Abstract
Objective
Klebsiella pneumoniae is a gram-negative pathogen common cause of nosocomial infections. Colistin is a last resort antibiotic to treat infections caused by K. pneumoniae. In recent years, the resistance rate to colistin has increased in K. pneumoniae. This study evaluated the prevalence of colistin resistance of K. pneumoniae isolates in Iran using a systematic review and meta-analysis.
Method
A systematic search was performed for relevant articles until August 2021 in the following database: PubMed, Scopus, SID and Google Scholar. The pooled prevalence of colistin resistance in clinical K. pneumoniae isolates analyzed using Comprehensive Meta-Analysis Software (CMA).
Results
Finally, 19 articles with appropriate criteria were included in the meta-analysis. Our results showed 6.9% of the pooled prevalence of colistin resistance in clinical K. pneumoniae isolates in Iran. The results of subgroup analysis demonstrated increase resistance of colistin from 4.8%; (95% CI 1.5–13.9%) in 2013–2018 to 8.2%; (95% CI 3.4–18.6%), in 2019–2021. Also, the results of our study showed a strong association between the carbapenem producing K. pneumoniae and increased resistance to colistin.
Conclusions
This study showed a high prevalence of colistin resistance in K. pneumoniae isolates. It is recommended that regular evaluation be performed to control colistin resistance.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12941-022-00520-8.
Keywords: Klebsiella pneumoniae, Antibiotic resistance, Colistin, Meta analysis, CMA
Introduction
Gram-negative bacterial (GNB) resistance to antimicrobials is increasing worldwide [1]. It is a significant public health problem and causes critical morbidity and mortality in hospitalized patients [2]. There is a relationship between antibiotic resistance and mortality rate of patients and length of hospital stay, and increased treatment costs [3]. Many gram-negative bacteria cause nosocomial infections, among which Klebsiella pneumoniae of the Enterobacteriaceae family plays a vital role due to its potent antibiotic resistance [4]. K. pneumoniae is a gram-negative, encapsulated, nonmotile, rod-shaped bacterium that is an actual cause of nosocomial infections that can lead to various infections, including respiratory, urinary tract and wound infections [5, 6]. Overuse of antibiotics have led to problems in the treatment of K. pneumoniae and limited options available for effective treatment of this bacterial infection [7]. The emergence of MDR K. pneumoniae has become very challenging due to their resistance to most antimicrobial drugs [8]. Polymyxin antibiotics such as colistin are one of the few antimicrobial agents that have activity against MDR-GNB and are considered the last line of treatment for MDR-GNB infections [9]. Polymyxins are cationic antimicrobial peptides that target the phosphate portion of the bacterial lipopolysaccharide (LPS), disrupts the negative charge of the outer membrane and causes cell death [10, 11]. Colistin resistance is mainly due to the covalent modification of LPS, resulting in decreased affinity between LPS and colistin [12]. Other possible mechanisms are overexpression of the efflux pump, increased capsule synthesis, and production of colistinase. The two-component system (TCS), consisting of PhoPQ and PmrAB, are regulatory systems that reduce the negative charge of lipid A and the binding affinity of colistin to LPS [13, 14]. Considering the importance of colistin as one of the last lines of treatment and the lack of a meta-analysis article on the resistance of K. pneumoniae to colistin, the present study aimed to investigate the prevalence of colistin resistance in K. pneumoniae isolates in Iran.
Material and method
Research strategy
Systematic research was performed on PubMed, Scopus, SID, and Google Scholar for published studies of prevalence colistin resistance K. pneumoniae isolates in Iran. All these databases were searched followed by this search strategy: (“Klebsiella pneumoniae” OR K.pneumoniae ) AND (Resistan* OR suscep*) AND (Colisticin OR “Polymyxin E” OR Colimycin OR colistin OR colistimethate) AND Iran (Additional file 1).
Inclusion and exclusion criteria for studies
The following studies were included in our study based on the following criteria: (1) original articles that reported the total number of K. pneumoniae isolates and the number of colistin resistant (2) Studies conducted in Iran (3) Studies were written in Persian or English.
Studies with insufficient information, reviews, comments, case report, studies not reporting K. pneumoniae isolates separately (total isolates and number of resistance) and studies that report the prevalence of colistin resistance of K. pneumoniae in animals were excluded.
Data extraction
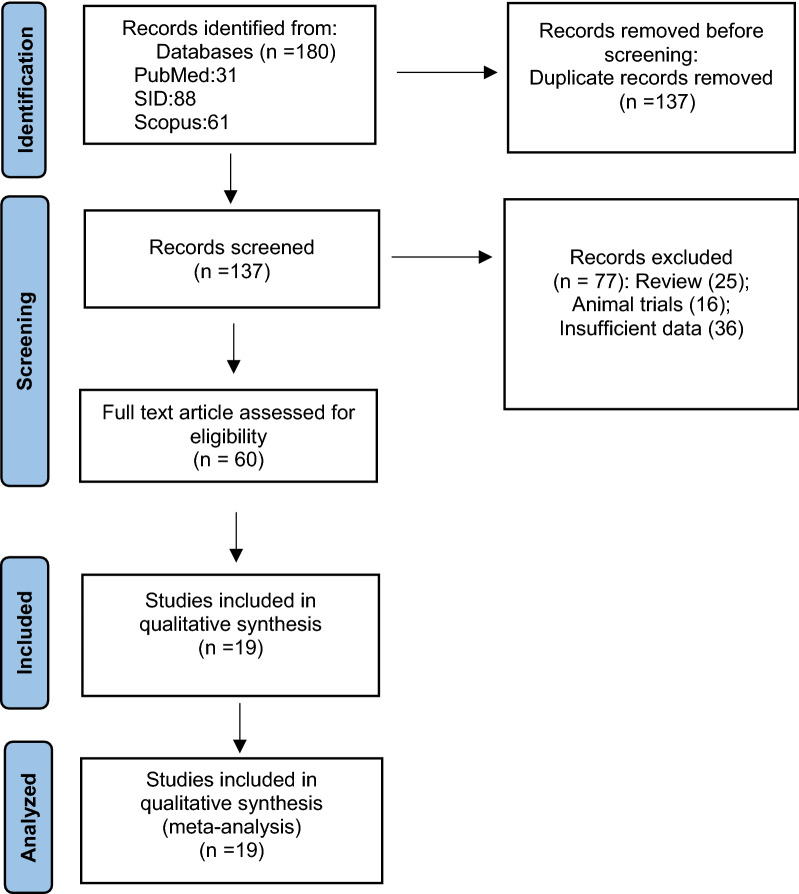
For data extraction, the abstracts and the full texts were independently searched according to the eligibility criteria by two researchers (Goodarzi F, Bavari Sh). The results were reviewed by a corresponding author (Narimia N), and any discrepancies between the researchers were resolved by a consensus and discussion. Data extraction format for the included article was: first author, publication year, study city, number of total isolates, number of colistin resistance isolates, detection method and guidelines for interrupting results (Table 1). This study selection process was presented in a Preferred Reporting Item for Systematic Reviews and Meta-Analyses (PRISMA) flowchart (Fig. 1).
Table 1.
Characteristics of included studies
| First author | Year | City | Method for assay | Guideline | Quality | Number of isolates | Number of resistance |
|---|---|---|---|---|---|---|---|
| Alizadeh [31] | 2021 | Isfahan | Broth microdilution | EUCAST | 8 | 240 | 10 |
| Heidary [32] | 2016 | Tehran | Disk diffusion | CLSI | 7 | 117 | 5 |
| Karimi [5] | 2021 | Hamadan | Broth microdilution | EUCAST | 9 | 83 | 0 |
| Mamishi [33] | 2019 | Tehran | – | CLSI | 6 | 6 | 0 |
| Mirshekar [34] | 2020 | Tehran | Broth microdilution | CLSI | 9 | 94 | 20 |
| Moosavian [35] | 2019 | Ahvaz | Broth microdilution | EUCAST | 9 | 119 | 26 |
| Saadatian [36] | 2018 | Tehran | Broth microdilution | CLSI | 7 | 81 | 14 |
| Shahraki-Zahedani [37] | 2016 | Zahedan | Disk diffusion | CLSI | 6 | 170 | 2 |
| Sharahi [38] | 2021 | Tehran | Broth microdilution | CLSI | 7 | 52 | 16 |
| Sheikh [39] | 2021 | Ahvaz | Disk diffusion | CLSI | 8 | 120 | 2 |
| Vaziri [40] | 2017 | Kermanshah | Disk diffusion | CLSI | 7 | 57 | 2 |
| Nejad [41] | 2017 | Arak | Disk diffusion | CLSI | 7 | 100 | 0 |
| Sheshblouki [42] | 2016 | Shiraz | Disk diffusion | CLSI | 6 | 111 | 4 |
| Darabi [43] | 2015 | Urmia | Disk diffusion | CLSI | 7 | 182 | 0 |
| Carbapenemase-producing K. pneumoniae | |||||||
| Bahrami [44] | 2021 | Isfahan | Disk diffusion | CLSI | 7 | 62 | 43 |
| Latifi [45] | 2020 | Bushehr | Broth microdilution | CLSI | 7 | 12 | 0 |
| Solgi [46] | Isfahan | Broth microdilution | EUCAST | 8 | 74 | 0 | |
| Jafari [47] | 2017 | Tehran | Disk diffusion | CLSI | 7 | 100 | 50 |
| Moghimi [48] | 2021 | Tabriz | Broth microdilution | CLSI | 5 | 5 | 2 |
Fig. 1.
The study Prisma flow diagram
Study quality assessment
The quality of studies was assessed using standard critical appraisal tools prepared by Joanna Briggs Institute (JBI) [15, 16]. This appraisal checklist for prevalence studies has nine essential questions. These questions focus on appropriate sampling frame, study subject and sufficient data analysis. Each item is graded yes, no or unclear. A score of 1 was given for the answer “yes”, while a score of 0 was given for the answer “no” and “unclear”. Finally, the mean score was independently calculated for each article independently with two authors (Goodarzi F, Bavari Sh) in consultation with a correspond author (Narimisa N). Studies with scores of 5 and above were rated as high quality.
Data analysis
Data Analysis of the prevalence of colistin resistance clinical K. pneumoniae isolates was calculated in Comprehensive Meta-Analysis Software (CMA). Subgroup analyses were done according to the publication year, study city, detection method and type of K. pneumoniae isolates. A random-effects model was applied to estimate the pooled prevalence of colistin resistance among clinical K. pneumoniae isolates in Iran at 95% CI.
Heterogeneity was checked using I2 test statistics. I2 ≤ 25% indicated low homogeneity, 25% < I2 ≤ 75% indicated moderate heterogeneity, and I2 > 75% indicated high heterogeneity. Funnel plot diagrams and Begg’s test were used to assess the existence of publication bias. The results were considered to have a publication bias at P < 0.05.
Results
Search results
A total of 180 studies were found, and after reviewing and removing duplicates through the Endnote software, 161 studies were excluded as they did not have the inclusion criteria. Finally, only 19 studies were potentially eligible, included in this meta-analysis (Fig. 1). In this study, 1532 isolates from 12 cities of Iran from 2015 to 2021 were examined. Disc diffusion and broth microdilution methods were used to detect antibiotic susceptibility testing of isolates. In 4 articles, the results were interpreted by EUCAST Guideline and in 15 articles by CLSI (Table 1).
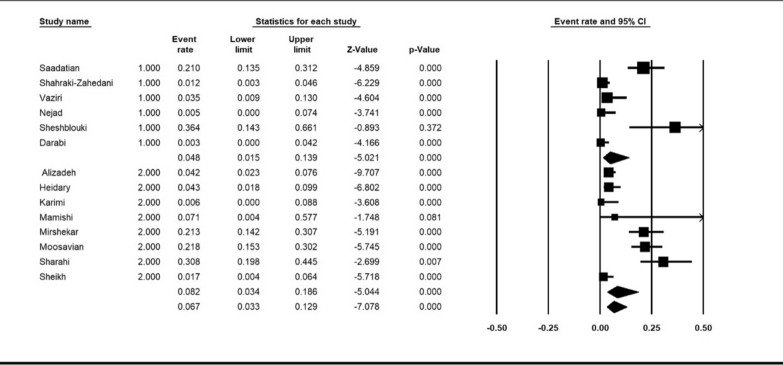
The pooled prevalence of colistin resistance in clinical K. pneumoniae isolates was estimated at 6.9% (95% CI; 3.6_ 12.8%; I2 = 87.12%; P < 0.001) (Fig. 2). Sensitivity analyses were performed, and no studies affected the prevalence of colistin-resistant isolates, as shown in the susceptibility analysis’s forest plot (Fig. 3). The result of publication bias was shown in the funnel plot; also, Begg’s tests was used to indicate the extent of bias (P = 0.352) (Fig. 4).
Fig. 2.
Forest plot of prevalence of colistin resistance of k. pneumoniae isolates in Iran
Fig. 3.
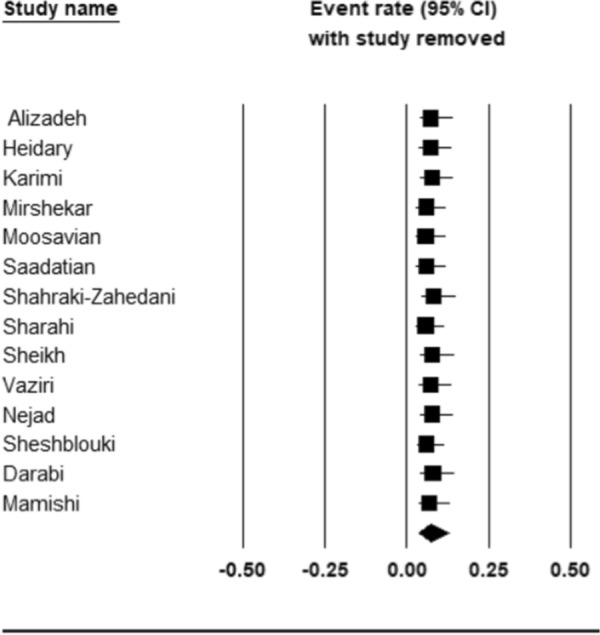
Forest plot of sensitivity analyses
Fig. 4.
Funnel plot for meta-analysis
Subgroup meta-analysis
Subgroup analysis was done according to the publication year, city of included studies, and detection method.
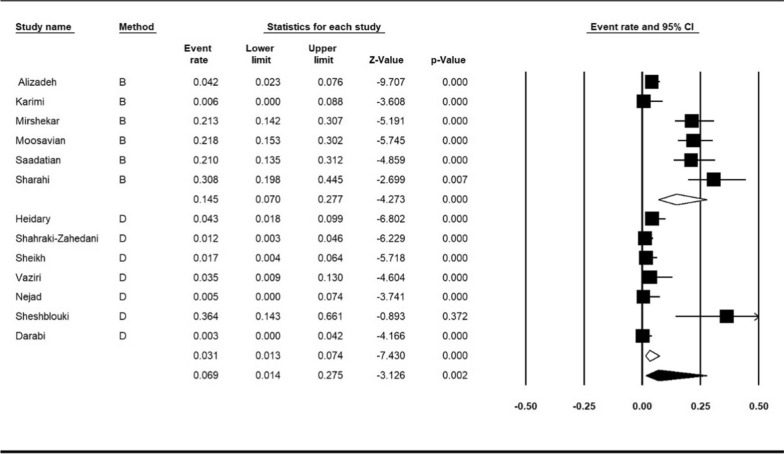
Subgroup meta-analysis for publish year was performed in two periods (2013–2018 and, 2019–2021). The year subgroup analysis indicated increase resistance of colistin from 4.8%; (95% CI 1.5–13.9%) in 2013–2018 to 8.2%; (95% CI 3.4–18.6%), (I2 = 87.12%; P < 0.001) in 2019–2021 (Fig. 5). Subgroup meta-analysis based on method of detection antimicrobial resistance revealed 14.5%; (95% CI 7–27%) for broth microdilution method and 3.1%; (95% CI 1.3–7.4%) for disk diffusion method (I2 = 88.08%; P < 0.001) (Fig. 6).
Fig. 5.
Subgroup meta-analysis for publish year (1: 2013–2018, and 2: 2019–2021)
Fig. 6.
Subgroup meta-analysis for detection method (B: broth microdilution method and D: disk diffusion method)
Subgroup meta-analysis of cites showed that Tehran with 16% (95% CI 7.3–31.5%) had the highest resistance to colistin and Urmia with 0.3% (95% CI 0.0–4.2%) had the lowest (I2 = 87.61%; P < 0.001) (Fig. 7).
Fig. 7.
Subgroup meta-analysis for cities
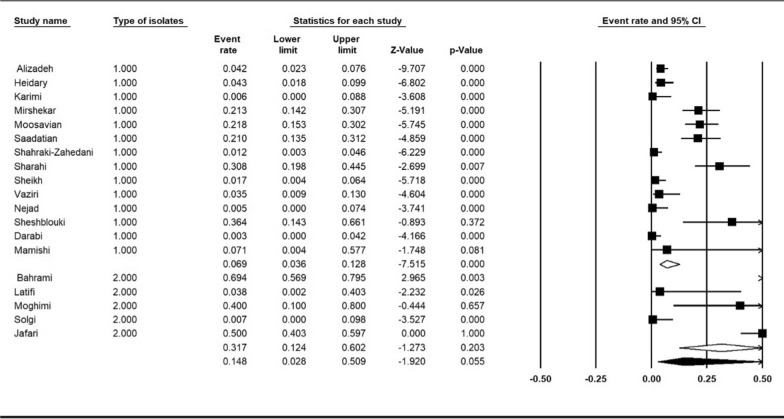
Five of the included studies reported the prevalence of colistin resistance in Carbapenemase-producing K. pneumoniae isolates. We compared the prevalence of colistin resistance in this group of isolates with sensitive isolates. The comparison of resistance in carbapenemase-producing K. pneumoniae isolates with sensitive isolates showed a higher level of resistance in carbapenemase-producing K. pneumoniae isolates 31.7% (95% CI 12.4–60.2%) compared to sensitive isolates 6.9% (95% CI 3.6–12.8%), (I2 = 92.15%; P < 0.001) (Fig. 8).
Fig. 8.
Subgroup meta-analysis based on isolates (1: sensitive K. pneumoniae isolates, 2: carbapenemase-producing K. pneumoniae isolates)
Discussion
Klebsiella pneumoniae is an important opportunistic bacterium that causes severe nosocomial infections [8]. Currently, excessive and inappropriate use of antibiotics against microbial infections has led to increased drug resistance [4, 17]. This resistance raises concerns about the choice of effective antibiotics to treat associated infections. In the past, the use of colistin was limited due to its toxicity, and in the 1970s it was replaced by antibiotics that were considered less toxic. Recently, colistin has been increasingly used as a rescue therapy alone or in combination with one or more other antimicrobials to treat carbapenem-resistant and MDR Gram-negative bacteria [18]. The use of colistin as an option to treat infections caused by carbapenem-resistant and MDR Gram-negative bacteria has led to increased resistance to this antibiotic in recent years [19].
In our research, the pooled prevalence of colistin resistance in clinical K. pneumoniae isolates was 6.9% in Iran. A study by Abdelhamid et al. reported that all 50 studied isolates from Egypt were sensitive to colistin [20]. A study by Bshabshe et al. from Saudi Arabia showed 34.5% resistance to colistin [21]. Poudyal et al. In Australia examined 21 MDR K. pneumoniae, of which 6 isolates were resistant to colistin (28.5%) [22].Other studies from India, Poland, Pakistan, and Spain reported 16, 0, 4.8 and 13% colistin resistance in K. pneumoniae isolates [23–26]. Although a similar meta-analysis is needed to compare the resistance of colistin to other countries, the study of resistance to this antibiotic in Iran showed a moderate resistance level compared to other countries. The results of our study also showed that the rate of colistin resistance increased significantly over time (2013–2018, 2019–2021), which may be due to the increased use of this antibiotic in recent years. Moreover, this increase emphasizes the need to design an accurate program to measure the resistance level to colistin. To measure the resistance to colistin CLSI guideline validates the broth microdilution, disk elution and agar dilution methods, and EUCAST guideline only suggests the microdilution method [27, 28]. Several included articles in this meta-analysis used the disk diffusion method to measure resistance, which is not recommended by EUCAST and CLSI guidelines. The results of the subgroup meta-analysis based on the measurement method showed that the rate of resistance in the disk diffusion method (3.1%) was lower than the broth microdilution method (14.5%), which may indicate that the disk diffusion method may not be sensitive enough to measure and identify resistant strains. Thus, it is essential to use the methods recommended by the standard guidelines.
Although colistin currently maintains a high activity level against most K. pneumoniae isolates, the decrease in activity against carbapenem-resistant isolates is worrisome. It has also been recognized that increased colistin use is responsible for outbreaks caused by species intrinsically resistant to polymyxins and the increasing isolation of colistin-resistant K. pneumoniae strains. Investigators have reported a correlation between the use of colistin to treat infections caused by carbapenem-resistant and the subsequent emergence of colistin-resistant strains [29]. Mansour et al. In Tunisia isolated 29 carbapenem-resistant K. pneumoniae, of which 7 (24.1%) were colistin-resistant isolates [30]. Our study showed a strong association between the carbapenem producing K. pneumoniae and increased resistance to colistin. Based on the higher percentage of colistin resistance observed among K. pneumoniae isolates producing a carbapenemase (31.7%), it will be necessary to monitor the use of colistin continually.
The present study had a few limitations. The evaluation of antibiotic resistance in K. pneumoniae was based on two methods, and this variety of methods can increase heterogeneity. Also, the heterogeneity between studies is relatively high. We used subgroup analysis to discover sources of heterogeneity and reduce the effect of heterogeneity on the results.
Conclusion
Given the recent use of colistin as a life-saving treatment for carbapenem-resistant and MDR K. pneumoniae, it is essential to know the prevalence of resistance to this antibiotic. Our study was the first study to investigate the resistance of colistin in K. pneumoniae isolates in Iran. The results of our meta-analysis showed a 6.9% prevalence of colistin among the clinical isolates of K. pneumoniae in Iran. Our findings support the idea that regulatory measures are essential for the use of colistin.
Supplementary Information
Author contributions
Conceptualization: NN. Methodology: FG and SB. Data curation, Writing—original draft preparation: NN, FGandSB. Visualization and investigation: NN, FGandSB. Supervision: NN. Writing—reviewing and editing: All authors. All authors read and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability
All the data in this review are included in the manuscript.
Declarations
Ethical approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Karakonstantis S, Kritsotakis EI, Gikas A. Pandrug-resistant Gram-negative bacteria: a systematic review of current epidemiology, prognosis and treatment options. J Antimicrob Chemother. 2020;75(2):271–282. doi: 10.1093/jac/dkz401. [DOI] [PubMed] [Google Scholar]
- 2.Bassetti M, et al. Treatment of bloodstream infections due to gram-negative bacteria with difficult-to-treat resistance. Antibiotics. 2020;9(9):632. doi: 10.3390/antibiotics9090632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosgrove SE. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis. 2006;42(Supplement_2):S82–S89. doi: 10.1086/499406. [DOI] [PubMed] [Google Scholar]
- 4.Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res Int. 2016 doi: 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karimi K, et al. Investigation of antibiotic resistance and biofilm formation in clinical isolates of Klebsiella pneumoniae. Int J Microbiol. 2021 doi: 10.1155/2021/5573388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narimisa N, et al. Type II toxin/antitoxin system genes expression in persister cells of Klebsiella pneumoniae. Rev Med Microbiol. 2020;31(4):215–220. doi: 10.1097/MRM.0000000000000232. [DOI] [Google Scholar]
- 7.Padmini N, et al. Extended spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae: critical tools for antibiotic resistance pattern. J Basic Microbiol. 2017;57(6):460–470. doi: 10.1002/jobm.201700008. [DOI] [PubMed] [Google Scholar]
- 8.Moradigaravand D, et al. Evolution and epidemiology of multidrug-resistant Klebsiella pneumoniae in the United Kingdom and Ireland. MBio. 2017;8(1):e01976-16. doi: 10.1128/mBio.01976-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Sayed Ahmed MA, et al. Colistin and its role in the Era of antibiotic resistance: an extended review (2000–2019) Emerg Microbes Infect. 2020;9(1):868–885. doi: 10.1080/22221751.2020.1754133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trimble MJ, et al. Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harbor Perspect Med. 2016;6(10):a025288. doi: 10.1101/cshperspect.a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narimisa N, et al. Biofilm establishment, biofilm persister cell formation, and relative gene expression analysis of type II toxin–antitoxin system in Klebsiella pneumoniae. Gene Rep. 2020;21:100846. doi: 10.1016/j.genrep.2020.100846. [DOI] [Google Scholar]
- 12.Khoshbayan A, et al. Mutation in mgrB is the major colistin resistance mechanism in Klebsiella pneumoniae clinical isolates in Tehran, Iran. Acta Microbiol Immunol Hungarica. 2022;69(1):61–67. doi: 10.1556/030.2022.01679. [DOI] [PubMed] [Google Scholar]
- 13.Aghapour Z, et al. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect Drug Resist. 2019;12:965. doi: 10.2147/IDR.S199844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamel M, Rolain J-M, Baron SA. The history of colistin resistance mechanisms in bacteria: progress and challenges. Microorganisms. 2021;9(2):442. doi: 10.3390/microorganisms9020442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng X, et al. The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: a systematic review. J Evid Based Med. 2015;8(1):2–10. doi: 10.1111/jebm.12141. [DOI] [PubMed] [Google Scholar]
- 16.Santos WM, Secoli SR, Püschel VA. The Joanna Briggs institute approach for systematic reviews. Revista latino-americana de enfermagem. 2018 doi: 10.1590/1518-8345.2885.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bungau S, et al. Aspects of excessive antibiotic consumption and environmental influences correlated with the occurrence of resistance to antimicrobial agents. Curr Opin Environ Sci Health. 2021;19:100224. doi: 10.1016/j.coesh.2020.10.012. [DOI] [Google Scholar]
- 18.Lim LM, et al. Resurgence of colistin: a review of resistance, toxicity, pharmacodynamics, and dosing. Pharmacotherapy. 2010;30(12):1279–1291. doi: 10.1592/phco.30.12.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peyclit L, Baron SA, Rolain J-M. Drug repurposing to fight colistin and carbapenem-resistant bacteria. Front Cell Infect Microbiol. 2019;9:193. doi: 10.3389/fcimb.2019.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdelhamid SM, et al. Genotyping and virulence analysis of drug resistant clinical Klebsiella pneumoniae isolates in Egypt. J Pure Appl Microbiol. 2020;14(3):1967–1976. doi: 10.22207/JPAM.14.3.36. [DOI] [Google Scholar]
- 21.Al Bshabshe A, et al. Rising Klebsiella pneumoniae infections and its expanding drug resistance in the intensive care unit of a tertiary Healthcare Hospital, Saudi Arabia. Cureus. 2020 doi: 10.7759/cureus.10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poudyal A, et al. In vitro pharmacodynamics of colistin against multidrug-resistant Klebsiella pneumoniae. J Antimicrob Chemother. 2008;62(6):1311–1318. doi: 10.1093/jac/dkn425. [DOI] [PubMed] [Google Scholar]
- 23.Azam M, et al. Colistin resistance among multiple sequence types of Klebsiella pneumoniae is associated with diverse resistance mechanisms: a report from India. Front Microbiol. 2021;12:215. doi: 10.3389/fmicb.2021.609840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baraniak A, et al. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008–2009. Antimicrob Agents Chemother. 2011;55(12):5493–5499. doi: 10.1128/AAC.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ejaz H, et al. Molecular epidemiology of extensively drug-resistant mcr encoded colistin-resistant bacterial strains co-expressing multifarious β-lactamases. Antibiotics. 2021;10(4):467. doi: 10.3390/antibiotics10040467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuster B, et al. Molecular epidemiology and drug-resistance mechanisms in carbapenem-resistant Klebsiella pneumoniae isolated in patients from a tertiary hospital in Valencia, Spain. J Glob Antimicrob Resist. 2020;22:718–725. doi: 10.1016/j.jgar.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Lei C-W, et al. Detection of mobile colistin resistance gene mcr-10.1 in a conjugative plasmid from Enterobacter roggenkampii of chicken origin in China. Antimicrob Agents Chemother. 2020;64(10):e01191-20. doi: 10.1128/AAC.01191-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jayol A, et al. Evaluation of three broth microdilution systems to determine colistin susceptibility of Gram-negative bacilli. J Antimicrob Chemother. 2018;73(5):1272–1278. doi: 10.1093/jac/dky012. [DOI] [PubMed] [Google Scholar]
- 29.Bradford PA, et al. Correlation of β-lactamase production and colistin resistance among Enterobacteriaceae isolates from a global surveillance program. Antimicrob Agents Chemother. 2015;60(3):1385–1392. doi: 10.1128/AAC.01870-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansour W, et al. Outbreak of colistin-resistant carbapenemase-producing Klebsiella pneumoniae in Tunisia. J Glob Antimicrob Resist. 2017;10:88–94. doi: 10.1016/j.jgar.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 31.Alizadeh H, et al. Molecular characteristics of carbapenem-resistant Klebsiella pneumoniae isolates producing blaVIM, blaNDM, and blaIMP in clinical centers in Isfahan, Iran. Jundishapur J Microbiol. 2021;14:e114473. doi: 10.5812/jjm.114473. [DOI] [Google Scholar]
- 32.Heidary M, et al. The prevalence of genes that encode quinolone resistance in Klebsiella pneumoniae strains isolated from hospitalized patients during 2013–2014. Arch Pediatr Infect Dis. 2016;10:e38343. [Google Scholar]
- 33.Mamishi S, et al. Antibiotic resistance and genotyping of gram-negative bacteria causing hospital-acquired infection in patients referred to Children’s Medical Center. Infect Drug Resist. 2019;12:3377. doi: 10.2147/IDR.S195126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirshekar M, et al. Analysis of mgrB gene mutations in colistin-resistant Klebsiella pneumoniae in Tehran, Iran. Gene Rep. 2020;21:100864. doi: 10.1016/j.genrep.2020.100864. [DOI] [Google Scholar]
- 35.Moosavian M, Emam N. The first report of emerging mobilized colistin-resistance (mcr) genes and ERIC-PCR typing in Escherichia coli and Klebsiella pneumoniae clinical isolates in southwest Iran. Infect Drug Resist. 2019;12:1001. doi: 10.2147/IDR.S192597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saadatian Farivar A, et al. RAPD PCR profile, antibiotic resistance, prevalence of armA gene, and detection of KPC enzyme in Klebsiella pneumoniae isolates. Can J Infect Dis Med Microbiol. 2018 doi: 10.1155/2018/6183162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shahraki-Zahedani S, et al. First report of TEM-104-, SHV-99-, SHV-108-, and SHV-110-producing Klebsiella pneumoniae from Iran. Revista da Sociedade Brasileira de Medicina Trop. 2016;49:441–445. doi: 10.1590/0037-8682-0114-2016. [DOI] [PubMed] [Google Scholar]
- 38.Sharahi JY, et al. Molecular characteristics of antibiotic-resistant Escherichia coli and Klebsiella pneumoniae strains isolated from hospitalized patients in Tehran, Iran. Ann Clin Microbiol Antimicrob. 2021;20(1):1–14. doi: 10.1186/s12941-021-00437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheikh AF, et al. Prevalence of carbapenemases and ESBL encoding genes among K. pneumoniae isolates obtained from an educational hospital in Ahvaz, Southwestern Iran. Gene Rep. 2021;23:101128. doi: 10.1016/j.genrep.2021.101128. [DOI] [Google Scholar]
- 40.وزیری, س. et al. بررسی فراوانی بتالاکتامازهای طیف گسترده در ایزوله های Klebsiella pneumonia جدا شده از نمونه های بیماران پنومونی وابسته به ونتیلاتور در کرمانشاه. مجله دانشکده پزشکي اصفهان, 1396. 35(444 #b0036): p.
- 41.ژاپونی نژاد, ع. et al. فراوانی ژن های کد کننده آنزیم های بتالاکتامازی AmpC با واسطه ی پلاسمید در ایزوله های بالینی کلبسیلا پنومونیه. مجله دانشکده پزشکي اصفهان, 1392. 31(249): p.
- 42.پورعلی شش بلوکی, غ., ج. مردانه, and ز. حسین زاده, تعیین مقاومت آنتیبیوتیکی متقاطع کلبسیلا پنومونیه و شناسایی سویه های با حساسیت وابسته به دوز نسبت به سفپیم. مجله دانشگاه علوم پزشكي فسا (J Adv Biomed Sci). 6(1):1395.
- 43.دارابی, ن., et al. الگوی حساسیت آنتی بیوتیکی ایزوله های بالینی کلبسیلا پنومونیه جداسازی شده از بیمارستان های آموزشی دانشگاه علوم پزشکی ارومیه و تعیین حداقل غلظت بازدارندگی ایزوله های مقاوم به ایمیپنم. مجله مطالعات علوم پزشکی (مجله پزشكي دانشگاه علوم پزشكي اروميه), 1394. 26(8): p.
- 44.Bahrami S, et al. Antimicrobial susceptibility pattern of carbapenemase-producing Gram-negative nosocomial bacteria at Al Zahra hospital, Isfahan, Iran. Iran J Microbiol. 2021;13(1):50. doi: 10.18502/ijm.v13i1.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Latifi B, et al. Phenotypic and genotypic characterization of carbapenemase-producing Klebsiella pneumoniae clinical isolates in Bushehr province, Iran. Gene Rep. 2020;21:100932. doi: 10.1016/j.genrep.2020.100932. [DOI] [Google Scholar]
- 46.Solgi H, et al. Molecular characterization of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae ST11 harbouring blaNDM-1 and blaOXA-48 carbapenemases in Iran. Microb Pathog. 2020;149:104507. doi: 10.1016/j.micpath.2020.104507. [DOI] [PubMed] [Google Scholar]
- 47.Jafari Z, et al. Molecular epidemiology and drug resistance pattern of carbapenem-resistant Klebsiella pneumoniae isolates from Iran. Microb Drug Resist. 2019;25(3):336–343. doi: 10.1089/mdr.2017.0404. [DOI] [PubMed] [Google Scholar]
- 48.Moghimi M, Haeili M, Mohajjel Shoja H. Characterization of tigecycline resistance among tigecycline non-susceptible Klebsiella pneumoniae isolates from humans, food-producing animals, and in vitro selection assay. Front Microbiol. 2021 doi: 10.3389/fmicb.2021.702006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data in this review are included in the manuscript.