Abstract
Background
Data suggest that immunomodulation induced by DNA hypomethylating agents can sensitize tumors to immune checkpoint inhibitors. We conducted a phase 1 dose-escalation trial (NCT02998567) of guadecitabine and pembrolizumab in patients with advanced solid tumors. We hypothesized that guadecitabine will overcome pembrolizumab resistance.
Methods
Patients received guadecitabine (45 mg/m2 or 30 mg/m2, administered subcutaneously on days 1–4), with pembrolizumab (200 mg administered intravenously starting from cycle 2 onwards) every 3 weeks. Primary endpoints were safety, tolerability and maximum tolerated dose; secondary and exploratory endpoints included objective response rate (ORR), changes in methylome, transcriptome, immune contextures in pre-treatment and on-treatment tumor biopsies.
Results
Between January 2017 and January 2020, 34 patients were enrolled. The recommended phase II dose was guadecitabine 30 mg/m2, days 1–4, and pembrolizumab 200 mg on day 1 every 3 weeks. Two dose-limiting toxicities (neutropenia, febrile neutropenia) were reported at guadecitabine 45 mg/m2 with none reported at guadecitabine 30 mg/m2. The most common treatment-related adverse events (TRAEs) were neutropenia (58.8%), fatigue (17.6%), febrile neutropenia (11.8%) and nausea (11.8%). Common, grade 3+ TRAEs were neutropaenia (38.2%) and febrile neutropaenia (11.8%). There were no treatment-related deaths. Overall, 30 patients were evaluable for antitumor activity; ORR was 7% with 37% achieving disease control (progression-free survival) for ≥24 weeks. Of 12 evaluable patients with non-small cell lung cancer, 10 had been previously treated with immune checkpoint inhibitors with 5 (42%) having disease control ≥24 weeks (clinical benefit). Reduction in LINE-1 DNA methylation following treatment in blood (peripheral blood mononuclear cells) and tissue samples was demonstrated and methylation at transcriptional start site and 5’ untranslated region gene regions showed enriched negative correlation with gene expression. Increases in intra-tumoural effector T-cells were seen in some responding patients. Patients having clinical benefit had high baseline inflammatory signature on RNAseq analyses.
Conclusions
Guadecitabine in combination with pembrolizumab is tolerable with biological and anticancer activity. Reversal of previous resistance to immune checkpoint inhibitors is demonstrated.
Keywords: drug therapy, combination; methylation; pembrolizumab; guadecitabine
Key messages.
DNA hypomethylating agents may sensitize tumors to immune checkpoint inhibitors.
This phase I/II trial established the recommended phase II dose of guadecitabine 30 mg/m2, days 1–4, and pembrolizumab 200 mg on day 1 every 3 weeks.
Thirty patients were evaluable for antitumor activity; 37% had disease control (progression-free survival) for ≥24 weeks including patients previously treated with immune checkpoint inhibitors.
On tumoral analysis reduction in LINE-1 methylation was seen and methylation at transcriptional start site and 5’ untranslated region gene regions showed enriched negative correlation with gene expression.
Guadecitabine in combination with pembrolizumab is tolerable with biological and anticancer activity.
Introduction
Epigenetic dysregulation is a key mechanism in oncogenic progression.1 A mechanism of epigenetic dysregulation is aberrant methylation, triggering chromatin condensation and gene silencing and leading to impairment of corresponding protein expression.2 3 DNA hypomethylating compounds (DHCs) reduce DNA methylation. DHCs cause an inflammatory response by several mechanisms (figure 1A): (1) Induction of gene promoter demethylation resulting in upregulation of tumor-associated antigens4 5 5–7; (2) Increased expression of Class I human leukocyte antigens (HLA) which are downregulated across a range of cancer types and associated with poor outcomes,8 9 with DHCs upregulating the expression of HLA class I antigens with resultant T-cell recognition10 11 and promotion of CD8 T-cells migration to tumor12; (3) DHCs can augment T-cell response; decitabine (a nucleoside analog that reduces DNA methyltransferases) induces CD80 expression on cancer cells via demethylation of the gene promoter, contributing to induction of cytotoxic T lymphocyte response.13 DHCs can also induce type 1 interferon responses,14 15 promoting T-cell proliferation and increased IFN-gamma T-cells.16
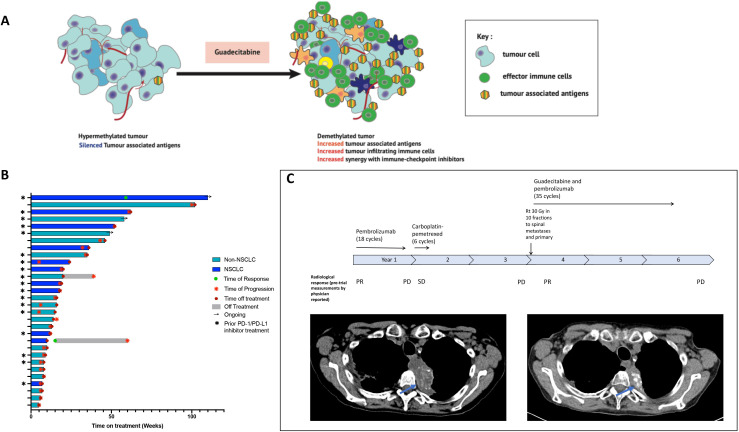
Figure 1.
(A) Proposed mechanism of action of guadecitabine and pembrolizumab based on preclinical evidence. (B) Swimmers plot of objective response (according to RECIST V.1.1) from start of treatment to disease progression. (C) A patient with adeno-NSCLC (PD-L1 greater than 50% TPS, EGFR wild-type, ALK rearrangement negative, was previously treated with pembrolizumab for 12 months followed by carboplatin and pemetrexed chemotherapy. On trial she achieved a partial response of −38% that lasted 110 weeks. C (upper panel): timeline of previous response to therapy. C (lower panel): computer tomography scan of thorax showing response in left upper lobe tumor (blue arrows) with 38% reduction in overall tumor burden by RECIST from baseline to cycle 19. ALK, anaplastic lymphoma kinase; EGFR, epidermal growth factor receptor; Gy, gray; NSCLC, non-small cell lung cancer; PD, progressive disease; PD-L1, programmed death ligand 1; PD-1, programmed death protein 1; PR, partial response; RECIST, Response Evaluation in Solid Tumors; Rt, radiotherapy; SD, stable disease; TPS, Tumor Proportion Score.
Demethylation of T cells occurs during the effector phase of chronic infection with remethylation occurring during exhaustion phase.17 Decitabine can reverse T-cell exhaustion improving T-cell responses to PD-1 (programmed cell death protein 1) inhibition with an increase in antigen specific and polyclonal T-cells in murine models.18 Demethylation of the PD-1 loci may be a mechanism of resistance to DHCs.19
PD-1 pathway blockade has led to major advances in the treatment of solid tumors. The PD-1 inhibitor pembrolizumab is licensed for treatment of malignancies including non-small cell lung cancer (NSCLC), melanoma and tumors with high tumor mutational burden.20–24 Challenges remain as single-agent activity is limited in many cancers and acquired resistance to PD-1 inhibitors an inevitability.25 We hypothesized that, given the immunostimulatory impacts of hypomethylation, the combination of DHC with pembrolizumab will enhance the efficacy of PD-1 inhibition and reverse resistance.
Methods
This open-label, dose-escalation phase I study, to determine the safety and tolerability of guadecitabine in combination with pembrolizumab, was conducted at two centers (Royal Marsden Hospital and University College London Hospitals, UK).
Eligibility criteria
Study inclusion criteria included written informed consent, age 18 years or older with histologically confirmed advanced solid tumors refractory to standard therapy or for which no conventional treatment exists, Eastern Co-operative Oncology Group performance status 0–1,26 RECIST (Response Evaluation in Solid Tumors) V.1.1 measurable disease and adequate bone marrow, renal and hepatic function. Exclusion criteria included radiotherapy, endocrine therapy, immunotherapy and chemotherapy in the 4 weeks prior to trial, brain metastases (unless, asymptomatic, treated and stable), active autoimmune disease, interstitial lung disease, history of grade 2 or higher immune-related toxicity and significant coexisting medical conditions.
Study design
Patients received guadecitabine daily on days 1–4 in 3-week cycles. Pembrolizumab 200 mg was administered every 3 weeks (Q3W). The study used a two-part design. The first part, a dose-escalation in a standard three-plus-three design with a guadecitabine starting dose level of 45 mg/m2. Dose-limiting toxicity (DLT) was defined as a drug-related toxicity occurring during the first two cycles including grade 3/4 neutropaenia or thrombocytopaenia for more than 7 days, and grade 3 or greater non-hematological toxicity. The maximum tolerated dose (MTD) was defined as the dose with a DLT rate of <33%. The expansion cohort, with a planned sample size of 20 patients, commenced once the recommended phase II dose (RP2D) of guadecitabine and pembrolizumab was established.
Safety
Safety assessments were performed at baseline, day 1, 8 and 15 of cycle 1 and 2 and day 1 of subsequent cycles including medical history and physical examination. ECG, hematology and chemistry blood analysis and urine analysis were performed. Adverse events (AEs) and laboratory parameters were assessed using Common Terminology Criteria for Adverse Events V.4.0.27
Tumor responses
Radiological assessment of disease was performed at baseline and every 6 weeks according to RECIST and iRECIST.28 29
Biomarker analysis
Paired tumor biopsies were taken at baseline, before the first dose of pembrolizumab at day 8 of cycle 2 (C2D8), and at end of treatment. Tissues were formalin-fixed and paraffin-embedded and intratumoral immune cell infiltration and PD-L1 (program death ligand-1) expression assessed by multiplexed immunohistochemistry (IHC) and immunofluorescence (online supplemental methods). Briefly, CD3 (cluster differentiation 3) IHC was performed using a rabbit anti-CD3 antibody (#A0452; rabbit polyclonal; Dako, Agilent Technologies) on the BOND RX automated staining platform (Leica Microsystems). PD-L1 IHC was performed using a rabbit anti-PD-L1 antibody (#13684; monoclonal (clone E1L3N); Cell Signaling Technology). A multiplex IF panel was performed on the BOND RX platform (Leica Microsystems) using antibodies against CD4 (#ab133616; Abcam), CD8 (#M7103, Dako, Agilent Technologies) FOXP3 (#13-4777-82, eBioscience) and PanCK (#4528S, Cell Signaling Technology).
jitc-2022-004495supp001.pdf (142.1KB, pdf)
Fresh tissue samples were snap frozen and sent for whole transcriptome sequencing (online supplemental methods). Briefly, Tumor RNA-Seq libraries were prepared using NEBNext Ultra II Directional RNA Library Prep Kit for Illumina NEB (#E7760) and ribo depletion using the NEBNext rRNA Depletion Kit (Human/Mouse/Rat) (NEB #6310). Sequencing was performed on the Illumina NextSeq 500 platform (Illumina) with 2×75 bp read length. FASTQ files were generated using BCL2FASTQ software. Transcriptomes reads were aligned to the human reference genome (GRCh37/hg19) using TopHat2 (V.2.0.7).
Methylation status by pyrosequencing of LINE-1 (long interspersed nuclear elements) and IL22RA1 (interleukin 22 receptor subunit alpha 1) was assessed in peripheral blood mononuclear cells (PBMCs) and tumor samples (online supplemental methods). LINE-1 refers to repetitive elements of DNA forming around 17% of the genome and used as a surrogate of global DNA methylation.30 Briefly, bisulfite modification of DNA using EZ DNA Methylation kit (Zymo Research) was performed followed by PCR amplification. Primers were designed using PyroMark Assay Design 2.0 Software (Qiagen). Paired two sample t-test was computed on samples for both baseline and on-treatment pyrovalues. Genome-wide DNA methylation at specific genomic loci was analyzed using Infinium Methylation EPIC BeadChip (Illumina) array, covering over 850,000 CpG sites (online supplemental methods).
The correlation of gene methylation levels of 135,047 methylation loci with RNA expression of corresponding 11,726 genes was assessed by Spearman’s correlation test. Genes with median gene expression level in the top 25th percentile and corresponding methylation loci with a methylation value SD of >0.1 were chosen for analysis.
Immunophenotyping was performed in whole blood (online supplemental methods). Lymphocytes were acquired on a FACSCanto II flow cytometer and analyzed using FACSDiva software (BD Biosciences, San Jose, California, USA).
Results
Thirty-four patients were treated into the study between January 31, 2017 and January 7, 2020 and included in the safety analysis (table 1). Dose escalation commenced at guadecitabine 45 mg/m2 days 1–4 with pembrolizumab 200 mg Q3W. Following a DLT in one of the initial three-patient cohort, a further three patients were recruited at this dose level. Following a further DLT the dose was de-escalated to 30 mg/m2 guadecitabine days 1–4. Six evaluable patients were included at this dose level with no DLTs. Twenty further patients were recruited to the expansion cohort of 30 mg/m2 guadecitabine days 1–4 in combination with pembrolizumab.
Table 1.
Demographics and clinical characteristics of all patients
| Characteristics | Escalation | Expansion |
| No of patients | 14 | 20 |
| Age (years), mean (IQR) | 52.3 (47.0–70.3) | 66.1 (56.9–73.5) |
| Sex | ||
| Male | 7 | 10 |
| Female | 7 | 10 |
| ECOG PS at baseline | ||
| 0 | 4 | 6 |
| 1 | 10 | 14 |
| Tumor type | ||
| Non-small cell lung cancer | 3 (21.4) | 11 (55.0) |
| Cervical cancer | 2 (14.3) | 0 (0) |
| Cholangiocarcinoma | 2 (14.3) | 1 (5.0) |
| Colorectal cancer | 1 (7.1) | 0 (0) |
| Breast cancer | 2 (14.3) | 0 (0) |
| Prostate cancer | 0 (0) | 2 (10.0) |
| Ovarian cancer | 1 (7.1) | 1 (0) |
| Mesothelioma | 3 (21.4) | 4 (20.0) |
| Renal cell cancer | 0 (0) | 1 (5.0) |
| Median no of prior lines of therapies and range | 2.5 (1–7) | 3 (1–8) |
ECOG PS, Eastern Co-operative group performance status; IQR, interquartile range.
DLTs and MTD
Two DLTS were observed: grade 3 febrile neutropaenia and grade 4 neutropaenia. Both events resolved within 14 days with the use of granulocyte-colony-stimulating factor. The observed DLT rate in cohort 1 of guadecitabine 45 mg/m2 days 1–4 was 33%. MTD and RP2D was established as 30 mg/m2 guadecitabine in combination with pembrolizumab 200 mg every 3 weeks.
Safety and tolerability
The most common all-grade treatment-related, treatment-emergent AEs were neutropaenia (58.8% (grade 3/4 38.2%)), fatigue (17.6% (no grade 3/4)), febrile neutropaenia (grade 3/4 11.8%), nausea (11.8% (no grade 3/4)), anemia (8.8% (no grade 3/4)) and thrombocytopaenia (8.8% (no grade 3/4)) (table 2).
Table 2.
Treatment related AEs
| TRAE | Total (N=34) | All AEs | Guadecitabine dose level | All AEs | All AEs | |||
| Escalation | All AEs | Escalation | Expansion | |||||
| 45 mg/m2 (N=6) | 30 mg/m2 (N=8) | 30 mg/m2 (N=20) | ||||||
| Grade | ≥Grade 3 | ≥Grade 3 | ≥Grade 3 | ≥Grade 3 | ||||
| Any TRAE | 18 (53%) | 53 | 6 (100%) | 16 | 5 (62.5%) | 10 | 8 (40%) | 27 |
| Neutropaenia | 13 | 20 | 4 | 6 | 3 | 5 | 6 | 9 |
| Fatigue | 0 | 6 | 0 | 1 | 0 | 1 | 0 | 4 |
| Febrile Neutropaenia | 4 | 4 | 2 | 2 | 1 | 1 | 1 | 1 |
| Anemia | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 3 |
| Nausea | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 3 |
| Thrombocytopaenia | 0 | 3 | 0 | 2 | 0 | 0 | 0 | 1 |
| Anemia | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 1 |
| Cough | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| Diarrhea | 1 | 2 | 0 | 0 | 1 | 2 | 0 | 0 |
| Fever | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 1 |
| Injection site reaction | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 1 |
| Rash | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 1 |
| Vomiting | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 1 |
AEs, adverse events; TRAE, treatment related AEs.
Antitumor activity
Thirty patients were evaluable for antitumor activity, having at least one postbaseline assessment of disease. Overall, 2 (2/30; 7%) patients achieved a confirmed RECIST 1.1 partial response (PR) and 15 (15/30; 50%) had a best response of RECIST 1.1 SD, with 11 (37%) achieving disease control of greater than 24 weeks. Of these, two patients had lack of progression observed after stopping IMP; one of these patients had initial progressive disease with subsequent PR for greater than 24 weeks (figure 1C). Of the two patients with PR both had NSCLC; one had not received previous PD-1/PD-L1 inhibitor previously and one had previously received pembrolizumab for 13 months with disease progression.
Eighteen patients had previously received prior PD-1/PDL-1 inhibitor (14 of whom experienced disease progression on prior PD-1/PDL-1 inhibitor) and were evaluable for response; of these, 7 (39%) patients had disease control of ≥24 weeks. Furthermore, 14 patients with confirmed prior disease progression on a PD-1/PD-L1 inhibitor were evaluable for response; interestingly, 7 (50%) of these patients had disease control of ≥24 weeks (figure 1). Of these seven benefiting patients, three were previously on PD-1/PD-L1 inhibition for <6 months before coming off drug for radiological disease progression, including one patient with colorectal cancer who had previously been treated with nivolumab for 8 weeks before disease progression and had clinical benefit lasting 58 weeks on trial. This patient had mismatch repair deficiency with loss of MLH1 and PMS2. A second of these patient had NSCLC and was on pembrolizumab for less than 2 months before radiological disease progression.
There were 12 evaluable patients with NSCLC recruited to this trial of whom 2 (17%) achieved a confirmed PR and 7 (58%) had SD with 5 (42%) NSCLC patients having disease control ≥24 weeks. Of these 12 evaluable patients with NSCLC, 10 had received prior PD-1 or PD-L1 inhibitor; 3 (30%) of these patients had disease control of ≥24 weeks (figure 1B).
Methylation modulation
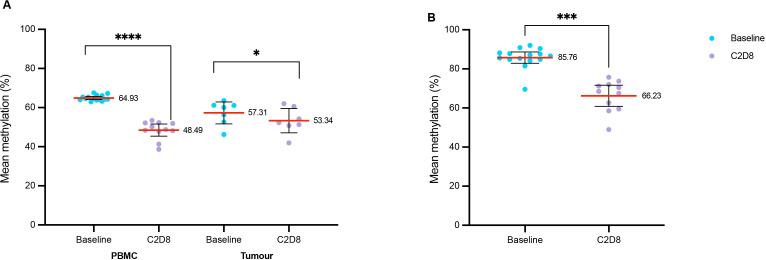
Serial blood samples from 15 treated patients were analyzed for PBMC methylation by pyrosequencing. DNA was also obtained from 7 patients with tumor biopsies at baseline and at C2D8. All samples passed in-house quality assurance criteria. The number of samples that passed quality control for these and other biomarker analyses are shown in online supplemental figure 1. LINE-1 showed a significant reduction in global methylation following treatment in PBMCs and tumor; being most pronounced in PBMC samples at C2D8 (median 48.7%, range 38.7%–53.5%) compared with baseline (median 64.3%, range 63%–66.4%) (p=5.8 × 10-7). In tumors, C2D8 global methylation (median 52.3%, range 42%–60.6%) was reduced compared with baseline (median 60%, range 46.3%–63.6%) (p=0.020). Demethylation was observed at IL22RA (single gene locus assay; highly methylated in PBMC) between blood samples at C2D8 (median 68.5, range 48.9%–75.7%), compared with baseline (median 86.6%, range 84.2%–92.1%) (p=4.54 × 10-6) (figure 2).
Figure 2.
Methylation status of LINE-1 pre-C2D8 (baseline) and post- (C2D8) guadecitabine. (A) methylation of LINE-1 in PBMC and tumor samples. (B) methylation of IL22RA1 in PBMC samples. *p<0.5, ***p<0.001, ****p<0.0001. C2D8, cycle 2 day 8; CD, cluster of differentiation; FOX-P3, forkhead box P3; IL22RA1, interleukin 22 receptor subunit alpha 1; LINE-1, long interspersed element-1; PBMC, peripheral blood mononuclear cell; PD-L1, programmed death ligand 1.
jitc-2022-004495supp002.pdf (271KB, pdf)
Selected loci of interest associated with immune responses were analyzed for change in methylation level using Illumina array. Six paired samples passed quality assurance; 64 genes involved in antigen presentation and immunomodulation were included. Differentially methylated positions with a biologically significant change in methylation were defined using a cut-off of delta-beta 0.1 in at least three of six patients. Loci demonstrating hypomethylation with guadecitabine included PRAME, PAX8 and GAGE2A. Some loci demonstrated hypermethylation including B2M (online supplemental table 1).
jitc-2022-004495supp003.pdf (90KB, pdf)
Transcriptome analysis
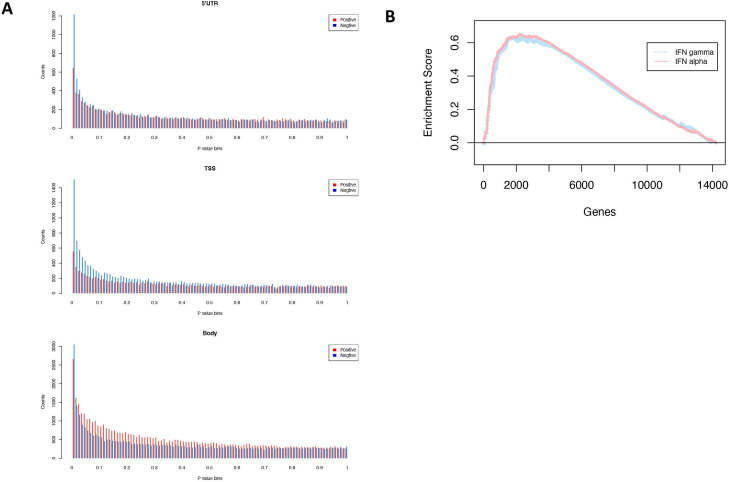
We performed RNAseq analysis for patients with paired biopsies at baseline and C2D8 and conducted an unbiased gene-set enrichment (GSEA) test to identify genes over-represented in benefiting patients; 16 paired biopsy samples passed quality control for RNAseq analysis. Patients with SD or PR for ≥24 weeks were assigned as achieving a clinical benefit (n=5), vs those who did not (n=11). GSEA test showed that biopsies from the clinical benefit group had a significantly higher general baseline inflammatory response signature (normalised enrichment score (NES)=1.9, false discovery rate (FDR) q value=1.4 × 10-5), and interferon alpha and gamma response signatures (NES=2.1 FDR q value=2.4 × 10-6 and NES=2.2 FDR q value=1.6 × 10-9) (figure 3B and online supplemental table 2).
Figure 3.
Methylation changes. (A) Correlation of p value distribution of gene methylation and its expression (Red bar—positive correlation; blue bar—negative correlation) in 5’ UTR, TSS and gene body. (B) Gene set enrichment test of IFN alpha and IFN gamma (HALLMARK) pathway in groups. Clinical benefit group versus non clinical benefit group baseline sample. C2D8, cycle 2 day 8; IFN, interferon; 5’UTR, 5’ untranslated region; TSS, transcriptional start site.
Integrated RNA and methylome analysis
To evaluate the tumor methylation profile impact on gene expression, we integrated methylation profile from the Illumina Array and RNAseq data from the four patients (baseline and C2D8 biopsies) in which both RNA and methylation data were available. Globally, methylation at transcriptional start site (TSS) and 5’ untranslated region (5’UTR) gene regions showed enriched negative correlation with expression (negative Spearman’s correlation p≤0.01 count of 1.9-fold and 2.7-fold comparing to positive test) but not gene body methylation (1.1-fold compared with positive test) (figure 3A). We then focused on PD-L1; the methylation of PD-L1 negatively associated with expression in individual samples (Pearson’s r value=−0.9, p=0.003); however, the methylation level of PD-L1 did not consistently change with guadecitabine treatment in these four patients (online supplemental figure 3).
Tumor infiltrating lymphocytes
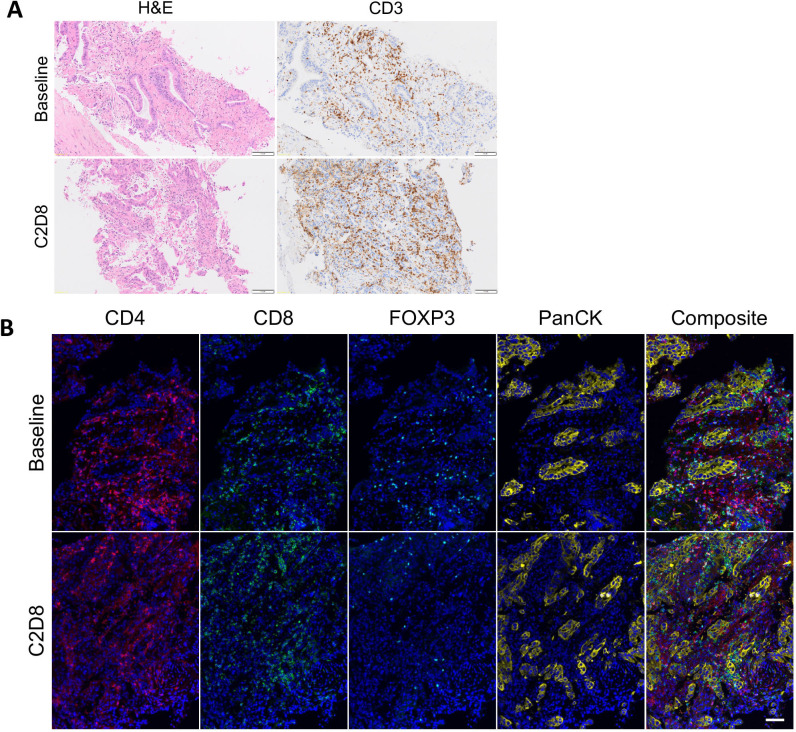
We next assessed immune cell populations by multiplex immunofluorescence for the 19 patients with paired tumor biopsies and by IHC for 18 patients with paired tumor biopsies. T-helper cells/mm2 (CD4 positive, FOXP3 negative) showed a statistically significant increase post-guadectabine, with a baseline median of 73.38 (range 0–375.5) vs 87.72 (range 0–805.9; p=0.043) at C2D8. An increase in CD3-positive cell/mm2 with guadecitabine was observed but this was not statistically significant, with a baseline median of 400.9 (range 8.65–2162) vs 575.6 (range 38.42–2881; p=0.899) at C2D8. Interestingly, three of the six patients achieving clinical benefit with paired biopsies available for analysis demonstrated an intratumoral increase of CD3 positive cells (range 0.34%–135.81% increase) (figure 4A), CD4 positive/FOXP3 negative cells (T-helper cells) (range 24.65%–503.34%), and CD8 positive cells in tumor (range 104.46%–120.7%) (figure 4B).
Figure 4.
A patient with adenosquamous NSCLC (EGFR wild-type, ALK negative and PD-L1 TPS 60%) had previously received treatment with carboplatin and gemcitabine followed by pembrolizumab for 17 months (with radiotherapy for oligometastatic progression in brain and lung during pembrolizumab course) and achieved stable disease lasting for 52 weeks on trial. (A) On IHC analysis of intratumoral T-cell subsets, C2D8 biopsy showed increase in CD3+ cells from 2161.58/mm2 to 2757.28/mm2 (increase of 27.55%) from baseline. (B) On immunofluorescence analysis of intratumoral T-cell subsets C2D8 biopsy showed an increase in CD4+/FOXP3- cells (T-helper cells) from 108.5/mm2 to 135.24/mm2 (increase of 24.65%), a decrease in CD4+ FOXP3+ cells (T-regulatory cells) from 79.57/mm2 to 22.97/mm2 (decrease of 71.13%) and an increase in CD8+ cells from 370.35/mm2 to 890.53/mm2 (increase of 140.46%) from baseline. Scale bar 100 µm. ALK, anaplastic lymphoma kinase; CD, cluster of differentiation; C2D8, cycle 2 day 8; EGFR, epidermal growth factor receptor; FOXP3, forkhead box P3; IHC, immunohistochemistry; NSCLC, non-small cell lung cancer; PanCK, pan cytokeratin; PD-L1, programmed death ligand 1; TPS, Tumor Proportion Score.
Peripheral blood immunophenotyping
On peripheral blood immunophenotyping, in 34 patients, a statistically significant increase in CD8-positive cells (1.4% increase in median percent CD8 positive cells; p=0.019) and natural killer (NK) cells (51% increase in median percent NK cells; p=0.023) was observed at cycle 2, day 15 compared with baseline following treatment.
Immune modulation: PD-L1
IHC was also performed for membranous PD-L1 Tumor Proportion Score (TPS) in 19 patients whose samples passed quality control; low levels of PD-L1 expression at baseline was observed with a median membranous TPS of 1 (range 0–70) with no change in median expression in the group at C2D8 (median expression of 1 at C2D8; p=0.852).
Discussion
To our knowledge, this is the one of the first reports evaluating guadecitabine in combination with pembrolizumab in patients with refractory solid tumors with embedded proof-of-mechanism and proof-of-concept biomarker studies in pursuit of the Pharmacological Audit Trail.31 Guadecitabine was chosen since it has advantageous pharmacokinetic properties over decitabine with data suggesting it results in favorable immunomodulation compared with other subcutaneous DHCs.14 32 The RP2D of guadecitabine in patients with hematological malignancies is 60 mg/m2 on days 1–5 of a 4-week cycle32; studies of guadecitabine in combination with chemotherapy reported MTDs of 30–45 mg/m2 in 3-weekly or 4-weekly cycles.33 34 We administered guadecitabine every 3 weeks; therefore, guadecitabine starting dose was adjusted to 45 mg/ m2 on days 1–4. Here, we established the MTD and RP2D as 30 mg/m2 of guadecitabine administered, in combination with pembrolizumab 200 mg every 3 weeks. Guadecitabine has been previously studied in combination with the CTLA4 targeting antibody ipilimumab, administered up to a dose of 60 mg/m2 on day 1–5 of a 3-week cycle without DLT.35 In this study, patients were mostly treatment-naïve, so possibly with higher bone marrow reserve than the heavily pretreated population recruited to our study. Eighty-eight per cent of patients in the 45 and 60 mg/m2 cohorts developing grade 3–4 neutropaenia, during treatment that was limited to a maximum of four cycles. A phase II trial in ovarian cancer investigated guadecitabine 30 mg/m2 on day 1–4 in combination with 200 mg intravenously every 3 weeks pembrolizumab.36
The antitumor activity observed in this trial is noteworthy, with 37% achieving disease control ≥24 weeks, for a population where 82% of patients had had ≥2 lines of prior therapy. Though a limitation of this trial in testing reversal of immunotherapy resistance was that not all patients included had experience of prior PD-1 or PD-L1 inhibitors, 47% of the patients had progressed on previous anti-PD-1/PD-L1 compounds. Five (42%) evaluable NSCLC patients experienced disease control for ≥24 weeks; 10 (83%) patients with NSCLC had progressed on previous anti-PD-1/PD-L1 therapy and the two PD-1/PD-L1 naïve patients had no expression of PD-L1 at baseline and would have been predicted to have primary resistance to PD-1 inhibition. Durable responses were observed in patients with primary resistance to PD-1 inhibitors namely two patients with colorectal cancer and NSCLC respectively who had previously progressed on PD-1 inhibition within 8 weeks of starting treatment. Rechallenging of pembrolizumab alone can produce a response; in trials of pembrolizumab and durvalumab, when patients were permitted to restart therapy having experienced disease response followed by progression after completion of the primary course of therapy (secondary resistance), disease control rates of 47.1%–83% were reported.37 To our knowledge, the response rate to rechallenging with PD-1 inhibition for tumors with primary resistance has not been previously described.
Global demethylation changes were seen in PBMCs and paired tumor biopsies, taken preguadecitabine and postguadecitabine administration, providing proof-of-mechanism. Globally, methylation of TSS and 5’UTR of genes showed enriched negative correlation with gene expression but not gene body methylation though this analysis was limited by data being only available from eight biopsies. The data herein are in keeping with existing data showing that methylation of promoter regions causes consistent negative effects on gene regulation in comparison to methylation of the gene body that may be positively correlated with gene regulation.38
Significant increases in effector T-cells were seen in some responding patients. The mechanism by which tumor inflammation and clinical response is achieved is likely to be complex and may include (1) upregulation of antigen presenting cells, (2) reversal of T-cell exhaustion, and (3) activation of T-cells. Methylation analysis of key genes involved in antigen presentation reveals variable methylation induced by guadecitabine with hypomethylation induced in some CTAs (Cancer Testis Antigens), though hypermethylation of other CTAs. In terms of T-cell exhaustion and activation; increased tumor infiltration of CD8, CD4 and T-helper cells was seen in responding patients suggesting T-cell activation. Data from this study is, however, limited by sample size, patient cohort heterogeneity, and biopsies being performed at an early time point after guadecitabine alone.
The dynamic changes reported here in circulating immune components including CD8 positive cell and NK cells may be attributable to immune stimulation; the observed changes in NK cells is worthy of further investigation given that NK cells undergo DNA methylation changes and play a role in immunosurveillance and cytotoxicity.39 To our knowledge, NK cell population changes with pembrolizumab alone have not been reported.40 41
Interestingly, baseline transcription in immune modulating pathways was more pronounced in those achieving clinical benefit; this may indicate a pre-existing inflammed phenotype (as opposed to an immune desert or immune excluded phenotype). This potential predictive biomarker of response will need to be further defined in future studies to assess utility for patient selection. Others have identified transcriptomic signatures as predictive of response to PD-1 inhibitors in NSCLC.42 43
In conclusion, the combination of guadecitabine and pembrolizumab is safe, tolerable, and has antitumor activity in patients previously treated with immune checkpoint inhibitors. Guadecitabine with the dosing schedule utilized induced robust pharmacodynamic modulation, with induction of circulating T-cell changes and T-cell infiltration into tumors in some patients, with baseline transcription signatures associating with clinical benefit and preliminary evidence of antitumor activity in NSCLC that merits further study.
jitc-2022-004495supp004.pdf (877.8KB, pdf)
Footnotes
Twitter: @ChristinaBYap, @Anna_Minchom
DP-P and WY contributed equally.
JdB and AM contributed equally.
Contributors: DP-P and AP: Data acquisition, manuscript writing, manuscript review. WY: Data analysis, statistical analysis, manuscript writing, manuscript review. MC, AF, BG, AC, CBe, CBa, SM, NM, WC, RP, IF, RM, RR, SC: Data analysis, manuscript review. TP, BJ: Data analysis, statistical analysis, manuscript review. MA, MCP, NT, UB, JL: Data acquisition, manuscript review. AZ, MP, AT, CY: Statistical analysis, manuscript review. RB: Data analysis, manuscript review. JdB: Funding acquisition, Data acquisition, manuscript review. AM: Data acquisition, manuscript writing, manuscript review, guarantor
Funding: This work was supported by Merck and Astex Pharmaceuticals. This study represents independent research supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at the Royal Marsden NHS Foundation Trust, the Institute of Cancer Research and Imperial College. The authors acknowledge funding support through a Cancer Centre grant from Cancer Research UK, and from the Experimental Cancer Medicine Centre (ECMC) Initiative to The Institute of Cancer Research and Royal Marsden.
Disclaimer: The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care.
Competing interests: DP-P: Has served on advisory boards for Takeda, Pfizer, Astra-Zeneca, Boehringer-Ingelheim, Roche. Has received honoraria from Boehringer-Ingelheim, Amgen, Pfizer, Astra-Zeneca, Takeda. Has received research funding (coapplicant) from Amgen. All unrelated to this work. CY: Has served as a consultant/independent contractor with Faron Pharmaceuticals, and as an honorarium recipient with Celgene. All unrelated to this work. MCP: Has served on advisory Board for BMS and Eisai, All unrelated to this work. RB: Has received funding from Cancer Research UK, Ovarian Cancer action and Astra Zeneca. All unrelated to this work. UB: Has received honoraria fron Astellas, Novartis, Karus Therapuetics, Pheonix Solutions, Eli Lilly, Astex, Vernalis, Boehringer IngelheimIs a recipient of an NIHR Research Professorship Award and has received CRUK funding: Cancer Research UK Scientific Executive Board, Cancer Research UK Centre Award. Cancer Research UK Drug Discovery Committee-Programme Award. All unrelated to this work JL: Research grant funding from Roche, Basilea, and Genmab unrelated to this workIs an editor for BJCJ de Bono: JdB has served on advisory boards and received fees from many companies including Astra Zeneca, Astellas, Bayer, Bioxcel Therapeutics, Boehringer Ingelheim, Cellcentric, Daiichi, Eisai, Genentech/Roche, Genmab, GSK, Janssen, Merck Serono, Merck Sharp & Dohme, Menarini/Silicon Biosystems, Orion, Pfizer, Qiagen, Sanofi Aventis, Sierra Oncology, Taiho, Vertex Pharmaceuticals. He is an employee of The ICR, which have received funding or other support for his research work from AZ, Astellas, Bayer, Cellcentric, Daiichi, Genentech, Genmab, GSK, Janssen, Merck Serono, MSD, Menarini/Silicon Biosystems, Orion, Sanofi Aventis, Sierra Oncology, Taiho, Pfizer, Vertex, and which has a commercial interest in abiraterone, PARP inhibition in DNA repair defective cancers and PI3K/AKT pathway inhibitors (no personal income). He was named as an inventor, with no financial interest, for patent 8,822,438. He has been the CI/PI of many industry sponsored clinical trials. JDB is a National Institute for Health Research (NIHR) Senior Investigator. AM: Has served on advisory boards for Janssen Pharmaceuticals, Merck Pharmaceuticals, Genmab Pharmaceuticals and Takeda Pharmaceuticals. Has received honoraria from Chugai Pharmaceuticals, Novartis Oncology, Faron Pharmaceuticals, Bayer Pharmaceuticals. Has received expenses from Amgen Pharmaceuticals and LOXO Oncology. All unrelated to this work.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplemental information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Institutional board: Royal Marsden Committee for Clinical Research CCR 4420, Ethics Committee: London - Surrey Borders Research Ethics Committee, 16/LO/1605.
References
- 1.Morel D, Jeffery D, Aspeslagh S, et al. Combining epigenetic drugs with other therapies for solid tumours - past lessons and future promise. Nat Rev Clin Oncol 2020;17:91–107. 10.1038/s41571-019-0267-4 [DOI] [PubMed] [Google Scholar]
- 2.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010;70:27–56. 10.1016/B978-0-12-380866-0.60002-2 [DOI] [PubMed] [Google Scholar]
- 3.Maio M, Covre A, Fratta E, et al. Molecular pathways: at the crossroads of cancer epigenetics and immunotherapy. Clin Cancer Res 2015;21:4040–7. 10.1158/1078-0432.CCR-14-2914 [DOI] [PubMed] [Google Scholar]
- 4.Fratta E, Coral S, Covre A, et al. The biology of cancer testis antigens: putative function, regulation and therapeutic potential. Mol Oncol 2011;5:164–82. 10.1016/j.molonc.2011.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sigalotti L, Fratta E, Coral S, et al. Intratumor heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation-regulated and functionally reverted by 5-aza-2'-deoxycytidine. Cancer Res 2004;64:9167–71. 10.1158/0008-5472.CAN-04-1442 [DOI] [PubMed] [Google Scholar]
- 6.Guo ZS, Hong JA, Irvine KR, et al. De novo induction of a cancer/testis antigen by 5-aza-2'-deoxycytidine augments adoptive immunotherapy in a murine tumor model. Cancer Res 2006;66:1105–13. 10.1158/0008-5472.CAN-05-3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coral S, Parisi G, Nicolay HJMG, et al. Immunomodulatory activity of SGI-110, a 5-aza-2'-deoxycytidine-containing demethylating dinucleotide. Cancer Immunol Immunother 2013;62:605–14. 10.1007/s00262-012-1365-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGranahan N, Rosenthal R, Hiley CT, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 2017;171:e11:1259–71. 10.1016/j.cell.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campoli M, Ferrone S. HLA antigen changes in malignant cells: epigenetic mechanisms and biologic significance. Oncogene 2008;27:5869–85. 10.1038/onc.2008.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonsatti E, Nicolay HJM, Sigalotti L, et al. Functional up-regulation of human leukocyte antigen class I antigens expression by 5-aza-2'-deoxycytidine in cutaneous melanoma: immunotherapeutic implications. Clin Cancer Res 2007;13:3333–8. 10.1158/1078-0432.CCR-06-3091 [DOI] [PubMed] [Google Scholar]
- 11.Coral S, Sigalotti L, Colizzi F, et al. Phenotypic and functional changes of human melanoma xenografts induced by DNA hypomethylation: immunotherapeutic implications. J Cell Physiol 2006;207:58–66. 10.1002/jcp.20540 [DOI] [PubMed] [Google Scholar]
- 12.Luo N, Nixon MJ, Gonzalez-Ericsson PI, et al. DNA methyltransferase inhibition upregulates MHC-I to potentiate cytotoxic T lymphocyte responses in breast cancer. Nat Commun 2018;9:248. 10.1038/s41467-017-02630-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L-X, Mei Z-Y, Zhou J-H, et al. Low dose decitabine treatment induces CD80 expression in cancer cells and stimulates tumor specific cytotoxic T lymphocyte responses. PLoS One 2013;8:e62924. 10.1371/journal.pone.0062924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fazio C, Covre A, Cutaia O, et al. Immunomodulatory properties of DNA hypomethylating agents: selecting the optimal epigenetic partner for cancer immunotherapy. Front Pharmacol 2018;9:1443. 10.3389/fphar.2018.01443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiappinelli KB, Strissel PL, Desrichard A, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 2015;162:974–86. 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Zhang Y, Chen M, et al. Increased IFNγ+ T cells are responsible for the clinical responses of low-dose DNA-demethylating agent decitabine antitumor therapy. Clin Cancer Res 2017;23:6031–43. 10.1158/1078-0432.CCR-17-1201 [DOI] [PubMed] [Google Scholar]
- 17.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15:486–99. 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghoneim HE, Fan Y, Moustaki A, et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 2017;170:e19:142–57. 10.1016/j.cell.2017.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H, Bueso-Ramos C, DiNardo C, et al. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia 2014;28:1280–8. 10.1038/leu.2013.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadgeel S, Rodríguez-Abreu D, Speranza G, et al. Updated analysis from KEYNOTE-189: pembrolizumab or placebo plus pemetrexed and platinum for previously untreated metastatic nonsquamous non-small-cell lung cancer. J Clin Oncol 2020;38:1505–17. 10.1200/JCO.19.03136 [DOI] [PubMed] [Google Scholar]
- 21.Antonarakis ES, Piulats JM, Gross-Goupil M, et al. Pembrolizumab for treatment-refractory metastatic castration-resistant prostate cancer: multicohort, open-label phase II KEYNOTE-199 study. J Clin Oncol 2020;38:395–405. 10.1200/JCO.19.01638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reck M, Rodríguez-Abreu D, Robinson AG, et al. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J Clin Oncol 2019;37:537–46. 10.1200/JCO.18.00149 [DOI] [PubMed] [Google Scholar]
- 23.Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 2020;21:1353–65. 10.1016/S1470-2045(20)30445-9 [DOI] [PubMed] [Google Scholar]
- 24.Robert C, Ribas A, Schachter J, et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol 2019;20:1239–51. 10.1016/S1470-2045(19)30388-2 [DOI] [PubMed] [Google Scholar]
- 25.Schoenfeld AJ, Hellmann MD. Acquired resistance to immune checkpoint inhibitors. Cancer Cell 2020;37:443–55. 10.1016/j.ccell.2020.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oken MM, Creech RH, Tormey DC, et al. Toxicity and response criteria of the eastern cooperative Oncology Group. Am J Clin Oncol 1982;5:649–56. 10.1097/00000421-198212000-00014 [DOI] [PubMed] [Google Scholar]
- 27.Diagnosis; NCIDoCTa . Common terminology criteria for adverse events (CTCAE), 2020. Available: https://ctep.cancer.gov/protocolDevelopment/electronic_applications/ctc.htm#ctc_40 [Accessed 19 Oct 2020].
- 28.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000;92:205–16. 10.1093/jnci/92.3.205 [DOI] [PubMed] [Google Scholar]
- 29.Seymour L, Bogaerts J, Perrone A, et al. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol 2017;18:e143–52. 10.1016/S1470-2045(17)30074-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet 2009;10:691–703. 10.1038/nrg2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerji U, Workman P. Critical parameters in targeted drug development: the pharmacological audit TRAIL. Semin Oncol 2016;43:436–45. 10.1053/j.seminoncol.2016.06.001 [DOI] [PubMed] [Google Scholar]
- 32.Issa J-PJ, Roboz G, Rizzieri D, et al. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: a multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol 2015;16:1099–110. 10.1016/S1470-2045(15)00038-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee V, Wang J, Zahurak M, et al. A phase I trial of a guadecitabine (SGI-110) and irinotecan in metastatic colorectal cancer patients previously exposed to irinotecan. Clin Cancer Res 2018;24:6160–7. 10.1158/1078-0432.CCR-18-0421 [DOI] [PubMed] [Google Scholar]
- 34.Matei D, Ghamande S, Roman L, et al. A phase I clinical trial of guadecitabine and carboplatin in platinum-resistant, recurrent ovarian cancer: clinical, pharmacokinetic, and pharmacodynamic analyses. Clin Cancer Res 2018;24:2285–93. 10.1158/1078-0432.CCR-17-3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Giacomo AM, Covre A, Finotello F, et al. Guadecitabine plus ipilimumab in unresectable melanoma: the NIBIT-M4 clinical trial. Clin Cancer Res 2019;25:clincanres.1335.2019. 10.1158/1078-0432.CCR-19-1335 [DOI] [PubMed] [Google Scholar]
- 36.Matei D, Pant A, Moroney JW, et al. Phase II trial of guadecitabine priming and pembrolizumab in platinum resistant recurrent ovarian cancer. Journal of Clinical Oncology 2020;38:6025–25. 10.1200/JCO.2020.38.15_suppl.6025 [DOI] [Google Scholar]
- 37.Yang K, Li J, Sun Z, et al. Retreatment with immune checkpoint inhibitors in solid tumors: a systematic review. Ther Adv Med Oncol 2020;12:1758835920975353. 10.1177/1758835920975353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jjingo D, Conley AB, Yi SV, et al. On the presence and role of human gene-body DNA methylation. Oncotarget 2012;3:462–74. 10.18632/oncotarget.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia M, Wang B, Wang Z, et al. Epigenetic regulation of NK cell-mediated antitumor immunity. Front Immunol 2021;12:672328. 10.3389/fimmu.2021.672328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pico de Coaña Y, Wolodarski M, van der Haar Àvila I, et al. Pd-1 checkpoint blockade in advanced melanoma patients: NK cells, monocytic subsets and host PD-L1 expression as predictive biomarker candidates. Oncoimmunology 2020;9:1786888. 10.1080/2162402X.2020.1786888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tietze JK, Angelova D, Heppt MV, et al. Low baseline levels of NK cells may predict a positive response to ipilimumab in melanoma therapy. Exp Dermatol 2017;26:622–9. 10.1111/exd.13263 [DOI] [PubMed] [Google Scholar]
- 42.Jang H-J, Lee H-S, Ramos D, et al. Transcriptome-based molecular subtyping of non-small cell lung cancer may predict response to immune checkpoint inhibitors. J Thorac Cardiovasc Surg 2020;159:1598–610. 10.1016/j.jtcvs.2019.10.123 [DOI] [PubMed] [Google Scholar]
- 43.Hwang S, Kwon A-Y, Jeong J-Y, et al. Immune gene signatures for predicting durable clinical benefit of anti-PD-1 immunotherapy in patients with non-small cell lung cancer. Sci Rep 2020;10:643. 10.1038/s41598-019-57218-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-004495supp001.pdf (142.1KB, pdf)
jitc-2022-004495supp002.pdf (271KB, pdf)
jitc-2022-004495supp003.pdf (90KB, pdf)
jitc-2022-004495supp004.pdf (877.8KB, pdf)
Data Availability Statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplemental information.