Plant cell surface receptors are at the forefront of defence against pathogens, involved in pathogen sensing through the detection of conserved molecules named pathogen‐associated molecular patterns (PAMP) and highly variable pathogen virulence (effector) proteins. The recently reported Brassica napus (canola, oilseed rape) disease resistance gene Rlm9 encodes a wall‐associated kinase‐like (WAKL) receptor which confers race‐specific resistance against races of the blackleg pathogen Leptosphaeria maculans carrying the corresponding effector gene, AvrLm5‐9 (Larkan et al., 2020). Rlm4 and Rlm7 are located on B. napus chromosome A07 and genetically tightly linked to Rlm9 (Larkan et al., 2016). The L. maculans effectors AvrLm4‐7 and AvrLm7 are small, secreted cysteine‐rich proteins encoded by a single locus, AvrLm4‐7. A single amino acid change in AvrLm4‐7 masks recognition by Rlm4 without affecting Rlm7 function. Here, we report the cloning of Rlm4 and Rlm7, both alleles of the Rlm9 WAKL locus.
Genomic sequencing data was generated for the B. napus introgression lines; Topas‐Rlm4 and Topas‐Rlm7 using Illumina HiSeq 2500. Close to 537 million reads were assembled using SOAPdenovo assembler. Contigs generated from each line were mapped to the B. napus reference genome ‘Darmor‐bzh’ using Bowtie2. Based on ~1.3 billion RNA sequence reads (using Illumina HiSeq 2500 2 × 125 bp, 3 biological replicates) mapped to the Rlm3‐4‐7‐9 gene cluster (Figure 1a), Rlm4 and Rlm7 genes were determined to be allelic, with each being 8507 bp in length consisting of three exons (Figure 1b). To prove the function of the predicted genes, the entire gene including introns and 5’ intergenic region (1750 bp) for each allele was synthesized (GenScript, USA) and cloned into the plant transformation vector pMDC123, modified to contain the nosT terminator sequence downstream of the cloning site using Gateway cloning technology. Rlm4 and Rlm7 genomic constructs were transferred into the blackleg‐susceptible B. napus line, Westar N‐o‐1. Regenerated transgenic (T0) plants that survived herbicide selection were screened via droplet digital PCR (ddPCR) to identify lines carrying insertions, then selfed to produce the T1 generation. The resulting transgenic lines (13 for Rlm4, 17 for Rlm7) were initially tested for resistance response using the L. maculans isolate v23.1.3 (avrLm3, AvrLm4‐7, avrLm9) using a standard cotyledon assay (Larkan et al., 2013) with all lines displaying hypersensitive response at the point of infection except for one Rlm4 line with poor germination. Additional ddPCR was conducted to identify homozygous, single insertion events in T1 plants, and one plant for each construct was selected and selfed to produce homozygous T2 lines for further characterization (hereafter referred to as Westar:Rlm4 and Westar:Rlm7). Further confirmation was obtained by utilizing transgenic L. maculans to demonstrate effector‐specific activation of resistance conferred by the Rlm4 and Rlm7 candidate genes. The L. maculans isolate 2367 (avrLm3, avrLm4‐7, avrLm9) and the transgenic isolates 2367:AvrLm4‐7, 2367:AvrLm7 (Larkan et al., 2016), and 2367:AvrLm5‐9 (Ghanbarnia et al., 2018) were used to inoculate Westar N‐o‐1, Westar:Rlm4, Westar:Rlm7 and Westar:Rlm9 (Larkan et al., 2020). Four seedlings of each line were inoculated with each isolate (performed in triplicate). No resistance reaction was induced in either Westar:Rlm4 or Westar:Rlm7 in response to AvrLm5‐9. A hypersensitive response was induced in Westar:Rlm4 only in response to AvrLm4‐7, while Westar:Rlm7 responded to both AvrLm4‐7 and AvrLm7, as expected, confirming the cloned genomic constructs as Rlm4 and Rlm7, respectively (Figure 1c).
Figure 1.
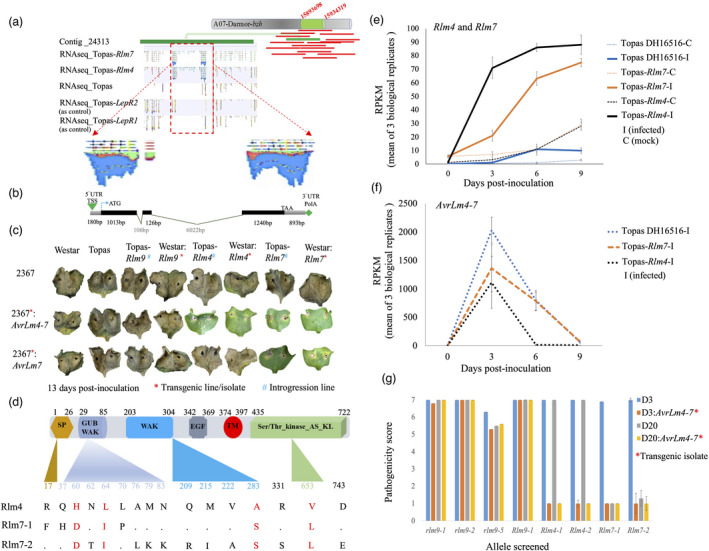
Cloning of B. napus genes; Rlm4 and Rlm7. (a) Identification of Rlm4 and Rlm7 through contig walking. (b) Rlm4 and Rlm7 gene structures. Exons are shown as black bar, intron black line valley, and UTR as grey line. (c) Transgenic B. napus expressing Rlm4 and Rlm7. Cotyledons of Westar, Topas, Topas‐Rlm9, Westar:Rlm9, Topas‐Rlm4, Topas‐Rlm7, Westar:Rlm4 and Westar:Rlm7 inoculated with isolate 2367 (virulent towards Rlm4, Rlm7 and Rlm9), 2367:AvrLm4‐7 (avirulent towards Rlm4 and Rlm7) and 2367:AvrLm7 (avirulent towards Rlm7). (d) Protein domains of Rlm4 and Rlm7. Protein structure is represented by the signal peptide (yellow bar), extracellular GUB_WAK (dark blue), C‐terminal WAK (blue bar), and EGF‐like Ca2+ (grey bar), transmembrane (red bar) and an intracellular serine/threonine Protein Kinase (green bar) domains. (e, f) Expression profile of Rlm4, Rlm7, and AvrLm4‐7. RNA‐seq analysis showed a significant upregulation of Rlm4, Rlm7 in the resistance lines and expression of AvrLm4‐7 peaked at 3 days post‐inoculation in both susceptible and resistant lines. (g) Confirmation of Rlm4 and Rlm7 alleles through pathogenicity phenotyping. B. napus lines harbouring susceptible alleles, Rlm9, Rlm4, and Rlm7 were screened using isolates D3, D20, D3:AvrLm4‐7 and D20:AvrLm4‐7.
Rlm4 and Rlm7 open reading frames are 2379 bp encoding proteins of 792 amino acids (aa). Sequence polymorphism between the two genes is limited to a total of 13 single nucleotide substitutions resulting in 4 synonymous and 9 non‐synonymous changes. InterPro predicts Rlm4 and Rlm7 as transmembrane proteins consisting of secretory signal peptide (SP), extracellular wall‐associated receptor kinase galacturonan‐ binding (WAK_GUB), WAK, epithelial growth factor (EGF) like calcium binding domains and a cytoplasmic kinase domain (Figure 1d). There are a total of 7 amino acid differences between Rlm4 and Rlm7 (Figure 1d). A further 128 lines that had been phenotyped for the presence of Rlm4 and Rlm7, using isolates with AvrLm4‐7 or AvrLm7, were investigated using whole genome sequencing leading to the identification of two additional resistant alleles (Rlm4‐2 and Rlm7‐2). The coding sequences of the Rlm4‐1 and Rlm4‐2 alleles were identical, with any polymorphisms limited to the intronic regions of the gene. The Rlm7‐2 allele, identified in the B. napus variety Caiman, contained 13 non‐synonymous SNPs; however, these amino acid changes do not affect the recognition of Rlm7. The Rlm7‐1 and Rlm7‐2 proteins differ from Rlm4 in conserved amino acids in the putative extracellular ligand‐binding domains, at positions 60, 64 (WAK_GUB), and 283 (WAK), which may explain the variation in recognition specificity between Rlm4 and Rlm7 towards AvrLm7. An additional conserved polymorphism is found in the kinase domain (at position 653), though this is unlikely to be involved in recognition of AvrLm4‐7 (Figure 1d). While Rlm4 and Rlm7 proteins are highly similar, Rlm9 protein is more diverse, with numerous SNPs mainly at the ectodomain (Figure S1).
The Topas‐Rlm4 and Topas‐Rlm7 were utilized to monitor the expression of Rlm4, Rlm7, and the corresponding Avr gene, AvrLm4‐7, during cotyledon infection by the reference isolate v23.1.3 (as previously described by Haddadi et al., 2019). Expression of Rlm4 and Rlm7 increased substantially in both Topas‐Rlm4 and Topas‐Rlm7 in response to infection (Figure 1e). AvrLm4‐7 expression peaked at 3 days post‐inoculation, both in susceptible and resistant lines, but rapidly declined at later time points (Figure 1f).
Additional pathology tests using lines harbouring both Rlm4 and Rlm7 alleles, as well as the Rlm9 and four of the susceptible alleles, were performed in triplicate with isolates transformed with AvrLm4‐7. Progenitor isolate D3 is virulent towards both Rlm4 and Rlm7 whilst D20 is only virulent towards Rlm4. Isolate D3:AvrLm4‐7 resulted in an avirulent reaction on lines harbouring each of the Rlm4 and Rlm7 alleles but remained virulent on all other lines as expected. Isolate D20:AvrLm4‐7 resulted in an avirulent reaction on lines harbouring both Rlm4 alleles but remained virulent on the lines harbouring the susceptible alleles or Rlm9 and remained avirulent on the Rlm7 lines as expected (Figure 1g).
The cloning of Rlm4 and Rlm7 will provide valuable information for canola breeding programmes worldwide and expands the toolbox for further study of the newly emerging WAKL class of plant R genes.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
NJL and HB conceived and designed the study, PH and HB extracted the Rlm4‐1 and Rlm7‐1 ORFs and designed the transgenic constructs, NJL and EB analysed transformants, AVdW characterized additional Rlm4 and Rlm7 lines, YZ and TXN characterized additional alleles, PH, PB, and DE provided bioinformatic analysis. PH, NJL, AVdW, JB, and HB prepared the manuscript.
Funding
This work was supported by funding provided by Agriculture and Agri‐Food Canada’s (AAFC) Canadian Agricultural Partnership (CAP, Canola Council of Canada), the canola industry (SaskCanola), and the Australian Grains Research and Development Corporation (UWA1905‐006RTX).
Supporting information
Figure S1 Alignment of Rlm4, Rlm7, and Rlm9 proteins.
Haddadi, P. , Larkan, N. J. , Van deWouw, A. , Zhang, Y. , Xiang Neik, T. , Beynon, E. , Bayer, P. , Edwards, D. , Batley, J. and Borhan, M. H. (2022) Brassica napus genes Rlm4 and Rlm7, conferring resistance to Leptosphaeria maculans, are alleles of the Rlm9 wall‐associated kinase‐like resistance locus. Plant Biotechnol. J., 10.1111/pbi.13818
References
- Ghanbarnia, K. , Ma, L. , Larkan, N.J. , Haddadi, P. , Fernando, W.G.D. and Borhan, M.H. (2018) Leptosphaeria maculans AvrLm9: A new player in the game of hide‐and‐seek with AvrLm4‐7. Mol. Plant Pathol. 19, 1754–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddadi, P. , Larkan, N.J. and Borhan, M.H. (2019) Dissecting R gene and host genetic background effect on the Brassica napus defense response to Leptosphaeria maculans . Sci. Rep. 9, 6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkan, N.J. , Lydiate, D.J. , Parkin, I.A.P. , Nelson, M.N. , Epp, D.J. , Cowling, W.A. , Rimmer, S.R. et al. (2013) The Brassica napus blackleg resistance gene LepR3 encodes a receptor‐like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 197, 595–605. [DOI] [PubMed] [Google Scholar]
- Larkan, N.J. , Ma, L. , Haddadi, P. , Buchwaldt, M. , Parkin, I.A.P. , Djavaheri, M. and Borhan, M.H. (2020) The Brassica napus wall‐associated kinase‐like (WAKL) gene Rlm9 provides race‐specific blackleg resistance. Plant J. 104, 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkan, N.J. , Yu, F. , Lydiate, D.J. , Rimmer, S.R. and Borhan, M.H. (2016) Single R gene introgression lines for accurate dissection of the Brassica ‐ Leptosphaeria pathosystem. Front. Plant Sci. 7, 1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Alignment of Rlm4, Rlm7, and Rlm9 proteins.


