Graphical abstract
Keywords: Stropharia rugosoannulata mushroom, Peptide distribution, Structural characteristics, Formation path, Molecular docking, ACE inhibition mechanism
Highlights
-
•
Nonapeptides and decapeptides were the main active peptides prepared by probe ultrasound.
-
•
Heptapeptides were the main active peptides prepared by bath ultrasound.
-
•
PV, RP, PL, D combined with hydrophobic amino acids were the structural characteristics of peptides.
-
•
Hydrogen bonding and cation-pi interaction were the main interaction modes between ACE and peptides.
Abstract
To reveal the structural characteristics and angiotensin-converting enzyme (ACE) inhibition mechanism of Stropharia rugosoannulata mushroom peptides prepared by multifrequency ultrasound, the peptide distribution, amino acid sequence composition characteristics, formation pathway, and ACE inhibition mechanism of S. rugosoannulata mushroom peptides were studied. It was found that the peptides in S. rugosoannulata mushroom samples treated by multifrequency ultrasound (probe ultrasound and bath ultrasound mode) were mainly octapeptides, nonapeptides, and decapeptides. Hydrophobic amino acids were the primary amino acids in the peptides prepared by ultrasound, and the amino acid dissociation of the peptide bonds at the C-terminal under the action of ultrasound was performed mainly to produce hydrophobic amino acids. Pro and Val (PV), Arg and Pro (RP), Pro and Leu (PL), and Asp (D) combined with hydrophobic amino acids were the characteristic amino acid sequence basis of the active peptides of the S. rugosoannulata mushroom. The docking results of active peptides and ACE showed that hydrogen bond interaction remained the primary mode of interaction between ACE and peptides prepared by ultrasound. The peptides can bind to the amino acid residues in the ACE active pocket, zinc ions, or key amino acids in the domain, and this results in inhibition of ACE activity. Cation–pi interactions also played an important role in the binding of mushroom peptides to ACE. This study explains the structural characteristics and ACE inhibition mechanism used by S. rugosoannulata mushroom peptides prepared by ultrasound, and it will provide a reference for the development and application of S. rugosoannulata mushroom peptides.
1. Introduction
Edible fungi are a healthy food with high protein and low fat. Their bioactive substances can be used to solve numerous problems in medicine and biotechnology. The development of health food, functional food, and drugs using edible fungi as raw materials can greatly improve the product value after processing compared with the raw materials [1]. The Stropharia rugosoannulata mushroom, also known as Tricholoma matsutake, is an edible fungus with a high straw utilization rate. The flavor of the S. rugosoannulata mushroom is unique, and it is rich in nutrients such as proteins, peptides, and amino acids [2], [3]. It is a high-quality raw material for the development of edible mushroom proteins, peptides, and their derivatives.
Peptides can be developed into new ‘natural, nutritious, and safe’ food ingredients and health products because of their important nutritional and bioactive effects. Peptides can be classified as endogenous or exogenous according to their sources. According to its function, a peptide can be a physiologically active peptide or a food sensory peptide. At present, researchers have conducted a great deal of research on physiologically active peptides from mushrooms, and have focused on the preparation and pharmacological activity of peptides with antioxidant [4], [5], antimicrobial [6], [7], antitumor [8], antihypertensive [4], [5], [9], [10], and immune regulation [8] effects.
Through previous research, we found that the free peptides in the S. rugosoannulata mushroom reached 112.84–125.56 mg/g dry weight. Therefore, the development of active peptides is a new research direction for the deep processing of the S. rugosoannulata mushroom. Through the optimization of the ultrasonic extraction process for the preparation of peptides from the S. rugosoannulata mushroom, we found that when prepared by 300 W/L ultrasonic power density and 20/28 kHz synchronous dual frequency (probe ultrasound and bath ultrasound mode) for 20 min, the content of peptides reached 343.52–385.29 mg/g dry weight, with satisfactory biological activities for the peptide matrixes. At the concentration of peptide matrix 2 mg/mL, the angiotensin-converting enzyme (ACE) inhibition rate was greater than 94%. The IC50 of peptide matrixes is 0.037–0.069 mg/mL, and the ACE inhibitory activity is 3.07 times higher than that of a peptide matrix without ultrasonic treatment, and the peptide matrixes also exhibited antioxidant activity [11]. Therefore, the structural characteristics of peptides, ultrasonic action site, and bioactivity mechanism of the S. rugosoannulata mushroom peptides prepared by ultrasound deserve further analysis.
As a green and pollution-free technology, the use of ultrasonics in food processing circumvents the problems of food contamination and environmental pollution due to excessive use of chemical solvents, and the destruction of food nutrients caused by high processing temperatures. Ultrasonic technology can significantly increase the processing efficiency and food quality by strengthening the mass transfer effect [12], [13], [14]. Ultrasound technology plays an important role in the preparation and increase in activity of peptides [15], [16], [17]. Ultrasound technology accelerates protein hydrolysis by inducing protein structure expansion and exposing protein cleavage sites, which provides a new method for deeply developing protein resources and assists in the preparation of bioactive peptides [18], [19], [20].
Therefore, we chose peptide matrixes obtained by our previous ultrasonic preparation process and (i) analyzed the peptide sequences using the peptidomics method to clarify the amino acid sequence in the peptide matrixes prepared by ultrasound; (ii) used proteomics to mine the precursor proteins of the peptide formation pathway under the action of an ultrasonic physical field and the specific action site of ultrasound; and (iii) used molecular docking to analyze binding sites to clarify the action mode between active peptides and ACE, and elucidate the ACE inhibitory mechanism. This study will increase our understanding of structural characteristics and the ACE inhibitory mechanism of S. rugosoannulata mushroom peptides produced by ultrasound.
2. Materials and methods
2.1. Preparation of the peptide matrix
Fresh S. rugosoannulata mushrooms were provided by the Shanghai Gulinyuan Cooperative (mushroom strain certificate 2004062, NCBI strain release No. SRR14469700). Fresh mushroom samples were dehydrated by hot air drying. The dried S. rugosoannulata mushrooms were crushed by a pulverizer (80 mesh), and a liquid concentrate of S. rugosoannulata mushroom was prepared at 50 g/L for ultrasonic treatment to obtain the mushroom peptide matrix. The probe ultrasound and bath ultrasound were developed by the Institute of Food Physical Processing of Jiangsu University, and the equipment used was the same as that reported by Chen [20]. Ultrasonic treatment was administered at 300 W/L ultrasonic power density, 20/28 kHz synchronous dual frequency, for 20 min. The samples were centrifuged at 8000× g for 10 min, and the supernatant was then filtered using the AKTA flux membrane filtration system equipped with a GE 3000 Da hollow fiber membrane (Glaser Life Technology Co., Ltd., Shanghai, China) to collect the ultrafiltrate, which was then freeze-dried at −70 °C for 48 h to obtain the peptide matrix of the S. rugosoannulata mushroom. The peptide matrixes prepared by probe ultrasound and bath ultrasound were named as PBUF and FPUF, respectively.
2.2. Methods
2.2.1. Peptide sequence analysis method
A Ziptip C18 column was used for desalting pretreatment of the peptide matrix. The samples were eluted with 60% ACN/0.1% TFA, and then dried in vacuum to obtain the pretreated peptide matrix.
Liquid chromatography tandem mass spectrometry (LC-MS/MS) (Thermo Easy NLC liquid chromatograph, Thermo Scientific Q-Exactive Orbitrap mass spectrometer, 75 µm I.D. ×150 mm Acclaim PepMap RSLC C18 Nanoviper chromatographic column, Thermo Fisher Scientific, Roskilde, Denmark) was used to analyze the peptide sequences of the FPUF and PBUF pretreated peptide matrixes. The pretreated peptide matrix was dissolved in 20 μL of 0.1% formic acid and 5% acetonitrile. Next, it was fully oscillated in a vortex, and then centrifuged at 8000× g and 4 °C for 20 min. The supernatant was transferred to a loading tube, and 8 μL was injected for mass spectrometry identification. For the liquid chromatography mobile phase A, 0.1% formic acid was used, and for mobile phase B, 0.1% formic acid and 80% acetonitrile were used. The LC-MS/MS setting parameters, identification parameters, and filtering parameters are shown in Supplementary Table 1. The peptide sequences of samples were mapped to the UniProtKB precursor protein sequence database. A total of 26 kinds of protein sequences were obtained by searching the keyword Stropharia rugosoannulata (https://www.uniprot.org/).
2.2.2. Analysis of peptide specific action sites
According to the peptide sequences identified by mass spectrometry, combined with peptigram online visual analysis software (https://bioware.ucd.ie/peptigram), the molecular weight distribution and amino acid information for peptides were extracted from the peptide database. Then, the specific action sites of peptides produced by ultrasound were analyzed.
2.2.3. Prediction of peptide activity
The BIOPEP-UWM website (https://biochemia.uwm.edu.pl/biopep-uwm/) was used to predict the activity of peptides identified by LC-MS/MS, and the peptide ranker score was obtained.
2.2.4. Inhibitory mechanism used by peptides against ACE
According to the peptide ranker score predicted by the BIOPEP-UWM website, combined with the peptide spectrum abundance data obtained by LC-MS/MS, the peptides with a high peptide ranker score or MS abundance were identified. The interaction between mushroom peptides and ACE was simulated by molecular docking, and the inhibitory mechanism against ACE used by peptides was predicted. The structure of ACE (PDB: 1O86) was optimized by molecular docking software. Semi-flexible docking between ACE and peptides was conducted, and the peptide-ACE complex structure with the highest affinity score was selected. The binding sites and action modes of peptides and ACE were analyzed, and the mechanism of peptides exerting ACE inhibitory activity was predicted.
2.2.5. Statistical analysis of the data
The data collected by LC-MS/MS was retrieved and analyzed by Peaks software. Microsoft Excel was used to process the data. A two-layer pie plot was used for peptide MS data analysis and visualization. Line maps for peptide and protein precursor analysis and visualization were plotted in Python.
3. Results
3.1. Peptide sequence assay
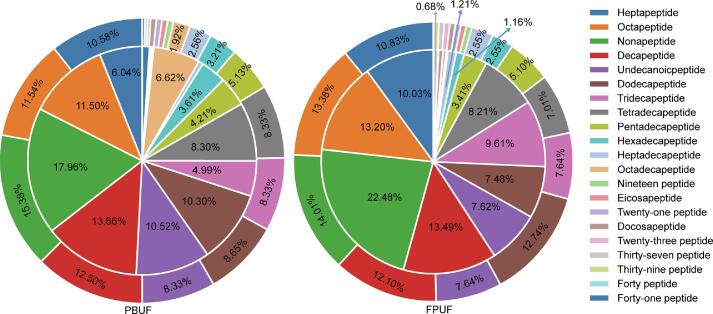
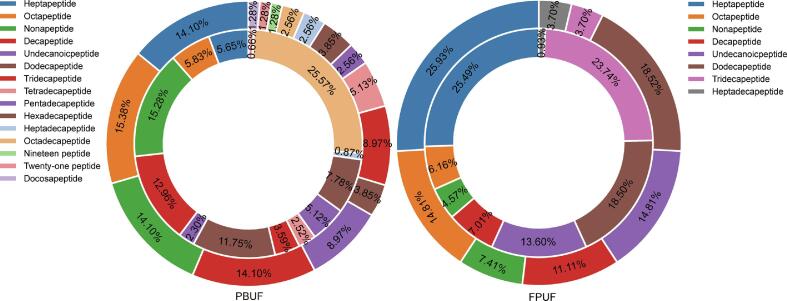
Under the filter setting parameters of peptide −10 lgp ≥ 15 and protein −10 lgp ≥ 20, a total of 310 peptides were identified in the peptide matrix PBUF, and a total of 155 peptides were identified in the peptide matrix FPUF. There were more types of peptides identified in the peptide matrix PBUF. The distribution of peptide number and peak area obtained from PBUF and FPUF is shown in Fig. 1. The peptide MS areas of octapeptides, nonapeptides, and decapeptides were relatively high, and the number of heptapeptides- pentadecapeptides was relatively rich.
Fig. 1.
Distribution of peptide number and peak area in PBUF and FPUF. Note: The internal pie plot is for peptide MS area, and the external pie plot is for peptide number.
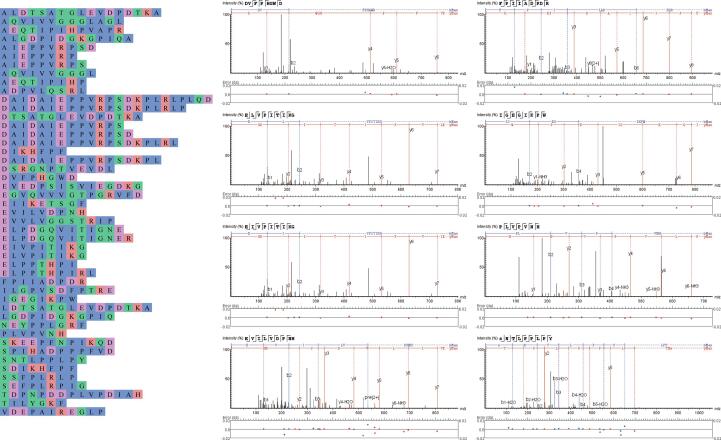
There were 47 common peptides identified in the PBUF and FPUF peptide matrixes (Supplementary Table 2). Among the common peptides, the peptide MS areas of the octapeptide, nonapeptide, and decapeptide were higher, with nonapeptides being the most abundant. The distribution of amino acids in common peptides was analyzed (Fig. 2). Among the common peptides, 17 hydrophobic amino acids were distributed at the N-terminal, of which Ala accounted for a large proportion (10); 21 acidic amino acids, composed of Asp (10) and Glu (11); 10 polar uncharged amino acids, in which Ser appeared 6 times. Among the C-terminal amino acids, hydrophobic amino acids accounted for a high proportion (51%), and consisted mainly of Phe, Leu, Pro, and Ala. C-terminal acidic amino acids (9) accounted for 19%, and Asp was the main acidic amino acid. C-terminal polar uncharged amino acids (8) accounted for 17%, and Gly was the main polar uncharged amino acid. C-terminal basic amino acids accounted for 13%.
Fig. 2.
MS diagram of common peptides and partial peptides in PBUF and FPUF. Note: Hydrophobic amino acids F, I, L, M, V, W, A, and P; basic amino acids R, K, and H; acidic amino acids D and E; and polar uncharged amino acids G, S, T, C, N, Q, and Y.
The above results indicate that there were more acidic amino acids at the N-terminal of the peptides in PBUF and FPUF, followed by hydrophobic amino acids. The proportion of hydrophobic amino acids at the C-terminal was dominant. The proportion of basic amino acids at the C-terminal and N-terminal was low. The frequency of hydrophobic amino acids in the peptide was higher (284), with the frequency of Pro being the highest (92), followed by Ile, Val, and Leu (55, 45, and 40 respectively). The frequency of P-I, P-V, and P-L combinations was also higher. Acidic amino acid Asp (56) was the second-highest frequency amino acid, and was incorporated mostly in D-P and D-A combinations.
Ultrasound technology has been proven to be conducive to the generation of hydrophobic amino acids at the peptide end. From the results of common peptides, there were 24 types of peptides that produced hydrophobic amino acids at the C-terminal, accounting for 51%, which is in accordance with the action law of ultrasound technology. There were similar sequences for some of the peptides produced by ultrasound, such as SDIKHFPF and DIKHFPF, AIEPPVRPSD, AIEPPVRPS and AIEPPVRP, DAIDAIEPPVRPSDKPLRLP, DAIDAIEPPVRPSDKPLRL, and DAIDAIEPPVRPSDKPL. The C-terminal and N-terminal amino acids of peptides can depolymerize under the action of ultrasound to form many peptides with similar amino acid sequences (Supplementary Table 2). The high proportion of hydrophobic amino acids (52%), polar uncharged amino acids (21%), and acidic amino acids (17%) was the basis of the characteristic amino acid sequences of the peptides in PBUF and FPUF.
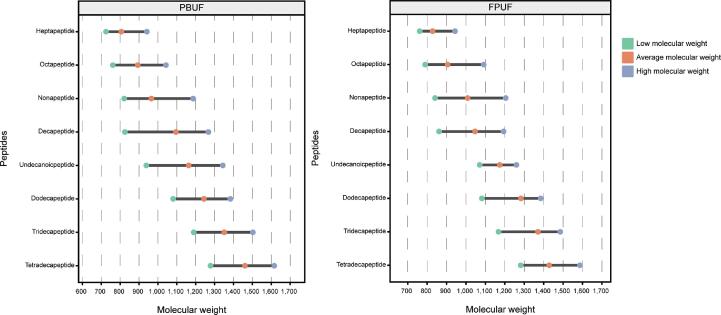
The molecular weight distributions of unique peptides in PBUF (263) and FPUF (108) were analyzed. The average molecular weight of the peptides with the same peptide chain length in the two peptide matrixes was similar. The molecular weight distribution ranges of decapeptides in PBUF and nonapeptides in FPUF were broader (Fig. 3).
Fig. 3.
Molecular weight of unique peptides in PBUF and FPUF.
Seqlogo was used to analyze the distribution of amino acids in the unique peptides in the PBUF and FPUF peptide matrixes of S. rugosoannulata mushroom. The amino acid characters represented the proportion. The larger the amino acid characters, the higher the proportion. Hydrophobic amino acids were still the primary amino acids in the unique peptides of PBUF and FPUF peptide matrixes. Among the unique peptides of PBUF, the proportion of basic amino acids in the second position at the C-terminal was high, and the proportion decreased with the increase of the peptide chain. The proportion of acidic amino acids D and E and polar uncharged amino acids S and G also increased with the rise of the peptide chain. The N-terminal and the second position of the N-terminal of the unique peptide in the FPUF peptide matrixes, D, E, S, and G, accounted for a relatively high proportion. Due to the limited number of statistical peptides in FPUF, the amino acid distribution characteristics in the identified peptides only represented some of the characteristic structures of the peptides of S. rugosoannulata mushroom prepared by bath ultrasound. By comparing the distribution characteristics of unique peptide amino acids in PBUF and FPUF, it can be seen that amino acids with a high proportion in FPUF can also be found in PBUF peptide matrixes. Due to the more abundant types of peptides in PBUF, the ratio of corresponding amino acids decreased. In a word, hydrophobic amino acids were the primary amino acids in the unique peptides of PBUF and FPUF peptide matrixes. The proportion of D, E, K, H, and R in oligopeptides (heptapeptides – decapeptides) was also high, and the ratio of S, G, and T increased with the increase of the peptide chain (Fig. 4).
Fig. 4.
Amino acid distribution of unique peptides in PBUF and FPUF. Note: A-H, the unique peptides in PBUF, a-h, the unique peptides in FPUF.
The Bio-UWM peptide ranker was used to predict the activity of common peptides (Supplementary Table 2). There are 13 common peptides with a score greater than 0.5. Among them, the MS area of peptides with a predicted score greater than 0.5 in PBUF accounted for 23% of the common peptides, and the MS area of peptides with a predicted score greater than 0.5 in FPUF accounted for 16%. All peptides exhibited satisfactory water solubility. Among the 13 predicted active peptides, there were 3 hydrophobic amino acids at the N-terminal, 4 polar uncharged amino acids, and 6 acidic amino acids. Most of the C-terminal amino acids were hydrophobic amino acids (9), acidic amino acids (2), basic amino acids, and polar uncharged amino acids (1). The amount of hydrophobic amino acids was the highest (83), including Pro (31), followed by Asp, Ile, and Leu (18, 15, and 11, respectively).
The scoring prediction rule from the BIO-UWM website is based on the distribution law of amino acids in the active peptide sequences reported in the literature, and hydrophobic amino acids have been proven to be closely related to the activity of peptides in many studies. From the above results, among the identified peptides of PBUF and FPUF, hydrophobic amino acids still appear most frequently in the peptides with a score greater than 0.5. Among the common peptides with a score greater than 0.5, the combination of PV, RP, and PL appeared more frequently. D was combined more often with hydrophobic amino acid residues, which may be the characteristic amino acid sequence basis of the active peptide of the S. rugosoannulata mushroom.
3.2. Ultrasound specific action site assay
The amide bonds dissociated by ultrasound were analyzed. Through peptide amino acid sequence alignment, and from the statistical results of amide dissociation sites of 263 unique peptides of PBUF, the probe ultrasound promoted the depolymerization of peptide C-terminal amino acids. The depolymerization of C-terminal amino acids occurring between hydrophobic amino acids and polar uncharged amino acids accounted for 30% of the amide bonds, between hydrophobic amino acids accounted for 22%, between hydrophobic and basic amino acids accounted for 15%, between hydrophobic and acidic amino acids accounted for 7%, and the dissociated amide bond between non-hydrophobic amino acids accounted for 26%. From the amount of amide bond dissociation of peptides by probe ultrasound, it was proven again that ultrasound can promote the dissociation and exposure of hydrophobic amino acids (Supplementary Table 3 shows 51 peptides with similar sequences in PBUF, and Supplementary Fig. S1 shows the protein sequences and peptide information for PBUF).
Based on the activity prediction of the BIO-UWM website, it was found that among the unique peptides of PBUF, the predicted activity score of 78 peptides was higher than 0.5, and the total MS area accounted for 35% (Fig. 5, Supplementary Table 4). The MS area for the nonapeptides and decapeptides was higher, and the numbers of heptapeptides and octapeptides were abundant. The peptide MS areas for dodecapeptides, hexadecapeptides, and octadecapeptides were also higher.
Fig. 5.
Distribution of unique activity peptide number and peak area in PBUF and FPUF. Note: An internal layer pie plot for unique peptide MS area is shown, as well as an external layer pie plot for unique peptide number.
The peptides with similar amino acid sequences among the 108 unique peptides in FPUF were analyzed. The amide dissociation sites produced by bath ultrasound indicated that the depolymerization of C-terminal amino acids occurring in the peptide bonds related to hydrophobic amino acids accounted for 86% of the amide bonds, polar uncharged amino acids accounted for 50%, acidic amino acids accounted for 29%, and basic amino acids accounted for 14%. It was proven again that ultrasound can promote the dissociation and exposure of hydrophobic amino acids (Supplementary Table 5 shows 24 peptides of similar sequence in FPUF). Based on the activity prediction from the BIO-UWM website, the predicted score of 27 peptides in FPUF was higher than 0.5, and the total proportion of the MS area was 29% (Fig. 5, Supplementary Table 6). Among the active peptides of FPUF, there were advantages for the heptapeptides in the MS area and the number of peptides.
The commonality between the unique peptides of PBUF and FPUF consisted of the ability of ultrasound to dissociate amide bonds at the C-terminal and mainly produce hydrophobic amino acids. The number of amide bonds associated with the ultrasonic dissociation of hydrophobic amino acids and non-polar amino acids was high. There were 34 hydrophobic amide bonds dissociated by probe ultrasound, and they were related to 14 basic amino acids, 10 acidic amino acids, and 23 non polar amino acids, while 12 hydrophobic amide bonds dissociated by bath ultrasound were related to 2 basic amino acids, 4 acidic amino acids, and 7 non polar amino acids. Owing to the different numbers of unique peptides, the number of dissociated amide bonds by probe ultrasound was higher than that by bath ultrasound. Based on the prediction score of active peptides from the BIO-UWM website, the MS area proportion of active peptides in unique peptides prepared by ultrasound was higher than 30%. Among the active peptides produced by ultrasound, a nonapeptide and a decapeptide in PBUF and heptapeptide in FPUF were the main peptides produced by the respective ultrasound modes.
The precursor protein sequence of the common peptide was analyzed. The precursor protein sequences for 45 common peptides were found in PBUF and FPUF. Some peptides were identified in several protein sequences. Through UniProt protein sequence alignment, it was found that there was high similarity (identity >70%) between protein sequences where the peptides were repeatedly identified. Considering the matching number of peptides in the protein, only one version of precursor protein information for the same peptide was retained. The statistical results are shown in Supplementary Fig. S2. The precursor protein sequence with the most common peptides was A0D2LMI9 (11), and this was also the protein sequence that was used to annotate the greatest number of peptides prepared by probe and bath ultrasound. In the PBUF prepared by probe ultrasound, 36 peptides were annotated (Fig. S1), and in the FPUF prepared by bath ultrasound, 15 peptides were annotated. The protein contained elongation factor 1-alpha function.
In the PBUF, among the 78 unique peptides with activity score greater than 0.5, 53 active peptides found a precursor protein. The statistical results were obtained through precursor protein sequence alignment and peptide matching, and are shown in Supplementary Fig. S3. A0D2LMI9 remained the precursor protein sequence with the greatest number of unique peptides (11) prepared by probe ultrasound. In the FPUF, among the 28 unique peptides with an activity score greater than 0.5, 20 active peptides successfully found precursor proteins. The statistical results are shown in Supplementary Fig. S4. The precursor protein sequence with the greatest number of unique peptides was A0A409X3S3 (4), which was prepared by bath ultrasound.
3.3. Mechanism used by peptides to inhibit ACE
Molecular docking was used to predict the ACE inhibitory activity of peptides. Molecular docking is a technology based on the principles of receptor structure, electrostatic matching between receptor and ligand molecules, chemical environment complementarity, and spatial complementarity to simulate and find the most optimal binding mode and evaluate the stability of the binding conformation. It is used to identify peptides that can interact with target proteins and explain their biological mechanism.
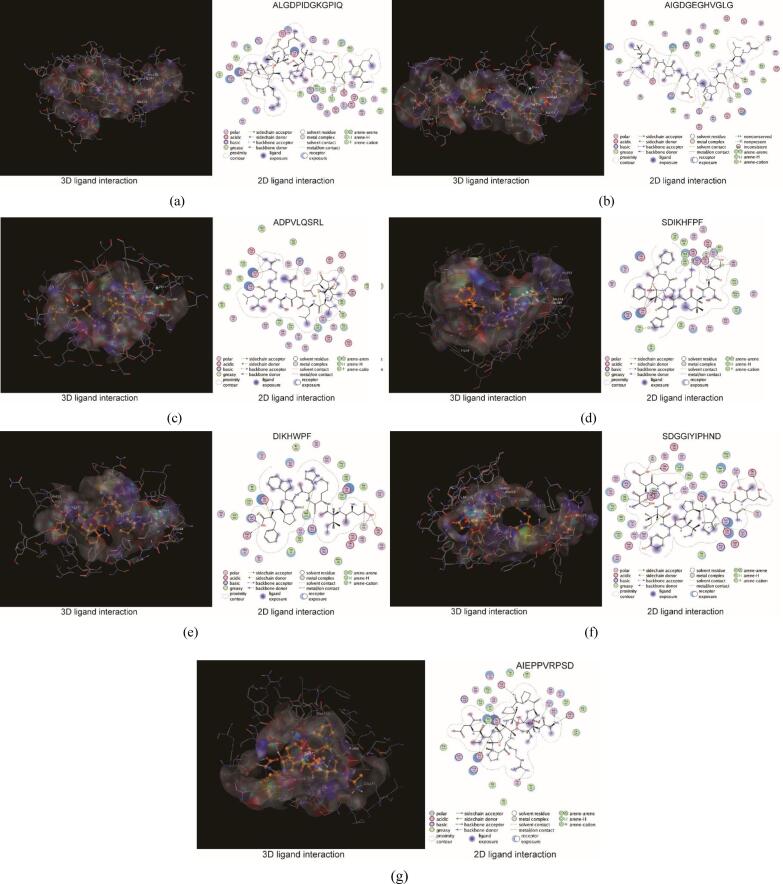
The results of possible amino acid binding sites in the ACE receptor protein cavity confirmed by molecular docking are shown in Fig. 6 (the amino acid binding sites are represented by yellow spheres). The amino acid binding sites in the cavity were composed of 133 amino acid residues, including three active pockets, S1 (Ala354, Glu384, and Tyr523), S2 (Gln281, His353, His513, Lys511, and Tyr520), and S1′ (Glu162) and Zn2+, as reported in the literature [21]. The docking results for several selected active peptides with ACE are shown in Fig. 7.
Fig. 6.
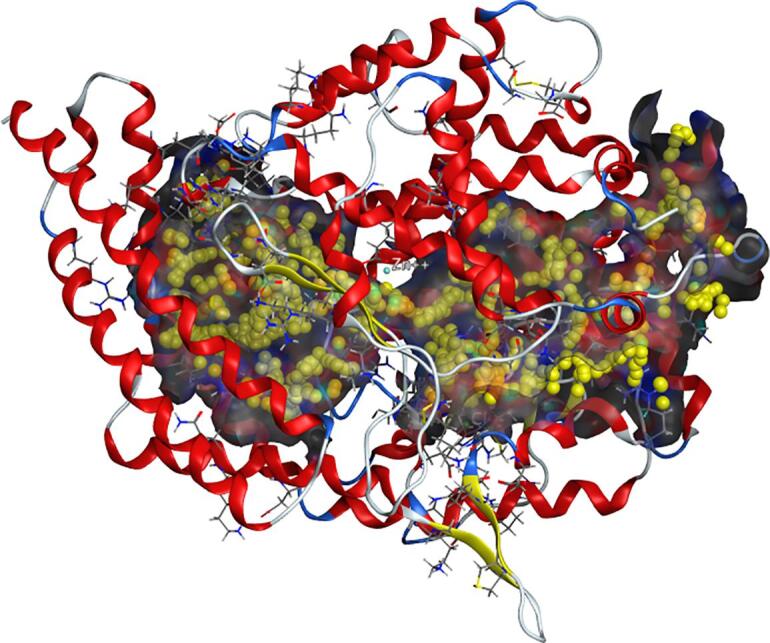
ACE docking cavity and amino acid binding sites.
Fig. 7.
3D and 2D ligand interactions between peptides and ACE amino acids.
The docking score for ALGDPIDGKGPIQ (tridecapeptide, molecular weight 1280, peptide ranker score 0.50) was −14.52 kcal/mol. The peptide participated in hydrogen bond interaction with amino acid residues Ala354, Glu384, and other amino acid residues of ACE (5). It also participated in ionic interaction with Glu123 and Glu403, and H-pi interaction with 5-ring His383. The oxygen atoms in the peptides and Zn2+ (2.04 Å) participated in metal receptor interactions (Fig. 7(a)). Based on the structural analysis of active peptides on the BIO-UWM website, the amino acids with ACE inhibitory activity contained in the peptide were GP, KG, GK, LG, Gd, DG, and KGP.
The docking score of AIGDGEGHVGLG (dodecapeptide, molecular weight 1081, peptide ranker score 0.55) was −16.38 kcal/mol. Nine hydrogen bonds with different distances were formed between the peptide and amino acid residues Ala354, Glu384, Gln281 and other amino acid residues of ACE. The hydrogen bond with S2 Gln281 was the shortest (2.96 Å), and Asp121 participated in pi–pi interaction with 5-ring His410. The oxygen atoms in peptides and Zn2+ (2.10 Å) participated in metal receptor interactions (Fig. 7(b)). The amino acids with ACE inhibitory activity contained in the peptide were the combinations of VG, IG, GL, GH, GE, AI, LG, GD, EG, and DG. Liu et al. [22] discovered that His was the main residue that interacted between peptides (YLVR and TLVGR, inhibitory peptides derived from the hazelnut) and ACE, and cation–pi interaction was crucial to the binding affinity between the peptide and ACE. Similarly, the pi–pi interaction between AIGDGEGHVGLG and His410 also promoted binding between the peptide and ACE.
The docking score of ADPVLQSRL (nonapeptide, molecular weight 998, peptide ranker score 0.54) was −12.72 kcal/mol. Eight hydrogen bonds with different distances were formed between the peptide and amino acid residues Ala354, Glu384, and other amino acid residues of ACE, and ionic interaction occurred between Glu123 and Asp121. The hydrogen bond between ADPVLQSRL and ACE active pocket S1 Ala354 is the shortest (2.69 Å). The oxygen atoms in the peptides and Zn2+ (2.06 Å) participated in metal receptor interactions (Fig. 7(c)). The amino acids with ACE inhibitory activity contained in the peptide were RL and LQ.
The docking score of SDIKHFPF (octapeptide, molecular weight 990, peptide ranker score 0.89) was −12.06 kcal/mol. The peptide participated in hydrogen bond interaction with amino acid residues Ala354, Glu384, and other amino acid residues of ACE (4 hydrogen bonds), and the hydrogen bond between peptide and ACE active pocket S1 Ala354 was the shortest (2.84 Å). Peptides participated in ionic interactions with amino acid residues such as active pocket S1 Glu384, and participated in cation-pi interaction with 5-ring His353 and His387. The oxygen atoms in the peptides and Zn2+ (2.04 Å) participated in metal receptor interactions (Fig. 7(d)). The amino acid with ACE inhibitory activity contained in the peptide was FP. Although the combination of amino acids in SDIKHFPF that inhibit ACE as predicted by BIO-UWM was only FP, the interaction between the peptide and ACE might be the main reason for its high activity.
The docking score of DIKHWPF (heptapeptide, molecular weight 941, peptide ranker score 0.90) was −12.27 kcal/mol. Hydrogen bond interaction (7 hydrogen bonds) occurred between the peptide and the amino acid residues Glu384 and other amino acid residues of ACE. Two oxygen atoms in the peptide interacted with Zn2+ (2.15 Å/2.60 Å), and metal receptor interactions resulted (Fig. 7(e)). Although the predicted DIKHWPF peptide ranker score based on BIO-UWM was very high, BIO-UWM did not show that the peptide exhibited ACE inhibitory activity. The website predicted that the peptides were the DPP-III inhibitor and DPP-IV inhibitor, and the active amino acid combinations were WP, HW, KH, and PF. Therefore, the predicted score was related to the DPP-III and DPP-IV inhibitors. Ashok et al. [23] identified heptapeptide GIPLPLI from the fat globulin membrane protein (FGMP) hydrolysate of buffalo colostrum, which can inhibit ACE (IC50, 74.27 mM) and DPP-IV (IC50, 3.83 mM). Previous studies indicated that peptides with ACE inhibitory activity usually also exhibit antioxidant activity [15], [24], and there may be multiple active synergistic effects of peptides [25].
It has been reported that hydrophobic aromatic amino acids usually lead to higher ACE activity. The last two amino acids at the C-terminal are the main amino acids affecting ACE inhibitory activity because they can be hydrolyzed by ACE. Studies have found that the presence of Pro at the C-terminal or penultimate position will promote the ACE inhibitory activity of peptides [26]. There were higher prediction scores for SDIKHFPF and DIKHWPF on the BIO-UWM website, while the C-terminal of the two peptides is PF, and therefore, the PF amino acid combination may confer inhibition of ACE.
The docking score of SDGGIYIPHND (undecanoicpeptide, molecular weight 1187, peptide ranker score 0.26) was −15.20 kcal/mol. The peptide participated in hydrogen bond interaction (8 hydrogen bonds) with amino acid residues Glu411, Glu403, Ala356, Asn66, and other amino acid residues of ACE, and ionically interacted with His387, with a cation-pi interaction between His412 and Lys118. The oxygen atoms in the peptides and Zn2+ (2.08 Å) participated in metal receptor interactions (Fig. 7(f)). The amino acids with ACE inhibitory activity contained in the peptide were IY, IP, GI, GG, DG, and PH.
The docking score of AIEPPVRPSD (decapeptide, molecular weight 1080, peptide ranker score 0.22) was −11.65 kcal/mol. The peptide participated in hydrogen bond interactions with Glu403, Asn66, Asp358, and other amino acid residues of ACE (11 hydrogen bonds), ionic interaction with Glu403 and other amino acid residues, cation-pi interaction with Trp59, and H-pi interaction with Phe570 (Fig. 7(g)). The amino acids with ACE inhibitory activity contained in the peptide were VRP, RP, AI, VR, IE, PP, and IEP.
ADPVLQSRL, AIGDGEGHVGLG, ALGDPIDGKGPIQ, DIKHWPF, and SDIKHFPF can occupy the active center of ACE, interact with the amino acid residues in the ACE active pocket, and strongly bind with zinc ions. All these interactions play an important role in ACE inhibition. Among the above peptides, the lowest docking energy was for AIGDGEGHVGLG, and it contained the highest number of amino acid combinations with ACE inhibitory activity. The more active amino acid combinations in the peptide, the more active peptides it can degrade when taken orally. Peptides may be degraded by gastrointestinal digestive enzymes, and the stability of peptides depends on the length, molecular size, and structural characteristics of peptides and their sensitivity to digestive enzymes. Obtaining greater gastrointestinal stability, a higher absorption rate, and longer in vivo activity time and bioavailability is important for the development of functional foods rich in peptides.
The docking results for AIEPPVRPSD and SDGGIYIPHND indicate that the two peptides did not bind to the S1, S2, or S1′ amino acid residues of the ACE active pocket. Liu [27] found that the key amino acid residues in the ACE N domain were Asp43, Ala334, and Asn494, while the key amino acid residues in the ACE C domain were Asn66, Ala356, Asp358, and Glu403. Natesh et al. [28] found that His383, His387, and Glu411 are responsible for the bioactivity of ACE, and were also the regions where ACE inhibitors bind. SDGGIYIPHND participated in hydrogen bond interactions with Glu411, Asn66, Ala356, and Glu403, and AIEPPVRPSD participated in hydrogen bond interactions with Glu403, Asn66, and Ala358. The two peptides also interacted with His and Trp to form a cation–pi interaction. The interaction between hydrogen bonds and cation-pi promoted the binding of peptides and ACE, which thus inhibited the activity of ACE.
Hydrogen bond interaction remains the primary mode of action between peptides and ACE. The combination of peptides with active pocket amino acid residues, zinc ions, or key amino acids in the appropriate domain were the main reasons for the peptide to exert ACE inhibitory activity. The cation-pi interaction also plays an important role in the binding of peptides to ACE, and the His residue of ACE was the key amino acid residue in the formation of the cation–pi interaction.
4. Discussion
Many studies have shown that ultrasonic treatment can significantly increase the ACE inhibitory activity of protein hydrolysates [29], [30], [31]. Ultrasonic treatment can also increase the proportion of hydrophobic amino acids in the released peptide [32]. The results of this study showed that hydrophobic amino acids accounted for a high proportion in the peptide matrix prepared by ultrasound, which was consistent with the above research results.
Many researchers use molecular docking, bioinformatics tools, and real-time updated protein databases to focus on the preparation of ACE inhibitory peptides and the relationship between peptide structure and activity, to deeply understand the mechanism used for reducing blood pressure. Li et al. [33] obtained RGLSK from Pixian broad bean paste, and the peptide exhibited strong ACE inhibitory activity. RGLSK forms hydrogen bonds with the S1 active sites (Ala354, Tyr523, Glu384) of ACE and coordination bonds with Zn2+. From the hydrolysates of Channa striatus, Ma et al. [34] identified the peptides EYFR and LPGP, which may function as ACE inhibitors. The two peptides bind to ACE S1 and S2 amino acid residues. Hydrogen bonds, electrostatic interaction, and hydrophobic interaction are the main methods for stabilizing the ACE-peptide complex. Wei et al. [35] identified seven ACE inhibitory peptides from distilled spent grain, among which Pro-Arg with satisfactory IC50 values and high content participated in competitive inhibition with ACE. The peptide interacted with the residues near the ACE active site to form six hydrogen bonds, and the binding bond length ranged from 1.7 to 2.6 Å. Lu et al [36] found four ACE inhibitory peptides from black tea, QTDEYGNPPR, AGFAGDDAPR, IQDKEGIPPDQQR, and SIDELR. The first three peptides inhibited ACE activity in an uncompetitive manner, which required the presence of substrate. IDESLRI inhibited ACE in a non-competitive manner, in which the presence of substrate is not necessary. The molecular docking results showed that the four peptides did not bind to the active sites of ACE, which indicated that the four peptides were allosteric inhibitors. Two bovine collagen-derived ACE inhibitor peptides, VGPV and GPRGF, were examined through a molecular docking study. It was found that hydrogen bond interaction was the main mode of action between collagen peptides and ACE. GPRGF formed two hydrogen bonds with zinc ions in the active center, which may be the reason that GPRGF exhibited higher ACE inhibitory activity as compared to VGPV. These two collagen peptides did not bind to the S1 or S2 active amino acid residues, and they inhibited ACE activity mainly through a noncompetitive mechanism [37].
It was found that the inhibitor competitively binds to ACE active sites, and noncompetitively binds when it interacts with the enzyme-substrate complex. In most cases, noncompetitive inhibitors are long-chain peptides. They bind to sites other than substrate binding sites on the enzyme and affect the binding between substrate and enzyme. In some cases, the ACE inhibitory activity of long-chain peptides is higher than that of short-chain peptides (competitive inhibitors) [38]. It can be inferred that among the molecular docking peptides in this study, except for DIKHWPF, other peptides inhibited ACE activity according to website prediction, and contained a variety of ACE inhibitory amino acid combinations. The peptide possessed the ability to bind to ACE active amino acid sites, zinc ions, or major amino acid residues in the domain.
Peptide chain length, amino acid composition, and structural characteristics will affect the bioactivity of peptides. A website predictive peptide ranker score can be used as an auxiliary reference for peptide activity prediction, and molecular docking involves structural binding and energy binding. Therefore, the results of molecular docking can also be used as a predictive means for identifying active peptides as well as molecular verification. Based on this study, the inhibitory type and ACE inhibitory activity of peptide monomers can be verified by solid-phase synthesis, molecular interaction, and cell and animal tests.
5. Conclusion
The peptides in PBUF and FPUF prepared by ultrasound were mainly octapeptides, nonapeptides, and decapeptides. The peptide bonds dissociated by ultrasound were mainly amide bonds that produce hydrophobic amino acids. There was a high proportion of hydrophobic amino acids in the C-terminal of the peptide, and there were many combinations of PV, RP, and PL. D was mostly combined with hydrophobic amino acid residues. The nonapeptide and decapeptide in PBUF and heptapeptide in FPUF are the main active peptides produced by ultrasound in their respective modes. The hydrogen bond interaction was the main force that the active peptides interact with the active amino acid residues of ACE. The interaction of cation-pi also played an important role in the binding of peptides to ACE. A His residue of ACE was the critical amino acid residue that enabled cation-pi interaction. This study provides a reference for understanding the structural characteristics and ACE inhibition mechanism of peptides prepared by ultrasound.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This research was funded by Shanghai Agriculture Applied Technology Development Program, China (No. 2020-02-08-00-12-F01484), National Natural Science Foundation of China (No. 31901812).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ultsonch.2022.106074.
Contributor Information
Haile Ma, Email: mhl@ujs.edu.cn.
Yan Yang, Email: yangyan@saas.sh.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Gargano M.L., van Griensven L.J.L.D., Isikhuemhen O.S., Lindequist U., Venturella G., Wasser S.P., Zervakis G.I. Medicinal mushrooms: valuable biological resources of high exploitation potential. Plant Biosystems. 2017;151(3):548–565. [Google Scholar]
- 2.Hu S., Feng X., Huang W., Ibrahim S.A., Liu Y. Effects of drying methods on non-volatile taste components of Stropharia rugoso-annulata mushrooms. LWT – Food Science and Technology. 2020;127(3) [Google Scholar]
- 3.Chen W., Li W., Wu D.i., Zhang Z., Chen H., Zhang J., Wang C., Wu T., Yang Y. Characterization of novel umami-active peptides from Stropharia rugoso-annulata mushroom and in silico study on action mechanism. Journal of Food Composition and Analysis. 2022;110:104530. [Google Scholar]
- 4.Geng X.R., Tian G.T., Zhang W.W., Zhao Y.C., Zhao L.Y., Wang H.X., Ng T.B. A Tricholoma matsutake peptide with angiotensin converting enzyme inhibitory and antioxidative activities and antihypertensive effects in spontaneously hypertensive rats. Scientific Reports. 2016;6:24130. doi: 10.1038/srep24130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang N., Ko S.C., Kim H.S., Yang H.W., Ahn G., Lee S.C., Lee T.G., Lee J.S., Jeon Y.J. Structural evidence for antihypertensive effect of an antioxidant peptide purified from the edible marine animal Styela clava. Journal of Medicinal Food. 2020;23(2):132–138. doi: 10.1089/jmf.2019.4415. [DOI] [PubMed] [Google Scholar]
- 6.Farzaneh P., Khanahamadi M., Ehsani M.R., Sharifan A. Bioactive properties of Agaricus bisporus and Terfezia claveryi proteins hydrolyzed by gastrointestinal proteases. LWT – Food Science and Technology. 2018;91:322–329. [Google Scholar]
- 7.Mishra J., Rajput R., Singh K., Puri S., Goyal M., Bansal A., Misra K. Antibacterial natural peptide fractions from Indian Ganoderma lucidum. International Journal of Peptide Research and Therapeutics. 2018;24(4):543–554. [Google Scholar]
- 8.Sun Y.N., Hu X.L., Li W.X. Antioxidant, antitumor and immunostimulatory activities of the polypeptide from Pleurotus eryngii mycelium. International Journal of Biological Macromolecules. 2017;97:323–330. doi: 10.1016/j.ijbiomac.2017.01.043. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q., Li Y., Peng K., Wang X.L., Ding Z.Y., Liu L.M., Xu P., Liu G.Q. Isolation and characterization of three antihypertension peptides from the mycelia of Ganoderma Lucidum (Agaricomycetes) Journal of Agricultural and Food Chemistry. 2019;67(29):8149–8159. doi: 10.1021/acs.jafc.9b02276. [DOI] [PubMed] [Google Scholar]
- 10.Lau C.C., Abdullah N., Shuib A.S., Aminudin N. Novel angiotensin I-converting enzyme inhibitory peptides derived from edible mushroom Agaricus bisporus (J.E. Lange) Imbach identified by LC-MS/MS. Food Chemistry. 2014;148:396–401. doi: 10.1016/j.foodchem.2013.10.053. [DOI] [PubMed] [Google Scholar]
- 11.Li W., Chen W.C., Ma H.L., Wu D., Zhang Z., Yang Y. Ultrasonic Preparation of Stropharia rugoso-annulata peptides and analysis of their taste characteristics and pharmacological activities. Acta Edulis Fungi. 2022;29(3):11–24. [Google Scholar]
- 12.Ashokkumar M. Applications of ultrasound in food and bioprocessing. Ultrasonics Sonochemistry. 2015;25:17–23. doi: 10.1016/j.ultsonch.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Jin J., Ma H.L., Wang B., Yagoub A.E.G.A., Wang K., He R.H., Zhou C.S. Effects and mechanism of dual-frequency power ultrasound on the molecular weight distribution of corn gluten meal hydrolysates. Ultrasonics Sonochemistry. 2016;30:44–51. doi: 10.1016/j.ultsonch.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 14.Bian C., Cheng H., Yu H., Mei J., Xie J. Effect of multi-frequency ultrasound assisted thawing on the quality of large yellow croaker (Larimichthys crocea) Ultrasonics Sonochemistry. 2022;82:105907. doi: 10.1016/j.ultsonch.2021.105907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou C.S., Ma H.L., Yu X.J., Liu B., Yagoub A.A., Pan Z.L. Pretreatment of defatted wheat germ proteins (by-products of flour mill industry) using ultrasonic horn and bath reactors: Effect on structure and preparation of ACE-inhibitory peptides. Ultrasonics Sonochemistry. 2013;20(6):1390–1400. doi: 10.1016/j.ultsonch.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Kadam S.U., Tiwari B.K., Alvarez C., O'Donnell C.P. Ultrasound applications for the extraction, identification and delivery of food proteins and bioactive peptides. Trends in Food Science & Technology. 2015;46(1):60–67. [Google Scholar]
- 17.Ruan S.Y., Luo J., Li Y.L., Wang Y.C., Huang S.F., Lu F., Ma H.L. Ultrasound-assisted liquid-state fermentation of soybean meal with Bacillus subtilis: Effects on peptides content, ACE inhibitory activity and biomass. Process Biochemistry. 2020;91:73–82. [Google Scholar]
- 18.Wu Q.Y., Zhang X.F., Jia J.Q., Kuang C., Yang H.S. Effect of ultrasonic pretreatment on whey protein hydrolysis by alcalase: Thermodynamic parameters, physicochemical properties and bioactivities. Process Biochemistry. 2018;67:46–54. [Google Scholar]
- 19.Ulug S.K., Jahandideh F., Wu J.P. Novel technologies for the production of bioactive peptides. Trends In Food Science & Technology. 2021;108:27–39. [Google Scholar]
- 20.Chen W., Ma H., Wang Y.-Y. Recent advances in modified food proteins by high intensity ultrasound for enhancing functionality: Potential mechanisms, combination with other methods, equipment innovations and future directions. Ultrasonics Sonochemistry. 2022;85:105993. doi: 10.1016/j.ultsonch.2022.105993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Q.Y., Jia J.Q., Yan H., Du J.J., Gui Z.Z. A novel angiotensin-I converting enzyme (ACE) inhibitory peptide from gastrointestinal protease hydrolysate of silkworm pupa (Bombyxmori) protein: Biochemical characterization and molecular docking study. Peptides. 2015;68:17–24. doi: 10.1016/j.peptides.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 22.Liu C.L., Fang L., Min W.H., Liu J.S., Li H.M. Exploration of the molecular interactions between angiotensin-I-converting enzyme (ACE) and the inhibitory peptides derived from hazelnut (Corylus heterophylla Fisch.) Food Chemistry. 2018;245:471–480. doi: 10.1016/j.foodchem.2017.10.095. [DOI] [PubMed] [Google Scholar]
- 23.Ashok A., Brijesha N., Aparna H.S. Discovery, synthesis, and in vitro evaluation of a novel bioactive peptide for ACE and DPP-IV inhibitory activity. European Journal of Medicinal Chemistry. 2019;180:99–110. doi: 10.1016/j.ejmech.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Wang X.M., Chen H.X., Fu X.G., Li S.Q., Wei J. A novel antioxidant and ACE inhibitory peptide from rice bran protein: Biochemical characterization and molecular docking study. LWT-Food Science and Technology. 2017;75:93–99. [Google Scholar]
- 25.Xue L., Yin R.X., Howe K., Zhang P.Z. Activity and bioavailability of food protein-derived angiotensin-I-converting enzyme-inhibitory peptides. Comprehensive Reviews in Food Science and Food Safety. 2021;20(2):1150–1187. doi: 10.1111/1541-4337.12711. [DOI] [PubMed] [Google Scholar]
- 26.Ibrahim H.R., Ahmed A.S., Miyata T. Novel angiotensin-converting enzyme inhibitory peptides from caseins and whey proteins of goat milk. Journal of Advanced Research. 2017;8(1):63–71. doi: 10.1016/j.jare.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C. Jilin University; Jilin Province, China: 2021. Study on the inhibitory mechanism of food-derived bioactive peptides on ACE N and C domains. Master Thesis. [Google Scholar]
- 28.Natesh R., Schwager S.L.U., Sturrock E.D., Acharya K.R. Crystal structure of the human angiotensin-converting enzyme- lisinopril complex. Nature. 2003;421(6922):551–554. doi: 10.1038/nature01370. [DOI] [PubMed] [Google Scholar]
- 29.Guerra-Almonacid C.M., Torruco-Uco J.G., Murillo-Arango W., Mendez-Arteaga J.J., Rodríguez-Miranda J. Effect of ultrasound pretreatment on the antioxidant capacity and antihypertensive activity of bioactive peptides obtained from the protein hydrolysates of Erythrina edulis. Emirates Journal of Food and Agriculture. 2019;31:288–296. [Google Scholar]
- 30.Wali A., Ma H.L., Aadil R.M., Zhou C.S., Rashid M.T., Liu X. Effects of multifrequency ultrasound pretreatment on the enzymolysis, ACE inhibitory activity, and the structure characterization of rapeseed protein. Journal of Food Processing and Preservation. 2017;41(6):1–12. [Google Scholar]
- 31.Jia J.Q., Ma H.L., Zhao W.R., Wang Z.B., Tian W.M., Luo L., He R.H. The use of ultrasound for enzymatic preparation of ACE-inhibitory peptides from wheat germ protein. Food Chemistry. 2010;119(1):336–342. [Google Scholar]
- 32.Liang Q., Ren X., Ma H., Li S., Xu K., Oladejo A.O. Effect of low-frequency ultrasonic-assisted enzymolysis on the physicochemical and antioxidant properties of corn protein hydrolysates. Journal of Food Quality. 2017;2017:1–10. [Google Scholar]
- 33.Li M., Fan W., Xu Y. Identification of angiotensin converting enzyme (ACE) inhibitory and antioxidant peptides derived from Pixian broad bean paste. LWT-Food Science and Technology. 2021;151:112221. [Google Scholar]
- 34.Ma T., Fu Q., Mei Q., Tu Z., Zhang L.u. Extraction optimization and screening of angiotensin-converting enzyme inhibitory peptides from Channa striatus through bioaffinity ultrafiltration coupled with LC-Orbitrap-MS/MS and molecular docking. Food Chemistry. 2021;354:129589. doi: 10.1016/j.foodchem.2021.129589. [DOI] [PubMed] [Google Scholar]
- 35.Wei D., Fan W.-L., Xu Y. Identification of water-soluble peptides in distilled spent grain and its angiotensin converting enzyme (ACE) inhibitory activity based on UPLC-Q-TOF-MS and proteomics analysis. Food Chemistry. 2021;353:129521. doi: 10.1016/j.foodchem.2021.129521. [DOI] [PubMed] [Google Scholar]
- 36.Lu, Y.T., Wang, Y., Huang, D.Y., Bian, Z., Lu, P., Fan, D.M., & Wang, X.C. (2021). Inhibitory mechanism of angiotensin-converting enzyme inhibitory peptides from black tea. Journal of Zhejiang University-Science B. 22(7). 575-589. [DOI] [PMC free article] [PubMed]
- 37.Fu Y., Young J.F., Rasmussen M.K., Dalsgaard T.K., Lametsch R., Aluko R.E., Therkildsen M. Angiotensin I-converting enzyme–inhibitory peptides from bovine collagen: insights into inhibitory mechanism and transepithelial transport. Food Research International. 2016;89:373–381. doi: 10.1016/j.foodres.2016.08.037. [DOI] [PubMed] [Google Scholar]
- 38.Xue L., Wang X.D., Hu Z.H., Wu Z., Wang L.J., Wang H., Yang M. Identification and characterization of an angiotensin-converting enzyme inhibitory peptide derived from bovine casein. Peptides. 2018;99:161–168. doi: 10.1016/j.peptides.2017.09.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.