Abstract
The recent spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) exemplifies the critical need for accurate and rapid diagnostic assays to prompt clinical and public health interventions. Currently, several quantitative reverse-transcription polymerase chain reaction (RT-qPCR) assays are being used by clinical, research, and public health laboratories. However, it is currently unclear if results from different tests are comparable. Our goal was to make independent evaluations of primer-probe sets used in four common SARS-CoV-2 diagnostic assays. From our comparisons of RT-qPCR analytical efficiency and sensitivity, we show that all primer-probe sets can be used to detect SARS-CoV-2 at 500 viral RNA copies per reaction. The exception for this is the RdRp-SARSr (Charité) confirmatory primer-probe set which has low sensitivity, likely due to a mismatch to circulating SARS-CoV-2 in the reverse primer. We did not find evidence for background amplification with pre-COVID-19 samples or recent SARS-CoV-2 evolution decreasing sensitivity. Our recommendation for SARS-CoV-2 diagnostic testing is to select an assay with high sensitivity and that is regionally used to ease comparability between outcomes.
SARS-CoV-2 was first identified as the cause of an outbreak of pneumonia in Wuhan, China, in December 2019, and rapidly spread around the world1–3, which exemplifies the critical need for accurate and rapid diagnostic assays to prompt clinical and public health interventions. In response, several molecular assays (i.e. RT-qPCR) were developed to detect COVID-19 cases4–7; however, it is not clear to many clinical, research, and public health laboratories which assay they should adopt or if the data are comparable. Independent evaluations of the designed primer-probe sets used in the primary SARS-CoV-2 RT-qPCR detection assays are necessary to compare findings across studies and select appropriate assays for in-house testing. Our goal was to compare the analytical efficiencies and sensitivities of the primer-probe sets used in four commonly used SARS-CoV-2 RT-qPCR assays developed by the China Center for Disease Control (China CDC)7, United States CDC (US CDC)6, Charité Institute of Virology, Universitätsmedizin Berlin (Charité)5, and Hong Kong University (HKU)4 (Supplementary Table 1). Importantly, we did not directly compare the assays per se, as that would involve many different variables. Here, we used the same (1) primer-probe concentrations (500 nM of forward and reverse primer, and 250 nM of probe); (2) PCR reagents (New England Biolabs Luna Universal Probe One-step RT-qPCR kit); and (3) thermocycler conditions (10 minutes at 55°C, 1 minute at 95°C, followed by 40 cycles [45 for clinical samples] of 10 seconds at 95°C and 30 seconds at 55°C) in all reactions.
Results
Generation of RNA transcript standards for RT-qPCR validation
A barrier to implementing and validating RT-qPCR molecular assays for SARS-CoV-2 detection was the availability of virus RNA standards. Using RNA from a SARS-CoV-2 isolate derived from an early COVID-19 case in the US8, we generated small RNA transcripts (704–1363 nt) from the non-structural protein 10 (nsp10), RNA-dependent RNA polymerase (RdRp), non-structural protein 14 (nsp14), envelope (E), and nucleocapsid (N) genes spanning the primer and probe sets of each assay (Extended Data Fig. 1; Supplementary Tables 2–3). By measuring PCR amplification using 10-fold serial dilutions of our RNA transcript standards, we found the efficiencies of each of the nine primer-probe sets to be above 90% (Extended Data Fig. 1), which match the criteria for an efficient RT-qPCR assay9. Our RNA transcripts can thus be used for assay validation, positive controls, and standards to quantify viral loads: critical steps for a diagnostic assay. Our protocol to generate the RNA transcripts is openly available10, and any clinical or research diagnostic lab can directly request them for free through our lab website (www.grubaughlab.com).
Analytical comparisons of RT-qPCR primer-probe sets
By testing each of the nine primer-probe sets using 10-fold dilutions of SARS-CoV-2 RNA derived from cell culture8 (Fig. 1a) or 10-fold dilutions of SARS-CoV-2 RNA spiked into RNA extracted from pooled nasopharyngeal swabs taken from patients in 2017 (SARS-CoV-2 RNA-spiked mocks; Fig. 1b), we again found that the PCR amplification efficiencies were near or above 90% (Fig. 1c). Our measured PCR efficiencies corresponded to an average of 3.5 cycle threshold (Ct) values between the 10-fold SARS-CoV-2 RNA dilutions (i.e. slope), with a range of 3.1–3.7 corresponding to the highest and lowest efficiencies, respectively (Fig. 1c; See Source Data for Ct values). These again match the criteria for efficient RT-qPCR9. To measure the analytical sensitivity of virus detection, we used the Ct value in which the expected linear dilution series would cross the y-intercept when tested with 1 viral RNA copy per μL of RNA. Our measured sensitivities (y-intercept Ct values) were similar among most of the primer-probe sets, except for the RdRp-SARSr (Charité) set (Fig. 1d). We found that the Ct values from the RdRp-SARSr set (using only RdRp_SARSr-P2 [probe 2]) were usually 6–10 Cts higher (lower virus detection) than the other primer-probe sets.
Fig. 1. Analytical efficiency and sensitivity of the nine primer-probe sets used in SARS-CoV-2 qRT-qPCR assays.
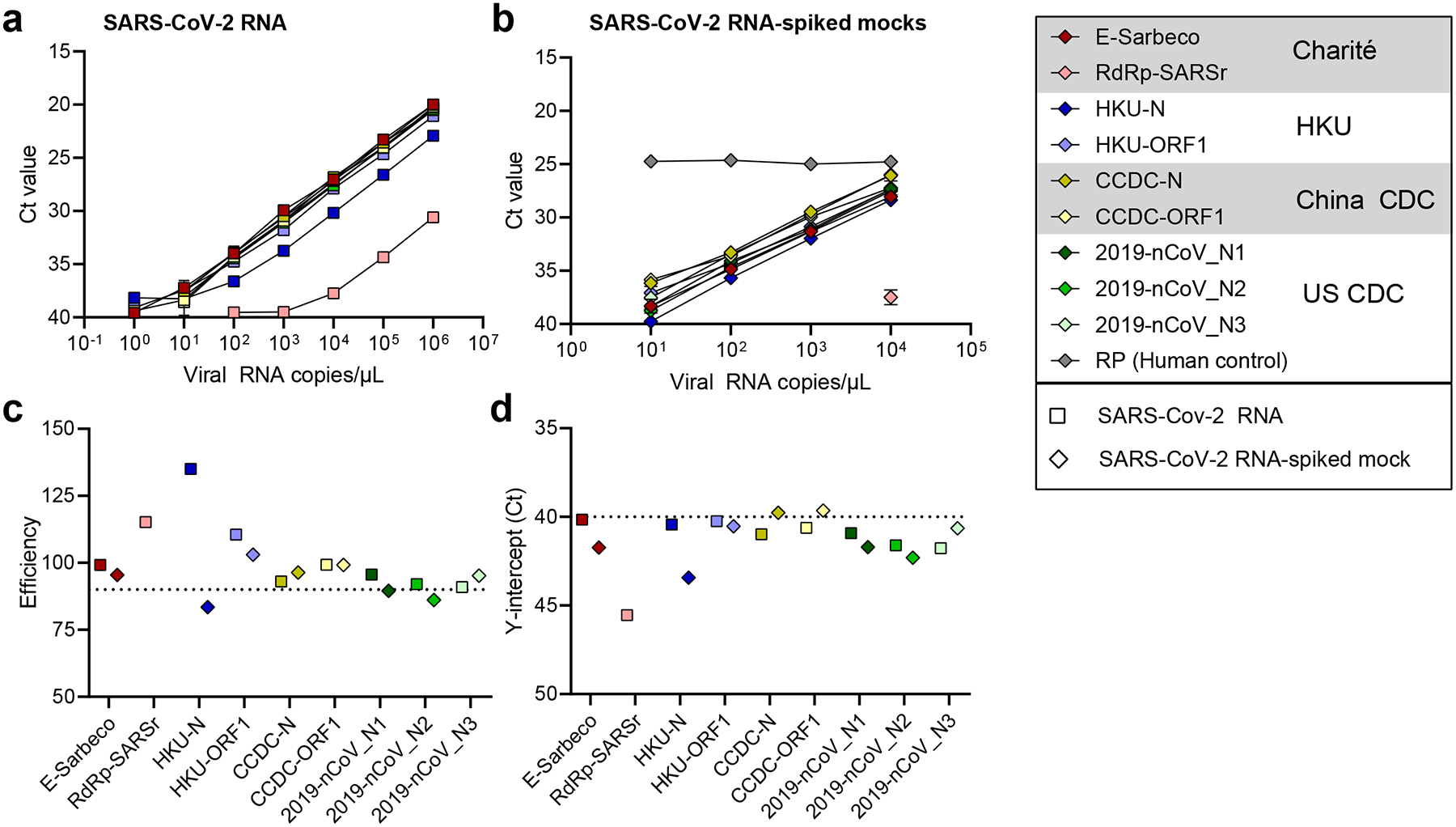
a,b, Mean Ct values for nine primer-probe sets and a human control primer-probe set targeting the human RNase P gene tested for 2 technical replicates with 10-fold dilutions of (a) full-length SARS-CoV-2 RNA and (b) pre-COVID-19 nasopharyngeal swabs spiked with known concentrations of SARS-CoV-2 RNA (SARS-CoV-2 RNA-spiked mocks). The CDC human RNase P (RP) assay was included as an extraction control. c,d, From the dilution curves in panels a and b, (c) PCR efficiency and (d) y-intercept Ct values (measured analytical sensitivity) were calculated for each of nine primer-probe sets. Symbols depict sample types: squares represent tests with SARS-CoV-2 RNA and diamonds represent SARS-CoV-2 RNA-spiked mock samples. Colors depict the nine tested primer-probe sets. The primer and probe sequences can be found in Supplementary Table 1. Data used to make this figure can be found in Source Data Fig. 1.
To determine the lower limit of detection and the occurrence of false positive or inconclusive detections, we tested the primer-probe sets using SARS-CoV-2 RNA spiked into RNA extracted from pooled nasopharyngeal swabs from respiratory disease patients during 2017 (pre-COVID-19). We made four independent pools of viral transport media from four nasopharyngeal swabs, and tested six technical replicates of each without virus (24 total replicates) or two replicates of each with 100, 101, or 102 viral RNA copies/μL of extracted nucleic acid concentrations (8 total replicates each). From the pooled nasopharyngeal swabs without viral RNA, we did not detect RT-qPCR amplification for any of the tested primer-probe sets (Fig. 2). These findings suggest that there is no cross-reactivity between the tested primer-probe sets and host or possible other microbial nucleic acid present in nasopharyngeal swabs from non-COVID-19 patients. At 100 and 101 viral RNA copies/μL, our results show that all primer-probe sets, except RdRp-SARSr and 2019-nCoV_N2, were able to partially detect (Ct values <40) SARS-CoV-2 from clinical sample (Fig. 2). At 102 viral RNA copies/μL, we could detect viral RNA and differentiate between the negative samples for all primer-probe sets, except for the RdRp-SARSr (Charité) set, which was negative (Ct values >40) for all 100–102 viral RNA copies/μL concentrations (Fig. 2). Our mock clinical samples demonstrated that all primer-probe sets, except RdRp-SARSr (Charité), are 100% sensitive to SARS-CoV-2 detection at 100 viral RNA copies/μL of extracted nucleic acid (500 copies/reaction), and between 0–50% sensitive at 1–10 viral RNA copies/μL (5–50 copies/reaction).
Fig. 2. Comparison of analytical sensitivity of SARS-CoV-2 primer-probe sets using pre-COVID-19 nasopharyngeal swabs.
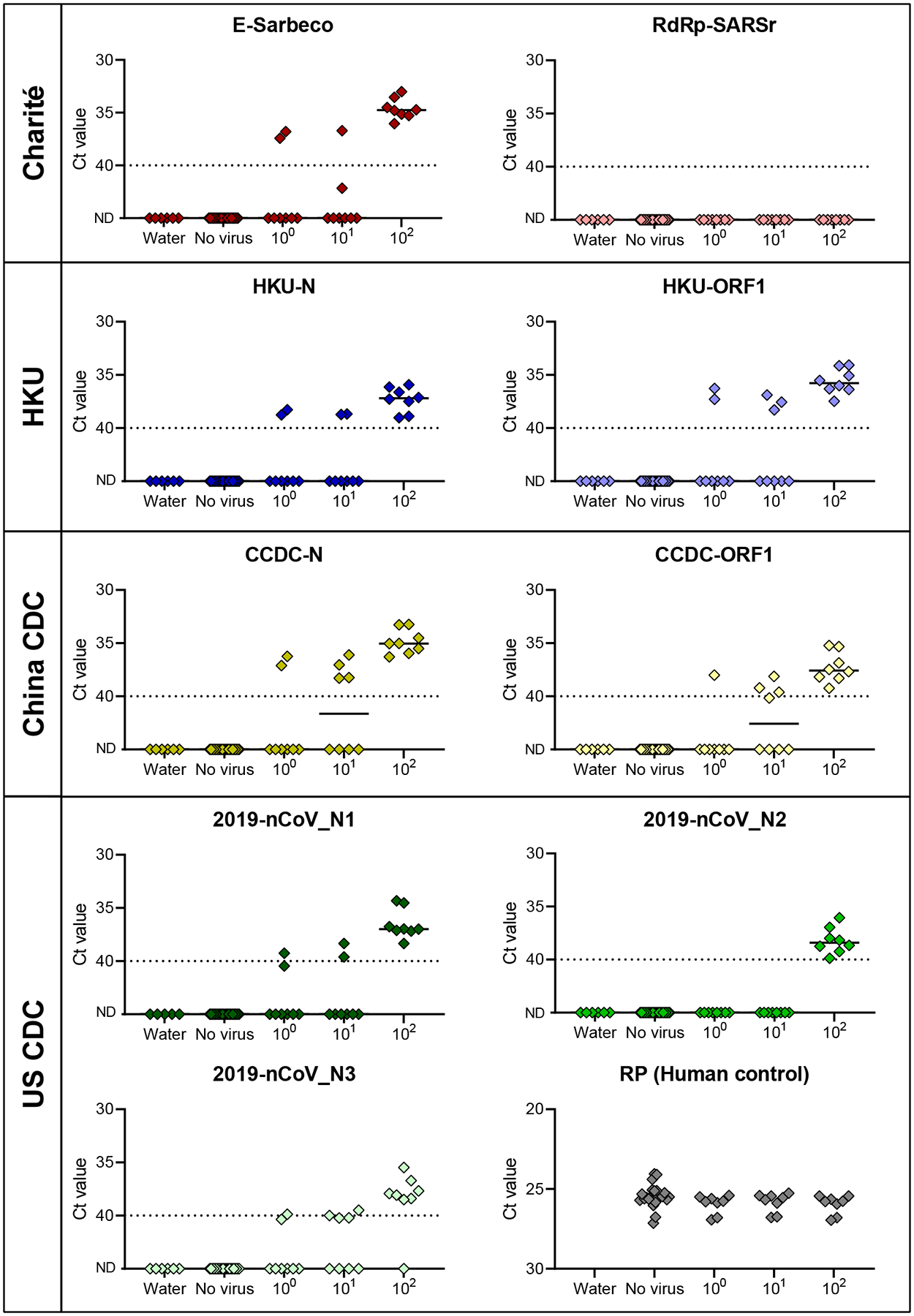
The lower detection limit of nine primer-probe sets as well as the human RNase P control from RNA extracted from nasopharyngeal swabs collected in 2017 spiked with known concentrations of SARS-CoV-2 RNA. Each primer-probe set was performed using 24 technical replicates of pooled swab RNA without spiking SARS-CoV-2 RNA (‘no virus’; 6 replicates with 4 independent pools of 4 swabs) and 8 replicates (2 replicates with 4 independent pools of each 4 nasopharyngeal swabs) spiked with 100–102 viral RNA copies/μL of SARS-CoV-2 RNA. ND = not detected. Black lines indicate the median and the dashed line indicates the detection limit. Data used to make this figure can be found in Source Data Fig. 2.
Clinical evaluation of US CDC primer-probe sets
For the US CDC assay, we found that the 2019-nCoV_N1 (N1) primer-probe set was more sensitive than the 2019-nCoV_N2 (N2) primer-probe set (Fig. 2). To investigate if the differences in analytical sensitivity between N1 and N2 would cause inconclusive results, we compared results from 172 clinical samples taken during the COVID-19 pandemic (Fig. 3). We tested RNA from nasopharyngeal swabs, saliva, urine, and rectal swabs from COVID-19 patients and healthcare workers enrolled in our research protocol at the Yale-New Haven Hospital. We found that more samples had lower Ct values (more efficient virus detection) using the N1 primer-probe set as compared to N2, again showing the N1 is more sensitive for SARS-CoV-2 detection (Fig. 3a). When the N2 set had lower Ct values, each instance was paired with N1 not detected (>45 Ct), indicating that the N1 set had a more distinct separation between positive and negative values (Fig. 3b). When we look at the US CDC assay outcomes, which take into account both the N1 and N2 results, only 1 out of 172 tests was deemed inconclusive due to N1 being negative (>40 Ct) and N2 being positive (<40 Ct; Table 1). We found more inconclusive results where N1 was the only positive set at both 40 Ct (3/172) and 38 Ct (5/172) cut-offs (Table 1), likely because the N1 primer-probe set is more sensitive. Overall, we found inconclusive results from less than 3% of the tested clinical samples that had low (35–40 Ct) or no (>40 Ct) virus detection using the US CDC primer-probe sets, indicating that the US CDC N1 and N2 primer-probe sets are consistent at differentiating between true negatives and positives.
Fig. 3. Low rate of inconclusive testing outcomes using the US CDC N1 and N2 primer-probe sets.
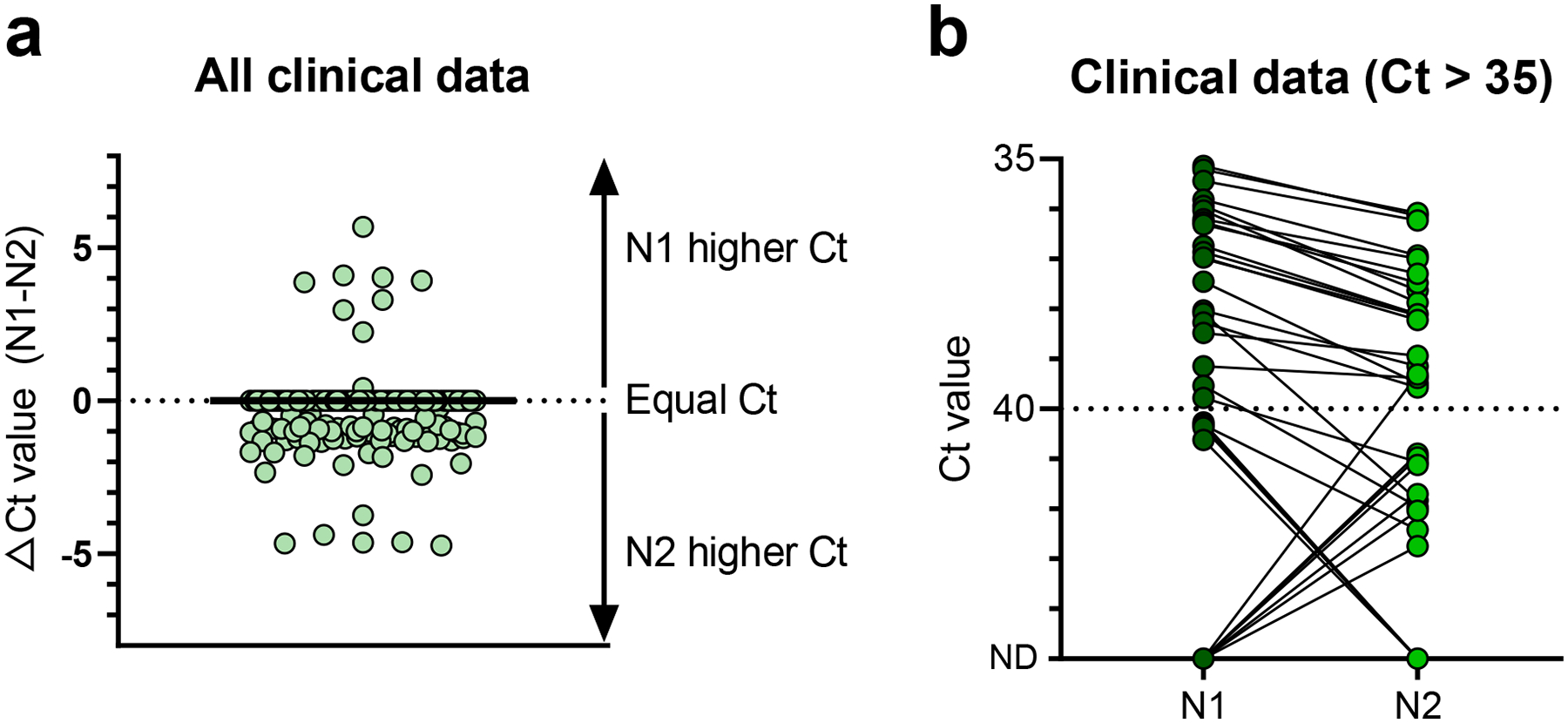
Clinical samples negative or low positive for SARS-CoV-2 were used to determine if differences between the analytical sensitivities of the US CDC N1 and N2 primers produced inconclusive results. a, Cycle threshold (Ct) values for the same 172 clinical samples testing using the N1 and N2 primer probe sets. b, We compared Ct values obtained with the two primer-probe sets for clinical samples with Ct values higher than 35. N1 = 2019-nCoV_N1, N2 = 2019-nCoV_N2, ND = not detected. Solid black line indicates the median, and dashed line indicates the detection limit. Data used to make this figure can be found in Source Data Fig. 3.
Table 1: Differences in sensitivity between N1 and N2 primer-probe sets do not affect performance of the US CDC assay.
We evaluated outcomes of the US CDC assay based on N1 and N2 at two different cut-off levels (Ct = 40 or 38). We found that N2 has a broader range of Ct values between 40–45, whereas N1 only detected Ct values just above 40. We conclude that these differences do not affect the overall performance of the US CDC assay as the percentage of inconclusive samples is below 3% for cut-off values of 40 or more strictly 38 Ct. N1 = 2019-nCoV_N1, N2 = 2019-nCoV_N2.
| Outcome | Cut-off 40 Ct | Cut-off 38 Ct |
|---|---|---|
| Positive | 61/172 (39.0%) | 58/172 (33.7%) |
| Negative | 101/172 (58.7%) | 109/172 (63.4%) |
| Inconclusive | ||
| N1 positive | 3/172 (1.7%) | 5/172 (2.9%) |
| N2 positive | 1/172 (0.6%) |
Lower sensitivity of RdRp-SARSr (Charité) primer-probe set
To further investigate the relatively low sensitivity of the RdRp-SARSr (Charité) primer-probe set, we compared our standardized primer-probe concentrations with the recommended concentrations in the confirmatory (containing both RdRp_SARSr-P1 [probe 1] and RdRp_SARSr-P2 [probe 2]) and discriminatory (probe 2 only, as performed in Figs. 1–2) RdRp-SARSr (Charité) assays. We deviated from the recommended concentrations in the original assays to make a fair comparison across primer-probe sets, using 500 nM of each primer and 250 nM of probe 2. To investigate the effect of primer-probe concentration on the ability to detect SARS-CoV-2, we made a direct comparison between (1) our standardized primer (500 nM) and probe 2 (250 nM) concentrations, (2) the recommended concentrations of 600 nM of forward primer, 800 nM of reverse primer, and 100 nM of probe 1 and 2 (confirmatory assay), and (3) the recommended concentrations of 600 nM of forward primer, 800 nM of reverse primer, and 200 nM of probe 2 (discriminatory assay) per reaction5. We found that adjusting the primer-probe concentrations or using the combination of probes 1 and 2 did not increase SARS-CoV-2 RNA detection when using 10-fold serial dilutions of our RdRp RNA transcripts, or full-length SARS-CoV-2 RNA from cell culture (Extended Data Fig. 2). The Charité Institute of Virology Universitätsmedizin Berlin assay is designed to use the E-Sarbeco primer-probes as an initial screening assay, and the RdRp-SARSr primer-probes as a confirmatory test5. Our data suggest that the RdRp-SARSr assay is not a reliable confirmatory assay at <1000 viral RNA copies/μL of extracted nucleic acid.
Mismatches in primer and probe binding regions
As viruses evolve during outbreaks, nucleotide substitutions can emerge in primer or probe binding regions and alter the sensitivity of PCR assays. To investigate whether this had already occurred during the early COVID-19 pandemic, we calculated the accumulated genetic diversity from 992 available SARS-CoV-2 genomes (released as of 22 March 2020; Fig. 4) and compared that to the primer and probe binding regions (Table 2). Thus far, we detected 12 primer-probe nucleotide mismatches that have occurred in at least two of the 992 SARS-CoV-2 genomes. The most potentially problematic mismatch is in the RdRp-SARSr reverse primer (Table 2), which likely explains the sensitivity issues with this set (Figs. 1–2). Oddly, the mismatch is not derived from a new variant that has arisen, but rather that the primer contains a degenerate nucleotide (S, binds with G or C) at position 12, and 990 of the 992 SARS-CoV-2 genomes encode for a T at this genome position (Table 2). This degenerate nucleotide appears to have been added to help the primer anneal to SARS-CoV and bat-SARS-related CoV genomes5, seemingly to the detriment of consistent SARS-CoV-2 detection. Earlier in the outbreak, before hundreds of SARS-CoV-2 genomes became available, non-SARS-CoV-2 data were used to infer genetic diversity that could be anticipated during the outbreak. As a result, several of the primers contain degenerate nucleotides (Supplementary Table 4). For RdRp-SARSr, adjusting the primer (S→A) may resolve its low sensitivity.
Fig. 4. Genetic diversity of available SARS-CoV-2 genomes.
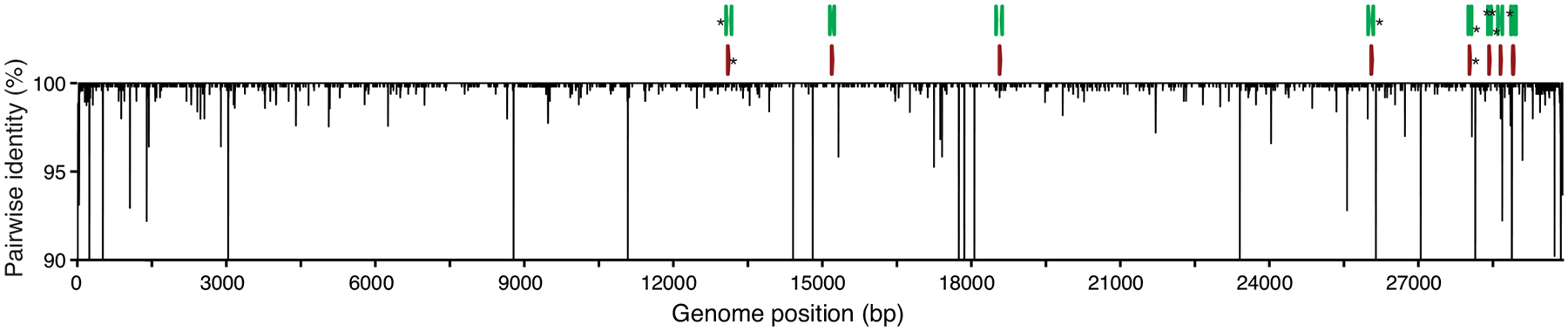
992 SARS-CoV-2 genomes available as of 22 March 2020 (listed in Source Data Fig. 4) were aligned to calculate nucleotide diversity and investigate mismatches with the nine primer-probe sets. Genetic diversity was measured using pairwise identity (%) at each position, disregarding gaps and ambiguous nucleotides. Asterisks (*) at the top indicate primers (green) and probes (red) targeting regions with one or more mismatches. Genomic plots were designed using DNA Features Viewer 3.0.1 in Python version 3.715.
Table 2: High frequency primer and probe mismatches may result in decreased sensitivity for SARS-CoV-2 detection.
Listed mismatched nucleotides with primers and probes with frequencies above 0.1% in 992 genomes inspected in this analysis. The last column highlights the various frequencies of mismatches, which would represent a mispairing upon binding of the primers listed above. A list of degenerate nucleotides incorporated into the primer and probe sequences can be found in Supplementary Table 4. Data used to make this Table can be found in Source Data Fig. 4.
| Institute | Primer/probe | Primer/probe position 5’-3’ | Genome position 5’-3’ | Primer/probenucleotide | Nucleotide in ref genome1 (RC) | Expected target nucleotide | Mismatch target in genomes2 (frequency) |
|---|---|---|---|---|---|---|---|
| China CDC | CCDC-N-F | 1 | 28,881 | G | G (C) | C | TRC
(126/992; 12.7%) |
| CCDC-N-F | 2 | 28,882 | G | G (C) | C | TRC (126/992; 12.7%) |
|
| CCDC-N-F | 3 | 28,883 | G | G (C) | C | GRC
(126/992; 12.7%) |
|
| CCDC-ORF1-F | 17 | 13,358 | C | C (G) | G | ARC (2/992; 0.2%) |
|
| CCDC-ORF1-P | 26 | 13,402 | T | T (A) | A | CRC
(4/992; 0.4%) |
|
| Charité | E_Sarbeco_R | 12 | 26,370 | G | C (G) | C | T (4/992; 0.4%) |
| RdRp-SARSr_R | 12 | 15,519 | S | T (A) | C or G |
T
(990/992; 99.8%) |
|
| HKU | HKU-N-F | 4 | 29,148 | T | T (A) | A | GRC (5/992; 0.5%) |
| US CDC | 2019-nCoV_N1-P | 3 | 28,311 | C | C (G) | G | ARC (2/992; 0.2%) |
| 2019-nCoV_N1-R | 15 | 28,344 | G | C (G) | C | A (4/992; 0.4%) |
|
| 2019-nCoV_N3-F | 8 | 28,688 | T | T (A) | A | GRC
(39/992; 3.9%) |
|
| 2019-nCoV_N3-R | 14 | 28,739 | C | G (C) | G | T (4/992; 0.4%) |
RC Reverse Complement
Nucleotide (DNA form) found in the reference genome (NC_045512) and its reverse complement (RC)
Mismatch target is the disagreement between the expected target nucleotide and the nucleotide in the genome
Of the variants that we detected in the primer-probe regions, we only found four in more than 30 of the 992 SARS-CoV-2 genomes (>3%, Table 2). Most notable was a stretch of three nucleotide substitutions (GGG→AAC) at genome positions 28,881–28,883, which occur in the three first positions of the CCDC-N forward primer binding site. While these substitutions define a large clade that includes ~13% of the available SARS-CoV-2 genomes released as of 22 March 2020, and has been detected in numerous countries11, their position on the 5’ location of the primer may not be detrimental to sequence annealing and amplification. The other high frequency variant that we detected was T→C substitution at the 8th position of the binding region of the 2019-nCoV_N3 forward primer, a substitution found in 39 genomes (position 28,688). While this primer could be problematic for detecting viruses with this variant, the CDC revised their assay on March 15, 2020, by removing the 2019-nCoV_N3 primer-probe set12. We found another seven variants in only five or fewer genomes (<0.5%, Table 2), and their minor frequency at present does not pose a major concern for viral detection. This scenario may change if those variants increase in frequency: most of them lie in the second half of the primer binding region, and may decrease primer sensitivity13. The WA1_USA strain8 (GenBank: MN985325) that we used as a reference for our comparisons only contains the mismatch with the RdRp reverse primer (T at position 15,519), and therefore we cannot directly assess the impact of the other variants. Continued monitoring of SARS-CoV-2 evolution (e.g. gisaid.org), and how arising variants may alter PCR detection, is needed.
Discussion
Our study provides a comprehensive and independent comparison of analytical performance of primer-probe sets for SARS-CoV-2 testing in various parts of the world. Our findings show a high similarity in the analytical sensitivities for SARS-CoV-2 detection, which indicates that outcomes of different assays are comparable. The primary exception to this is the RdRp-SARSr (Charité) primer-probe set which had the lowest sensitivity, as also shown by an independent study14, likely stemming from a mismatch in the reverse primer. In the US, we recommend using the US CDC SARS-CoV-2 assay as: (1) we found similar analytical sensitivity as compared to the other three assays; (2) we detected a low rate of inconclusive results with low-virus clinical samples; (3) it includes a human RNase P primer-probe set (RP) that allows for quality control of RNA extraction methods; and (4) its wide-spread use in the US makes it easier to compare results. In other regions of the world, however, a different test may be preferable based on existing usage.
Our study has limitations to consider. We standardized the concentration of primers and probes, PCR kits, and thermocycler conditions to directly compare primer-probes sets used in four common RT-qPCR assays for detection of SARS-CoV-2. By standardizing the PCRs, we deviated from some of the recommended conditions, which means that not all of our results can be directly transferable to how the assays were intended in clinical diagnostic settings. For instance, we selected an annealing temperature of 55°C which was lower than recommended for the assays developed by Charité (58°C)5 and HKU (60°C)4, but similar to the assay developed by US CDC (55°C)6. No specific PCR conditions were reported for the assay developed by the China CDC7. We found that the two assays with higher annealing temperatures (Charité and HKU) had high analytical sensitivity and no background amplification, which suggests that our standardized annealing temperature likely did not have a large effect on our findings. In addition, we selected one RT-qPCR kit (Luna Universal Probe One-step RT-qPCR) for all comparisons. We selected this kit specifically because it was not approved by the US Federal Drug Administration for SARS-CoV-2 diagnostics and thus our research would not compete with clinical diagnostic laboratories for resources. In doing so, we provide an alternative protocol for SARS-CoV-2 RT-qPCR for research testing (Supplementary File 1), which is especially helpful as more resources are required to expand testing around the world. Finally, we performed all of our RT-qPCR tests on one thermocycler (BioRad CFX). It is possible that our standardization methods may have influenced analytical performance of the tested primer-probe sets, and our results may not directly apply to other PCR kits or thermocyclers9. Thus, we strongly urge that each laboratory should locally validate analytical sensitivities and positive-negative cut-off values when establishing these assays, which can be performed using our RNA transcripts and study framework.
Methods
Ethics
Residual de-identified nasopharyngeal samples collected during 2017 (pre-COVID-19) were obtained from the Yale-New Haven Hospital Clinical Virology Laboratory. In accordance with the guidelines of the Yale Human Investigations Committee (HIC), this work with de-identified samples is considered non-human subjects research. These samples were used to create the mock substrate for the SARS-CoV-2 spike-in experiments. Collection of clinical samples from COVID-19 patients and healthcare workers at the Yale New Haven Hospital was approved by the Institutional Review Board of the Yale Human Research Protection Program (FWA00002571, Protocol ID. 2000027690). Written consent was obtained from all patients and healthcare workers. These samples were used to test the US CDC 2019-nCoV_N1 and 2019-nCoV_N2 primer-probe sets.
Generation of RNA transcript standards
We generated RNA transcript standards for each of the five genes targeted by the diagnostic RT-qPCR assays using T7 transcription. A detailed protocol can be found here10. Briefly, cDNA was synthesized from full-length SARS-CoV-2 RNA (WA1_USA strain from UTMB; GenBank: MN985325). Using PCR, we amplified the nsp10, RdRp, nsp14, E, and N genes with specifically designed primers (Supplementary Table 2). We purified PCR products using the Mag-Bind TotalPure NGS kit (Omega Bio-tek, Norcross, GA, USA) and quantified products using the Qubit High Sensitivity DNA kit (ThermoFisher Scientific, Waltham, MA, USA). We determined fragment sizes using the DNA 1000 kit on the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). After quantification, we transcribed 100–200 ng of each purified PCR product into RNA using the Megascript T7 kit (ThermoFisher Scientific). Although RNA transcripts were DNase treated with TURBO DNase, low concentrations of residual DNA may still be present. We quantified RNA transcripts using the Qubit High sensitivity RNA kit (ThermoFisher Scientific) and checked quality using the Bioanalyzer RNA pico 6000 kit. For each of the RNA transcript standards (Supplementary Table 3), we calculated the number of viral RNA copies per μL using Avogadro’s number. We generated a genomic annotation plot with all newly generated RNA transcript standards and the nine tested primer-probe sets based on the NC_045512 reference genome using the DNA Features Viewer 3.0.1 in Python version 3.7 (Extended Data Fig. 1)15. We generated standard curves for each combination of primer-probe set with its corresponding RNA transcript standard, using standardized RT-qPCR conditions as described below.
RT-qPCR conditions
To make a fair comparison between nine primer-probe sets (Supplementary Table 1), we used the same RT-qPCR reagents and conditions for all comparisons. We used the Luna Universal Probe One-step RT-qPCR kit (New England Biolabs, Ipswich, MA, USA) with 5 μL of RNA and standardized primer and probe concentrations of 500 nM of forward and reverse primer, and 250 nM of probe for all comparisons. PCR cycler conditions were reverse transcription for 10 minutes at 55°C, initial denaturation for 1 min at 95°C, followed by 40 cycles (45 cycles for clinical samples) of 10 seconds at 95°C and 30 seconds at 55°C on the Biorad CFX96 qPCR machine (Biorad, Hercules, CA, USA). We applied the fluorescence drift correction for plates with autofluorescence and refrained from manual adjustment of the threshold. A detailed protocol can be found in Supplementary File 1. We calculated analytical efficiency of RT-qPCR assays tested with corresponding RNA transcript standards using the following formula16,17:
Validation with SARS-CoV-2 RNA and pre-COVID-19 samples
We prepared mock samples by extracting RNA from de-identified nasopharyngeal swabs collected in 2017 (pre-COVID-19) from hospital patients with respiratory disease using the MagMAX Viral/Pathogen Nucleic Acid Isolation kit (ThermoFisher Scientific) following manufacturer’s protocol. We used 300 μL of sample and eluted in 75 μL. We compared analytical efficiency and sensitivity of primer-probe sets by testing 10-fold dilutions (106–100 viral RNA copies/μL) of SARS-CoV-2 RNA as well as the SARS-CoV-2 mock samples spiked with RNA after extraction (eluates pooled from 12 individuals), in duplicate. In addition, we pooled eluates from 4 patients to create 4 independent pools (16 individuals total) and spiked these mock samples with 10-fold dilutions of SARS-CoV-2 RNA (100–102 viral RNA copies/μL) to determine the lower detection limit of each primer-probe set. We tested RNA-spiked mock samples from each of the four independent pools in duplicate (in total 8 reps). Lastly, we tested mock samples (no spiked-in virus) from each pool for six replicates (in total 24 reps per primer-probe set) to test for potential background amplification.
Clinical samples
Clinical samples from COVID-19 diagnosed patients and healthcare workers were obtained from the Yale New Haven Hospital. We extracted nucleic acid from nasopharyngeal swabs, saliva, urine, and rectal swabs using the MagMax Viral/Pathogen Nucleic Acid Isolation kit following manufacturer’s protocol. We used 300 μL of each sample and eluted in 75 μL. We used the Luna Universal Probe One-step RT-qPCR kit with standardized primer and probe concentrations of 500 nM of forward and reverse primer, and 250 nM of probe for the 2019-nCoV_N1, 2019-nCoV_N2, and RP (human control) primer-probe sets to detect SARS-CoV-2 in each sample. PCR cycler conditions were reverse transcription for 10 minutes at 55°C, initial denaturation for 1 min at 95°C, followed by 45 cycles of 10 seconds at 95°C and 30 seconds at 55°C on the Biorad CFX96 qPCR machine (Biorad, Hercules, CA, USA). All figures were made with GraphPad Prism 8.3.0.
Mismatches in primer and probe binding regions
We investigated mismatches in primer binding regions by calculating pairwise identities (%) for each nucleotide position in binding sites of assay primers and probes. Ignoring gaps and ambiguous bases, we compared all possible pairs of nucleotides in all columns of a multiple sequence alignment including all available SARS-CoV-2 genomes from GISAID (as of 22 March 2020; Source Data Fig. 4). We assigned a score of 1 for each identical pair of bases, and divided the final score by the total number of valid nucleotide pairs, to finally express pairwise identities as percentages. Pairwise identity of less than 100% indicates mismatches between primers or probes and some SARS-CoV-2 genomes. We calculated mismatch frequencies and reported absolute and relative frequencies for mismatches with frequency higher than 0.1%. The DNA Features Viewer 3.0.1 package in Python version 3.7 was used to generate the diversity plot (Fig. 4)15.
Extended Data
Extended Data Fig. 1. Generation of RNA transcript standards for validation of SARS-CoV-2 RT-qPCR assays.
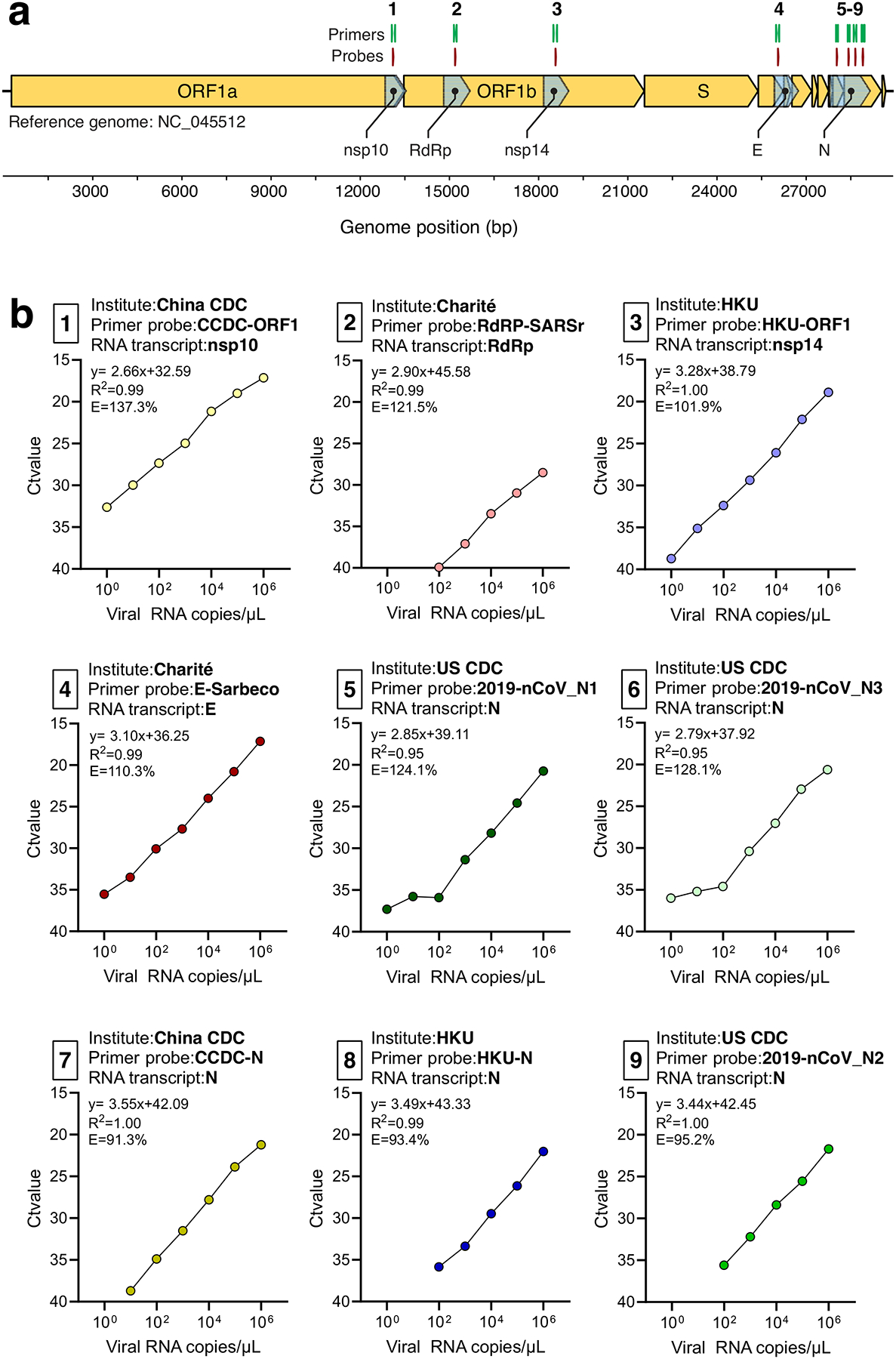
a, SARS-CoV-2 genome locations of generated RNA transcript standards for the non-structural protein 10 (nsp10), RNA-dependent RNA polymerase (RdRp), non-structural protein 14 (nsp14), envelope (E), and nucleocapsid (N) genes and the nine primer-probe sets used RT-qPCR assays. b, The slope, intercept, R2, and efficiency of RT-qPCR using tenfold dilutions (100-106 viral RNA copies/μL) of RNA transcript standards with the corresponding primer-probe sets. Shown are mean Ct values based on 2 technical replicates. The primer-probe sets are numbered as shown in panel A. The RNA transcript primers and sequences can be found in Supplementary Table 2 and Supplementary Table 3, respectively. Data used to make this figure can be found in Source Data Extended Data Fig. 1.
Extended Data Fig. 2. No effect of different concentrations of RdRp-SARSr primers and probes on analytical sensitivity.
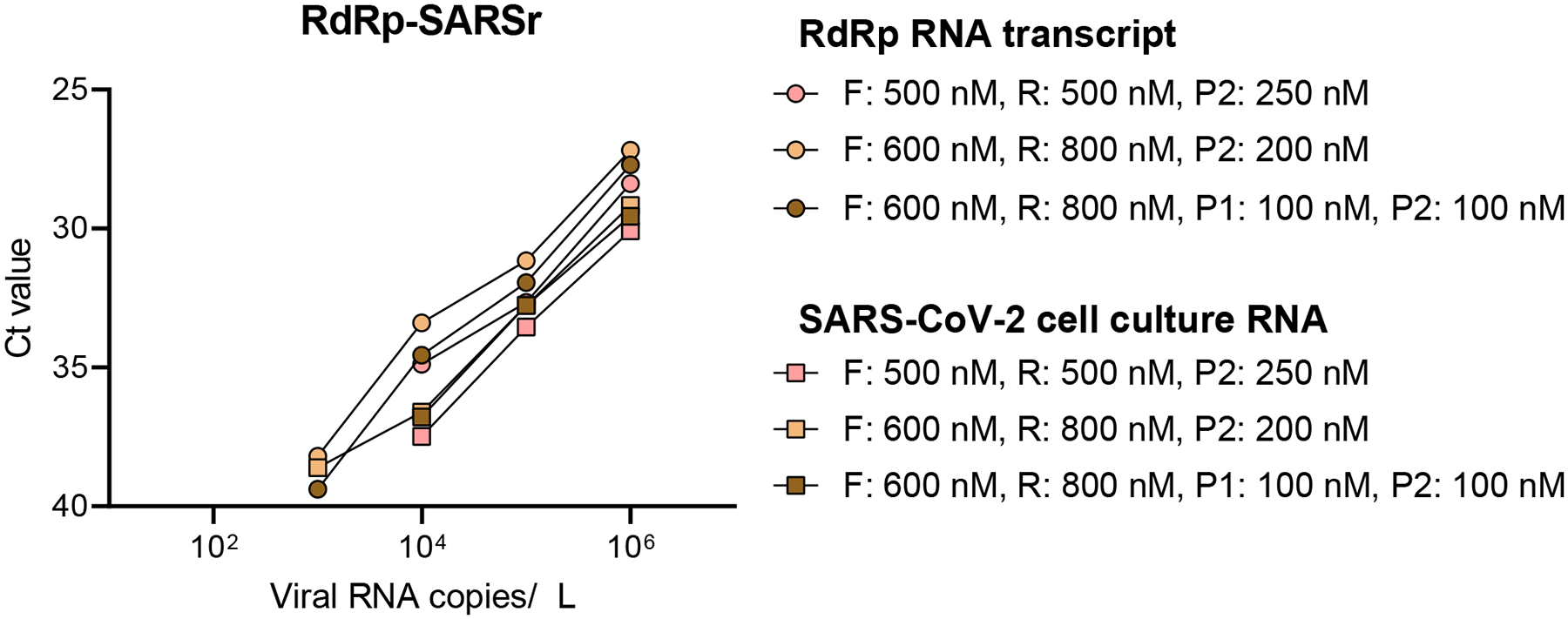
Low performance of the standardized RdRp-SARSr primer-probe set triggered us to further investigate the effect of primer concentrations. We compared our standardized primer-probe concentrations (500 nM of forward and reverse primers, and 250 nM of probe) with the recommended concentrations in the confirmatory assay (600 nM of forward primer, 800 nM of reverse primer, 100 nM of probe 1, and 100 nM of probe 2), and the discriminatory assay (600 nM of forward primer, 800 nM of reverse primer, and 200 nM of probe 2) as developed by the Charité Institute of Virology Universitätsmedizin Berlin. Standard curves for both RdRp-transcript standard and full-length SARS-CoV-2 RNA are similar, which indicates that higher primer concentrations did not improve the performance of the RdRp-SARSr set. Symbol indicates tested sample type (circles = RdRp transcript standard, and squares = full-length SARS-CoV-2 RNA from cell culture) and colors indicate the different primer and probe concentrations. Data used to make this figure can be found in Source Data Extended Data Fig 2.
Supplementary Material
Acknowledgements
We thank K. Plante and the University of Texas Medical Branch World Reference Center for Emerging Viruses for providing SARS-CoV-2 RNA, the Yale COVID-19 Laboratory Working Group for technical support, P. Jack and S. Taylor for discussions, and A. Greninger and X. Lu for feedback on a previous version of this manuscript. We also thank scientists from around the world that openly shared their SARS-CoV-2 genomic data on GISAID, and they are acknowledged in Source Data Fig. 4. This research was funded by the generous support from the Yale Institute for Global Health and the Yale School of Public Health start-up package provided to NDG. CBFV is supported by NWO Rubicon 019.181EN.004.
Competing interests
ALW has received research funding through grants from Pfizer to Yale and has received consulting fees for participation in advisory boards for Pfizer. The other authors declare no competing interests.
Data availability
All data are included in this article, the supplementary files, and the Source Data.
References
- 1.Gorbalenya AE et al. Severe acute respiratory syndrome-related coronavirus: The species and its viruses – a statement of the Coronavirus Study Group. bioRxiv 2020.02.07.937862 (2020) doi: 10.1101/2020.02.07.937862. [DOI] [Google Scholar]
- 2.Wu F et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu DKW et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem (2020) doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corman VM et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC. Coronavirus Disease 2019 (COVID-19). Centers for Disease Control and Prevention; https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (2020). [Google Scholar]
- 7.Institute of Viral Diseases. China CDC. National Institute for Viral Disease Control and Prevention; http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html (2020). [Google Scholar]
- 8.Harcourt J et al. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with 2019 Novel Coronavirus Disease, United States. Emerg. Infect. Dis 26, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svec D, Tichopad A, Novosadova V, Pfaffl MW & Kubista M How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol Detect Quantif 3, 9–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogels CBF, Fauver JR, Ott I & Grubaugh ND Generation of SARS-COV-2 RNA transcript standards for qRT-PCR detection assays v1 (protocols.io.bdv6i69e) (2020) doi: 10.17504/protocols.io.bdv6i69e. [DOI] [Google Scholar]
- 11.Genomic epidemiology of novel coronavirus. Nextstrain https://nextstrain.org/ncov?c=gt-ORF14_50. [Google Scholar]
- 12.CDC Revises SARS-CoV-2 Assay Protocol; Surveillance Testing on Track to Start Next Week. 360Dx https://www.360dx.com/pcr/cdc-revises-sars-cov-2-assay-protocol-surveillance-testing-track-start-next-week. [Google Scholar]
- 13.Bru D, Martin-Laurent F & Philippot L Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example. Appl. Environ. Microbiol 74, 1660–1663 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casto AM et al. Comparative Performance of SARS-CoV-2 Detection Assays using Seven Different Primer/Probe Sets and One Assay Kit. medRxiv (2020) doi: 10.1101/2020.03.13.20035618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zulkower V & Rosser S DNA Features Viewer, a sequence annotations formatting and plotting library for Python. bioRxiv 2020.01.09.900589 (2020) doi: 10.1101/2020.01.09.900589. [DOI] [PubMed] [Google Scholar]
- 16.Broeders S et al. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol 37, 115–126 (2014). [Google Scholar]
- 17.Ginzinger DG Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp. Hematol 30, 503–512 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in this article, the supplementary files, and the Source Data.


