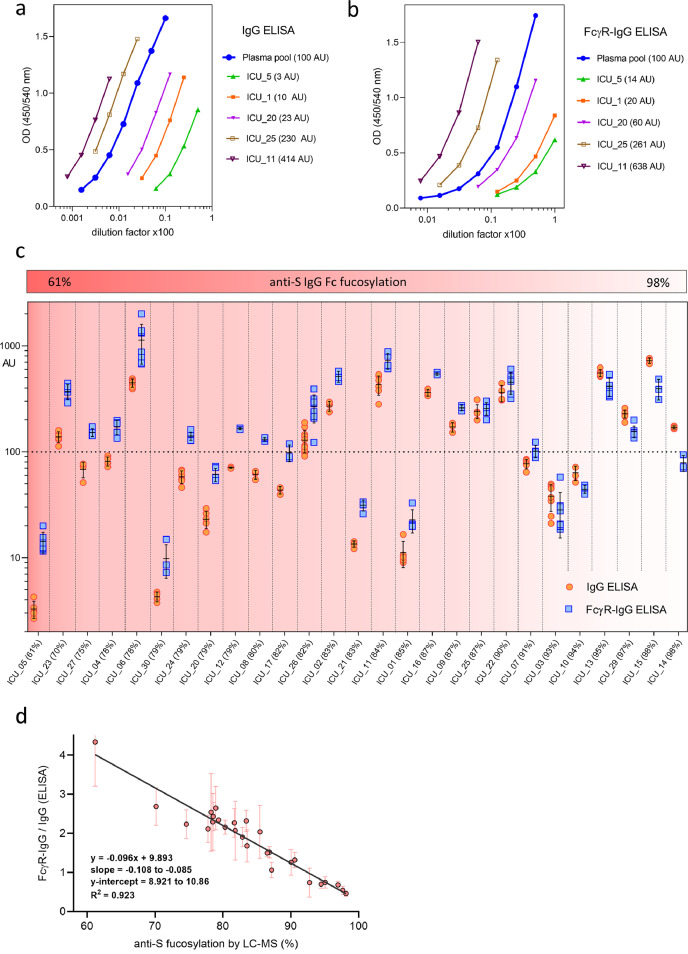
To develop a model for quantification of IgG Fc fucosylation directly from human plasma samples, we used 27 previously collected samples from severely ill COVID-19 patients after their admission to the ICU and measured the glycosylation of anti-S IgG with LC-MS.31 Anti-S IgG Fc fucosylation ranged from 61% to 98% (Table S2). These plasma samples were subsequently tested with FEASI.
In both the IgG and the FcγR-IgG ELISA, pooled plasma from ∼150 COVID-19 convalescent blood donors47 was used as a calibrator set to 100 arbitrary units (AU). To estimate the absolute concentration of spike-reactive IgG antibodies, serial dilutions of the plasma pool were compared with recombinant anti-S IgG and 100 AU were found to correspond to approximately 21 µg/ml (Figure S2a). Limit of detection was defined entirely by the FcγR-IgG ELISA and determined as 31.6(±8.7) ng/ml (Figure S2b). Consequently, the IgG ELISA required higher dilutions compared to the FcγR-IgG ELISA (Figure 3a, b). The differences among individual samples were primarily due to the differences in anti-S levels (for both ELISAs), and secondly due to different levels of fucosylation of the anti-S IgG (for the FcγR-IgG ELISA), (Figure 3a, b). The results for all 27 samples were converted to AU for both the IgG and the FcγR-IgG ELISA, mean values for each sample were calculated with average relative standard deviation (RSD) of 11.4 (±6.3) for the IgG and 15.4 (±9.9) for the FcγR-IgG ELISA (Table S2). Mean AU values of the FcγR-IgG ELISA were higher compared to the mean AU values of the IgG ELISA for samples with lower anti-S IgG Fc fucosylation (Figure 3c, Table S2), demonstrating that this dual ELISA approach (FEASI) provides information about the IgG Fc fucosylation status.
Figure 3.
Evaluation of clinical samples by FEASI. a, Measurement of representative COVID-19 ICU plasma samples with IgG- and b, FcγR-IgG ELISA. In both ELISAs, a serially diluted COVID-19 plasma pool was used as a calibrator set at 100 arbitrary units (AU). c, anti-S levels of severely ill (ICU-admitted) COVID-19 patients measured by the IgG and the FcγR-IgG ELISA. Samples (n = 27) are organized from lower anti-S fucosylation to the left to higher anti-S fucosylation to the right, as measured by LC-MS, according to the indicated colour scale. d, Linear regression model showing the ratio of FcγR-IgG and IgG ELISA readout compared to anti-S IgG Fc fucosylation percentage as determined by LC-MS. All ELISA measurements were done a minimum of three times independently for each sample and converted to AU relative to the plasma pool used. Error bars represent standard deviation of the ratio itself, after error propagation done under the assumption of independence between the two ELISA.
To obtain a quantitative relationship between the ELISA and the LC-MS data, we calculated the ratios between the mean AU values for the FcγR-IgG and the IgG ELISA. These ratios strongly correlated with anti-S IgG Fc fucosylation levels obtained by the LC-MS (R2 = 0.923), by a simple linear regression defined as y = -0.096(±0.011)x + 9.893(±0.970), (Figure 3d).
Validating the FEASI model
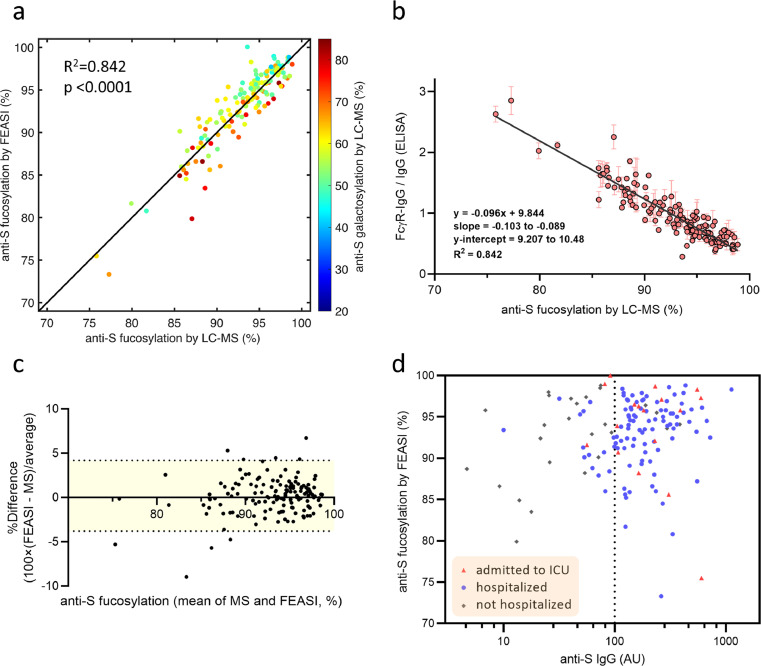
We tested this model on another cohort consisting of 145 convalescent COVID-19 plasma donors, who recovered from varying degrees of disease severity (Tables S3 and S4). We analyzed anti-S IgG Fc fucosylation by both LC-MS and FEASI in an independent and blinded fashion. To obtain the anti-S IgG Fc fucosylation percentages from the ratios of the FcγR-IgG and IgG ELISA, we used the linear regression model defined above (Figure 3d). A strong significant correlation was found between FEASI and the LC-MS-based fucosylation data (R2 = 0.845), (Figure 4a, Table S4). A linear regression model of the FcγR-IgG and IgG ELISA ratio and the anti-S IgG Fc fucosylation percentages measured by LC-MS, defined as y = -0.096(±0.007)x + 9.844(±0.637), (Figure 4b), closely replicated the linear regression model previously obtained with the ICU samples, as both the slope and the y-intercept fall within the 95% confidence interval of the linear regression model for the ICU samples. This confirms that FEASI can be applied to quickly and reliably screen for samples with decreased fucosylation of antigen-specific IgG, in different patient populations sampled at different times. However, slight deviations were observed between the anti-S IgG Fc fucosylation measured by FEASI and the LC-MS (Figure 4c). Interestingly, highly galactosylated (>60%) anti-S IgG gave slightly elevated FcγR-IgG/IgG ratios and therefore a minor underestimation of fucosylation (Figure 4a, Figure S3, Table S4). This confirmed previous findings that increased galactosylation slightly enhances binding to FcγRIIIa, especially for afucosylated IgG.18
Figure 4.
Evaluation of anti-S IgG Fc fucosylation in plasma of convalescent COVID-19 blood donors. a, Pearson's correlation of anti-spike IgG Fc fucosylation measured by FEASI and LC-MS. All samples (n = 145) were tested in both the IgG and the FcγR-IgG ELISA with two independent replicate measurements. Red and orange colours indicate samples with high galactose (>60%), according to the indicated colour scale. b, Linear regression model for the ratio of FcγR-IgG and IgG ELISA done on plasma from COVID-19 convalescent blood donors compared to anti-S IgG Fc fucosylation percentage as determined by LC-MS. Shown are 95% confidence intervals for both the slope and the y-intercept. c, Comparison between fucosylation percentage measurements by FEASI and LC-MS expressed as Bland-Altman plot. Horizontal dotted lines represent 95% limits of agreement ranging from 4.15 to -3.82% difference. The x-axis represents the geometric mean of FEASI and LC-MS measured anti-S fucosylation. d, Anti-spike IgG Fc fucosylation in COVID-19 convalescent plasma samples (n = 145) compared to the anti-spike IgG quantity of each sample. Samples obtained from blood donors treated at the ICU are shown with red triangles, samples from blood donors who have been hospitalized are shown with blue dots, and grey squares represent samples from blood donors who have not been admitted to the hospital. The vertical dotted line indicates anti-spike IgG level equal to 100 AU.
Multiple linear regression analysis showed that FEASI outcome correlated significantly and independently with both IgG Fc fucosylation (p < 0.0001) and galactosylation (p = 0.0381). However, IgG Fc fucosylation correlated positively with FEASI, while galactosylation had a slight negative contribution, as indicated by the negative value of its parameter estimate (Table S5), suggesting that high galactose content of the IgG Fc glycan can result in slightly lower fucosylation estimate by FEASI. Together, these results confirm that fucose is the primary sensing residue for FEASI, with an additional minor contribution of galactose. Sialylation and bisection did not significantly contribute to the FEASI outcome (Table S5), in line with previous findings that IgG Fc bisection has no influence on IgG binding to FcγR.18
Importantly, FEASI identified some samples with high titres (>100 AU47) and low anti-S IgG Fc fucosylation levels in the cohort of COVID-19 convalescent blood donors (Figure 4d), showing the continued presence of low fucosylated anti-S IgG in serum of individuals who recovered from COVID-19. These results were independently confirmed by the LC-MS analysis (Table S4).
To assess the validity of the FEASI approach, we analysed recombinant IgG using a general anti-IgG capturing method but with the same detection reagents. No difference was observed in the anti-IgG ELISA, while the FcγR-IgG ELISA showed a pronounced difference in signal between high and low fucosylated variants of anti-RhD IgG1 (Figure S4).
Discussion
Immunoglobulin-bound glycans affect protein folding, flexibility and its interactions with other biological macromolecules. As such, they encode information influencing both the intracellular signal transduction as well as systemic immune responses.7 Previous work from our group and others, has shown that the presence or absence of core fucose on the IgG N297-linked glycan is one of the key determinants of IgG binding affinity to its cognate receptors of the FcγRIII family.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 For this reason, afucosylated therapeutic antibodies are being tested in clinical trials and some were already approved for treatment of various haematological malignancies.48 In human serum, afucosylated IgG can be present transiently, for a few weeks or days, as in COVID-19,31,49,50 or be maintained over a decade, as in malaria35 and alloimmunizations.24, 25, 26, 27, 28 Interestingly, afucosylated IgG seems to be crucial to mount an efficient protective response in some contexts, such as HIV and malaria infections,34,35 while in others it could be responsible for exaggerated immune responses with lethal outcomes as seen in dengue fever29 and COVID-19.31, 32, 33 Further work is needed to better understand the clinical implications of decreased IgG Fc fucosylation in COVID-19 and in other diseases mediated by enveloped viruses. Until now, no widely accessible tools have been available to investigate IgG Fc fucosylation.
Here, we describe the development of FEASI, a two-tier immunoassay that measures the IgG affinity for FcγRIIIa and thus quantifies the prevalence of afucosylated antigen-specific IgG directly from patient sera or plasma. Since human FcγRIIIa is the receptor responsible for triggering enhanced immune activation when encountering pathogens opsonized with afucosylated IgG,13 we used it as a sensing tool to measure IgG Fc fucosylation. Unlike biophysical methods such as LC-MS that delineate glycan structure, FEASI offers a functional readout which is primarily sensitive to IgG afucosylation, but also influenced by other functionally relevant glycan residues.18 Importantly, FEASI can only detect afucosylated IgG1 and IgG3, as FcγRIIIa does not bind IgG2 and IgG4.51 However, our previous work has shown that only IgG1 and IgG3 are produced as a primary response to protein antigens likely to result in afucosylated IgG.31 By contrast, IgG2 and IgG4 are not generated to protein antigens1 and IgG2 responses are almost fully fucosylated in humans.36,37 The least abundant IgG subclass in humans, IgG4, can contain a minor afucosylated fraction, however the biological importance of this remains uncertain.1 Furthermore, previous studies strongly suggest that afucosylated IgG1 responses are also reflected in IgG3 within the same individual,52 and both IgG1 and IgG3 bind to fucosylation-sensitive FcγRIIIa with high avidity.51 Limitation of the FEASI method is the potential loss of sensitivity to extreme afucosylation beyond 50%. That is expected considering that asymmetrical IgG with only one fucose in either of its heavy chains behaves virtually identical to fully afucosylated variants in functional assays,53 suggesting that changes beyond 50% afucosylation are likely to be less clinically relevant than changes in the range that can be confidently measured by FEASI. Moreover, for diseases such as COVID-19 and Dengue, that is not a concern as changes less than 40% afucosylation have not been observed.
Another limitation of this study is the dependence on standard samples with known IgG Fc fucosylation to define the relationship between the ELISA ratio and the IgG Fc fucosylation. However, even without prior knowledge of fucosylation of a specific calibrator, the described dual ELISA setup holds the potential to stratify samples according to the ratio between the two ELISAs. More precisely, samples with the FEASI ratio higher than 1 would have fucosylation percentage lower than that of the chosen calibrator, and vice versa.
The FEASI method makes it possible to quickly yet reliably identify high titre and low IgG Fc fucosylation plasma in large cohort studies. This screening approach could be especially important for convalescent plasma used in the treatment of COVID-19 patients, where it may be beneficial to select plasma containing afucosylated IgG. A recent clinical trial studying the efficacy of COVID-19 treatment with convalescent plasma concluded that transfusion of convalescent plasma containing IgG with high ADCC potential associated with better clinical outcomes.54 This suggests that strong effector functions can be beneficial even in COVID-19, as supported by studies in animal models,55 and that they associate with severely exaggerated pathologies only in critically ill patients.31, 32, 33 Our results show that some plasma donors continue to produce low fucosylated anti-spike IgG after recovery. Further work is needed to find out if continued production of afucosylated anti-spike IgG has clinical consequences. This is particularly important as afucosylated IgG responses seem to elevate symptoms in secondary infections with heterotypic serotypes of dengue viruses.30 In COVID-19, the full spectrum of clinical implications of the IgG fucosylation remains to be investigated for current and emerging variants of concern of SARS-CoV-2, as well as in response to vaccines.
The importance of the Fc domain characteristics of antigen-specific antibodies in vaccine-mediated protection is being increasingly recognized and different FcγR binding-based methodologies have been established to evaluate antigen-specific antibody effector profiles.56, 57, 58, 59, 60 For example, high-throughput assays using antigen-coated beads to capture antigen-specific IgG have been used in systems serology studies to assess FcγR binding.59 While the FcγR binding profile reflects the abundance of afucosylated glycoforms, none of the established assays offer quantitative assessment of the IgG Fc fucosylation. Moreover, unlike these assays, FEASI does not require fluorescent probes and flow cytometry platform making it more accessible to research settings with limited equipment. In addition, while previously established assays rely on dimeric or multimeric constructs, here we demonstrate the applicability of a monomeric FcγRIIIa-based construct whose affinity to IgG is sufficient to discriminate between afucosylated and fucosylated IgG glycoforms. We previously identified the affinity range of monomeric FcγRIIIa to IgG (FcγRIIIa-V158 KD of ∼100–200 nM for fucosylated and 5–40 nM for afucosylated IgG18,51), and hypothesized that this affinity range is more discriminative to the presence or absence of fucose in IgG1 (and IgG3) compared to higher avidity dimeric and multimeric constructs, whose binding to IgG might be more reflective of the quantity of antigen-specific IgG in given sample, but that remains to be verified. Likewise, FcγRIIIa-F158 may offer even better discrimination between fucosylated and afucosylated IgG1 than V158, but will suffer from the approximately 5-fold lower affinity to IgG1,18,51,61 reducing sensitivity of the assay at low IgG concentrations.
Importantly, FEASI can be adapted for quantifying Fc fucosylation of other antigen-specific antibodies, by introducing the FcγR-IgG ELISA as an additional assay in existing ELISA-based platforms. As a proof of concept, we applied the same principle for total IgG ELISA, which demonstrated the generalizability of the approach. Furthermore, wide accessibility and applicability of this method can boost the interest in both the biological and clinical relevance of the IgG Fc N-glycan variation and provide data necessary to advance our understanding of the regulation of antibody effector functions in immune responses. This goes for both immune responses to naturally occurring viral infections as well as vaccine-induced responses, where FEASI can be applied to identify plasma with low fucosylated IgG as a biomarker of an ongoing inflammatory process.
Contributors
T.S., I.R. and N.I.L.D.: - investigation (ELISA), J.V.C, M.D.L., J.N., W.W., T.D. and M.W.: - investigation (LC-MS); T.S., J.V.C., M.D.L., and T.D.: - Data curation, Methodology, Visualization, Z.S., F.L., R.V., J.Y.M., D.M.G., W.J.E.v.E., S.W.d.T., M.J.v.G., and L.v.d.W.: - Resources, T.S., T.R. and G.V.: - Conceptualization, Writing - original draft, T.S., J.V.C., C.E.v.d.S., M.W., T.R. and G.V.: - Validation, Formal analysis, Writing - review & editing, C.E.v.d.S., M.W., T.R. and G.V.: - Supervision, G.V. - Funding acquisition. All authors read and approved the final version of the manuscript. T.S, J.V.C., M.W., T.R. and G.V. verified the data.
Data sharing statement
The datasets generated for this study are available on request from the corresponding author.
Declaration of interests
The authors declare no conflicts of interest.
Acknowledgements
We thank Amsterdam UMC COVID-19 biobank study group for their assistance in collecting the blood samples. We thank Michael Cloesmeijer, Nikolina Šoštarić, Maurice Steenhuis and Josip Kličinović for their advice on data interpretation and statistics. This work was funded by the Stichting Sanquin Bloedvoorziening (PPOC 19–08 and SQI00041) and ZonMW 10430 01 201 0021.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104109.
Appendix. Supplementary materials
References
- 1.Vidarsson G, Dekkers G, Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front Immunol. 2014;5:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nimmerjahn F, Ravetch JV. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science. 2005;310(5753):1510–1512. doi: 10.1126/science.1118948. [DOI] [PubMed] [Google Scholar]
- 3.Nagelkerke SQ, Schmidt DE, de Haas M, Kuijpers TW. Genetic variation in low-to-medium-affinity fcgamma receptors: functional consequences, disease associations, and opportunities for personalized medicine. Front Immunol. 2019;10:2237. doi: 10.3389/fimmu.2019.02237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lighaam LC, Rispens T. The immunobiology of immunoglobulin G4. Semin Liver Dis. 2016;36(3):200–215. doi: 10.1055/s-0036-1584322. [DOI] [PubMed] [Google Scholar]
- 5.Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313(5787):670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 6.Parekh RB, Dwek RA, Sutton BJ, et al. Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature. 1985;316(6027):452–457. doi: 10.1038/316452a0. [DOI] [PubMed] [Google Scholar]
- 7.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 8.Gudelj I, Lauc G, Pezer M. Immunoglobulin G glycosylation in aging and diseases. Cell Immunol. 2018;333:65–79. doi: 10.1016/j.cellimm.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Kobata A. The N-linked sugar chains of human immunoglobulin G: their unique pattern, and their functional roles. Biochim Biophys Acta. 2008;1780(3):472–478. doi: 10.1016/j.bbagen.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Shields RL, Lai J, Keck R, et al. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem. 2002;277(30):26733–26740. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- 11.Shinkawa T, Nakamura K, Yamane N, et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J Biol Chem. 2003;278(5):3466–3473. doi: 10.1074/jbc.M210665200. [DOI] [PubMed] [Google Scholar]
- 12.Ferrara C, Grau S, Jager C, et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc Natl Acad Sci USA. 2011;108(31):12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falconer DJ, Subedi GP, Marcella AM, Barb AW. Antibody fucosylation lowers the FcgammaRIIIa/CD16a affinity by limiting the conformations sampled by the N162-Glycan. ACS Chem Biol. 2018;13(8):2179–2189. doi: 10.1021/acschembio.8b00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lippold S, Knaupp A, de Ru AH, et al. Fc gamma receptor IIIb binding of individual antibody proteoforms resolved by affinity chromatography-mass spectrometry. MAbs. 2021;13(1) doi: 10.1080/19420862.2021.1982847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Sethuraman N, Stadheim TA, et al. Optimization of humanized IgGs in glycoengineered Pichia pastoris. Nat Biotechnol. 2006;24(2):210–215. doi: 10.1038/nbt1178. [DOI] [PubMed] [Google Scholar]
- 16.Siberil S, de Romeuf C, Bihoreau N, et al. Selection of a human anti-RhD monoclonal antibody for therapeutic use: impact of IgG glycosylation on activating and inhibitory Fc gamma R functions. Clin Immunol. 2006;118(2–3):170–179. doi: 10.1016/j.clim.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Liu SD, Chalouni C, Young JC, Junttila TT, Sliwkowski MX, Lowe JB. Afucosylated antibodies increase activation of FcgammaRIIIa-dependent signaling components to intensify processes promoting ADCC. Cancer Immunol Res. 2015;3(2):173–183. doi: 10.1158/2326-6066.CIR-14-0125. [DOI] [PubMed] [Google Scholar]
- 18.Dekkers G, Treffers L, Plomp R, et al. Decoding the human immunoglobulin G-glycan repertoire reveals a spectrum of Fc-receptor- and complement-mediated-effector activities. Front Immunol. 2017;8:877. doi: 10.3389/fimmu.2017.00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Temming AR, de Taeye SW, de Graaf EL, et al. Functional attributes of antibodies, effector cells, and target cells affecting NK Cell-mediated antibody-dependent cellular cytotoxicity. J Immunol. 2019;203(12):3126–3135. doi: 10.4049/jimmunol.1900985. [DOI] [PubMed] [Google Scholar]
- 20.Subedi GP, Barb AW. The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc gamma receptor. MAbs. 2016;8(8):1512–1524. doi: 10.1080/19420862.2016.1218586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei B, Gao X, Cadang L, et al. Fc galactosylation follows consecutive reaction kinetics and enhances immunoglobulin G hexamerization for complement activation. MAbs. 2021;13(1) doi: 10.1080/19420862.2021.1893427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Osch TLJ, Nouta J, Derksen NIL, et al. Fc Galactosylation promotes hexamerization of human IgG1, leading to enhanced classical complement activation. J Immunol. 2021;207(6):1545–1554. doi: 10.4049/jimmunol.2100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Haan N, Reiding KR, Driessen G, van der Burg M, Wuhrer M. Changes in healthy human IgG Fc-glycosylation after birth and during early childhood. J Proteome Res. 2016;15(6):1853–1861. doi: 10.1021/acs.jproteome.6b00038. [DOI] [PubMed] [Google Scholar]
- 24.Kapur R, Kustiawan I, Vestrheim A, et al. A prominent lack of IgG1-Fc fucosylation of platelet alloantibodies in pregnancy. Blood. 2014;123(4):471–480. doi: 10.1182/blood-2013-09-527978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapur R, Della Valle L, Sonneveld M, et al. Low anti-RhD IgG-Fc-fucosylation in pregnancy: a new variable predicting severity in haemolytic disease of the fetus and newborn. Br J Haematol. 2014;166(6):936–945. doi: 10.1111/bjh.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonneveld ME, Natunen S, Sainio S, et al. Glycosylation pattern of anti-platelet IgG is stable during pregnancy and predicts clinical outcome in alloimmune thrombocytopenia. Br J Haematol. 2016;174(2):310–320. doi: 10.1111/bjh.14053. [DOI] [PubMed] [Google Scholar]
- 27.Wuhrer M, Porcelijn L, Kapur R, et al. Regulated glycosylation patterns of IgG during alloimmune responses against human platelet antigens. J Proteome Res. 2009;8(2):450–456. doi: 10.1021/pr800651j. [DOI] [PubMed] [Google Scholar]
- 28.Kapur R, Della Valle L, Verhagen OJ, et al. Prophylactic anti-D preparations display variable decreases in Fc-fucosylation of anti-D. Transfusion. 2015;55(3):553–562. doi: 10.1111/trf.12880. [DOI] [PubMed] [Google Scholar]
- 29.Wang TT, Sewatanon J, Memoli MJ, et al. IgG antibodies to dengue enhanced for FcgammaRIIIA binding determine disease severity. Science. 2017;355(6323):395–398. doi: 10.1126/science.aai8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bournazos S, Vo HTM, Duong V, et al. Antibody fucosylation predicts disease severity in secondary dengue infection. Science. 2021;372(6546):1102–1105. doi: 10.1126/science.abc7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen MD, de Graaf EL, Sonneveld ME, et al. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science. 2021;371(6532):8378. doi: 10.1126/science.abc8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakraborty S, Gonzalez J, Edwards K, et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat Immunol. 2021;22(1):67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoepel W, Chen HJ, Geyer CE, et al. High titers and low fucosylation of early human anti-SARS-CoV-2 IgG promote inflammation by alveolar macrophages. Sci Transl Med. 2021;13(596):8654. doi: 10.1126/scitranslmed.abf8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ackerman ME, Crispin M, Yu X, et al. Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity. J Clin Invest. 2013;123(5):2183–2192. doi: 10.1172/JCI65708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larsen MD, Lopez-Perez M, Dickson EK, et al. Afucosylated Plasmodium falciparum-specific IgG is induced by infection but not by subunit vaccination. Nat Commun. 2021;12(1):5838. doi: 10.1038/s41467-021-26118-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wuhrer M, Stam JC, van de Geijn FE, et al. Glycosylation profiling of immunoglobulin G (IgG) subclasses from human serum. Proteomics. 2007;7(22):4070–4081. doi: 10.1002/pmic.200700289. [DOI] [PubMed] [Google Scholar]
- 37.Falck D, Jansen BC, de Haan N, Wuhrer M. High-throughput analysis of IgG Fc glycopeptides by LC-MS. Methods Mol Biol. 2017;1503:31–47. doi: 10.1007/978-1-4939-6493-2_4. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez-Quintela A, Alende R, Gude F, et al. Serum levels of immunoglobulins (IgG, IgA, IgM) in a general adult population and their relationship with alcohol consumption, smoking and common metabolic abnormalities. Clin Exp Immunol. 2008;151(1):42–50. doi: 10.1111/j.1365-2249.2007.03545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dekkers G, Bentlage AEH, Plomp R, et al. Conserved FcgammaR- glycan discriminates between fucosylated and afucosylated IgG in humans and mice. Mol Immunol. 2018;94:54–60. doi: 10.1016/j.molimm.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Brouwer PJM, Caniels TG, van der Straten K, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dekkers G, Bentlage AEH, Stegmann TC, et al. Affinity of human IgG subclasses to mouse Fc gamma receptors. MAbs. 2017;9(5):767–773. doi: 10.1080/19420862.2017.1323159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Taeye SW, Bentlage AEH, Mebius MM, et al. FcgammaR Binding and ADCC Activity of Human IgG Allotypes. Front Immunol. 2020;11:740. doi: 10.3389/fimmu.2020.00740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Temming AR, Tammes Buirs M, Bentlage AEH, et al. C-Reactive protein enhances IgG-mediated cellular destruction through IgG-Fc receptors in vitro. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.594773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vink T, Oudshoorn-Dickmann M, Roza M, Reitsma JJ, de Jong RN. A simple, robust and highly efficient transient expression system for producing antibodies. Methods. 2014;65(1):5–10. doi: 10.1016/j.ymeth.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 45.MacLean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogelzang EH, Loeff FC, Derksen NIL, et al. Development of a SARS-CoV-2 total antibody assay and the dynamics of antibody response over time in hospitalized and nonhospitalized patients with COVID-19. J Immunol. 2020;205(12):3491–3499. doi: 10.4049/jimmunol.2000767. [DOI] [PubMed] [Google Scholar]
- 47.Steenhuis M, van Mierlo G, Derksen NI, et al. Dynamics of antibodies to SARS-CoV-2 in convalescent plasma donors. Clin Transl Immunol. 2021;10(5):e1285. doi: 10.1002/cti2.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pereira NA, Chan KF, Lin PC, Song Z. The “less-is-more” in therapeutic antibodies: Afucosylated anti-cancer antibodies with enhanced antibody-dependent cellular cytotoxicity. MAbs. 2018;10(5):693–711. doi: 10.1080/19420862.2018.1466767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pongracz T, Vidarsson G, Wuhrer M. Antibody glycosylation in COVID-19. Glycoconj J. 2022;39(3):335–344. doi: 10.1007/s10719-022-10044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pongracz T, Nouta J, Wang W, et al. Immunoglobulin G1 Fc glycosylation as an early hallmark of severe COVID-19. EBioMedicine. 2022;78 doi: 10.1016/j.ebiom.2022.103957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruggeman CW, Dekkers G, Bentlage AEH, et al. Enhanced effector functions due to antibody defucosylation depend on the effector cell fcgamma receptor profile. J Immunol. 2017;199(1):204–211. doi: 10.4049/jimmunol.1700116. [DOI] [PubMed] [Google Scholar]
- 52.Sonneveld ME, Koeleman CAM, Plomp HR, Wuhrer M, van der Schoot CE, Vidarsson G. Fc-Glycosylation in human IgG1 and IgG3 is similar for both total and anti-red-blood cell anti-K antibodies. Front Immunol. 2018;9:129. doi: 10.3389/fimmu.2018.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shatz W, Chung S, Li B, et al. Knobs-into-holes antibody production in mammalian cell lines reveals that asymmetric afucosylation is sufficient for full antibody-dependent cellular cytotoxicity. MAbs. 2013;5(6):872–881. doi: 10.4161/mabs.26307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Begin P, Callum J, Jamula E, et al. Convalescent plasma for hospitalized patients with COVID-19: an open-label, randomized controlled trial. Nat Med. 2021;27(11):2012–2024. doi: 10.1038/s41591-021-01488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schafer A, Muecksch F, Lorenzi JCC, et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J Exp Med. 2021;218(3) doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wines BD, Vanderven HA, Esparon SE, Kristensen AB, Kent SJ, Hogarth PM. Dimeric FcgammaR ectodomains as probes of the Fc receptor function of anti-influenza virus IgG. J Immunol. 2016;197(4):1507–1516. doi: 10.4049/jimmunol.1502551. [DOI] [PubMed] [Google Scholar]
- 57.Brown EP, Dowell KG, Boesch AW, et al. Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J Immunol Methods. 2017;443:33–44. doi: 10.1016/j.jim.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Selva KJ, van de Sandt CE, Lemke MM, et al. Systems serology detects functionally distinct coronavirus antibody features in children and elderly. Nat Commun. 2021;12(1):2037. doi: 10.1038/s41467-021-22236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karsten CB, Mehta N, Shin SA, et al. A versatile high-throughput assay to characterize antibody-mediated neutrophil phagocytosis. J Immunol Methods. 2019;471:46–56. doi: 10.1016/j.jim.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Natarajan H, Xu S, Crowley AR, et al. Antibody attributes that predict the neutralization and effector function of polyclonal responses to SARS-CoV-2. BMC Immunol. 2022;23(1):7. doi: 10.1186/s12865-022-00480-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruhns P, Iannascoli B, England P, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113(16):3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.