ABSTRACT
Candida auris is an urgent threat to human health due to its rapid spread in health care settings and its repeated development of multidrug resistance. Diseases that increase risk for C. auris infection, such as diabetes, kidney failure, or immunocompromising conditions, are associated with elevated levels of methylglyoxal (MG), a reactive dicarbonyl compound derived from several metabolic processes. In other Candida species, expression of MG reductase enzymes that catabolize and detoxify MG are controlled by Mrr1, a multidrug resistance-associated transcription factor, and MG induces Mrr1 activity. Here, we used transcriptomics and genetic assays to determine that C. auris MRR1a contributes to MG resistance, and that the main Mrr1a targets are an MG reductase and MDR1, which encodes a drug efflux protein. The C. auris Mrr1a regulon is smaller than Mrr1 regulons described in other species. In addition to MG, benomyl (BEN), a known Mrr1 stimulus, induces C. auris Mrr1 activity, and characterization of the MRR1a-dependent and -independent transcriptional responses revealed substantial overlap in genes that were differentially expressed in response to each compound. Additionally, we found that an MRR1 allele specific to one C. auris phylogenetic clade, clade III, encodes a hyperactive Mrr1 variant, and this activity correlated with higher MG resistance. C. auris MRR1a alleles were functional in Candida lusitaniae and were inducible by BEN, but not by MG, suggesting that the two Mrr1 inducers act via different mechanisms. Together, the data presented in this work contribute to the understanding of Mrr1 activity and MG resistance in C. auris.
IMPORTANCE Candida auris is a fungal pathogen that has spread since its identification in 2009 and is of concern due to its high incidence of resistance against multiple classes of antifungal drugs. In other Candida species, the transcription factor Mrr1 plays a major role in resistance against azole antifungals and other toxins. More recently, Mrr1 has been recognized to contribute to resistance to methylglyoxal (MG), a toxic metabolic product that is often elevated in different disease states. MG can activate Mrr1 and its induction of Mdr1 which can protect against diverse challenges. The significance of this work lies in showing that MG is also an inducer of Mrr1 in C. auris, and that one of the major pathogenic C. auris lineages has an activating Mrr1 mutation that confers protection against MG.
KEYWORDS: Candida auris, Candida, methylglyoxal, benomyl, Mrr1, RNA-seq, transcriptomics
INTRODUCTION
Although Candida albicans has historically been the most prominent Candida species associated with both superficial and invasive fungal infections, worldwide incidence of non-albicans Candida (NAC) species is increasing (1–10). Of particular concern is Candida auris, which the CDC classifies as an urgent threat due to its relatively high frequency of resistance to multiple different classes of drugs including amphotericin B, echinocandins, and azoles (reviewed in reference 11). Since its recognition as a novel Candida species in 2009, C. auris, has been reported in at least 40 countries (12–14). Whole-genome sequencing (WGS) analyses of C. auris isolates collected from across the globe indicate the concurrent emergence of four genetically distinct clades (15) with a potential fifth clade defined more recently (16). C. auris is thought to primarily colonize the skin (17–19) in addition to a diverse array of body sites, and most clinical isolates to date have been isolated from blood (20). Once C. auris has disseminated to the bloodstream, it can cause potentially fatal candidemia which has an estimated global mortality rate ranging from about 30% to 60% (15, 21, 22).
The resistance to azoles in C. auris is multifactorial; it has been shown that certain mutations in ERG11 (15, 23–31) and overproduction of Cdr1 (32–36) contribute to resistance to fluconazole (FLZ). In multiple Candida species, the transcriptional regulator Mrr1 also plays a role in FLZ resistance (37–45). Moreover, Mayr and colleagues (46) found three C. auris homologs of the transcriptional regulator Mrr1, and showed that one, MRR1a, modestly affected fluconazole resistance. Previously, we demonstrated that in Candida (Clavispora) lusitaniae, which is more closely related to C. auris relative to other well-studied Candida species (12, 47), Mrr1 regulates the expression of MDR1, and overexpression of MDR1 confers resistance to FLZ (40, 48–55), the host antimicrobial peptide histatin-5 (40, 56), bacterially produced phenazines (40), and other toxic compounds (57) in multiple Candida species. C. lusitaniae Mrr1 also regulates dozens of other genes with two of the most strongly regulated genes encoding methylglyoxal (MG) reductase enzymes, MGD1 and MGD2 (37, 40, 58). Mrr1 contributes to C. lusitaniae resistance to MG (58), which is a spontaneously formed dicarbonyl electrophile generated as a by-product of several metabolic processes by all living cells (reviewed in reference 59). Via its carbonyl groups, MG reacts non-enzymatically with biomolecules, which can lead to cellular stress and toxicity (reviewed in reference 59). Some of the risk factors (60–69) for candidiasis caused by C. auris or other Candida spp., such as diabetes (70–72), kidney disease (73–76), or septic shock (77), are associated with elevated MG in human serum. MG resistance across clinical isolates of the same Candida species, including C. auris, can vary (58).
Through specific regulators, MG and other reactive electrophiles induce stress responses in bacteria (78–80), plants (reviewed in reference 81), mammals (reviewed in reference 82), and the yeasts Saccharomyces cerevisiae (83–87) and Schizosaccharomyces pombe (88, 89) at subinhibitory concentrations. We found in C. lusitaniae, MG induces expression of MGD1 and MGD2 as well as MDR1, through a mechanism that involved Mrr1 (58), and that MG increased FLZ resistance. C. auris displays nosocomial transmission (61–63, 65–69), in part due to its resistance to high temperatures (90) and common surface antiseptics (91), and persistence on abiotic surfaces including latex and nitrile gloves (92), plastics (90), and axillary temperature probes (93). The factors that control C. auris stress resistance are not yet known.
In the present study, we show that C. auris MRR1a regulates resistance to MG and that MG is an inducer of Mrr1-regulated gene expression. Mrr1a regulates the gene orthologous to the methylglyoxal reductase genes C. lusitaniae MGD1 in addition to MDR1, which regulates FLZ efflux, but the Mrr1a regulon is smaller than that described for other species. Furthermore, we characterize Mrr1a in both clade I and clade III isolates and show that the Mrr1 variant in clade III is constitutively active. Transcriptomics analysis shows that MG elicits a large transcriptional response that is similar in both clade I and clade III, and that there are commonalities in the responses elicited by MG and the Mrr1 inducer benomyl. These data support the model that Mrr1 is a regulator of MG resistance in coordination with efflux proteins such as Mdr1 and provides the basis for future studies on the roles of Mrr1 and MG in survival of C. auris in hospital settings.
RESULTS
Mrr1a regulates expression of orthologs to MDR1 and MGD1 in C. auris strain B11221 and is involved in MG resistance.
To determine whether the C. auris MRR1 orthologs MRR1a, MRR1b, and MRR1c contributed to resistance to MG, we performed growth kinetic assays in yeast extract-peptone-dextrose (YPD) +/– 5 mM, 10 mM, or 15 mM MG. At MG concentrations of 10 mM (Fig. 1A) and 15 mM (Fig. S1), the mrr1aΔ mutant displayed a substantial growth defect relative to the parental isolate B11221 (WT), while the mrr1bΔ and mrr1cΔ mutants exhibited growth comparable to WT. None of the mutants (mrr1aΔ, mrr1bΔ, or mrr1cΔ) differed from the parental isolate B11221 (WT) in YPD alone or in the presence of 5 mM MG (Fig. S1). Like C. lusitaniae, the C. auris genome encodes multiple putative MG reductases; the closest orthologs to MGD1 and MGD2 were CJI97_000658 and CJI97_004624, respectively, in the B11221 genome assembly (58) and we will henceforth refer to these genes as MGD1 and MGD2. For reference, MGD1 and MGD2 correspond to B9J08_000656 and B9J08_004828, respectively, in the genome assembly of the C. auris reference strain B8441. By quantitative real-time PCR (qRT-PCR), basal expression of MGD1 was significantly decreased 24-fold in the mrr1aΔ mutant relative to B11221 WT (Fig. 1B), and expression of MGD2 trended lower in the mrr1aΔ mutant (∼1.2-fold) but this difference did not reach statistical significance (Fig. 1C). MGD1 was also more highly expressed than MGD2 in the WT B11221 as in C. lusitaniae (58). Consistent with the transcriptional patterns, C. auris Mgd1 shares slightly more identity with C. lusitaniae Mgd1 than does C. auris Mgd2 (63% identity versus 61% identity).
FIG 1.
Mrr1a regulates expression of MGD1 and MDR1 in C. auris isolate B11221. (A) Growth curves of B11221 WT (blue) and its mrr1aΔ (red), mrr1bΔ (green), and mrr1cΔ (purple) derivatives in YPD + 10 mM MG. Data shown represent the mean ± SD for three independent experiments. (B to C) qRT-PCR assessment of MGD1 (B) and MDR1 (C) expression in B11221 WT (blue) and mrr1aΔ (red) cultures grown to exponential phase in YPD at 37°C. Data shown represent the mean ± SD for three independent experiments. Ratio paired t test was used for statistical evaluation; * P < 0.05. (D) Volcano plot of all quantified genes in B11221 WT versus mrr1aΔ in the control condition. Each point represents a single gene; blue points indicate genes significantly more highly expressed in WT; red points indicate genes significantly more highly expressed in mrr1aΔ. Numbers adjacent to each colored point indicate the log2FC in mrr1aΔ versus WT.
The mrr1aΔ mutant has a growth defect in high concentrations of MG, but not at 5 mM MG or in the YPD control. Growth curves of B11221 WT (blue) and its mrr1aΔ (red), mrr1bΔ (green), and mrr1cΔ (purple) derivatives in YPD (left), or YPD supplemented with 5 mM (middle), or 15 mM (right) MG. Data shown represent the mean ± SD for three independent experiments. Download FIG S1, TIF file, 0.3 MB (269.2KB, tif) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In the C. auris B11221 background, expression of MDR1, another target of Mrr1 in other species including C. lusitaniae, also depended on Mrr1a, as the mrr1aΔ mutant exhibited a significant 21-fold decrease in MDR1 expression compared to the WT parent (Fig. 1D). These results indicate that in C. auris MDR1 and MGD1 are co-regulated, as has been reported in C. albicans (44, 45, 94–96), C. parapsilosis (97), and C. lusitaniae (37, 39, 40, 58, 98), and that higher expression of MGD1 and/or MDR1 contributes to growth in high concentrations of MG (Fig. 1A).
In C. lusitaniae and other Candida species, Mrr1 regulates dozens of genes in addition to MDR1 and MGD1 (37, 40). To further elucidate the Mrr1a regulon in C. auris isolate B11221, we performed an RNA-seq analysis of in B11221 WT and its mrr1aΔ derivative in cells from exponential phase cultures grown at 37°C in YPD. In the control condition (YPD + dH2O), only four genes, including MDR1 and MGD1, were differentially expressed between the two strains with the cutoff of a log2 fold change (log2FC) ≥ 1.00 or ≤ -1.00 and a P-value less than 0.05 (Fig. 1E and Data Set S1 for all data). MGD1 and MDR1 showed a 22- and 24-fold decrease, respectively, in mrr1aΔ compared with WT, consistent with our qRT-PCR data. CJI97_005632, which was 2.25-fold lower in mrr1aΔ, is orthologous to the C. albicans genes RIM11 and C2_04280W_A, both of which are predicted to encode proteins with serine/threonine kinase activity, though it is worth noting that levels of the transcript were much lower than levels of MDR1 and MGD1. CJI97_000852, which was 2.77-fold higher in mrr1aΔ than in WT, has 16 orthologs of diverse predicted or known functions in C. albicans, including USO5, USO6, and RBF1 (Fig. 1E and Data Set S1). Notably, MGD2 was not differentially expressed between B11221 WT and the mrr1aΔ mutant in our RNA-seq data (Data Set S1), consistent with our qRT-PCR results described above.
RNA-seq data, in the form of normalized counts per million (CPM), for all tested strains and conditions. Download Data Set S1, XLSX file, 5.7 MB (5.7MB, xlsx) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mrr1a regulates only MDR1 and MGD1 in response to MG and benomyl.
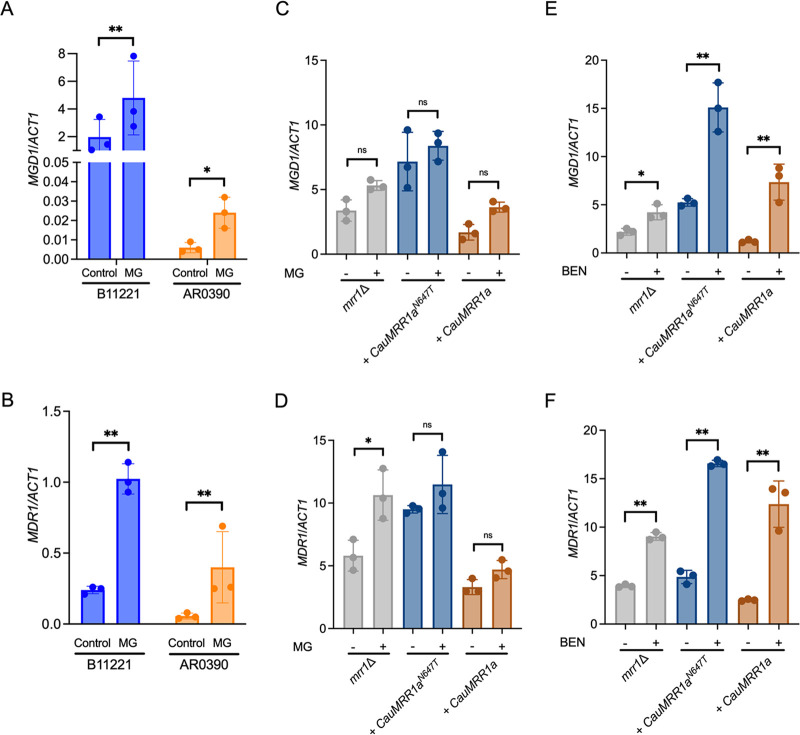
We have previously shown in C. lusitaniae that MG induces expression of the Mrr1-regulated genes MGD1 and MGD2 in an Mrr1-dependent manner, and MDR1 in a partially Mrr1-dependent manner (58). To determine if MG would induce expression of MGD1, MGD2, and/or MDR1 in C. auris, we purified RNA for qRT-PCR from exponential-phase cultures of B11221 WT and mrr1aΔ treated with 5 mM MG or an equal volume of dH2O for 15 min. We found that MG treatment significantly enhanced expression of MGD1 in WT by 2.4-fold but not in mrr1aΔ (Fig. 2A). MGD1 was also induced by a 30-min treatment with 25 μg/mL benomyl (BEN), a known inducer of Mrr1-regulated genes in other Candida species (37, 41, 43, 95, 99–104), by 7.5-fold in the WT (Fig. 2A). The different treatment times for MG and BEN were used to be consistent with previous studies using either compound in the related species C. lusitaniae (37, 58). Expression of MDR1 was also more highly induced by treatment with either MG or BEN in WT compared with the mrr1aΔ mutant by 6- and 14.5-fold, respectively (Fig. 2B). Although MDR1 expression was significantly induced by MG and BEN in the mrr1aΔ, transcript levels of MDR1 were approximately 20-fold higher in the WT than in the mrr1aΔ under these conditions (Fig. 2B), suggesting that Mrr1a is required for maximum expression of MDR1 in response to stimuli.
FIG 2.
MG and BEN both lead to a vast transcriptional response in C. auris B11221, which includes upregulation of MDR1 and MGD1. (A to B) qRT-PCR analysis for expression of MGD1 (A) and MDR1 (B) in exponential-phase cultures of B11221 WT (blue) or mrr1aΔ (red) treated with MG or BEN as indicated. Data shown represent the mean ± SD for three independent experiments. Ratio paired t test was used for statistical evaluation; ns P > 0.05; * P < 0.05; ** P < 0.01. (C to D) Volcano plots of all quantified genes in B11221 WT treated with either MG (C) or BEN (D). Each point represents a single gene; magenta points indicate genes that were significantly upregulated compared with the control condition, teal points indicate genes that were significantly downregulated compared to the control condition. MDR1 and MGD1 are shown along with the two most up- and downregulated genes in each condition. (E to F) Scatterplots of the average CPMs of all quantified genes in mrr1aΔ versus B11221 WT treated with MG (E) or BEN (F). Each point represents a single gene. Points below the dotted line indicate genes that were more highly expressed in the WT, and points above the dotted line indicated genes that were more highly expressed in the mrr1aΔ mutant. MDR1 and MGD1 are shown with red dots for reference.
To describe the complete Mrr1-dependent MG- and BEN-response regulon under our test conditions in C. auris, we also performed RNA-seq on exponential-phase cultures of B11221 WT and mrr1aΔ treated with MG or BEN as described above. In B11221 WT, MG led to the upregulation of 319 genes and downregulation of 133 genes compared with the control condition (Fig. 2C and Data Set S1). In the mrr1aΔ mutant, MG led to the upregulation of 349 genes and downregulation of 143 genes compared with the control condition (Fig. S2A and Data Set S1). Consistent with our qRT-PCR data in Fig. 2A, MG induced expression of MGD1 in the WT but not in the mrr1aΔ mutant (Table S1 and Data Set S1). Although expression of MDR1 was significantly induced by MG in both the WT and the mrr1aΔ mutant (Table S1 and Data Set S1), levels of MDR1 were substantially lower in the mrr1aΔ mutant even in the presence of MG (Fig. 2D and Data Set S1), also in agreement with our qRT-PCR data. MGD1 and MDR1 strongly stood out as the only two genes in the MG response that were strongly dependent on Mrr1a (Fig. 2D).
Select genes differentially expressed in response to MG and/or BEN in the C. auris B11221 background. Differentially expressed genes were determined using a cutoff of |Log2FC| ≥ 1.00 and P-value < 0.05. Download Table S1, DOCX file, 0.06 MB (60.7KB, docx) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The transcriptional response of mrr1aΔ to either MG or BEN is overall similar to that of the B11221 WT parent strain. Volcano plots of all quantified genes in the mrr1aΔ mutant treated with either MG (A) or BEN (B). Each point represents a single gene; magenta points indicate genes that were significantly upregulated compared to the control condition, teal points indicate genes that were significantly downregulated compared to the control condition. MDR1 is shown along with the two most up- and downregulated genes in each condition. Download FIG S2, TIF file, 0.3 MB (276.8KB, tif) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Treatment with BEN led to upregulation of 160 genes and downregulation of 163 genes in the WT (Fig. 2E and Data Set S1). In the mrr1aΔ mutant, 181 genes were upregulated, and 229 genes were downregulated in response to BEN (Fig. S2B and Data Set S1). Like MG, induction of MGD1 by BEN was completely dependent on Mrr1a (Table S1 and Data Set S1) and MGD2 expression was not induced by BEN (Data Set S1). Expression of MDR1 was also induced by BEN in both the WT and the mrr1aΔ mutant, but as with MG, MDR1 levels in the mrr1aΔ mutant did not reach that of the WT even with BEN treatment (Fig. 2F and Data Set S1). Again, MGD1 and MDR1, appear to be the only genes in C. auris whose induction of expression by either MG or BEN is dependent on Mrr1a. The Mrr1a-independent responses to MG and BEN are discussed further below.
B11221 has higher basal expression of MDR1 and of putative MG reductase genes compared with the clade I isolate AR0390.
Many clade III isolates, including B11221, contain an N647T single nucleotide polymorphism (SNP) in MRR1a (25, 105). In Iyer et al., this SNP was proposed to be a gain-of-function mutation due to the resistance of clade III isolates against azoffluxin, a novel antifungal compound that inhibits expression and activity of C. auris efflux pumps (105). As a first step to determine whether there were differences in activity between the Mrr1a protein encoded by the N647T allele found in clade III and the variant encoded by the allele found in clades I, II, and IV, we compared MG sensitivity of B11221 to that of clade I isolate AR0390. Interestingly, AR0390 grew substantially better than B11221 in the YPD control but showed a greater reduction in growth in YPD with 5 mM MG than did B11221 (Fig. S3). At concentrations of 10 mM (Fig. 3A) and 15 mM MG (Fig. S3), AR0390 exhibited a profound growth defect compared with B11221. To determine if differences in MG sensitivity were due to differences in MGD1 expression, we measured basal expression of MGD1 and its co-regulated gene MDR1 in B11221 and AR0390 using qRT-PCR. Both genes were significantly more highly expressed in B11221 by 42- and 4.2-fold, respectively (Fig. 3B and C).
FIG 3.
MDR1 and MGD1 are among the genes significantly more highly expressed in isolate B11221 compared with isolate AR0390. (A) Growth curves of B11221 (blue) and AR#0390 (orange) in YPD + 10 mM MG. Data shown represent the mean ± SD for three independent experiments. (B to C) qRT-PCR assessment of MGD1 (B) and MDR1 (C) expression in B11221 (blue) and AR0390 (orange) grown to exponential phase in YPD at 37°C. Data shown represent the mean ± SD for three independent experiments. Ratio paired t test was used for statistical evaluation; * P < 0.05; **** P < 0.0001. (D) Volcano plot of all quantified genes, matched by syntenic ortholog, in B11221 and AR0390 in the control condition (YPD). Each point represents a single gene; blue points indicate genes significantly more highly expressed in B11221; orange points indicate genes significantly more highly expressed in AR0390. (E to F) qRT-PCR expression analysis for MGD1 (E) and MDR1 (F) in C. lusitaniae U04 mrr1Δ (gray) and its derivatives expressing CauMRR1aN647T (dark blue) or CauMRR1a (brown). Data shown represent the mean ± SD for three independent experiments. One-way ANOVA was used for statistical evaluation; * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. (G) Growth curves of C. lusitaniae U04 mrr1Δ (gray) and its derivatives expressing CauMRR1aN647T (dark blue) or CauMRR1a (brown) in YPD + 15 mM MG. One representative experiment of three independent experiments is shown; error bars represent the standard deviation of technical replicates within the experiment.
C. auris strain AR0390 has a growth advantage over B11221 in YPD but loses that advantage in the presence of increasing concentrations of MG. Growth curves of B11221 (blue) and AR0390 (orange) in YPD (left), or YPD supplemented with 5 mM (middle), or 15 mM (right) MG. Data shown represent the mean ± SD for three independent experiments. Download FIG S3, TIF file, 0.2 MB (192.7KB, tif) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To gain a deeper understanding of the broader transcriptional differences between B11221 and AR0390, we compared the basal global gene expression in YPD of the two strains using RNA-seq. First, we matched the 5,227 syntenic orthologs between the genomes of B11221 and the clade I reference strain B8441 to compare expression of each gene under the control condition. Of these, 755 genes were differentially expressed between B11221 and AR0390 in the control condition (|log2FC| ≥ 1.00, FDR-corrected P < 0.05) (Fig. 3D, Data Set S1). The top 20 differentially expressed genes whose orthologs have known or predicted functions in C. albicans are reported in Table S2. Strikingly, the two genes which exhibited the largest difference in expression between B11221 and AR0390 were MGD2 (log2FC = 11.29) and MGD1 (log2FC = 8.53) (Fig. 3D, Table S2, and Data Set S1). A third gene with homology to MG reductases, CJI97_001800/B9J08_002257, was also more highly expressed in B11221, although the log2FC in expression of this gene in B11221 versus AR0390 was only 1.41 (Data Set S1). Low expression of MGD1, MGD2, and/or B9J08_002257 may contribute to the severe growth defect of AR0390 in the presence of MG. Consistent with our qRT-PCR data, MDR1 was also significantly more highly expressed in B11221 relative to AR0390 (log2FC = 4.42) (Fig. 3D and Table S2). Although MGD2 and B9J08_002257 do not appear to be regulated by Mrr1a in our studies, it is nonetheless interesting to note the elevated expression of three putative MG reductases in the MDR1-overexpressing C. auris isolate B11221, as the co-expression of MDR1 with at least one MG reductase has been reported in numerous studies in other Candida species (37, 40, 44, 45, 58, 94–97).
Top 20 genes with predicted functions differentially expressed between C. auris isolates B11221 and AR0390 in the control condition. Differentially expressed genes were determined using a cutoff of |Log2FC| ≥ 1.00 and P-value < 0.05. Download Table S2, DOCX file, 0.02 MB (25.3KB, docx) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Clade III Mrr1aN647T exhibits a gain-of-function phenotype compared with clade I Mrr1a when expressed in C. lusitaniae.
To compare the activities of the proteins encoded by the MRR1a alleles of B11221 and AR0390 more directly, we heterologously expressed each allele, henceforth referred to as CauMRR1aN647T and CauMRR1a, respectively, independently in a C. lusitaniae mrr1Δ mutant previously generated and characterized by our lab (37, 40, 58). All three C. lusitaniae clones expressing CauMRR1aN647T which we tested exhibited a 4-fold increase in FLZ MIC relative to the U04 mrr1Δ parent (16 μg/mL versus 4 μg/mL), confirming that C. auris clade III MRR1a can complement MRR1-dependent FLZ resistance in C. lusitaniae and adding support to the hypothesis that the N647T substitution in clade III MRR1a confers increased activity. However, the FLZ MIC of the three tested C. lusitaniae clones expressing CauMRR1a did not differ from that of U04 mrr1Δ (4 μg/mL), so FLZ MIC alone could not indicate whether this allele is functional in C. lusitaniae. One clone expressing each C. auris MRR1a allele was chosen at random for the remaining experiments described in this paper: clone #1 for CauMRR1aN647T and clone #5 for CauMRR1a. Using qRT-PCR, we then examined basal expression levels of C. lusitaniae MGD1 (CLUG_01281) and MDR1 (CLUG_01938/CLUG_01939) in the heterologous complements and the U04 mrr1Δ parent. Complementation with CauMRR1aN647T conferred a significant increase in basal expression of both MGD1 (Fig. 3E) and MDR1 (Fig. 3F) compared with the mrr1Δ parent, while complementation with CauMRR1a led to a small, but significant, decrease in expression of both genes relative to mrr1Δ (Fig. 3E and F). These results are consistent with our previous observations that C. lusitaniae strains expressing certain Mrr1 variants with low basal activity demonstrate lower expression of some Mrr1-regulated genes, including MDR1 and MGD1, compared with an isogenic mrr1Δ strain suggesting that Mrr1 has both repressing and activating roles (37, 58). Finally, we assessed the relative MG resistance of the isogenic C. lusitaniae strains expressing CauMRR1aN647T or CauMRR1a and the U04 mrr1Δ parent. The CauMRR1aN647T complement grew markedly better in 15 mM MG compared with U04 mrr1Δ whereas the CauMRR1a complement grew substantially worse than U04 mrr1Δ (Fig. 3G), consistent with the pattern of MGD1 expression we observed in these strains via qRT-PCR. None of the C. lusitaniae strains demonstrated growth differences in the YPD control, or in the presence of MG at concentrations of 5 mM or 10 mM (Fig. S4).
C. lusitaniae strains complemented with CauMRR1aN647T or CauMRR1a do not differ in growth from the mrr1Δ parent at MG concentrations below 15 mM. Growth curves of C. lusitaniae U04 mrr1Δ (grey) and its derivatives expressing CauMRR1aN647T (dark blue) or CauMRR1a (brown) in YPD (left) or YPD supplemented with 5 mM (middle), or 10 mM (right) MG. Data shown represent the mean ± SD for three independent experiments. Download FIG S4, TIF file, 0.2 MB (241.8KB, tif) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MG induces expression of MGD1 and MDR1 in C. auris B11221 and AR0390, but not in C. lusitaniae strains expressing C. auris MRR1a alleles.
Next, we compared induction of MGD1 and MDR1 by MG in the C. auris strains B11221 and AR0390 via qRT-PCR. MG significantly induced expression of MGD1 by 2.4-fold in C. auris strain B11221 and by 4.0-fold in C. auris strain AR0390 (Fig. 4A) and expression of MDR1 by 6.0-fold in B11221 and 9.3-fold in AR0390 (Fig. 4B). AR0390 displayed lower expression of both genes in MG, but a higher fold change compared to B11221, further supporting the hypothesis that the N647T allele is gain-of function.
FIG 4.
MG induces expression of MGD1 and MDR1 in C. auris isolates B11221 and AR0390, but C. auris MRR1a is not inducible by MG when heterologously expressed in C. lusitaniae. (A to B) qRT-PCR analysis for expression of MGD1 (A) and MDR1 (B) in exponential-phase cultures of B11221 (blue) or AR0390 (orange) treated with MG as indicated. Data shown represent the mean ± SD for three independent experiments. Ratio paired t test was used for statistical evaluation; ns P > 0.05; * P < 0.05; ** P < 0.01. (C to F) qRT-PCR analysis for expression of MGD1 (C, E) and MDR1 (D, F) in exponential-phase cultures of C. lusitaniae U04 mrr1Δ (gray) and its derivatives expressing CauMRR1aN647T (dark blue) or CauMRR1a (brown) treated with 5 mM MG for 15 min (C, D) or 25 μg/mL BEN for 30 min (E, F). Data shown represent the mean ± SD for three independent experiments. Ratio paired t test was used for statistical evaluation; ns P > 0.05; * P < 0.05; ** P < 0.01.
Finally, we compared induction of MGD1 and MDR1 by MG in the isogenic C. lusitaniae strains expressing either CauMRR1aN647T or CauMRR1a and the mrr1Δ parent. Additionally, we tested induction by BEN in these strains as a control. While the mrr1Δ parent exhibited a significant 1.8-fold induction of MDR1, neither C. lusitaniae strain expressing a C. auris Mrr1a allele demonstrated a significant change in MGD1 or MDR1 expression in response to MG (Fig. 4C and D), indicating that C. auris Mrr1a may repress MRR1-independent MG induction of MDR1 in C. lusitaniae and that induction of MGD1 by MG in C. lusitaniae requires a functional MRR1 allele from its own species. Treatment with BEN led to significant increase in expression of MGD1 (Fig. 4E) and MDR1 (Fig. 4F) in all three C. lusitaniae strains. In response to BEN, MGD1 was induced by 1.9-fold in mrr1Δ, 2.9-fold in the CauMRR1aN647T complement, and 6.1-fold in the CauMRR1a complement (Fig. 4E). Likewise, expression of MDR1 was induced by 2.3-fold in mrr1Δ, 3.5-fold in the CauMRR1aN647T complement, and 5.0-fold in the CauMRR1a complement in response to BEN (Fig. 4F). The striking difference in the ability of the C. lusitaniae strains expressing C. auris MRR1a alleles to respond to BEN versus MG suggests that there are differences in the mechanisms by which BEN and MG induce Mrr1-dependent transcriptional activation and that MG induction of C. auris Mrr1a is not supported by C. lusitaniae factors. These potential differences are a topic of future study and may shed light on mechanisms of Mrr1 activation in Candida species.
MG and BEN induced Mrr1a-independent transcriptional responses in C. auris.
We have previously observed heterogeneity in MG resistance as well as MG-induced FLZ resistance among several C. auris isolates from different clades (58), and thus we were interested in whether the overall transcriptional response to MG was more similar or different in B11221 and AR0390. AR0390 had greater number of genes differentially expressed by MG compared with B11221; 438 genes were significantly upregulated, and 242 genes were significantly downregulated by MG (see Fig. S5 for the volcano plot of all genes). More genes had a larger fold change in response to MG in AR0390 compared with B11221, including MGD1 and MDR1 (Fig. 5A), consistent with the qRT-PCR results in Fig. 4A and B. However, there was a large overlap of 254 genes which were induced by MG in both strains (Fig. 5B), suggesting a common response across these two genetically distinct clades. These commonly induced genes include many with putative roles in amino acid biosynthesis; transmembrane transport; or acquisition and usage of sulfur (Fig. 5C and Table S3). The complete comparison is available in Data Set S1.
FIG 5.
MG induces and represses common pathways across B11221 and AR0390. (A) Venn diagram of genes with syntenic orthologs between B11221 and AR0390 that were significantly induced (indicated by “up” arrows) or repressed (indicated by “down” arrows) by MG in either or both strains. (B) Scatterplot of the log2FC of genes significantly induced by MG in AR0390 versus the log2FC of genes induced by MG in B11221. Only genes with syntenic orthologs between the two strains are shown. Each point represents a single gene; points above the dotted line indicate genes which exhibited a greater Log2FC in AR0390, and points below the dotted line indicate genes which exhibited a greater log2FC in B11221. MGD1 and MDR1 are indicated with red dots for reference. (C) Graphic summary of major groups of genes that were significantly up- or downregulated in response to MG in both B11221 and AR0390. Genes in bold text were also up- or downregulated in response to BEN in B11221.
Comparison of select genes differentially expressed in response to MG in C. auris isolates B11221 and/or AR0390. Differentially expressed genes were determined using a cutoff of |Log2FC| ≥ 1.00 and P-value < 0.05. Download Table S3, DOCX file, 0.04 MB (43.7KB, docx) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Treatment with 5 mM MG leads to the differential expression of more genes in AR0390 than in B11221. Volcano plot of all quantified genes in AR0390 treated with MG. Each point represents a single gene; magenta points indicate genes that were significantly upregulated compared with the control condition, teal points indicate genes that were significantly downregulated compared with the control condition. MDR1 and MGD1 are shown for reference. Download FIG S5, TIF file, 0.2 MB (255KB, tif) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Only 68 genes with syntenic orthologs across both strains were commonly repressed by MG (Fig. 5B). These genes include some with putative roles in metal transport or carbohydrate uptake and metabolism (Fig. 5C and Table S3). We did not observe obvious patterns in genes that were only induced or repressed in one strain, and some genes that are listed as only induced or repressed in one strain were close to the cutoff in the other strain.
The groups of genes that were differentially expressed in response to MG in both B11221 and AR0390 were also evident in the response of B11221 to BEN as well as the response of the mrr1aΔ mutant in response to MG and BEN. In B11221, a total of 46 genes exhibited significant induction by both MG and BEN, including MGD1 and MDR1. Many of the 44 other genes have predicted roles in assimilation and biosynthesis of sulfur-containing compounds or xenobiotic transport (Fig. 5C and Table S1). MG also induced expression of many genes with predicted roles in the biosynthesis of amino acids. The two genes most highly upregulated upon MG treatment, in terms of fold change, in this strain were orthologous to the arginine biosynthesis genes ARG3 (log2FC = 4.77) and ARG1 (log2FC = 4.72) (Fig. 2C and Table S1). Conversely, BEN had a limited effect on expression of amino acid biosynthesis genes (Table S1). There were also common themes among the genes that were significantly repressed by both MG and BEN in B11221. Genes that were repressed by both MG and BEN included four orthologs of the HGT glucose transporter family, five genes with a predicted role in uptake of iron and/or copper, and ERG6, which encodes an enzyme in the ergosterol biosynthesis pathway (Fig. 5C and Table S1). The genes that were repressed by only one stimulus, MG or BEN, also included those involved in ergosterol biosynthesis and the uptake of iron, copper, or glucose (Fig. 5C, Table S1). In general, the transcriptional response of the mrr1aΔ mutant to MG and BEN was similar to that of B11221 WT (Fig. S2 and Table S1). The complete data sets for MG and BEN responses are available in Data Set S1.
DISCUSSION
In this work, we have demonstrated that in C. auris, the zinc-cluster transcription factor Mrr1a, which is orthologous to Mrr1 in other Candida species, strongly regulates expression of a putative MG reductase MGD1 in addition to MDR1, and that Mrr1a plays a role in MG resistance, highlighting a function of Mrr1 that is distinct from antifungal resistance. We also compared basal global gene expression in B11221 and AR0390 and found that MDR1, MGD1, and MGD2 were among the genes significantly more highly expressed in B11221, consistent with the higher MG resistance of this isolate relative to AR0390. These differences were explained by our finding that MRR1a from B11221 encoded a higher activity variant than that from AR0390 as evidenced by a higher FLZ MIC, higher expression of MDR1 and MGD1, and higher MG resistance in the strain expressing CauMRR1aN647T compared with the isogenic strain expressing CauMRR1a. The allele from B11221 contains an N647T amino acid substitution (25, 105) which is in the central region of the regulator where other gain of function substitutions have been found. Both alleles result in induction of MDR1 and MGD1 in response to BEN but not to MG in C. lusitaniae, suggesting that these two compounds activate Mrr1-dependent transcription through different mechanisms.
Under the conditions tested, Mrr1a regulation in the C. auris B11221 background was mainly of MGD1 and MDR1. Homologs of MDR1 and at least one gene encoding a known or predicted MG reductase are co-regulated by Mrr1 in C. albicans (44, 45, 94–96), C. parapsilosis (97), and C. lusitaniae (37, 40, 58), suggesting that the co-regulation of these two genes has been conserved throughout multiple Candida species. Gaining a deeper understanding of the evolutionary and biochemical relationship between methylglyoxal reductases and efflux pumps, particularly Mdr1, may shed light on how Candida species sense and respond to environmental or physiological stresses, evade host defense mechanisms, and develop antifungal resistance. In all other Candida species with published Mrr1 regulons, however, Mrr1 appears to regulate expression of many more genes than the four we have described here in the C. auris strain B11221 (37, 40, 44, 45, 97). The surprisingly small number of C. auris genes whose expression was significantly altered by genetic deletion of MRR1a may be due to possible redundancy between MRR1a and the other two MRR1 orthologs in C. auris, MRR1b, and MRR1c, although further studies would be necessary to test this hypothesis. It is striking, however, that MRR1a alone seems to be necessary for expression and induction of MGD1, which is further supported by our observation that only the mrr1aΔ mutant had a growth defect in MG compared with parental B11221 (Fig. 1A).
Our demonstration of increased basal activity of the CauMRR1aN647T allele compared with the allele from AR0390 supports the hypothesis put forth by Iyer et al. (105) that the N647T substitution found in many clade III isolates is a gain-of-function mutation. Furthermore, this may explain why deletion of MRR1a leads to a mild decrease in azole resistance in B11221, but not in the clade IV isolate B11243 (46). In C. albicans, knocking out gain-of-function MRR1 causes a significant decrease in FLZ resistance, but knocking out MRR1 with wild-type transcriptional activity does not alter FLZ resistance (41, 44, 45, 106). Similarly, knocking out gain-of-function MRR1 in C. lusitaniae also decreases FLZ resistance, although knocking out MRR1 alleles that do not encode a constitutively active protein generally leads to increased FLZ resistance (37).
Although Mrr1a does not appear to play a major role in C. auris azole resistance (46), our findings suggest that it contributes to resistance against MG, which may be encountered in the host environment. We have previously shown that Mrr1 also contributes to MG resistance in C. lusitaniae in a manner that is partially dependent on MGD1 and MGD2 (58). Indeed, gain-of-function mutations in MRR1 may arise in various Candida species due to selective pressures other than azoles. In C. lusitaniae, we have reported the emergence of gain-of-function mutations in MRR1 among isolates from a patient with no history of clinical antifungal use (40). In C. auris, most sequenced clade III isolates exhibit both the MRR1aN647T allele and the ERG11F126L allele (25), the latter of which has been shown to be a major contributor to azole resistance (31). Although it is not known whether the MRR1a or ERG11 mutation occurred first in the clade III lineage, it seems plausible that if the ERG11 mutation did occur first, evolution of the MRR1aN647T allele in C. auris is likely to be the result of selection for MGD1 expression and/or an unknown role for Mdr1 that is unrelated to azole resistance. Therefore, we hypothesize that Mrr1 may act, either directly or indirectly, as a response regulator for carbonyl stress in Candida species, and future studies will investigate a possible role for Mrr1 in resistance against other physiologically relevant reactive carbonyl compounds.
Curiously, although both variants of C. auris Mrr1a were inducible by BEN when expressed in C. lusitaniae, they were not inducible by MG under the conditions tested (Fig. 4E and F). One possible hypothesis for this observation is that Mrr1 must interact with at least one particular binding partner to induce transcription in response to MG, and that C. auris Mrr1a does not bind efficiently to this C. lusitaniae Mrr1-binding protein or complex. Differential requirements for Mrr1-dependent transcriptional activation by chemical stressors have reported in C. albicans. For example, the transcription factor Mcm1 is required for Mrr1-dependent induction of MDR1 in response to BEN but not to H2O2 (101), and the redox-sensing transcription factor Cap1 is required for MDR1 induction by H2O2 and may play a role in MDR1 induction by BEN (44). Furthermore, gain-of-function Mrr1 in C. albicans requires the Swi/Snf chromatin remodeling complex to maintain promoter occupancy, and the kinase Ssn3, which is a subunit of the Mediator complex, may act in opposition to Mrr1 or its coactivators (38). Thus, although C. auris Mrr1a can complement Mrr1-dependent basal and BEN-induced expression of MDR1 and MGD1 in C. lusitaniae, it may be incompatible with certain elements of the C. lusitaniae MG-responsive transcriptional machinery. Further studies on the differences between C. lusitaniae and C. auris Mrr1, particularly in the presence of MG, may elucidate more detailed mechanisms of Mrr1 activation.
In general, we observed substantial upregulation of genes with predicted roles in transmembrane transport, sulfur metabolism, and amino acid biosynthesis in response to MG in all three strains tested. Many genes downregulated in response to MG in all three strains have predicted roles in metal acquisition, particularly iron, and carbohydrate metabolism. In both B11221 WT and mrr1aΔ, BEN treatment led to differential expression of similar groups of genes as MG in addition to induction of genes with predicted roles in oxidative stress response. Our studies of the transcriptional response of C. auris to MG and BEN contribute to the understanding of how Candida species may adapt to oxidative and/or carbonyl stress, two types of stress that a pathogen is likely to encounter in the host environment. In humans, elevated serum MG has been reported in diabetes as well as in renal failure, which are both risk factors for Candida infection (107, 108). There is also evidence that neutrophils (109) and macrophages (110, 111) generate MG during the inflammatory response, consistent with elevated levels of MG in sepsis patients (77). In our transcriptomics analysis of three C. auris strains exposed to 5 mM MG for 15 min, upregulation of numerous genes involved in amino acid uptake, metabolism, and biosynthesis was one of the most striking responses to MG (Table S1 for comparison of MG and BEN in B11221 WT and mrr1aΔ and Table S2 for the comparison of genes induced by MG in B11221 and/or AR0390). In particular, induction of ARG genes is interesting considering the report that C. albicans upregulates expression of arginine biosynthesis genes when phagocytosed by macrophages or in response to sublethal concentrations of hydrogen peroxide, tert-butyl hydroperoxide, or menadione in vitro (112). This induction of ARG genes in C. albicans by macrophages is dependent on the gp91phox subunit of the macrophage oxidase, and thus is likely a direct response to oxidative stress rather than arginine depletion (112). In our data set, ARG3 and ARG1 exhibited the highest log2FC in response to MG in the B11221 background, independently of MRR1a (Table S1). We also observed, in all three C. auris strains, induction of several MET genes, which are involved in methionine synthesis and are an important branch of sulfur assimilation in yeast. Other genes involved in sulfur acquisition and assimilation that were induced by MG include the sulfate importer SUL2, a gene orthologous to both CYS3 and STR3 of S. cerevisiae, and numerous genes associated with iron-sulfur cluster formation (Table S1). A gene orthologous to MUP1 of S. cerevisiae and C. albicans was induced by MG in B11221 WT and AR0390 but fell short of the log2FC ≥ 1.00 cutoff in mrr1aΔ (Table S1 and Data Set S1). Induction of genes involved in sulfur metabolism, including the MET pathway, SUL2, CYS3, STR3, and MUP1, has previously been observed in Saccharomyces cerevisiae exposed to 1g/L acetaldehyde (113), another reactive aldehyde metabolite that is structurally similar to MG. Thus, sulfur acquisition and metabolism may be an important part of the carbonyl stress response in yeast.
In the B11221 background, we observed modest overlap in the genes and groups of genes that were up- or downregulated in response to either MG or BEN. MDR1 and MGD1 were among the genes induced by both compounds, and induction of MGD1 by either MG or BEN was completely dependent on MRR1a. Although BEN, which originated as an agricultural fungicide, is widely recognized as an inducer of expression of Mrr1-regulated genes in Candida species (37, 41, 43, 95, 99–104), the mechanism by which this induction occurs is not yet known. BEN is thought to cause oxidative stress in yeast (114, 115), which is consistent with our observation of an upregulation of genes with a predicted role in oxidative stress response in BEN-treated C. auris cultures (Table S1). Additionally, in mammalian cells, BEN exposure has been shown to inhibit aldehyde dehydrogenase enzymes (116–119), which may lead to an accumulation of reactive aldehydes, although this possible mechanism has not yet been investigated in fungi.
We also note similarities between the results of our study of MG- and BEN-treated C. auris and the recently published transcriptional analysis of the clade I C. auris strain NCPF 8973 exposed to 75 μM farnesol (120). In response to farnesol, the authors reported upregulation of many genes with predicted roles in transmembrane transport, such as MDR1 and CDR1, and downregulation of numerous genes predicted to be involved in metal acquisition and homeostasis, including multiple ferric reductases and iron permeases (120). As farnesol may cause oxidative stress in Candida species (120–123) and in S. cerevisiae (124, 125), the overlap in transcriptional changes in response to MG, BEN, and farnesol likely provides valuable insight into how C. auris and other Candida species sense and adapt to physiologically relevant stressors. In fact, MG itself may serve as a stress signal in various organisms. In plants, for example, intracellular MG increases in response to drought (126, 127), salinity (126, 128–131), cold stress (126), heavy metals (128), or phosphorous deficiency (131), and overexpression of certain genes involved in MG detoxification has been shown to enhance salt tolerance in tobacco (126) and in Brassica juncea (132). Investigating whether MG detoxification is linked to abiotic stressors such as salt, temperature, or desiccation in Candida species would be an interesting avenue of future research, particularly in C. auris due to its persistence on hospital surfaces and high salt tolerance.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The sources of all strains used in this study are listed in Table S4. All strains were stored long term in a final concentration of 25% glycerol at −80°C and freshly streaked onto YPD agar (10 g/L yeast extract, 20 g/L peptone, 2% glucose, 1.5% agar) once every 7 days and maintained at room temperature. Unless otherwise noted, all overnight cultures were grown in 5 mL YPD liquid medium (10 g/L yeast extract, 20 g/L peptone, 2% glucose) on a rotary wheel at 30°C. Media was supplemented with 25 μg/mL BEN (stock 10 mg/mL in DMSO) or 5 mM, 10 mM, or 15 mM MG (Sigma-Aldrich, 5.55 M) as noted. Escherichia coli strains were grown in LB with 15 μg/mL gentamicin (gent).
Strains and oligonucleotides used in this study. Download Table S4, DOCX file, 0.04 MB (43.7KB, docx) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids for complementation of C. auris MRR1a.
Plasmids for complementing C. auris MRR1a into C. lusitaniae were created as follows: the open reading frame of MRR1a was amplified from the genomic DNA of C. auris isolates B11221 (for CauMRR1aN647T) and AR0390 (for CauMRR1a) using a forward primer with homology to the 5′ flank of C. lusitaniae MRR1 and a reverse primer with homology to the 3′ flank of C. lusitaniae MRR1 for recombination into the C. lusitaniae MRR1 complementation plasmid pMQ30MRR1-L1191H+Q1197* (58). Plasmid pMQ30MRR1-L1191H+Q1197* was digested with AscI (New England BioLabs) and AgeI-HF (New England BioLabs). The PCR products and digested plasmid were cleaned using the Zymo DNA Clean & Concentrator kit (Zymo Research) and assembled using the S. cerevisiae recombination technique described in (133). Recombined plasmids were isolated from S. cerevisiae using a yeast plasmid miniprep kit (Zymo Research) before transformation into NEB®5-alpha competent E. coli (New England BioLabs). E. coli containing pMQ30-derived plasmids were selected for on LB containing 15 μg/mL gentamicin. Plasmids from E. coli were isolated using a Zyppy Plasmid Miniprep kit (Zymo Research) and subsequently verified by Sanger sequencing with the Dartmouth College Genomics and Molecular Biology Shared Resources Core. MRR1a complementation plasmids containing the correct sequences were linearized with NotI-HF (New England BioLabs), cleaned up with the Zymo DNA Clean & Concentrator kit (Zymo Research) and eluted in molecular biology grade water (Corning) before transformation of 1.5 μg into C. lusitaniae strain U04 mrr1Δ as described below. All plasmids and primers used and created in this study are listed in Table S4.
Transformation of C. lusitaniae with C. auris MRR1a complementation constructs.
Mutants in C. lusitaniae were generated using an expression-free CRISPR-Cas9 method as previously described (37, 58, 134). In brief, cells suspended in 1M sorbitol were electroporated immediately following the addition of 1.5 μg of C. auris MRR1a complementation plasmid that had been previously linearized with NotI-HF (New England BioLabs) and Cas9 ribonucleoprotein containing crRNA targeting the NAT1 gene. Transformants were selected on YPD agar containing 600 μg/mL hygromycin B (HygB). Successful transformants were identified via PCR of the C. lusitaniae MRR1 locus as previously described (37, 58). CRISPR RNAs (crRNAs; IDT) and primers used to validate transformants are listed in Table S4.
MIC assay.
MIC assays for FLZ were performed in RPMI 1640 medium (Sigma, containing l-glutamine, 165 mM MOPS, 2% glucose at pH 7) as described in Demers et al. (40) and Biermann et al. (58) using the broth microdilution method. The final concentration of FLZ in each well ranged from 64 μg/mL to 0.125 μg/mL. Plates were incubated at 35°C and scored for growth at 24 h and 48 h; the results are reported in Table S4. The MIC was defined as the drug concentration that abolished visible growth compared with a drug-free control.
Growth kinetics.
Growth kinetic assays were performed as previously described in Biermann et al. (58). In brief, exponential-phase cultures of C. auris or C. lusitaniae were washed and diluted in dH2O to an OD600 of 1; 60 μL of each diluted cell suspension was added to 5 mL fresh YPD. To each well of a clear 96-well flat-bottom plate (Falcon) was added 100 μL of YPD or YPD with MG at twice the desired final concentration and 100 μL of cell inoculum in YPD. Plates were arranged in technical triplicate for each strain and condition and incubated in a Synergy Neo2 Microplate Reader (BioTek, USA) according to the following protocol: heat to 37°C, start kinetic, read OD600 every 60 min for 36 h, end kinetic. Results were calculated in Microsoft Excel and plotted in GraphPad Prism 9.0.0 (GraphPad Software).
Quantitative real-time PCR.
Overnight cultures of C. auris or C. lusitaniae were diluted 1:50 into 5 mL fresh YPD, and grown to for 4 h at 37°C. To each culture was added MG to a final concentration of 5 mM (4.5 μL stock), BEN to a final concentration of 25 μg/mL (12.5 μL stock), or 4.5 μL molecular biology grade dH2O. Cultures were returned to the roller drum at 37°C for 15 min (MG or dH2O) or 30 min (BEN), then centrifuged at 5,000 rpm for 5 min. The differences in time of exposure in the experimental scheme was used to maintain consistency with published experiments in other species, and not because of known differences in kinetics of activity for the two inducers. RNA isolation, gDNA removal, cDNA synthesis, and quantitative real-time PCR were performed as previously described (40). Transcripts were normalized to C. auris or C. lusitaniae ACT1 expression as appropriate. Results were calculated in Microsoft Excel and plotted in GraphPad Prism 9.0.0 (GraphPad Software). Primers are listed in Table S4.
RNA sequencing.
Overnight cultures of C. auris were diluted to an OD600 of 0.1 in 5 mL fresh, pre-warmed YPD, and incubated on a roller drum at 37°C for five to six doublings (approximately 6 h). Cultures were diluted once more to an OD600 of 1 in 5 mL fresh, pre-warmed YPD and returned to the roller drum at 37°C for another five to six doublings. To each culture was added MG to a final concentration of 5 mM (4.5 μL), BEN to a final concentration of 25 μg/mL (12.5 μL), or 4.5 μL molecular biology grade dH2O. Cultures were returned to the roller drum at 37°C for 15 min (MG or dH2O) or 30 min (BEN), then centrifuged at 5,000 rpm for 5 min. Supernatants were discarded and RNA isolation was performed on cell pellets as described above for qRT-PCR. gDNA was removed from RNA samples as described above. DNA-free RNA samples were sent to the Microbial Genome Sequencing Center (https://www.migscenter.com/) for RNA sequencing.
Analysis of RNA-seq.
RNA-seq data were analyzed by the Microbial Genome Sequencing Center (https://www.migscenter.com/) as follows: Quality control and adapter trimming was performed with bcl2fastq (https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html). Read mapping was performed with HISAT2 (135). Read quantification was performed using Subread’s featureCounts (136) functionality. Read counts were loaded into R (https://www.R-project.org/) and normalized using edgeR’s (137) Trimmed Mean of M values (TMM) algorithm. Subsequent values were then converted to counts per million (cpm). Differential expression analysis was performed using edgeR’s Quasi Linear F-Test. In the supplementary file, the sheet named “All Quantified Genes” contain the results of the exact test for all genes in addition to the normalized counts per million for all samples. Differentially expressed genes were determined using the cutoff of |log2FC| > 1 and P < 0.05.
Identification of orthologs.
Orthologs of C. auris genes in C. albicans, C. lusitaniae, and S. cerevisiae, as well as orthologs between B11221 and the clade I reference strain B8441, were identified using FungiDB (https://fungidb.org) (138, 139).
Generation of Venn diagrams.
Venn diagrams of differentially expressed genes across different strains and conditions were computed using the Venn diagram tool from UGent Bioinformatics & Evolutionary Genomics, which is accessible at https://bioinformatics.psb.ugent.be/webtools/Venn/.
Statistical analysis and figure preparation.
All graphs were prepared with GraphPad Prism 9.0.0 (GraphPad Software). Ratio paired t-tests and one-way ANOVA tests were performed in Prism; details on each test are described in the corresponding figure legends. All P-values were two-tailed and P < 0.05 were considered significant for all analyses performed and are indicated with asterisks in the text: * p <0.05; ** p <0.01; *** p <0.001; **** p <0.0001.
Data availability.
The data supporting the findings in this study are available within the paper and its supplemental material and are also available from the corresponding author upon request. The raw sequence reads from the RNA-seq analysis have been deposited into NCBI sequence read archive under BioProject PRJNA801628 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA801628).
ACKNOWLEDGMENTS
We thank Joachim Morschhäuser and the FDA-CDC Antimicrobial Resistance Isolate Bank for providing strains. We thank Judith Berman for the pGEM-URA3 plasmid used for yeast cloning. We thank Elora Demers for primers.
A.R.B. and D.A.H. conceived and designed the experiments and wrote the paper; A.R.B. performed the experiments; A.R.B. and D.A.H. analyzed the data.
This study was supported by grants R01 5R01 AI127548 to D.A.H. Core services were provided by STANTO19R0 to CFF RDP, P30-DK117469 to DartCF, and P20-GM113132 to BioMT. Sequencing services and specialized equipment were provided by the Genomics and Molecular Biology Shared Resource Core at Dartmouth, NCI Cancer Center Support Grant 5P30 CA023108-41. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
The authors have declared that no competing interests exist.
Contributor Information
Deborah A. Hogan, Email: dhogan@dartmouth.edu.
Aaron P. Mitchell, University of Georgia
REFERENCES
- 1.Pfaller MA, Andes DR, Diekema DJ, Horn DL, Reboli AC, Rotstein C, Franks B, Azie NE. 2014. Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: data from the Prospective Antifungal Therapy (PATH) registry 2004–2008. PLoS One 9:e101510. doi: 10.1371/journal.pone.0101510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quindos G, Marcos-Arias C, San-Millan R, Mateo E, Eraso E. 2018. The continuous changes in the aetiology and epidemiology of invasive candidiasis: from familiar Candida albicans to multiresistant Candida auris. Int Microbiol 21:107–119. doi: 10.1007/s10123-018-0014-1. [DOI] [PubMed] [Google Scholar]
- 4.Abe M, Kinjo Y, Ueno K, Takatsuka S, Nakamura S, Ogura S, Kimura M, Araoka H, Sadamoto S, Shinozaki M, Shibuya K, Yoneyama A, Kaku M, Miyazaki Y. 2018. Differences in ocular complications between Candida albicans and non-albicans Candida infection analyzed by epidemiology and a mouse ocular candidiasis model. Front Microbiol 9:2477. doi: 10.3389/fmicb.2018.02477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leeyaphan C, Bunyaratavej S, Foongladda S, Rujitharanawong C, Maneeprasopchoke P, Surawan T, Muanprasat C, Matthapan L. 2016. Epidemiology, clinical characteristics, sites of infection and treatment outcomes of mucocutaneous Candidiasis caused by non-albicans species of Candida at a dermatologic clinic. J Med Assoc Thai 99:406–411. [PubMed] [Google Scholar]
- 6.Quindos G. 2014. Epidemiology of candidaemia and invasive candidiasis. a changing face. Rev Iberoam Micol 31:42–48. doi: 10.1016/j.riam.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Patel PK, Erlandsen JE, Kirkpatrick WR, Berg DK, Westbrook SD, Louden C, Cornell JE, Thompson GR, Vallor AC, Wickes BL, Wiederhold NP, Redding SW, Patterson TF. 2012. The changing epidemiology of oropharyngeal candidiasis in patients with HIV/AIDS in the era of antiretroviral therapy. AIDS Res Treat 2012:262471. doi: 10.1155/2012/262471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hachem R, Hanna H, Kontoyiannis D, Jiang Y, Raad I. 2008. The changing epidemiology of invasive candidiasis: Candida glabrata and Candida krusei as the leading causes of candidemia in hematologic malignancy. Cancer 112:2493–2499. doi: 10.1002/cncr.23466. [DOI] [PubMed] [Google Scholar]
- 9.Redding SW, Kirkpatrick WR, Dib O, Fothergill AW, Rinaldi MG, Patterson TF. 2000. The epidemiology of non-albicans Candida in oropharyngeal candidiasis in HIV patients. Spec Care Dentist 20:178–181. doi: 10.1111/j.1754-4505.2000.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 10.Abi-Said D, Anaissie E, Uzun O, Raad I, Pinzcowski H, Vartivarian S. 1997. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin Infect Dis 24:1122–1128. doi: 10.1086/513663. [DOI] [PubMed] [Google Scholar]
- 11.Bravo Ruiz G, Lorenz A. 2021. What do we know about the biology of the emerging fungal pathogen of humans Candida auris? Microbiol Res 242:126621. doi: 10.1016/j.micres.2020.126621. [DOI] [PubMed] [Google Scholar]
- 12.Du H, Bing J, Hu T, Ennis CL, Nobile CJ, Huang G. 2020. Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog 16:e1008921. doi: 10.1371/journal.ppat.1008921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 14.Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, Jang HC. 2011. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. 2019. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis 25:1780–1781. doi: 10.3201/eid2509.190686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Proctor DM, Dangana T, Sexton DJ, Fukuda C, Yelin RD, Stanley M, Bell PB, Baskaran S, Deming C, Chen Q, Conlan S, Park M, Mullikin J, Thomas J, Young A, Bouffard G, Barnabas B, Brooks S, Han J, Ho S-l, Kim J, Legaspi R, Maduro Q, Marfani H, Montemayor C, Riebow N, Schandler K, Schmidt B, Sison C, Stantripop M, Black S, Dekhtyar M, Masiello C, McDowell J, Thomas P, Vemulapalli M, Welsh RM, Vallabhaneni S, Chiller T, Forsberg K, Black SR, Pacilli M, Kong HH, Lin MY, Schoeny ME, Litvintseva AP, Segre JA, Hayden MK, NISC Comparative Sequencing Program . 2021. Integrated genomic, epidemiologic investigation of Candida auris skin colonization in a skilled nursing facility. Nat Med 27:1401–1409. doi: 10.1038/s41591-021-01383-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton MV, Johnson CJ, Kernien JF, Patel TD, Lam BC, Cheong JZA, Meudt JJ, Shanmuganayagam D, Kalan LR, Nett JE. 2020. Candida auris forms high-burden biofilms in skin niche conditions and on porcine skin. mSphere 5. doi: 10.1128/mSphere.00910-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uppuluri P. 2020. Candida auris biofilm colonization on skin niche conditions. mSphere 5. doi: 10.1128/mSphere.00972-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osei Sekyere J. 2018. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen 7:e00578. doi: 10.1002/mbo3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taori SK, Khonyongwa K, Hayden I, Athukorala GDA, Letters A, Fife A, Desai N, Borman AM. 2019. Candida auris outbreak: mortality, interventions and cost of sustaining control. J Infect 79:601–611. doi: 10.1016/j.jinf.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Arensman K, Miller JL, Chiang A, Mai N, Levato J, LaChance E, Anderson M, Beganovic M, Dela Pena J. 2020. Clinical outcomes of patients treated for Candida auris infections in a multisite health system, Illinois, USA. Emerg Infect Dis 26:876–880. doi: 10.3201/eid2605.191588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.AlJindan R, AlEraky DM, Mahmoud N, Abdalhamid B, Almustafa M, AbdulAzeez S, Borgio JF. 2020. Drug resistance-associated mutations in ERG11 of multidrug-resistant Candida auris in a tertiary care hospital of Eastern Saudi Arabia. J Fungi (Basel) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carolus H, Pierson S, Munoz JF, Subotic A, Cruz RB, Cuomo CA, Van Dijck P. 2021. Genome-wide analysis of experimentally evolved Candida auris reveals multiple novel mechanisms of multidrug resistance. mBio 12. doi: 10.1128/mBio.03333-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chow NA, Munoz JF, Gade L, Berkow EL, Li X, Welsh RM, Forsberg K, Lockhart SR, Adam R, Alanio A, Alastruey-Izquierdo A, Althawadi S, Arauz AB, Ben-Ami R, Bharat A, Calvo B, Desnos-Ollivier M, Escandon P, Gardam D, Gunturu R, Heath CH, Kurzai O, Martin R, Litvintseva AP, Cuomo CA. 2020. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 11. doi: 10.1128/mBio.03364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, Yadav P, Singh PK, Khillan V, Sachdeva N, Perlin DS, Meis JF. 2018. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 73:891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 27.Healey KR, Kordalewska M, Jimenez Ortigosa C, Singh A, Berrio I, Chowdhary A, Perlin DS. 2018. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother 62. doi: 10.1128/AAC.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Coste AT, Liechti M, Bachmann D, Sanglard D, Lamoth F. 2021. Novel ERG11 and TAC1b mutations associated with azole resistance in Candida auris. Antimicrob Agents Chemother. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munoz JF, Gade L, Chow NA, Loparev VN, Juieng P, Berkow EL, Farrer RA, Litvintseva AP, Cuomo CA. 2018. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun 9:5346. doi: 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rybak JM, Sharma C, Doorley LA, Barker KS, Palmer GE, Rogers PD. 2021. Delineation of the direct contribution of Candida auris ERG11 mutations to clinical triazole resistance. Microbiol Spectr 9:e0158521. doi: 10.1128/Spectrum.01585-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williamson B, Wilk A, Guerrero KD, Mikulski TD, Elias TN, Sawh I, Cancino-Prado G, Gardam D, Heath CH, Govender NP, Perlin DS, Kordalewska M, Healey KR. 2022. Impact of Erg11 amino acid substitutions identified in Candida auris Clade III isolates on triazole drug susceptibility. Antimicrob Agents Chemother 66:e0162421. doi: 10.1128/AAC.01624-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rybak JM, Doorley LA, Nishimoto AT, Barker KS, Palmer GE, Rogers PD. 2019. Abrogation of triazole resistance upon deletion of CDR1 in a clinical isolate of Candida auris. Antimicrob Agents Chemother 63. doi: 10.1128/AAC.00057-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SH, Iyer KR, Pardeshi L, Munoz JF, Robbins N, Cuomo CA, Wong KH, Cowen LE. 2019. Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SH, Iyer KR, Pardeshi L, Munoz JF, Robbins N, Cuomo CA, Wong KH, Cowen LE. 2019. Erratum for Kim et al., Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rybak JM, Munoz JF, Barker KS, Parker JE, Esquivel BD, Berkow EL, Lockhart SR, Gade L, Palmer GE, White TC, Kelly SL, Cuomo CA, Rogers PD. 2020. Mutations in TAC1B: a novel genetic determinant of clinical fluconazole resistance in Candida auris. mBio 11. doi: 10.1128/mBio.00365-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wasi M, Khandelwal NK, Moorhouse AJ, Nair R, Vishwakarma P, Bravo Ruiz G, Ross ZK, Lorenz A, Rudramurthy SM, Chakrabarti A, Lynn AM, Mondal AK, Gow NAR, Prasad R. 2019. ABC transporter genes show upregulated expression in drug-resistant clinical isolates of Candida auris: a genome-wide characterization of ATP-binding cassette (ABC) transporter genes. Front Microbiol 10:1445. doi: 10.3389/fmicb.2019.01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demers EG, Stajich JE, Ashare A, Occhipinti P, Hogan DA. 2021. Balancing positive and negative selection: in vivo evolution of Candida lusitaniae MRR1. mBio 12. doi: 10.1128/mBio.03328-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, Myers LC. 2017. Candida albicans Swi/Snf and Mediator complexes differentially regulate Mrr1-induced MDR1 expression and fluconazole resistance. Antimicrob Agents Chemother 61. doi: 10.1128/AAC.01344-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kannan A, Asner SA, Trachsel E, Kelly S, Parker J, Sanglard D. 2019. Comparative genomics for the elucidation of multidrug resistance in Candida lusitaniae. mBio 10. doi: 10.1128/mBio.02512-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demers EG, Biermann AR, Masonjones S, Crocker AW, Ashare A, Stajich JE, Hogan DA. 2018. Evolution of drug resistance in an antifungal-naive chronic Candida lusitaniae infection. Proc Natl Acad Sci USA 115:12040–12045. doi: 10.1073/pnas.1807698115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schubert S, Popp C, Rogers PD, Morschhauser J. 2011. Functional dissection of a Candida albicans zinc cluster transcription factor, the multidrug resistance regulator Mrr1. Eukaryot Cell 10:1110–1121. doi: 10.1128/EC.05100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schubert S, Rogers PD, Morschhauser J. 2008. Gain-of-function mutations in the transcription factor MRR1 are responsible for overexpression of the MDR1 efflux pump in fluconazole-resistant Candida dubliniensis strains. Antimicrob Agents Chemother 52:4274–4280. doi: 10.1128/AAC.00740-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider S, Morschhauser J. 2015. Induction of Candida albicans drug resistance genes by hybrid zinc cluster transcription factors. Antimicrob Agents Chemother 59:558–569. doi: 10.1128/AAC.04448-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert S, Barker KS, Znaidi S, Schneider S, Dierolf F, Dunkel N, Aid M, Boucher G, Rogers PD, Raymond M, Morschhauser J. 2011. Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob Agents Chemother 55:2212–2223. doi: 10.1128/AAC.01343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morschhauser J, Barker KS, Liu TT, Bla BWJ, Homayouni R, Rogers PD. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog 3:e164. doi: 10.1371/journal.ppat.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayr EM, Ramirez-Zavala B, Kruger I, Morschhauser J. 2020. A zinc cluster transcription factor contributes to the intrinsic fluconazole resistance of Candida auris. mSphere 5. doi: 10.1128/mSphere.00279-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spivak ES, Hanson KE. 2018. Candida auris: an emerging fungal pathogen. J Clin Microbiol 56. doi: 10.1128/JCM.01588-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin L, Cao Z, Wang Q, Wang Y, Wang X, Chen H, Wang H. 2018. MDR1 overexpression combined with ERG11 mutations induce high-level fluconazole resistance in Candida tropicalis clinical isolates. BMC Infect Dis 18:162. doi: 10.1186/s12879-018-3082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liston SD, Whitesell L, Kapoor M, Shaw KJ, Cowen LE. 2020. Enhanced efflux pump expression in Candida mutants results in decreased manogepix susceptibility. Antimicrob Agents Chemother 64. doi: 10.1128/AAC.00261-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wirsching S, Moran GP, Sullivan DJ, Coleman DC, Morschhauser J. 2001. MDR1-mediated drug resistance in Candida dubliniensis. Antimicrob Agents Chemother 45:3416–3421. doi: 10.1128/AAC.45.12.3416-3421.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.You L, Qian W, Yang Q, Mao L, Zhu L, Huang X, Jin J, Meng H. 2017. ERG11 gene mutations and MDR1 upregulation confer pan-azole resistance in Candida tropicalis causing disseminated candidiasis in an acute lymphoblastic leukemia patient on posaconazole prophylaxis. Antimicrob Agents Chemother 61. doi: 10.1128/AAC.02496-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wirsching S, Michel S, Morschhauser J. 2000. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol Microbiol 36:856–865. doi: 10.1046/j.1365-2958.2000.01899.x. [DOI] [PubMed] [Google Scholar]
- 53.Berkow EL, Manigaba K, Parker JE, Barker KS, Kelly SL, Rogers PD. 2015. Multidrug transporters and alterations in sterol biosynthesis contribute to azole antifungal resistance in Candida parapsilosis. Antimicrob Agents Chemother 59:5942–5950. doi: 10.1128/AAC.01358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Souza AC, Fuchs BB, Pinhati HM, Siqueira RA, Hagen F, Meis JF, Mylonakis E, Colombo AL. 2015. Candida parapsilosis resistance to fluconazole: molecular mechanisms and in vivo impact in infected Galleria mellonella Larvae. Antimicrob Agents Chemother 59:6581–6587. doi: 10.1128/AAC.01177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grossman NT, Pham CD, Cleveland AA, Lockhart SR. 2015. Molecular mechanisms of fluconazole resistance in Candida parapsilosis isolates from a U.S. surveillance system. Antimicrob Agents Chemother 59:1030–1037. doi: 10.1128/AAC.04613-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hampe IAI, Friedman J, Edgerton M, Morschhauser J. 2017. An acquired mechanism of antifungal drug resistance simultaneously enables Candida albicans to escape from intrinsic host defenses. PLoS Pathog 13:e1006655. doi: 10.1371/journal.ppat.1006655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hiller D, Sanglard D, Morschhauser J. 2006. Overexpression of the MDR1 gene is sufficient to confer increased resistance to toxic compounds in Candida albicans. Antimicrob Agents Chemother 50:1365–2591. doi: 10.1128/AAC.50.4.1365-1371.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biermann AR, Demers EG, Hogan DA. 2021. Mrr1 regulation of methylglyoxal catabolism and methylglyoxal-induced fluconazole resistance in Candida lusitaniae. Mol Microbiol 115:116–130. doi: 10.1111/mmi.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chakraborty S, Karmakar K, Chakravortty D. 2014. Cells producing their own nemesis: understanding methylglyoxal metabolism. Iubmb Life 66:667–678. doi: 10.1002/iub.1324. [DOI] [PubMed] [Google Scholar]
- 60.Tian S, Rong C, Nian H, Li F, Chu Y, Cheng S, Shang H. 2018. First cases and risk factors of super yeast Candida auris infection or colonization from Shenyang, China. Emerg Microbes Infect 7:128. doi: 10.1038/s41426-018-0131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sayeed MA, Farooqi J, Jabeen K, Mahmood SF. 2020. Comparison of risk factors and outcomes of Candida auris candidemia with non-Candida auris candidemia: A retrospective study from Pakistan. Med Mycol 58:721–729. doi: 10.1093/mmy/myz112. [DOI] [PubMed] [Google Scholar]
- 62.Shastri PS, Shankarnarayan SA, Oberoi J, Rudramurthy SM, Wattal C, Chakrabarti A. 2020. Candida auris candidaemia in an intensive care unit - prospective observational study to evaluate epidemiology, risk factors, and outcome. J Crit Care 57:42–48. doi: 10.1016/j.jcrc.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 63.Rudramurthy SM, Chakrabarti A, Paul RA, Sood P, Kaur H, Capoor MR, Kindo AJ, Marak RSK, Arora A, Sardana R, Das S, China D, Patel A, Xess I, Tarai B, Singh P, Ghosh A. 2017. Candida auris candidaemia in Indian ICUs: analysis of risk factors. J Antimicrob Chemother 72:1794–1801. doi: 10.1093/jac/dkx034. [DOI] [PubMed] [Google Scholar]
- 64.Ruiz-Gaitan A, Martinez H, Moret AM, Calabuig E, Tasias M, Alastruey-Izquierdo A, Zaragoza O, Mollar J, Frasquet J, Salavert-Lleti M, Ramirez P, Lopez-Hontangas JL, Peman J. 2019. Detection and treatment of Candida auris in an outbreak situation: risk factors for developing colonization and candidemia by this new species in critically ill patients. Expert Rev Anti Infect Ther 17:295–305. doi: 10.1080/14787210.2019.1592675. [DOI] [PubMed] [Google Scholar]
- 65.Al-Rashdi A, Al-Maani A, Al-Wahaibi A, Alqayoudhi A, Al-Jardani A, Al-Abri S. 2021. Characteristics, risk factors, and survival analysis of Candida auris cases: results of one-year national surveillance data from Oman. J Fungi (Basel) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caceres DH, Rivera SM, Armstrong PA, Escandon P, Chow NA, Ovalle MV, Diaz J, Derado G, Salcedo S, Berrio I, Espinosa-Bode A, Varon C, Stuckey MJ, Marino A, Villalobos N, Lockhart SR, Chiller TM, Prieto FE, Jackson BR. 2020. Case-case comparison of Candida auris versus other Candida species bloodstream infections: results of an outbreak investigation in Colombia. Mycopathologia 185:917–923. doi: 10.1007/s11046-020-00478-1. [DOI] [PubMed] [Google Scholar]
- 67.Khan Z, Ahmad S, Benwan K, Purohit P, Al-Obaid I, Bafna R, Emara M, Mokaddas E, Abdullah AA, Al-Obaid K, Joseph L. 2018. Invasive Candida auris infections in Kuwait hospitals: epidemiology, antifungal treatment and outcome. Infection 46:641–650. doi: 10.1007/s15010-018-1164-y. [DOI] [PubMed] [Google Scholar]
- 68.Pandya N, Cag Y, Pandak N, Pekok AU, Poojary A, Ayoade F, Fasciana T, Giammanco A, Caskurlu H, Rajani DP, Gupta YK, Balkan II, Khan EA, Erdem H. 2021. International Multicentre Study of Candida auris Infections. J Fungi (Basel) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rossow J, Ostrowsky B, Adams E, Greenko J, McDonald R, Vallabhaneni S, Forsberg K, Perez S, Lucas T, Alroy KA, Jacobs Slifka K, Walters M, Jackson BR, Quinn M, Chaturvedi S, Blog D, New York Candida auris Investigation Workgroup . 2021. Factors associated with Candida auris colonization and transmission in skilled nursing facilities with ventilator Units, New York, 2016–2018. Clin Infect Dis 72:e753–e760. doi: 10.1093/cid/ciaa1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang XJ, Ma SB, Liu ZF, Li H, Gao WY. 2019. Elevated levels of alpha-dicarbonyl compounds in the plasma of type II diabetics and their relevance with diabetic nephropathy. J Chromatography B-Analytical Technologies in the Biomedical and Life Sciences 1106:19–25. [DOI] [PubMed] [Google Scholar]
- 71.McLellan AC, Thornalley PJ, Benn J, Sonksen PH. 1994. Glyoxalase system in clinical diabetes mellitus and correlation with diabetic complications. Clin Sci (Lond) 87:21–29. doi: 10.1042/cs0870021. [DOI] [PubMed] [Google Scholar]
- 72.Lu J, Randell E, Han Y, Adeli K, Krahn J, Meng QH. 2011. Increased plasma methylglyoxal level, inflammation, and vascular endothelial dysfunction in diabetic nephropathy. Clin Biochem 44:307–311. doi: 10.1016/j.clinbiochem.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 73.Odani H, Shinzato T, Usami J, Matsumoto Y, Brinkmann Frye E, Baynes JW, Maeda K. 1998. Imidazolium crosslinks derived from reaction of lysine with glyoxal and methylglyoxal are increased in serum proteins of uremic patients: evidence for increased oxidative stress in uremia. FEBS Lett 427:381–385. doi: 10.1016/S0014-5793(98)00416-5. [DOI] [PubMed] [Google Scholar]
- 74.Karg E, Papp F, Tassi N, Janaky T, Wittmann G, Turi S. 2009. Enhanced methylglyoxal formation in the erythrocytes of hemodialyzed patients. Metabolism 58:976–982. doi: 10.1016/j.metabol.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 75.Lapolla A, Flamini R, Lupo A, Arico NC, Rugiu C, Reitano R, Tubaro M, Ragazzi E, Seraglia R, Traldi P. 2005. Evaluation of glyoxal and methylglyoxal levels in uremic patients under peritoneal dialysis. Ann N Y Acad Sci 1043:217–224. doi: 10.1196/annals.1333.027. [DOI] [PubMed] [Google Scholar]
- 76.Mukhopadhyay S, Ghosh A, Kar M. 2008. Methylglyoxal increase in uremia with special reference to snakebite-mediated acute renal failure. Clin Chim Acta 391:13–17. doi: 10.1016/j.cca.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 77.Brenner T, Fleming T, Uhle F, Silaff S, Schmitt F, Salgado E, Ulrich A, Zimmermann S, Bruckner T, Martin E, Bierhaus A, Nawroth PP, Weigand MA, Hofer S. 2014. Methylglyoxal as a new biomarker in patients with septic shock: an observational clinical study. Crit Care 18. doi: 10.1186/s13054-014-0683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Juarez P, Jeannot K, Plesiat P, Llanes C. 2017. Toxic electrophiles induce expression of the multidrug efflux pump MexEF-OprN in Pseudomonas aeruginosa through a novel transcriptional regulator, CmrA. Antimicrob Agents Chemother 61. doi: 10.1128/AAC.00585-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ozyamak E, de Almeida C, de Moura AP, Miller S, Booth IR. 2013. Integrated stress response of Escherichia coli to methylglyoxal: transcriptional readthrough from the nemRA operon enhances protection through increased expression of glyoxalase I. Mol Microbiol 88:936–950. doi: 10.1111/mmi.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee C, Shin J, Park C. 2013. Novel regulatory system nemRA-gloA for electrophile reduction in Escherichia coli K-12. Mol Microbiol 88:395–412. doi: 10.1111/mmi.12192. [DOI] [PubMed] [Google Scholar]
- 81.Mostofa MG, Ghosh A, Li ZG, Siddiqui MN, Fujita M, Tran LP. 2018. Methylglyoxal - a signaling molecule in plant abiotic stress responses. Free Radic Biol Med 122:96–109. doi: 10.1016/j.freeradbiomed.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 82.Kosmachevskaya OV, Shumaev KB, Topunov AF. 2017. Signal and regulatory effects of methylglyoxal in eukaryotic cells (review). Appl Biochem Microbiol 53:273–289. doi: 10.1134/S0003683817030103. [DOI] [Google Scholar]
- 83.Maeta K, Izawa S, Okazaki S, Kuge S, Inoue Y. 2004. Activity of the Yap1 transcription factor in Saccharomyces cerevisiae is modulated by methylglyoxal, a metabolite derived from glycolysis. Mol Cell Biol 24:8753–8764. doi: 10.1128/MCB.24.19.8753-8764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aguilera J, Prieto JA. 2004. Yeast cells display a regulatory mechanism in response to methylglyoxal. FEMS Yeast Res 4:633–641. doi: 10.1016/j.femsyr.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 85.Roy A, Hashmi S, Li Z, Dement AD, Cho KH, Kim JH. 2016. The glucose metabolite methylglyoxal inhibits expression of the glucose transporter genes by inactivating the cell surface glucose sensors Rgt2 and Snf3 in yeast. Mol Biol Cell 27:862–871. doi: 10.1091/mbc.E15-11-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Anonymous. 2016. Correction for The glucose metabolite methylglyoxal inhibits expression of the glucose transporter genes by inactivating the cell surface glucose sensors Rgt2 and Snf3 in yeast. Mol Biol Cell 27:3178–3179. doi: 10.1091/mboc.27.20.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maeta K, Izawa S, Inoue Y. 2005. Methylglyoxal, a metabolite derived from glycolysis, functions as a signal initiator of the high osmolarity glycerol-mitogen-activated protein kinase cascade and Calcineurin/Crz1-mediated pathway in Saccharomyces cerevisiae. J Biological Chemistry 280:253–260. doi: 10.1074/jbc.M408061200. [DOI] [PubMed] [Google Scholar]
- 88.Takatsume Y, Izawa S, Inoue Y. 2007. Modulation of Spc1 stress-activated protein kinase activity by methylglyoxal through inhibition of protein phosphatase in the fission yeast Schizosaccharomyces pombe. Biochem Biophys Res Commun 363:942–947. doi: 10.1016/j.bbrc.2007.09.071. [DOI] [PubMed] [Google Scholar]
- 89.Takatsume Y, Izawa S, Inoue Y. 2006. Methylglyoxal as a signal initiator for activation of the stress-activated protein kinase cascade in the fission yeast Schizosaccharomyces pombe. J Biol Chem 281:9086–9092. doi: 10.1074/jbc.M511037200. [DOI] [PubMed] [Google Scholar]
- 90.Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJ, Litvintseva AP. 2017. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol 55:2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rutala WA, Kanamori H, Gergen MF, Sickbert-Bennett EE, Weber DJ. 2019. Susceptibility of Candida auris and Candida albicans to 21 germicides used in healthcare facilities. Infect Control Hosp Epidemiol 40:380–382. doi: 10.1017/ice.2019.1. [DOI] [PubMed] [Google Scholar]
- 92.Jabeen K, Mal PB, Tharwani A, Hashmi M, Farooqi J. 2020. Persistence of Candida auris on latex and nitrile gloves with transmission to sterile urinary catheters double dagger. Med Mycol 58:128–132. doi: 10.1093/mmy/myz033. [DOI] [PubMed] [Google Scholar]
- 93.Eyre DW, Sheppard AE, Madder H, Moir I, Moroney R, Quan TP, Griffiths D, George S, Butcher L, Morgan M, Newnham R, Sunderland M, Clarke T, Foster D, Hoffman P, Borman AM, Johnson EM, Moore G, Brown CS, Walker AS, Peto TEA, Crook DW, Jeffery KJM. 2018. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med 379:1322–1331. doi: 10.1056/NEJMoa1714373. [DOI] [PubMed] [Google Scholar]
- 94.Hoehamer CF, Cummings ED, Hilliard GM, Morschhauser J, Rogers PD. 2009. Proteomic analysis of Mrr1p- and Tac1p-associated differential protein expression in azole-resistant clinical isolates of Candida albicans. Proteomics Clin Appl 3:968–978. doi: 10.1002/prca.200800252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Karababa M, Coste AT, Rognon B, Bille J, Sanglard D. 2004. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob Agents Chemother 48:3064–3079. doi: 10.1128/AAC.48.8.3064-3079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rogers PD, Barker KS. 2003. Genome-wide expression profile analysis reveals coordinately regulated genes associated with stepwise acquisition of azole resistance in Candida albicans clinical isolates. Antimicrob Agents Chemother 47:1220–1227. doi: 10.1128/AAC.47.4.1220-1227.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Silva AP, Miranda IM, Guida A, Synnott J, Rocha R, Silva R, Amorim A, Pina-Vaz C, Butler G, Rodrigues AG. 2011. Transcriptional profiling of azole-resistant Candida parapsilosis strains. Antimicrob Agents Chemother 55:3546–3556. doi: 10.1128/AAC.01127-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kannan A, Asner SA, Trachsel E, Kelly S, Parker J, Sanglard D. 2020. Erratum for Kannan et al., Comparative genomics for the elucidation of multidrug resistance in Candida lusitaniae. mBio 11. doi: 10.1128/mBio.03403-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rognon B, Kozovska Z, Coste AT, Pardini G, Sanglard D. 2006. Identification of promoter elements responsible for the regulation of MDR1 from Candida albicans, a major facilitator transporter involved in azole resistance. Microbiology (Reading) 152:3701–3722. doi: 10.1099/mic.0.29277-0. [DOI] [PubMed] [Google Scholar]
- 100.Ramirez-Zavala B, Mogavero S, Scholler E, Sasse C, Rogers PD, Morschhauser J. 2014. SAGA/ADA complex subunit Ada2 is required for Cap1- but not Mrr1-mediated upregulation of the Candida albicans multidrug efflux pump MDR1. Antimicrob Agents Chemother 58:5102–5110. doi: 10.1128/AAC.03065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mogavero S, Tavanti A, Senesi S, Rogers PD, Morschhauser J. 2011. Differential requirement of the transcription factor Mcm1 for activation of the Candida albicans multidrug efflux pump MDR1 by its regulators Mrr1 and Cap1. Antimicrob Agents Chemother 55:2061–2066. doi: 10.1128/AAC.01467-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hiller D, Stahl S, MorschhäUser J. 2006. Multiple cis-acting sequences mediate upregulation of the MDR1 efflux pump in a fluconazole-resistant clinical Candida albicans isolate. Antimicrob Agents Chemother 50:2300–2308. doi: 10.1128/AAC.00196-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Harry JB, Oliver BG, Song JL, Silver PM, Little JT, Choiniere J, White TC. 2005. Drug-induced regulation of the MDR1 promoter in Candida albicans. Antimicrob Agents Chemother 49:2785–2792. doi: 10.1128/AAC.49.7.2785-2792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gupta V, Kohli A, Krishnamurthy S, Puri N, Aalamgeer SA, Panwar S, Prasad R. 1998. Identification of polymorphic mutant alleles of CaMDR1, a major facilitator of Candida albicans which confers multidrug resistance, and its in vitro transcriptional activation. Curr Genet 34:192–199. doi: 10.1007/s002940050385. [DOI] [PubMed] [Google Scholar]
- 105.Iyer KR, Camara K, Daniel-Ivad M, Trilles R, Pimentel-Elardo SM, Fossen JL, Marchillo K, Liu Z, Singh S, Munoz JF, Kim SH, Porco JA, Jr, Cuomo CA, Williams NS, Ibrahim AS, Edwards JE, Jr, Andes DR, Nodwell JR, Brown LE, Whitesell L, Robbins N, Cowen LE. 2020. An oxindole efflux inhibitor potentiates azoles and impairs virulence in the fungal pathogen Candida auris. Nat Commun 11:6429. doi: 10.1038/s41467-020-20183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dunkel N, Blass J, Rogers PD, Morschhauser J. 2008. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol Microbiol 69:827–840. doi: 10.1111/j.1365-2958.2008.06309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rodrigues CF, Rodrigues ME, Henriques M. 2019. Candida sp. infections in patients with diabetes mellitus. J Clin Med 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pyrgos V, Ratanavanich K, Donegan N, Veis J, Walsh TJ, Shoham S. 2009. Candida bloodstream infections in hemodialysis recipients. Med Mycol 47:463–467. doi: 10.1080/13693780802369332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang MM, Ong CL, Walker MJ, McEwan AG. 2016. Defence against methylglyoxal in group A Streptococcus: a role for Glyoxylase I in bacterial virulence and survival in neutrophils? Pathog Dis 74. [DOI] [PubMed] [Google Scholar]
- 110.Rachman H, Kim N, Ulrichs T, Baumann S, Pradl L, Nasser Eddine A, Bild M, Rother M, Kuban RJ, Lee JS, Hurwitz R, Brinkmann V, Kosmiadi GA, Kaufmann SH. 2006. Critical role of methylglyoxal and AGE in mycobacteria-induced macrophage apoptosis and activation. PLoS One 1:e29. doi: 10.1371/journal.pone.0000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Prantner D, Nallar S, Richard K, Spiegel D, Collins KD, Vogel SN. 2021. Classically activated mouse macrophages produce methylglyoxal that induces a TLR4- and RAGE-independent proinflammatory response. J Leukoc Biol 109:605–619. doi: 10.1002/JLB.3A0520-745RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jimenez-Lopez C, Collette JR, Brothers KM, Shepardson KM, Cramer RA, Wheeler RT, Lorenz MC. 2013. Candida albicans induces arginine biosynthetic genes in response to host-derived reactive oxygen species. Eukaryot Cell 12:91–100. doi: 10.1128/EC.00290-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aranda A, del Olmo ML. 2004. Exposure of Saccharomyces cerevisiae to acetaldehyde induces sulfur amino acid metabolism and polyamine transporter genes, which depend on Met4p and Haa1p transcription factors, respectively. Appl Environ Microbiol 70:1913–1922. doi: 10.1128/AEM.70.4.1913-1922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lucau-Danila A, Lelandais G, Kozovska Z, Tanty V, Delaveau T, Devaux F, Jacq C. 2005. Early expression of yeast genes affected by chemical stress. Mol Cell Biol 25:1860–1868. doi: 10.1128/MCB.25.5.1860-1868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lelandais G, Tanty V, Geneix C, Etchebest C, Jacq C, Devaux F. 2008. Genome adaptation to chemical stress: clues from comparative transcriptomics in Saccharomyces cerevisiae and Candida glabrata. Genome Biol 9:R164. doi: 10.1186/gb-2008-9-11-r164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Staub RE, Quistad GB, Casida JE. 1998. Mechanism for benomyl action as a mitochondrial aldehyde dehydrogenase inhibitor in mice. Chem Res Toxicol 11:535–543. doi: 10.1021/tx980002l. [DOI] [PubMed] [Google Scholar]
- 117.Fitzmaurice AG, Rhodes SL, Lulla A, Murphy NP, Lam HA, O'Donnell KC, Barnhill L, Casida JE, Cockburn M, Sagasti A, Stahl MC, Maidment NT, Ritz B, Bronstein JM. 2013. Aldehyde dehydrogenase inhibition as a pathogenic mechanism in Parkinson disease. Proc Natl Acad Sci USA 110:636–641. doi: 10.1073/pnas.1220399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Casida JE, Ford B, Jinsmaa Y, Sullivan P, Cooney A, Goldstein DS. 2014. Benomyl, aldehyde dehydrogenase, DOPAL, and the catecholaldehyde hypothesis for the pathogenesis of Parkinson's disease. Chem Res Toxicol 27:1359–1361. doi: 10.1021/tx5002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Leiphon LJ, Picklo MJ. Sr, 2007. Inhibition of aldehyde detoxification in CNS mitochondria by fungicides. Neurotoxicology 28:143–149. doi: 10.1016/j.neuro.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 120.Jakab A, Balla N, Ragyak A, Nagy F, Kovacs F, Sajtos Z, Toth Z, Borman AM, Pocsi I, Baranyai E, Majoros L, Kovacs R. 2021. Transcriptional profiling of the Candida auris response to exogenous farnesol exposure. mSphere 6:e0071021. doi: 10.1128/mSphere.00710-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nagy F, Vitalis E, Jakab A, Borman AM, Forgacs L, Toth Z, Majoros L, Kovacs R. 2020. In vitro and in vivo effect of exogenous farnesol exposure against Candida auris. Front Microbiol 11:957. doi: 10.3389/fmicb.2020.00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shirtliff ME, Krom BP, Meijering RA, Peters BM, Zhu J, Scheper MA, Harris ML, Jabra-Rizk MA. 2009. Farnesol-induced apoptosis in Candida albicans. Antimicrob Agents Chemother 53:2392–2401. doi: 10.1128/AAC.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hasim S, Vaughn EN, Donohoe D, Gordon DM, Pfiffner S, Reynolds TB. 2018. Influence of phosphatidylserine and phosphatidylethanolamine on farnesol tolerance in Candida albicans. Yeast 35:343–351. doi: 10.1002/yea.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Machida K, Tanaka T, Fujita K, Taniguchi M. 1998. Farnesol-induced generation of reactive oxygen species via indirect inhibition of the mitochondrial electron transport chain in the yeast Saccharomyces cerevisiae. J Bacteriol 180:4460–4465. doi: 10.1128/JB.180.17.4460-4465.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fairn GD, MacDonald K, McMaster CR. 2007. A chemogenomic screen in Saccharomyces cerevisiae uncovers a primary role for the mitochondria in farnesol toxicity and its regulation by the Pkc1 pathway. J Biol Chem 282:4868–4874. doi: 10.1074/jbc.M610575200. [DOI] [PubMed] [Google Scholar]
- 126.Yadav SK, Singla-Pareek SL, Ray M, Reddy MK, Sopory SK. 2005. Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem Biophys Res Commun 337:61–67. doi: 10.1016/j.bbrc.2005.08.263. [DOI] [PubMed] [Google Scholar]
- 127.Nahar K, Hasanuzzaman M, Alam MM, Fujita M. 2015. Glutathione-induced drought stress tolerance in mung bean: coordinated roles of the antioxidant defence and methylglyoxal detoxification systems. AoB Plants 7:plv069. doi: 10.1093/aobpla/plv069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Melvin P, Bankapalli K, D'Silva P, Shivaprasad PV. 2017. Methylglyoxal detoxification by a DJ-1 family protein provides dual abiotic and biotic stress tolerance in transgenic plants. Plant Mol Biol 94:381–397. doi: 10.1007/s11103-017-0613-9. [DOI] [PubMed] [Google Scholar]
- 129.Vemanna RS, Babitha KC, Solanki JK, Amarnatha Reddy V, Sarangi SK, Udayakumar M. 2017. Aldo-keto reductase-1 (AKR1) protect cellular enzymes from salt stress by detoxifying reactive cytotoxic compounds. Plant Physiol Biochem 113:177–186. doi: 10.1016/j.plaphy.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 130.Mahmud JA, Hasanuzzaman M, Khan MIR, Nahar K, Fujita M. 2020. Beta-aminobutyric acid pretreatment confers salt stress tolerance in Brassica napus L. by modulating reactive oxygen species metabolism and methylglyoxal detoxification. Plants (Basel) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rohman MM, Islam MR, Monsur MB, Amiruzzaman M, Fujita M, Hasanuzzaman M. 2019. Trehalose protects maize plants from salt stress and phosphorus deficiency. Plants (Basel) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Veena Reddy VS, Sopory SK. 1999. Glyoxalase I from Brassica juncea: molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress. Plant J 17:385–395. doi: 10.1046/j.1365-313x.1999.00390.x. [DOI] [PubMed] [Google Scholar]
- 133.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Grahl N, Demers EG, Crocker AW, Hogan DA. 2017. Use of RNA-Protein complexes for genome editing in non-albicans Candida Species. mSphere 2:e00218-17. doi: 10.1128/mSphere.00218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 137.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Basenko EY, Pulman JA, Shanmugasundram A, Harb OS, Crouch K, Starns D, Warrenfeltz S, Aurrecoechea C, Stoeckert CJ, Jr, Kissinger JC, Roos DS, Hertz-Fowler C. 2018. FungiDB: an integrated bioinformatic resource for fungi and oomycetes. J Fungi (Basel) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF, Stoeckert CJ, Jr, Roos DS. 2012. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res 40:D675–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The mrr1aΔ mutant has a growth defect in high concentrations of MG, but not at 5 mM MG or in the YPD control. Growth curves of B11221 WT (blue) and its mrr1aΔ (red), mrr1bΔ (green), and mrr1cΔ (purple) derivatives in YPD (left), or YPD supplemented with 5 mM (middle), or 15 mM (right) MG. Data shown represent the mean ± SD for three independent experiments. Download FIG S1, TIF file, 0.3 MB (269.2KB, tif) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA-seq data, in the form of normalized counts per million (CPM), for all tested strains and conditions. Download Data Set S1, XLSX file, 5.7 MB (5.7MB, xlsx) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Select genes differentially expressed in response to MG and/or BEN in the C. auris B11221 background. Differentially expressed genes were determined using a cutoff of |Log2FC| ≥ 1.00 and P-value < 0.05. Download Table S1, DOCX file, 0.06 MB (60.7KB, docx) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The transcriptional response of mrr1aΔ to either MG or BEN is overall similar to that of the B11221 WT parent strain. Volcano plots of all quantified genes in the mrr1aΔ mutant treated with either MG (A) or BEN (B). Each point represents a single gene; magenta points indicate genes that were significantly upregulated compared to the control condition, teal points indicate genes that were significantly downregulated compared to the control condition. MDR1 is shown along with the two most up- and downregulated genes in each condition. Download FIG S2, TIF file, 0.3 MB (276.8KB, tif) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
C. auris strain AR0390 has a growth advantage over B11221 in YPD but loses that advantage in the presence of increasing concentrations of MG. Growth curves of B11221 (blue) and AR0390 (orange) in YPD (left), or YPD supplemented with 5 mM (middle), or 15 mM (right) MG. Data shown represent the mean ± SD for three independent experiments. Download FIG S3, TIF file, 0.2 MB (192.7KB, tif) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Top 20 genes with predicted functions differentially expressed between C. auris isolates B11221 and AR0390 in the control condition. Differentially expressed genes were determined using a cutoff of |Log2FC| ≥ 1.00 and P-value < 0.05. Download Table S2, DOCX file, 0.02 MB (25.3KB, docx) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
C. lusitaniae strains complemented with CauMRR1aN647T or CauMRR1a do not differ in growth from the mrr1Δ parent at MG concentrations below 15 mM. Growth curves of C. lusitaniae U04 mrr1Δ (grey) and its derivatives expressing CauMRR1aN647T (dark blue) or CauMRR1a (brown) in YPD (left) or YPD supplemented with 5 mM (middle), or 10 mM (right) MG. Data shown represent the mean ± SD for three independent experiments. Download FIG S4, TIF file, 0.2 MB (241.8KB, tif) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of select genes differentially expressed in response to MG in C. auris isolates B11221 and/or AR0390. Differentially expressed genes were determined using a cutoff of |Log2FC| ≥ 1.00 and P-value < 0.05. Download Table S3, DOCX file, 0.04 MB (43.7KB, docx) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Treatment with 5 mM MG leads to the differential expression of more genes in AR0390 than in B11221. Volcano plot of all quantified genes in AR0390 treated with MG. Each point represents a single gene; magenta points indicate genes that were significantly upregulated compared with the control condition, teal points indicate genes that were significantly downregulated compared with the control condition. MDR1 and MGD1 are shown for reference. Download FIG S5, TIF file, 0.2 MB (255KB, tif) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and oligonucleotides used in this study. Download Table S4, DOCX file, 0.04 MB (43.7KB, docx) .
Copyright © 2022 Biermann and Hogan.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The data supporting the findings in this study are available within the paper and its supplemental material and are also available from the corresponding author upon request. The raw sequence reads from the RNA-seq analysis have been deposited into NCBI sequence read archive under BioProject PRJNA801628 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA801628).