Abstract
Introduction
Pathogenic variants in the pyruvate carboxylase (PC) gene cause a wide spectrum of recessive phenotypes, ranging from the early-onset fatal encephalopathy to the adult-onset benign form.
Results
Patient 1 is a 6 y.o. boy with ataxia, hypoglycemia and episodes of lactic acidosis. WGS revealed the novel heterozygous missense variant c.1372A > G (p.Asn458Asp) in the PC gene. Additional analysis revealed discordant reads mapped to chromosomes 11 and 1, so a reciprocal translocation disrupted the PC gene was suspected. The translocation was validated via FISH-analysis and Sanger sequencing of its boundaries.
Patient 2 is a 13 y.o. girl with psychomotor delay, episodes of lactic acidosis and ketonuria. WES revealed the novel homozygous intronic variant c.1983-116C > T. The PC's mRNA analysis demonstrated the exonization of several intron 16 sequences and some residual amount of WT mRNA isoform.
Two other patients had more severe course of the disease. Their genotype represents missense variants in compound heterozygous and homozygous state (c.1876C > T (p.Arg626Trp), c.2606G > C (p.Gly869Ala), c.2435C > A (p.Ala812Asp).
Conclusion
In patients with metabolic crises, lactic acidosis and hypoglycemia analysis of PC gene is recommended. WGS with deep bioinformatic analysis should be taken into consideration when none or the only one pathogenic variant in the PC gene is found.
Keywords: Pyruvate carboxylase deficiency, Reciprocal translocations, Deep intronic variants, WES, WGS
Highlights
-
•
In patients with metabolic crises with lactic acidosis and hypoglycemia WGS with deep bioinformatic analysis should be taken when none or the only one heterozygous pathogenic variant in the PC gene is found.
-
•
PC gene may have more deep intronic pathogenic variants than included in known mutation databases.
1. Introduction
Pyruvate carboxylase deficiency (PCD, MIM #262150) is a rare autosomal recessive metabolic disease. Pyruvate carboxylase (PC) is an enzyme which works in mitochondrial matrix and converts pyruvate to oxaloacetate, which serves as a substrate for phosphoenolpyruvate carboxykinase and for the Krebs cycle. The PC plays an important role in gluconeogenesis, where it is considered to be the major regulatory enzyme, and is also involved in lipogenesis and biosynthesis of neurotransmitters. PC forms a homotetramer with each subunit having one molecule of biotin covalently attached.
PCD has three major partially overlapping phenotypes [1,2]. The B or “French” phenotype is a neonatal form with severe biotin-unresponsive lactic acidosis, hyperammonemia, hypercitrullinemia, and fatal outcome in the first few months of life. The A form or “North American phenotype” is characterized by infantile onset, mild to moderate lactic acidemia and longer survival. The third phenotype C is rare and is characterized by mild lactic acidemia with normal psychomotor development. [3,4]
PC is encoded by the PC gene located on chromosome 11. The PC gene contains 22 exons (including 2 non-coding) which encode 1178 amino acid polypeptide with three domains: biotin carboxylation domain (BC), transcarboxylation domain (CT) and biotin carboxyl carrier domain (BCCP), and two spacer regions. PC functions as a homotetramer and is localized in the mitochondrial matrix.
Currently (12.06.2022) 62 variants in the PC gene are published in the HGMD (http://www.hgmd.cf.ac.uk/): 42 missense/nonsense, 7 splicing, 7 small deletions, 5 small insertions, 1 small duplication. No large structural or deep intronic variants are reported at the moment of publication.
Several hereditary metabolic diseases share the common features with PCD: biotin disorders, disorders of pyruvate metabolism and the tricarboxylic acids cycle (TAC), respiratory chain disorders, mitochondrial fatty acid oxidation disorders, gluconeogenic defects. Detecting the decreased PC activity in fibroblasts and lymphocytes is definitive for the diagnosis of patients with suspected PCD. However, the residual enzymatic activity is not strongly correlated with the disease severity. The reported genotype-phenotype correlations in PCD forms (infantile, neonatal and benign) are inconsistent [5,6].
Modern diagnostic approach implies targeted genes sequencing or whole exome sequencing (WES) for rapid analysis.
PC-deficiency has an incidence of around 1 in 250,000 and was initially reported in 1976 [7] but since then only small groups of patients have been published in literature. In the present work we describe four patients with PCD, two of which have rare types of genetic variants, that could be identified only with WES/WGS methods.
2. Methods
2.1. Editorial policies and ethical considerations
The work has been carried out in accordance with The Code of Ethics of the World Medical Association Declaration of Helsinki) for experiments involving humans. Informed consent was obtained from all patients. The study was approved by the local ethics committee of the Federal State Budgetary Institution “Research Centre for Medical Genetics”. The approval number 2015–5/3. Date 2015.05.03.
2.2. Patients
Samples from four patients (1 male, 3 females) were collected from several hospitals and based on the clinical and biochemical signs of PCD. The PC activity assay was not provided due to technical issues.
Briefly, clinical symptoms, biochemical and molecular data are summarized in Table 1.
Table 1.
Clinical, laboratory and molecular data of 4 patients with PCD.
| Patient 1 (Type A) | Patient 2 (Type C) | Patient 3 (Type A) | Patient 4 (Type B) | |
|---|---|---|---|---|
| Gender | M | F | F | F |
| Consanguinity | No | Yes | No | No |
| Ethnic origin | russian | osetian | Mixed (russian+azerbajdzhan) | belarusian |
| Pregnancy and birth | uneventful | Anemia, chronic fetus hypoxia | placental insufficiency, urgent cesarean section | Anemia, chronic fetus hypoxia, cesarean section |
| GA at birth | 39–40 | 40 | 31–32 | 41 |
| Birth weight | 3200 | 2990 | 2020 | 2650 |
| Age at onset | 13 months | 1 day | 1 day | 1 day |
| Age at last visit | 6 years | 13 years | 6 months | 3 months |
| Main clinical symptoms/signs | Ataxia, dysarthria, hepatosplenomegaly, recurrent episodes of vomiting, floppiness and pallor | Recurrent episodes of vomiting, floppiness and pallor, hepatomegaly | Epileptic encephalopathy, respiratory failure, coma, jaundice | Respiratory failure, hyporeflexia, hepatomegaly |
| Brain imaging | ND | Periventricular leukopathy | Cysts in the anterior horns of the lateral ventricles | Asymmetric ventricular enlargement, periventricular cyst from left, retrocerebelar cyst, small cyst of septum pellucidum |
| aminoacids in blood | N | ↑ AC C0 | N | N |
| organic acids in urine | 2-OH-butyrate - 378 mmol/mol CRE (ref. 0–2) 3-OH-butyrate – 681 mmol/mol CRE (ref. 0–3) 3-Methyl-2-oxovaleric acid – 1442 mmol/mol CRE (ref. 0–2) Lactate – 4332 mmol/mol CRE (ref. 0–25) Pyruvate – 135 mmol/mol CRE (ref. 0–12) Acetoacetate - 43 mmol/mol CRE (ref. 0–2) fumaric acid – 15 mmol/mol CRE (ref. 0–2) 4-OH-phenylpyruvate – 10 mmol/mol CRE (ref. 0–2) |
3-OH-butyrate – 502 mmol/mol CRE (ref. 0–3) Lactate – 1115 mmol/mol CRE (ref. 0–25) Acetoacetate – 115 mmol/mol CRE (ref. 0–2) 2-OH-butyrate - 378 mmol/mol CRE (ref. 0–2) |
2-OH-butyrate – 6707 mmol/mol CRE (ref. 0–2) 3-OH-butyrate – 10,187 mmol/mol CRE (ref. 0–3) Lactate – 22,568 mmol/mol CRE (ref. 0–25) Pyruvate – 181 mmol/mol CRE (ref. 0–12) 2-OH-isovalerianic acid – 848 mmol/mol CRE (ref. 0–46) 3-OH-propionic acid – 113 mmol/mol CRE (ref. 3–10) 2-oxoglutaric acid – 4126 mmol/mol CRE (ref. 0–152) Fumaric acid – 180 mmol/mol CRE (ref. 0–2) Acetoacetate – 631 mmol/mol CRE (ref. 0–2) 4-OH-phenylpyruvate – 830 mmol/mol CRE (ref. 0–2) Glycolic acid – 709 mmol/mol CRE (ref. 11–103) 4-OH-phenyllactate – 462 mmol/mol CRE (ref. 6–28) |
2-OH-butyrate – 736 mmol/mol CRE (ref. 0–2) Lactate – 8690 mmol/mol CRE (ref. 0–25) 3-OH-butyrate – 1271 mmol/mol CRE (ref. 0–3) Acetoacetate – 1342 mmol/mol CRE (ref. 0–2) Pyruvate – 150 mmol/mol CRE (ref. 0–12) Fumaric acid – 23 mmol/mol CRE (ref. 0–2) 2-OH-isovalerianic acid – 50 mmol/mol CRE (ref. 0–2) 4-OH-phenylacetate – 420 mmol/mol CRE (ref. 6–28) 2-oxoglutaric acid – 504 mmol/mol CRE (ref. 0–152) |
| Routine laboratory tests | pH = 7,13 (ref. 7,35–7,45), BE = −6 to −19 mmol/L (ref. −2 to +2 mEq/L), blood lactate = 9-15 mmol/L (ref. < 2.1) glucoseb = 1,5–2,9 mmol/L (ref.3–5) |
pH = 6,94 (ref. 7,35–7,45), blood lactate = 2,7–10 mmol/L (ref. 〈2,1) glucoseb = 2,2–2,6 mmol/L (ref.3–5) BEa = −7,6 to - 27,3 mmol/L |
pH = 6,92 (ref. 7,35–7,45), blood lactate = 13 mmol/L (ref. <2,1) glucoseb = 2,2 mmol/L (ref. 3–5 mmol/L) |
pH = 7,18–7,57 (ref. 7,35–7,45), blood lactate = 6,8–20 mmol/L (ref. <2,1), total bilirubin = 203/286/166 mkm/L (ref. 3–21), K = 4,6–2,3 mmol/L (ref. 3, 8), Na = 146-155 mmol/L (ref. 135–150) Mg = 0,41–0,47 mmol/L (ref. 0,75-1,5) Hb = 73 (ref. 90–130 g/L) protein in blood = 40 g/L (ref. 51–73) AST = 115 U/L (ref. <82) ALT = 86 U/L (ref. <54) |
| Genotype | [c.1372A > G] + [t(1;11) (p.36.32; q13.2)] | [c.1983-116C > T] + [c.1983-116C > T] | [c.1876C > T] + [c.2606G > C] | [c.2435C > A] + [c.2435C > A] |
Base excess.
Glucose concentration in blood at the metabolic crises before the glucose infusions.
2.3. DNA and RNA extraction and Sanger sequencing
Genomic DNA was extracted from blood samples with the use of QiaAMP DNA-mini kit (Qiagen, USA), following the manufacturer's protocol.
Total RNA was isolated from whole blood cells using Leukocyte RNA Purification Plus Kit (Norgene, Thorold, ON, Canada). The first strand of cDNA was synthesized using ImProm-II™ Reverse Transcriptase (Promega, Madison, WI, USA) and oligo(dT) primers. The fragment of the PC gene cDNA spanning exons 15–18 was amplified by PCR with primers 5’-GTTGCCCACAACTTCAGCAAG and 5’-ATGCTGGGCTGTGAAGTCATC.
Sanger sequencing was performed using an ABI Prism 3500XL (Thermo Fisher Scientific, USA), following the manufacturer's protocol. Exons of the PC gene and genetic variants are named according to the NM_022172.3 reference sequence and GRCh37 (hg19) genome assembly.
2.4. Metabolite analyses
MS/MS analysis of acylcarnitines in dried blood spots was performed using «NeoGram Amino Acids and Acylcarnitines TandemMass Spectrometry Kit» (Perkin Elmer, Finland). The urinary organic acids were extracted by diethyl ether/ethyl acetate, derivatized and analysed by GC/MS 7890А/5975С(Agilent Technologies, USA) withНР-5МS [8].
2.5. WES and WGS
Whole genome sequencing was performed with TruSeq DNA PCR-Free sample preparation kit on NovaSeq 6000 (Illumina, San Diego, CA, USA).
Bioinformatic pipeline: sequence reads were aligned to human reference genome GRCh37 (hg19) using Burrows-Wheeler Aligner (http://bio-bwa.sourceforge.net/). Single-nucleotide variants and small insertions and deletions (indels) were called with Strelka2 Small Variant Caller (https://github.com/Illumina/strelka) and the Genome Analysis Toolkit v.4 (https://gatk.broadinstitute.org/). Structural variants were identified with Manta [https://github.com/Illumina/manta]. The reported variants were annotated with their genomic coordinates, allele frequency (gnomAD database, http://gnomad.broadinstitute.org/), functional consequence and Impact level on the gene product using SnpEff v5 (http://pcingola.github.io/SnpEff/). Variants were prioritized by the consensus score of the set of bioinformatic tools, which predict the pathogenicity of the variant and the deleterious effect on protein (SIFT, SIFT4G, Polyphen2, MutationAssessor, FATHMM, PROVEAN, DEOGEN2, LRT, PrimateAI, MetaSVM, MetaLR, SpliceAI, MMsplice, SPiP, Spidex). Data analysis was performed with in-home NGSData-Genome interface.
The PC variants revealed by massive parallel sequencing were validated by the Sanger sequencing in all patients and both their parents, confirming the biallelic state.
Variants were named according to the HGVS nomenclature and validated using VariantValidator (https://variantvalidator.org/). The novel variants were classified according to ACMG recommendations [9] and submitted to the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) (accession numbers: SCV002003956 - SCV002003961).
2.6. Cytogenetics
Cytogenetic analysis of patient 1 and his parents was performed on GTG-banded metaphase chromosomes obtained from cultured peripheral blood lymphocytes according to standard procedures. The translocation was confirmed by FISH-analysis using WCP probes for chromosomes 1 and 11 (Whole chromosome 1, Whole chromosome 11; KREATECH, The Netherlands) according to manufactures protocols. The analysis was performed using an AxioImager M.1 epifluorescence microscope (Carl Zeiss, Germany) and an Isis digital image processing computer program (MetaSystems, Germany).
3. Results
3.1. Clinical, biochemical, electrophysiological, and neuroradiological features of the patients
3.1.1. Patient 1
The patient is a 6-year-old boy from non-consanguineous marriage. He presented with delayed motor skills from birth. At the age of 1 year and 2 months he developed the first episode of indomitable, severe vomiting, hypoglycemia, lactic acidosis (Table 1). The disease developed acutely and was not provoked by infection or other external factors. He underwent mechanical ventilation during a month. At the age of 2 years he was able to walk with support, but with profound hypotonia. At the moment of the last examination the patient was intellectually normal, demonstrated spasticity with poor walking skills, delayed speech with dysarthria. Severe metabolic crises with hypoglycemia and acidosis (рН 7,13, ВЕ = −19,4 mmol/L) attack him 1–2 times a year.
3.1.2. Patient 2
The patient is a 9-year-old girl from consanguineous marriage. The girl suffered from recurrent crises of lactic acidosis, hypoglycemia and acetonuria since birth. At the first day of life the disease manifested with respiratory failure and metabolic acidosis. From the age of 1 year 2 months she developed symptoms of metabolic decompensation (vomiting, hypoglycemia, metabolic acidosis, lactic acidosis, neurological deterioration). The first episode was diagnosed like a generalized bacterial infection of unknown etiology associated with EBV infection. During hospitalization at 9 years of age the patient presented the diffuse muscle hypotonia, ataxia with polyneuropathy, pathological bilateral reflexes, clonus of feet and the Gowers's sings. She walked independently and speak sentences but was intellectually delayed. At the time of last visit she had 3 years remission of metabolic crises and presented with good neurological condition with slight cognitive deficiency. The patient has a brother, 1 m. of age, with persistent lactatemia, pH = 7,32, BE = − 9,2 mmol/L.
3.1.3. Patient 3
The patient is the first child from healthy parents and non-consanguineous marriage. Shortly after birth she developed respiratory distress syndrome and was transferred to the intensive care unit with respiratory insufficiency and CNS depression syndrome. She was on СРАР for 12 h. Later, at 2 months of age, her mother noted paroxysms with clones of fingers and aversion of the eyeballs (5 s duration). The patient was hospitalized to a neurological department. EEG showed epileptiform activity in the both right and left fronto-central areas. Valproic acid and Levetiracetam were started. MRI did not show any specific damages. At the age of 5 months after few days of cold without fever, the child presented with dyspnea and was admitted to the hospital with depression of consciousness. During 2 weeks the patient had been on mechanical ventilation. She had a development delay, but started to acquire new motor skills. After receiving genetic tests results, antiepileptic therapy was modified (valproic acid was canceled). Until the age of 2y 7 m the girl was stable, she received metabolic, antiepileptic treatment and physical therapy. At 2y7m she developed symptoms of flu (3 days fever) and was admitted to the intensive care unit because of a lethargy and dyspnea. Metabolic acidosis pH 7,31 (ref. 7,35–7,45) and hyperlactatemia (9 mmol/L, ref. <2.1 mmol/L) and X-ray pictures of bilateral pneumonia were noted. After a few days, she developed a stroke-like episode with brainstem damage, tracheo- and gastro-tubes were installed. She received sodium bicarbonate daily because of metabolic acidosis, however lactate level is always kept at 4–6 (ref. <2.1 mmol/L). The girl died at 3 years of age.
3.1.4. Patient 4
The patient was the first child of healthy non-consanguineous marriage. On the first day of life she had respiratory failure, acrocyanosis and hyporeflexia. On the sixth day she was moved to the intensive care unit with respiratory distress and cerebral depression, the mechanical ventilation was initiated. The results of blood tests are given in Table 1.
At the age of 2 months she demonstrated a severe developmental delay, muscle hypotonia, hyporeflexia, seizures, anemia – Hb = 73 (ref. 90–130 g/L) and hypoproteinemia (protein level in blood was 40 g/L (ref. 51–73 g/L), AST = 115 U/L (ref. <82 U/L), ALT = 86 U/L (ref. <54 U/L), despite of bicarbonate infusion the lactate concentration in blood exceeded 5,0–20,0 (ref. <2.1 mmol/L).
The girl died at the age of 85 days with symptoms of increasing respiratory and hepatic failure. Pathology examination identified enlarged liver, histological - balloon dystrophy of hepatocytes.
3.2. Molecular genetics
3.2.1. Patient 1
Initially, the large targeted gene panel sequencing (587 genes of metabolic diseases) was performed to search for variants in the genes responsible for pyruvate metabolism. The novel missense variant c.1372A > G (p.Asn458Asp) in exon 11 of the PC gene was revealed in heterozygous state. The variant is located in the biotin binding domain and leads to the substitution of the highly conserved and neutrally charged Asparagine to the negatively charged Aspartic acid. No other changes in the PC gene were revealed on the panel sequencing. Subsequent analysis of the patient's fibroblasts-derived mRNA demonstrated the monoallelic expression of the r.1372A > G allele, which may indicate the presence of a large deletion, deep intronic or regulatory region mutations on the second allele.
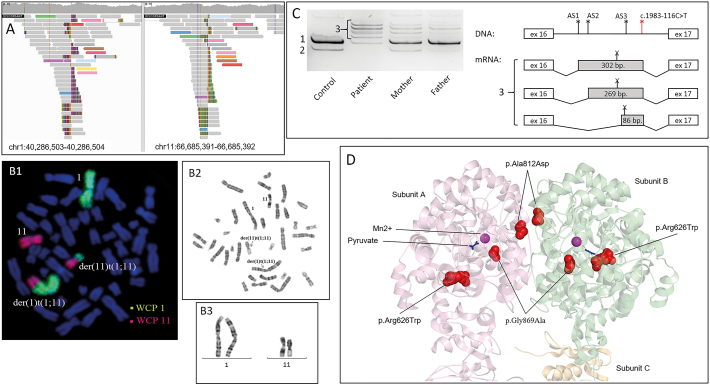
To identify the second pathogenic variant, WGS was performed. The structural variant analysis identified the discordant reads deep in the intron 2 of the PC gene (chromosome 11) whose mates were mapped to the chromosome 1 (Fig. 1A). The translocation between two chromosomes was suspected and the subsequent PCR and Sanger sequencing validated the position of the chromosomal breaks (NC_000011.10:g.66917921 and NC_000001.11:g.39820831). Karyotype analysis confirmed the presence of the balanced translocation between chromosomes 1 and 11 - t(1;11)(p.36.32; q13.2)dmat (Fig. 1B). Analysis of the breakpoint junction also revealed the 3-bp microhomology (CTC), which is a frequent feature in balanced translocations. Previously, Nilsson D et al. reported that microhomology (2–6 nt) was present in 21 of the 74 chromosomal junctions (28%) and in 16 of the 37 translocation events (43%) in their series [10]. The translocation disrupts the 5′ end of the PC gene which includes exons 1, 2 and the promoter region, thus explaining the monoallelic expression in patient's mRNA. Segregation analysis in the family showed that translocation was inherited from the patient's mother and the missense-variant – from the father.
Fig. 1.
A – the IGV browser window demonstrating two groups of discordant reads at chromosomes 1 and 11, which indicate the translocation breakpoints for patient 1. B -. C – the results of blood derived mRNA analysis for patient 2 and his parents. The cDNA fragment spanning exons 15–17 of the PC gene was amplified and visualized by polyacrylamide gel electrophoresis: 1 – the WT isoform, 2 – skipping of exon 16, 3 – several isoforms with cryptic exons. Cryptic exons are activated by the c.1983-116C > T variant, creating strong donor splice site (indicated as red asterisk) and several acceptor splice sites in intron 16 (AS1-AS3, indicated as black asterisks). All of the cryptic exons contains the premature stop-codon (indicated as X). D – the 3D model of the PC dimer (pdb 3BG9, amino acids 482–1178) and location of the novel missense-variants.
3.2.2. Patient 2
WES revealed the novel deep intronic variant c.1983-116C > T in the PC gene in homozygous state. Although such deep intronic regions are not usually catched by WES 19× coverage was obtained in this region in the patient's sample. According to the SpliceAI (https://spliceailookup.broadinstitute.org/#), this variant creates the strong donor splice site altering the PC gene's splicing. The results of the patient's blood-derived mRNA analysis demonstrated the exonization of several intron 16 sequences with some residual amount of WT mRNA isoform (Fig. 1C). All of the identified aberrant mRNA isoforms contain premature stop-codons. The presence of some residual amount of WT RNA isoform suggests, that c.1983-116C > T variant is a leaky splicing variant, which could lead to mild phenotypes, which is consistent with patient's presentations. The segregation analysis of the variant confirmed recessive inheritance and the homozygous state could be explained by the consanguineous marriage.
3.2.3. Patient 3
In patient 3, two novel compound-heterozygous missense-variants were identified by massive parallel sequencing (587 genes of inherited metabolic diseases): c.1876C > T (p.Arg626Trp) in exon 15 and c.2606G > C (p.Gly869Ala) in exon 18.
The c.1876C > T (p.Arg626Trp) residue is highly conserved and is located in the core of the carboxyl transferase domain. The missense variant p.Arg626Gln in the same codon was described earlier, enzymatic activity of pyruvate carboxylase measured in skin fibroblasts was 0.09 nm·min−1·mg−1 protein (6% of control sample; control = 1.5 nm·min−1·mg−1) and PC/CS ratio was 0.18 (4% of control sample) [11].
The variant c.2606G > C (p.Gly869Ala) is located in the highly conserved region of the carboxyl transferase domain's active site (Fig. 1D). The variant p.Gly869Asp in the same codon was identified in homozygous state in a patient with the severely decreased activity of PC [12]. As demonstrated earlier, the torsion angles for Gly869 are unusual: only glycine is flexible enough to make these angles and substitution into another residue will force the local backbone into an incorrect conformation and will disturb the local structure of active site [13].
The revealed variants in patient 3 seem to cause infantile sever form of PCD, as the patient died at 3 years of age. The variants in the same codons also led to severe PCD in literature (11,12).
3.2.4. Patient 4
In patient 3 a novel missense change c.2435C > A (p.Ala812Asp) in homozygous state in exon 17 of the PC gene was identified by massive parallel sequencing (587 genes of inherited metabolic diseases). The segregation analysis confirmed the heterozygous state of the variant in parents. The Ala812 is located at the interface of 2 neighbor pyruvate carboxyltransferase domains (Fig. 1D). Neutrally charged alanine is substituted by the negatively charged aspartic acid, which probably alters the interaction between the subunits. Considering also the patient's phenotype and the homozygous state of the variant, we can suggest that c.2435C > A (p.Ala812Asp) is a severe mutation, which lead to destabilization of the PC's tetrameric structure.
4. Discussion
In this study we demonstrate at first time the rare types of genomic alterations in PCD patients. Clinical and biochemical findings allowed to suspect a mitochondrial disorder in all four patients, but in 2 of 4 cases targeted genes sequencing did not reveal the genetic cause of the disease: in patient 1 we only revealed one likely pathogenic allele and in patient 2 no suspicious variants were detected. Further whole genome sequencing in patient 1 showed the second allele being large reciprocal translocation that comprises almost the half of the p arm of chromosome 1 and whole q arm of chromosome 11, including 5′ end of the PC gene. The breakpoint junction is located deep in the 2nd intron of the PC gene, so the transcription of the PC RNA is totally disrupted. Importantly, such deep intronic alterations could be missed when using whole exome sequencing. This fact should be taken into account in cases of rare genetic disorders where the diagnosis is highly suspicious by clinical and biochemical data. The chromosomal abnormalities are rare but well established phenomenon among other rare monogenic diseases, for example 13 cases of the Hunter disease in females are known due to structural anomalies, a non-random chromosome X inactivation or chromosome X monosomy [[14], [15], [16]]. The similar cases with translocations that disrupt known disease genes were described for the other X-linked disorders, for example OTC-deficiency and Menkes disease [17,18]. Use of chromosomal rearrangements as signposts for genes important in human disease is well documented [10,[19], [20], [21], [22]].
Overall, reciprocal translocations are observed in 0.09% of newborns; the majority of which are inherited from either parent. Approximately 20% of translocations occur de novo with an estimated mutational rate of 2.7 × 10–4 per gamete per generation [23]. Obviously, balanced translocations could be the cause of the dominant inherited disorder if it affects the one allele in known genes with dominant phenotypes and recessive monogenic disorder when found in trans with a point pathogenic alteration in the genes causing recessive phenotypes. Disruption of recessive alleles has to be taken into consideration concerning reproduction and risk for disease in the offspring as structural variation can contribute to recessive disease carrier states [24].
Deep intronic variant in patient 2 c.1983-116C > T is the deepest splicing change in the PC gene being described to date. Despite the fact that truncated transcripts with stop codons are formed due to the nucleotide transition, the patient's phenotype corresponds to a mild form C. This can be explained by the presence of a residual amount of normally spliced PC transcripts in the cells (Fig. 1C left). Earlier 5 splicing variants were published in the HGMD database, including branch point mutation c.1369-29A > G [12]. At least 2 cases with only one possible causative PC allele found in compound heterozygous with unknown intronic variant supposed to disrupt pre-mRNA splicing are published [11]. So, the frequency of deep intronic variants in the PC gene could be underestimated. Hereby, wide-scale genomic search (WES, WGS) is necessary in cases when only one heterozygous allele is found in a patient during targeted genetic search. The difficulties in the investigation of intronic abnormalities are associated with the methods of their functional characterization. Here we used mRNA from a patient's blood to show aberrant transcripts, but this method is only suitable for genes that have expression in blood cells, so more labor-consuming methods such as mini-genes may be required in other cases.
All four novel missense variants in our patients p.Asn458Asp, p.Arg626Trp, p.Gly869Ala, p.Ala812Asp are located in conservative regions and functional domains of the protein and are predicted to alter PC function. The classification of all identified changes in the PC gene is given in Table 2. The variant p.Asn458Asp is present in patient 1 in compound with the LoF variant, so as the clinical picture of the patient is consistent with type A of PCD this missense variant can be classified as moderate. The variants p.Arg626Trp, p.Gly869Ala could be classified as moderate based on type A phenotype in patient 3 but the pathogenic variants in the same codons were described in patients with fatal neonatal form of PCD. Patient 4 has homozygous missense variant p.Ala812Asp which has severe effect on the protein based on the clinical picture of the patient.
Table 2.
Classification of the identified variants.
| Variant | Zygosity | Type of PCD | gnomAD frequency | SpliceAI (delta score) | Consensuns computional verdict for missense variants | ACMG criteria | Classification |
|---|---|---|---|---|---|---|---|
| c.1372A > G (p.Asn458Asp) | het | A | 0 | US | P | PM2, PM3, PP3 | Uncertain significance |
| c.1876C > T (p.Arg626Trp) | het | A | 0 | US | P | PM2, PM3 (sup), PM5, PP3 | Likely pathogenic |
| c.1983-116C > T | hom | C | 0 | 0.89 for Donor Gain | PM2, PS3, PM3 (sup) | Likely pathogenic | |
| c.2435C > A (p.Ala812Asp) | hom | B | 0 | US | P | PM2, PM3 (sup), PP3 | Uncertain significance |
| c.2606G > C (p.Gly869Ala) | het | A | 0 | US | P | PM2, PM3 (sup), PM5, PP3 | Likely pathogenic |
| Translocation | het | A | PVS1, PM2, PM3 (sup) | Pathogenic |
The enzyme activity measurement was not available in all patients, but as it was established earlier, the PC activity often poor correlates with type of PCD [5, 6].
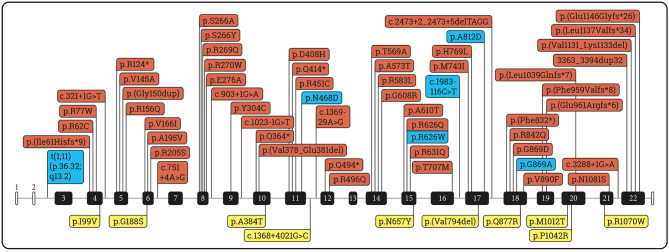
This paper adds six additional variants to the mutation spectrum in PCD deficiency. Among 62 variants in HGMD there are 42 missense/nonsense, 7 splicing, 7 small deletions, 5 small insertions, 1 duplication of 32 b.p. (Fig. 2). Two missense variants (p.Asn458Asp, p.Ala812Asp) from this work are classified according ACMG as variants of uncertain significance (VOUS) nevertheless a vivid and rather specific clinical picture of the patients allows to accept the variants as causative.
Fig. 2.
PC gene variants published in HGMD professional (ver.2022.2) and novel variants revealed in this study. Red color – damaging mutations according to HGMD; yellow – uncertain variants according to HGMD; blue – novel variants revealed in patients from our study.
5. Conclusion
PCD is variable pathology, clinically and molecularly. We describe novel rare types of the PC gene mutations that could be found only using WES/WGS methods - reciprocal translocation and deep intronic nucleotide change.
This study improves our understanding of molecular mechanisms of PCD and emphasizes the value of whole genome sequencing in cases when pyruvate metabolism defect is clinically and biochemically suspected.
Funding
The research was carried out within the state assignment of the Ministry of Science and Higher Education of the Russian Federation for RCMG.
Author statement
P Tsygankova Data curation, Writing- Original draft preparation, Sanger sequencing, WES and WGS analysis, supervision.
I Bychkov RNA analysis, figures preparing, review of the manuscript.
M Minzhenkova karyotyping and FISH-analysis.
N Pechatnikova Patients 2,3 descriptions and the review of the manuscript.
L Bessonova consulted on the diagnostics algorithm for patient 1.
G Buyanova Patients 1 description and the review of the manuscript.
I Naumchik Patients 4 description and the review of the manuscript.
N Beskorovainiy WES/WGS pipeline and raw data analysis.
V Tabakov cells cultivation for FISH and karyotype analysis.
Y Itkis panel sequencing and Sanger sequencing.
N Shilova design, analyzing the FISH and karyotype for patient 1.
E Zakharova Conceptualization.
Declaration of Competing Interest
All authors have seen and approved the manuscript.
All authors declare no conflict of interest.
Acknowledgements
The research was carried out within the state assignment of the Ministry of Science and Higher Education of the Russian Federation for RCMG.
Data availability
Data will be made available on request.
References
- 1.Carbone M.A., Applegarth D.A., Robinson B.H. Intron retention and frameshift mutations result in severe pyruvate carboxylase deficiency in two male siblings. Hum. Mutat. 2002;20:48–56. doi: 10.1002/humu.10093. [DOI] [PubMed] [Google Scholar]
- 2.Marin-Valencia I., Roe C.R., Pascual J.M. Pyruvate carboxylase deficiency: mechanisms, mimics and anaplerosis. Mol. Genet. Metab. 2010;101:9–17. doi: 10.1016/j.ymgme.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton J., Rae M.D., Logan R.W., Robinson P.H. A case of benign pyruvate carboxylase deficiency with normal development. J. Inherit. Metab. Dis. 1997;20:401–403. doi: 10.1023/a:1005350600278. [DOI] [PubMed] [Google Scholar]
- 4.Van Coster R.N., Fernhoff P.M., De Vivo D.C. Pyruvate carboxylase deficiency: a benign variant with normal development. Pediatr. Res. 1991;30:1–4. doi: 10.1203/00006450-199107000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Wang D., Yang H., De Braganca K.C., Lu J., Yu Shih L., Briones P., Lang T., De Vivo D.C. The molecular basis of pyruvate carboxylase deficiency: mosaicism correlates with prolonged survival. Mol. Genet. Metab. 2008;95:31–38. doi: 10.1016/j.ymgme.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monnot S., Serre V., Chadefaux-Vekemans B., Aupetit J., Romano S., De Lonlay P., Rival J.M., Munnich A., Steffann J., Bonnefont J.P. Structural insights on pathogenic effects of novel mutations causing pyruvate carboxylase deficiency. Hum. Mutat. 2009;30:734–740. doi: 10.1002/humu.20908. [DOI] [PubMed] [Google Scholar]
- 7.Saudubray J.M., Marsac C., Cathelineau C.L., Besson Leaud M., Leroux J.P. Neonatal congenital lactic acidosis with pyruvate carboxylase deficiency in two siblings. Acta Paediatr. Scand. 1976;65:717–724. doi: 10.1111/j.1651-2227.1976.tb18009.x. [DOI] [PubMed] [Google Scholar]
- 8.Kurkina M.V., Mihaylova S.V., Baydakova G.V., Saifullina E.V., Korostelev S.A., Pyankov D.V., Kanivets I.V., Yunin M.A., Pechatnikova N.L., Zakharova E.Y. Molecular and biochemical study of glutaric aciduria type 1 in 49 Russian families: nine novel mutations in the GCDH gene. Metab. Brain Dis. 2020;35:1009–1016. doi: 10.1007/s11011-020-00554-x. [DOI] [PubMed] [Google Scholar]
- 9.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H.L. ACMG laboratory quality assurance committee, standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsson D., Pettersson M., Gustavsson P., Förster A., Hofmeister W., Wincent J., Zachariadis V., Anderlid B.-M., Nordgren A., Mäkitie O., Wirta V., Käller M., Vezzi F., Lupski J.R., Nordenskjöld M., Syk Lundberg E., Carvalho C.M.B., Lindstrand A. Whole-genome sequencing of cytogenetically balanced chromosome translocations identifies potentially pathological gene disruptions and highlights the importance of microhomology in the mechanism of formation. Hum. Mutat. 2017;38:180–192. doi: 10.1002/humu.23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coci E.G., Gapsys V., Shur N., Shin-Podskarbi Y., Groot B.L., Miller K., Vockley J., Sondheimer N., Ganetzky R., Freisinger P. Pyruvate carboxylase deficiency type A and type C: characterization of five novel pathogenic variants in PC and analysis of the genotype–phenotype correlation. Hum. Mutat. 2019;40:816–827. doi: 10.1002/humu.23742. [DOI] [PubMed] [Google Scholar]
- 12.Ostergaard E., Duno M., Møller L.B., Kalkanoglu-Sivri H.S., Dursun A., Aliefendioglu D., Leth H., Dahl M., Christensen E., Wibrand F. 2012. Novel Mutations in the PC Gene in Patients with Type B Pyruvate Carboxylase Deficiency; pp. 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venselaar H., te Beek T.A., Kuipers R.K., Hekkelman M.L., Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semyachkina A.N., Voskoboeva E.Y., Zakharova E.Y., Nikolaeva E.A., Kanivets I.V., Kolotii A.D., Baydakova G.V., Kharabadze M.N., Kuramagomedova R.G., Melnikova N.V. Case report: a rare case of hunter syndrome (type II mucopolysaccharidosis) in a girl. BMC Med. Genet. 2019;20:66. doi: 10.1186/s12881-019-0807-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sohn Y.B., Kim S.J., Park S.W., Park H.-D., Ki C.-S., Kim C.H., Huh S.W., Yeau S., Paik K.-H., Jin D.-K. A mother and daughter with the p.R443X mutation of mucopolysaccharidosis type II: genotype and phenotype analysis. Am. J. Med. Genet. Part A. 2010;152A:3129–3132. doi: 10.1002/ajmg.a.33589. [DOI] [PubMed] [Google Scholar]
- 16.Lonardo F., Di Natale P., Lualdi S., Acquaviva F., Cuoco C., Scarano F., Maioli M., Pavone L.M., Di Gregorio G., Filocamo M., Scarano G. Mucopolysaccharidosis type II in a female patient with a reciprocal X;9 translocation and skewed X chromosome inactivation. Am. J. Med. Genet. Part A. 2014;164:2627–2632. doi: 10.1002/ajmg.a.36667. [DOI] [PubMed] [Google Scholar]
- 17.Abusaad I., Mohammed S.N., Ogilvie C.M., Ritchie J., Pohl K.R., Docherty Z. Clinical expression of Menkes disease in a girl with X;13 translocation. Am. J. Med. Genet. 1999;87:354–359. (doi:10588844) [PubMed] [Google Scholar]
- 18.Zenker M., Wermuth B., Trautmann U., Knerr I., Kraus C., Rauch A., Reis A. Severe, neonatal-onset OTC deficiency in twin sisters with a de novo balanced reciprocal translocation t(X;5)(p21.1;q11) Am. J. Med. Genet. Part A. 2005;132A:185–188. doi: 10.1002/ajmg.a.30414. [DOI] [PubMed] [Google Scholar]
- 19.Zaki M., Shehab M., El-Aleem A.A., Abdel-Salam G., Koeller H.B., Ilkin Y., Ross M.E., Dobyns W.B., Gleeson J.G. Identification of a novel recessiveRELN mutation using a homozygous balanced reciprocal translocation. Am. J. Med. Genet. Part A. 2007;143A:939–944. doi: 10.1002/ajmg.a.31667. [DOI] [PubMed] [Google Scholar]
- 20.Ray P.N., Belfall B., Duff C., Logan C., Kean V., Thompson M.W., Sylvester J.E., Gorski J.L., Schmickel R.D., Worton R.G. Cloning of the breakpoint of an X;21 translocation associated with Duchenne muscular dystrophy. Nature. 1985;318:672–675. doi: 10.1038/318672a0. [DOI] [PubMed] [Google Scholar]
- 21.Viskochil D., Buchberg A.M., Xu G., Cawthon R.M., Stevens J., Wolff R.K., Culver M., Carey J.C., Copeland N.G., Jenkins N.A., White R., O’Connell P. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-A. [DOI] [PubMed] [Google Scholar]
- 22.Spinner N.B., Rand E.B., Fortina P., Genin A., Taub R., Semeraro A., Piccoli D.A. Cytologically balanced t(2;20) in a two-generation family with alagille syndrome: cytogenetic and molecular studies. Am. J. Hum. Genet. 1994;55:238–243. http://www.ncbi.nlm.nih.gov/pubmed/8037203 [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs P.A., Browne C., Gregson N., Joyce C., White H. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J. Med. Genet. 1992;29:103–108. doi: 10.1136/jmg.29.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boone P.M., Campbell I.M., Baggett B.C., Soens Z.T., Rao M.M., Hixson P.M., Patel A., Bi W., Cheung S.W., Lalani S.R., Beaudet A.L., Stankiewicz P., Shaw C.A., Lupski J.R. Deletions of recessive disease genes: CNV contribution to carrier states and disease-causing alleles. Genome Res. 2013;23:1383–1394. doi: 10.1101/gr.156075.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.