ABSTRACT
In mammalian cells, DNA double-strand breaks (DSBs) are mainly repaired by nonhomologous end joining (NHEJ) pathway. Ku (a heterodimer formed by Ku70 and Ku80 proteins) and DNA ligase IV are the core NHEJ factors. Ku could also be involved in other cellular processes, including telomere length regulation, DNA replication, transcription, and translation control. Leishmania, an early branching eukaryote and the causative agent of leishmaniasis, has no functional NHEJ pathway due to its lack of DNA ligase IV and other NHEJ factors but retains Ku70 and Ku80 proteins. In this study, we generated Leishmania donovani Ku70 disruption mutants and Ku70 and Ku80 double gene (Ku70/80) disruption mutants. We found that Leishmania Ku is still involved in DSB repair, possibly through its binding to DNA ends to block and slowdown 5′ end resections and Ku-Ku or other protein interactions. Depending on location of a DSB between the direct repeat genomic sequences, Leishmania Ku could have an inhibiting effect, no effect or a promoting effect on the DSB repair mediated by single strand annealing (SSA), the most frequently used DSB repair pathway in Leishmania. Ku70/80 proteins are also required for the healthy proliferation of Leishmania cells. Interestingly, unlike in Trypanosoma brucei and L. mexicana, Ku70/80 proteins are dispensable for maintaining the normal lengths of telomeres in L. donovani. We also show it is possible to reconstitute the two components (Ku and Ligase D) NHEJ pathway derived from Mycobacterium marinum in Leishmania. This improved DSB repair fidelity and efficiency in Leishmania and sets up an example that the bacterial NHEJ pathway can be successfully reconstructed in an NHEJ-deficient eukaryotic parasite.
IMPORTANCE Nonhomologous end joining (NHEJ) is the most efficient double-stranded DNA break (DSB) repair pathway in mammalian cells. In contrast, the protozoan parasite Leishmania has no functional NHEJ pathway but retains the core NHEJ factors of Ku70 and Ku80 proteins. In this study, we found that Leishmania Ku heterodimers are still participating in DSB repair possibly through blocking 5′ end resections and Ku-Ku protein interactions. Depending on the DSB location, Ku could have an inhibiting or promoting effect on DSB repair mediated by the single-strand annealing repair pathway. Ku is also required for the normal growth of the parasite but surprisingly dispensable for maintaining the telomere lengths. Further, we show it is possible to introduce Mycobacterium marinum NHEJ pathway into Leishmania. Understanding DSB repair mechanisms of Leishmania may improve the CRISPR gene targeting specificity and efficiency and help identify new drug targets for this important human parasite.
KEYWORDS: Leishmania donovani, CRISPR-Cas9, gene targeting, gene deletion, double-strand DNA break, DSB repair, single-strand annealing, SSA, nonhomologous end joining, NHEJ, Ku, Ku70, Ku80, telomere maintenance, Mycobacterium marinum, Ligase D
INTRODUCTION
In order to maintain genome integrity and survive, living cells must repair DNA double-strand breaks (DSBs) generated by various endogenous or exogenous stresses, including CRISPR gene targeting. Depending on the organism, cell type, and cell cycle phase, DSBs can be repaired through one of the following ends joining pathways, namely, nonhomologous end joining (NHEJ), microhomology-mediated end joining (MMEJ), single-strand annealing (SSA), and homologous recombination (HR) (1–3). Although those pathways may share some repair proteins and steps, each of those pathways requires specific repair factors. The presence of those specific factors, their relative activity, and availability of homologous repair sequences determine the repair pathway choice for a DSB. Due to differences of repair mechanisms, those pathways can produce very different repair outcomes from faithful repair to extensive deletions (1–3).
In human cells, DSBs are mainly repaired by NHEJ (>70% of repair events) and HR (<30% of repair events, mainly during S and G2 cell cycle phases when the sister chromatid is available for recombination) (4–6). The Ku70/80 heterodimer (Ku) formed by the abundant Ku70 and Ku80 nuclear proteins plays a central role in NHEJ. Ku, a ring-like structure, has very high affinity for DSB ends. Ku rapidly recognizes and threads onto broken DNA ends (within seconds) in the cell nucleus. Ku subsequently recruits other NHEJ effectors, including DNA-PKcs (DNA-dependent protein kinase catalytic subunit), nucleases, polymerases, and the DNA Ligase IV complex which form a DNA synapse to bridge the ends. After minor processing of the ends by nucleases or polymerases to expose short complementary sequences (1 to 4 nucleotides [nt]), the ligatable termini are then rejoined by Ligase IV, usually within 30 min after DSB. NHEJ typically attempts to repair DSBs first, often resulting in small deletions or insertions and is much faster than HR (7 h or longer is required for HR) (4–8).
If NHEJ fails to repair the DSB, the Ku heterodimer rings will be displaced from the DNA ends by the Mre11, Rad50, and Nbs1 (MRN) complex which initiates 5′-end resections to generate 3′-single-stranded DNA (ssDNA) overhangs suitable for repair with MMEJ, SSA, or HR pathways (9–13). If the DSB occurs in cell cycle S and G2 phases, HR is active as sister chromatids are present in the cell. The nucleoprotein filaments formed from the 3′-ssDNA overhangs, replication protein A (RPA), and Rad51 recombinase protein will search for and invade the adjacent homology sequence in the sister chromatid as the HR repair template. As a result, DNA is often repaired faithfully by HR with no insertion or deletions (5). Because Ku blocks 5′-end resections, MMEJ is per se inefficient and SSA requires the existence of long repeat sequences flanking the DSB and extensive resections, MMEJ and SSA are rarely used for DSB repair in mammalian cells. They are the backup pathways when the NHEJ and HR pathways are absent or interrupted, such as in some cancer cells (10–15). MMEJ uses the microhomology sequences (5 to 25 nt) in the 3′-overhangs to anneal the two single-stranded DNA and causes deletion between the microhomology sequences. DNA polymerase θ helps anneal the microhomology sequences and plays a central role in MMEJ (12, 13). SSA mediated repair can take place if the DSBs occur between two direct repeat sequences (>26 bp). SSA usually requires extensive resections to reach the homologous repeat sequences flanking the DSB. With the assistance of Rad52 and other repair proteins, the single-strand 3′-overhangs are annealed between the complementary sequences of direct repeats located on opposite sides of the DSB, followed by the nucleolytic removal of remaining tails, DNA synthesis to fill in gaps, and ligation. Depending on the distance between the repeats, SSA often results in large deletion mutations and causes the most severe damage to the genome (15).
Unlike prokaryotic Escherichia coli which has only the HR DSB repair pathway, mycobacteria possess HR as well as NHEJ and SSA pathways (16). However, NHEJ in mycobacteria such as Mycobacterium tuberculosis and Mycobacterium marinum is mainly facilitated by two proteins, Ku and Ligase D (17–19). In vitro and in vivo studies have shown that mycobacterial Ku forms a homodimer which has high affinity for DNA ends and interacts physically with Ligase D and promotes its ligation activity (17, 20). Unlike Ligase IV in mammalian cells, Ligase D also contains nucleotidyl transferase activities, including gap-filling polymerase, terminal transferase, and primase, and the 3′- to 5′-exonuclease activity. Thus, Ligase D from mycobacteria can carry out DNA ends processing and join incompatible DSB ends (17). It has been demonstrated that the mycobacterial NHEJ pathway could be successfully reconstituted to repair DSBs in yeast and in Escherichia coli by introducing the Ku and Ligase D proteins (17–19).
Leishmania protozoan, an early branching eukaryote, is the causative agent of human leishmaniasis, one of the major neglected tropical diseases. Visceral leishmaniasis, caused by L. donovani is the second most deadly parasitic infection after malaria. There are no approved vaccines for leishmaniasis. Treatment of Leishmania infection relies on chemical drugs with side effects (21). Contrary to mammalian cells, Leishmania lacks the highly efficient NHEJ pathway due to absence of several key NHEJ factors, including DNA-PKcs, Ligase IV, and XRCC4, though, it still retains the core NHEJ factor Ku70 and Ku80 proteins (22–24). Like in many other organisms, homologous chromosomes are rarely used as the HR template for repairing DSB (25). Instead, Leishmania mainly relies on SSA (>95% of events if the direct repeat sequences flanking the DSBs exist) and MMEJ to repair DSBs, which involve extensive resections, prolonged repair time, and often result in large sequence deletions (up to 77 kb was detected) and frequently fail (26, 27). It was estimated that more than half of the DSBs could not be repaired in Leishmania, leading to cell death (25, 26, 28). Because of lack of a functional NHEJ DSB repair pathway, compared with other organisms, CRISPR gene targeting is not very efficient in Leishmania and antibiotic selection marker encoding donor DNAs (a kind of SSA mediated repair) are often required to isolate the CRISPR edited mutants (28–31).
As indicated above, the Ku dimer has strong affinity for DSB ends (7). If NHEJ could not be completed due to lack of other key NHEJ factors, Ku70/80 binding could block the 5′-end resections to prevent or slowdown other DSB repair pathways which rely on using the 3′-single-stranded DNA overhangs (32–36). For example, it has been recently reported that Ku70/80 heterodimer suppresses MMEJ in Ligase IV-deficient progenitor B cells, deletion of Ku70 in this B cell line significantly increases MMEJ (33). In addition, Ku has also been shown to be involved in other cellular processes such as telomere maintenance, DNA replication, transcription, and translation control (7, 8, 37–45). In this study, to investigate whether Leishmania Ku70 and Ku80 proteins actively participate in DSB repair in the absence of key NHEJ factors Ligase IV and XRCC4, we generated Leishmania donovani Ku70 null mutants and Ku70/80 double gene null mutants. It was observed that depending on location of a DSB between the direct repeat sequences, L. donovani Ku70/80 heterodimer could inhibit, have no effect, or assist DSB repair by SSA. In addition, Ku70/80 proteins are required for the normal healthy cell proliferation. However, unlike in other organisms (43–45), the normal lengths of the telomeres are maintained in Ku-deficient L. donovani cells. We also introduced Mycobacterium marinum two components (Ku and Ligase D) NHEJ pathway into Leishmania. This improved CRISPR gene targeting specificity and efficiency in Leishmania and represents the first example that the bacterial NHEJ pathway can be successfully reconstructed for DSB repair in an NHEJ-deficient eukaryotic parasite.
RESULTS AND DISCUSSION
Generation of L. donovani Ku70 null mutants and Ku70 and Ku80 double null mutants.
As noted in the Introduction, Leishmania retains Ku proteins but lacks other key NHEJ effectors, including DNA Ligase IV, DNA PKcs, and XRCC 4 (24). It was therefore interesting to compare Leishmania Ku to Ku proteins of human and mycobacteria that possess a functional NHEJ. L. donovani (Ld) Ku70 and Ku80 proteins retain the conserved domains present in human Ku proteins, including the N-terminal von Willebrand A domain, the central core DNA-binding domain and a divergent C-terminal region. Ld Ku70, however, has about 27% amino acid identity with human Ku70 and is larger containing 967 amino acids with molecular weight of 103,023 Dalton (Fig. 1; Fig. S1). Ld Ku80 protein has a similar size (798 amino acids) as human Ku80 protein (732 amino acids) and they share 22% amino acid sequence identity (Fig. 1; Fig. S1). Like human Ku proteins, Ld Ku70 and Ku80 proteins also share some sequence similarity with 21% amino acid identity over 660 amino acids alignment (Fig. S1). In contrast to eukaryotes, mycobacteria such as Mycobacteria marinum possess only a single small Ku protein of 291 amino acids which forms a Ku homodimer for binding DNA ends and NHEJ repair (19). Interestingly, Ld Ku70 and Ku80 also share 20% to 24% amino acid identities in their DNA binding domain with M. marinum Ku used in this study (Fig. 1; Fig. S1; see below).
FIG 1.
Schematics of Ku70 and Ku80 proteins of human and Leishmania donovani and Ku protein of Mycobacterium marinum. Like human Ku proteins, Leishmania Ku proteins also possess the Ku von Willebrand A (vWA) domain, DNA-binding domain, and SAF-A/B, Acinus, and PIAS (SAP) or C-terminal domain (Cter). While Ld Ku80 is only slightly larger than human Ku80, Ld Ku70 contains 967 amino acids and has a molecular weight of 103 kDa which is 30 kDa larger than Hu Ku70. Unlike eukaryotic cells, the prokaryote Mycobacterium marinum has only a single small Ku protein of 33 kDa mainly composed of the DNA binding domain and a short C-terminal domain (Cter). The Mm Ku Cter domain is involved in binding and activating Mm Ligase D and may also has some nonspecific DNA binding activity.
Amino acids alignments of Ku70 and Ku80 proteins of human and Leishmania donovani and Ku protein of Mycobacterium marinum. (A) Amino acids alignment of human Ku70 and Leishmania donovani Ku70. (B) Amino acids alignment of human Ku80 and Leishmania donovani Ku80. (C) Amino acids alignment of Leishmania donovani Ku70 and Ku80. (D) Amino acids alignment of Leishmania donovani Ku70 and Mycobacterium marinum Ku. (E) Amino acids alignment of Leishmania donovani Ku80 and Mycobacterium marinum Ku. Download FIG S1, DOCX file, 0.1 MB (110.9KB, docx) .
Copyright © 2022 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To investigate whether Leishmania Ku proteins are involved in DSB repair, we generated L. donovani Ku null mutants using the CRISPR method we previously described (46). To facilitate generating double gene knock out mutants and save the antibiotic selection markers for succeeding studies, oligonucleotides with stop codons were used as donors (repair templates) following DSB to disrupt both L. donovani Ku70 (LdBPK_291140) and Ku80 (LdBPK_300340) genes (Fig. 2A). To generate L. donovani Ku70 null mutants [Ku70(-)], the Leishmania CRISPR plasmid pLdSaCN-Ld291140, expressing neomycin (G418) resistance gene, SaCas9 nuclease and a gRNA targeting the middle in first half of the Ku70 gene (Fig. 2A) was transfected into L. donovani promastigotes. The G418 resistant cells were subsequently transfected with a 70 nt oligonucleotides donor with stop codons in all frames. PCR screening with a donor specific primer and DNA sequencing confirmed that both alleles of L. donovani Ku70 gene were disrupted as planned with the stop codons containing oligo donor (Fig. 2B). Likewise, to generate L. donovani Ku70 and Ku80 double gene knock out mutants [Ku70/80(-)], Ku70 null mutants [Ku70(-)] were retransfected with CRISPR plasmid pLdSaCH-Ld300340 which expresses hygromycin resistance gene, SaCas9 nuclease and a gRNA targeting the first half of Ku80 gene (Fig. 2C; Fig. S2A). The hygromycin resistant cells were then transfected with the stop codons containing oligonucleotides. PCR screening and DNA sequencing revealed that both alleles of L. donovani Ku80 gene were disrupted with the introduced stop codons (Fig. 2D; Fig. S2B). The CRISPR plasmids pLdSaCN and pLdSaCH were later removed from the Ku70(-) and Ku70/80(-) mutants by culturing in G418- and Hygromycin-free medium. Thus, with CRISPR gene editing, we generated the Ku70 single gene knock out mutants and Ku70/80 double gene knock out mutants with no remaining antibiotic selection markers. A total four Ku70(-) clones and three Ku70/80(-) clones were isolated. To avoid the potential clonal variation effect, all the Ku70(-) and Ku70/80(-) clones were pooled for subsequent characterization studies (see below).
FIG 2.
Generation of L. donovani Ku70 gene disruption mutant and Ku70 and Ku80 double gene disruption mutant. (A) The CRISPR strategy used to disrupt both alleles of Ld Ku70 genes with stop codons containing oligo donors. pLdSaCN CRISPR vector expresses Staphylococcus aureus Cas9 and a gRNA targeting Ld Ku70 gene. The gRNA targeting site is highlighted as green, the SaCas9 PAM site highlighted as purple. (B) Chromatogram showing both Ld Ku70 gene alleles were disrupted as planned with the stop codons containing oligo donors. (C) The same CRISPR strategy was used to further disrupt both alleles of Ld Ku80 gene in the Ld Ku70 disruption mutant to generate the Ku70 and Ku80 double gene disruption mutant (please see details in Fig. S2). (D) Chromatogram showing both Ld Ku80 gene alleles were disrupted as planned with the stop codons containing oligo donors.
Generation of L. donovani Ku70 and Ku80 double gene disruption mutant. (A) The CRISPR strategy used to disrupt both alleles of Ld Ku80 genes with stop codons containing oligo donors in Ku70 gene disruption mutant. pLdSaCH CRISPR vector expresses Staphylococcus aureus Cas9 and a gRNA targeting Ld Ku80 gene. The gRNA targeting site is highlighted as green, the SaCas9 PAM site highlighted as purple. (B) Chromatogram showing both Ld Ku80 gene alleles were disrupted as planned with the stop codons containing oligo donors. Download FIG S2, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2022 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Leishmania Ku proteins bind DSB ends and influence DSB repair.
Given that Leishmania lacks DNA Ligase IV and several other key NHEJ components, using the Ku mutants generated above, we determined whether Leishmania Ku proteins are still involved in DSB repair by binding DNA ends and interfering the 5′- to 3′-end resections. We have previously shown that L. donovani Miltefosine Transport (MT) gene (LdBPK_131590) is an ideal target for studying CRISPR gene targeting and DSB repair efficiency (25, 26, 28). Because the MT located on the cell membrane is responsible for importing Miltefosine (MLF) which is toxic to Leishmania parasites, deletion mutations of MT gene resulting from CRISPR targeting and successful DSB repair will lead to Leishmania resistance to MLF (25, 26, 28). Wild-type (WT), Ku70(-), and Ku70/80(-) L. donovani promastigotes were transfected with a series of CRISPR vectors which express gRNAs targeting various sites of the MT gene (Fig. 3B) (26). The MLF resistance rates were determined by placing the transfectants (7-weeks posttransfection) in 96-well tissue culture plates containing MLF.
FIG 3.
Leishmania Ku is still involved in DSB repair. (A) Comparison of MLF resistant rates of WT, Ld Ku70- and Ku70/80-deficient Leishmania cells 7-weeks posttransfection of CRISPR vectors expressing various gRNAs targeting MT gene. As explained in text and (B), MLF resistant rate also represents the frequency of successful repairs of DSBs generated on both alleles of MT gene following CRISPR targeting. The MLF resistance rates shown are mean value obtained from three experiments with standard error of the mean, SEM. (B) A speculative model explaining why the DSB repair efficiencies vary among different gRNAs in WT and Ku-deficient Leishmania cells. L. donovani MT gene (Ld131590) together with Ld131600 gene are flanked by two 460 bp direct repeats (blue arrows). The positions of those genes and direct repeats in L. donovani chromosome 13 are indicated. The geomatic center between the two repeats is marked with a vertical black line. The seven gRNA targeting sites in MT gene are marked with red arrows. Note, the gRNAh and 5′-targeting sites are very close to the middle line, the gRNAi site is near the left repeat but away from the right repeat. After CRISPR gene targeting and successful SSA mediated DSB repair, both MT and Ld131600 gene are deleted. (i) In WT cells, following gRNAi targeting and Cas9 cleavage, Ku heterodimers immediately bind the DSB ends and hold them together by Ku-Ku interactions, and the cell initiates 5′- to 3′-end resections. With progression of the end resections, Ku translocate inward and continue holding on the two broken chromosomal ends. In gRNAi targeting, because the left single-strand complementary direct repeat sequence is formed much earlier than the right single-strand repeat, Ku binding do not really help anneal these repeats. However, in gRNAh targeting (ii), since the left and the right single-strand repeats are formed at almost the same time and in proximity, Ku binding greatly help anneal these repeats. In the absence of Ku binding in the Ku-deficient cells (iii, iv), the 5′- to 3′-end resections can progress more rapidly to accelerate the DSB repair in the case of gRNAi targeting (iii) compared with in WT cells (i). However, in the case of gRNAh targeting (iv), without Ku binding, the left and the right single-strand repeats are formed at a distance and can move freely so delay the DSB repair compared with in WT cells (ii). See the detailed explanation in the text.
If the Ku proteins are not involved in DSB repair in Leishmania, the Ku null mutants would display similar MLF resistance rates as WT Leishmania cells. In contrast, if the Ku null mutants show much higher or lower MLF resistance rates, it would suggest that Leishmania Ku proteins are still involved in DSB repair by either an inhibition or promotion mechanism. Interestingly, depending on the gRNA targeting location, Leishmania Ku proteins could have opposite effects on DSB repair in Leishmania. As shown in Fig. 3A, a total of seven gRNAs were tested. In gRNAi targeted cells, the MLF resistance rates were found to be much higher (~5-fold) in Ku70(-) and Ku70/80(-) mutants than in WT cells, suggesting as expected, Ku70/80 in WT cells are able to bind DSB ends to slow the 5′- to-3′-end resections required for SSA mediated DSB repair. In contrast, a 3- to 7-fold reduction of MLF resistance rates were observed in the Ku null mutants transfected with gRNAh or gRNA5’. Addback of Ku70 in the Ku70(-) mutant restored the MLF resistance rates to the levels observed from WT cells, indicating Ku proteins promote DSB repair in these later circumstances despite obstructing the repair of DSB resulting from gRNAi targeting. However, MLF resistance rates using gRNAe, f, g, and j were similar between WT Leishmania cells and Ku null mutants. It is important to note that gRNA activity could vary among gRNAs due to the guard sequence differences, which explains why gRNA e and f had very different MLF resistant rates (26).
Interestingly, further analysis revealed that the targeting sites for those two gRNAs (h and 5′), which displayed much higher MLF resistance rates in WT cells, are near in the middle point between the two direct repeats used for SSA DSB repair (Fig. 3B). In contrast, the targeting site for gRNAi showed a much lower MLF resistance rate in WT cells near the left direct repeat but away from the right repeat. The remaining four gRNA targeting sites are between gRNAi and gRNAh sites (Fig. 3B). Therefore, this suggests that like in other organisms (32–36), Leishmania Ku proteins are able to bind the DSB DNA ends, inhibit and slow down the extended 5′- to 3′-end resections required for SSA or MMEJ DSB repair (see diagram Fig. 3B, panel i). However, like the resection-dependent NHEJ pathway recently discovered in mammalian cells (47, 48), with the progress of 5′- to 3′-end resections, the Ku heterodimers could also translocate inward (away from the DNA ends) while keep holding the two 3′-overhangs together through Ku-Ku and other protein interactions (7, 8). In the case of gRNAh and gRNA5’ targeting, the DSBs occurred in the middle between the two direct repeats, thus, the complementary 3′-overhangs would be formed from the two direct repeats around the same time and in close proximity, and thus be easily annealed, so as to speed up the SSA DSB repair process (Fig. 3B, panel ii). However, in the absence of Ku binding, the two 3′-overhangs would be separated and move freely, which could significantly slow down the annealing between the homologous 3′-overhangs although deficiency of Ku proteins could accelerate the 5′- to 3′-end resections (Fig. 3B, panel iv). However, in gRNAi targeting, because the homologous single-strand sequences were formed at a different time and at a distance, even the binding of Ku might hold the two 3′-overhangs together at the single to double-strand DNA transition site, Ku binding in this case would not promote 3′-overhang homologous sequences annealing but delay the 5′- to 3′-end resections (Fig. 3B, panel i and iii). For the remaining four gRNAs, their targeting sites are between gRNAi and gRNAh, and the slight annealing advantage of Ku binding could be repressed by the Ku blocking activity of 5′- to 3′-end resections. As a result, no significant changes in DSB repair efficiency could be observed for the four gRNAs between WT and Ku null mutants. Together, these data indicate that depending on the location of a DSB between two direct repeats, Ku protein binding could have an inhibition effect, no effect, or a promotion effect on the DSB repair by single-strand annealing. Ku proteins accelerate repair if the DSB occurs in the central region between two direct repeats by promoting annealing. In contrast, Ku proteins could impede repair by blocking the 5′- to 3- end resections if the DSB occurs near one direct repeat but away from the other. With the targeting site moving toward the middle, the Ku inhibition effect slowly changes to the promotion effect for SSA mediated DSB repair.
The gRNAi targeting data indicate, as reported in other organisms (32–36), the Leishmania Ku70/80 heterodimer could bind and compete for DSB ends against the MRN complex, and block or delay the 5′-end resections. However, the gRNAh and gRNA5’ targeting data in WT- and Ku-deficient cells also suggest that Leishmania Ku could translocate inward with progress of 5-′ to 3′-end resections and maintain a hold on the two 3′-single-strand overhangs. Indeed, limited inward Ku translocation was observed in recently discovered resection-dependent NHEJ in mammalian cells (47, 48). Although this Ku translocation model explains well the different DSB repair frequencies detected in WT and Ku-deficient L. donovani cells, human Ku heterodimers bounded on the DNA ends (if the NHEJ cannot be completed) are usually promptly removed by the MRN complex. Human Ku heterodimers can also be removed through Ku70 protein phosphorylation during 5′- to 3′-end resections to initiate the homologous recombination pathway (5, 9, 35, 36). It is surprising that Leishmania Ku might be able to translocate inward and hold together the two 3′-overhangs during extensive 5′- to 3′-end resections (more than 4 kb) required for SSA mediated repair in the MT gene locus. This could be due to several factors. First, DNA PKcs, DNA Ligase IV, and other NHEJ factors which limit human Ku translocation by forming a synapse between the DNA ends are not present in Leishmania. Second, although Ld Ku proteins possess all the main domains of human Ku, they share only 25% amino acid identity. Ld Ku70 contains extra amino acid sequences and is 30 kDa larger than human Ku70, and the phosphorylation sites required for human Ku displacement are not identified in Ld Ku70. The differences in sequence and size may facilitate Leishmania Ku translocation and Ku-Ku interactions. Third, the MRN complex required for 5-′ to 3′-end resections may function somewhat differently in Leishmania and could be more tolerant to the presence of Ku on the DNA ends. It may, together with other unknown factors or proteins, push L. donovani Ku to slide inward extensively during SSA mediated DSB repair in Leishmania. While this translocation model best elucidates the DSB repair rate differences between WT and Ku-deficient cells for gRNAh and gRNA5’ targeting, other mechanisms could also be possible. For example, the 3′-single-strand DNA overhangs may form stem-loop structures, through binding the stem loops, Leishmania Ku may hold the two 3′-overhangs together and assist their annealing and repair. Lastly, this Ku translocation model may also suggest it is better to select a gRNA targeting site near the middle between two direct repeat sequences in the genome. Because Leishmania mainly depends on SSA to repair DSB, this will likely improve the CRISPR gene deletion efficiency if a selection marker donor is not used.
L. donovani Ku proteins are dispensable for maintaining the normal lengths of telomeres.
Telomeres are repetitive DNA sequences (commonly TTAGGG repeats) found at the ends of chromosomes that are required to protect chromosome ends, prevent chromosome fusion, and maintain genomic integrity. In mammals, telomeres form a loop (T-loop) together with the six-protein telomere binding shelterin complex to protect chromosomal ends (49). Ku has been shown to directly bind the telomere sequences, interact with the telomere proteins, and help recruit telomerase through associating with the RNA component stem-loop structure. Therefore, Ku is involved in telomere length regulation in variety of organisms. For instance, deletion of the yeast Ku70 or Ku80 gene resulted in decreased telomere length (50, 51). Ku-deficient mice displayed abnormal telomere lengths with telomeric fusions (52). Human cells with a knockdown of Ku80 were reported to have shortened telomeres and increased cell lethality (53). In particular, progressive telomere shortening was observed in Ku-deficient Trypansoma brucei (43, 44) which like Leishmania also belongs to Trypanosomatida. In T. brucei, the telomere sequences ranging from 5 to 14 kb were progressively increased in WT cells but significantly shortened in the Ku80-deficient cells with an increasing or shortening rate of ~100 bp per week (44). In contrast, abnormally long telomeres were observed in the Ku knockout Drosophila cells (54). Notably, it has been recently reported that Leishmania mexicana telomeres were elongated from 250 to 850 bp to 250 to 3,000 bp upon deletion of the Ku70 or Ku 80 genes, and the lengths of the elongated telomeres then remain stable (unchanged) in the Ku-deficient L. mexicana cells (45).
To determine whether Ku is also involved in telomere maintenance in L. donovani, we compared the telomeres’ lengths in WT and Ku-deficient L. donovani cells by terminal restriction fragment (TRF) Southern blot and semiquantitative PCR analysis as described in methods (43–45, 55). L. donovani promastigotes were harvested from culture every 3 weeks for total 9 weeks. The genomic DNA prepared from those cell cultures were digested to completion with restriction enzyme Msp I (with CCGG cut site) and subjected to Southern blot analysis with a biotin labeled telomere oligo probe (TTAGGG)4. As shown in Fig. 4A, unlike what were reported in T. brucei and L. mexicana (43–45), no progressive telomere elongation or shortening was observed in WT or Ku70-deficient L. donovani cells. All the analyzed L. donovani cells retained similar lengths of telomeres over the 9-week culture period. Depending on the organism, telomere lengths can vary greatly ranging from several hundreds to more than 10,000 bp (56). Large telomere length variation was also observed in different Leishmania species and strains; for example, L. mexicana M379 telomeres (250 to 850 bp) are much shorter than the telomeres of L. major LV39 (550 to 13,300 bp) (45, 56). According to the Southern blot shown in Fig. 4A, we estimated the telomere lengths for this L. donovani 1S/Cl2D strain are between 1.5 to 3 kb. In addition, our semiquantitative PCR analysis with telomere specific primers revealed L. donovani Ku70/80-deficient cells also displayed no telomere shortening and contained the similar sizes of telomeres as WT L. donovani cells (Fig. 4B). Taken together, these data indicate L. donovani Ku proteins are dispensable for maintaining the normal telomere lengths.
FIG 4.
Normal telomere lengths are maintained in Ku-deficient L. donovani. (A) TRF Southern blot analysis. Genomic DNA from wild-type (LdWT), Ku70-deficient cells [Ku70(-)], and from the Ku70 addback cell line [Ku70(-) +Ku70], which expressed an ectopic copy of Ld Ku70 was prepared every 3 weeks for a period of 9 weeks. The DNA (10 μg for each sample) was digested with Msp I and separated by agarose gel electrophoresis, Southern-blotted, and probed with a biotin-labeled telomeric (TTAGGG)4 probe. Note, because L. donovani is a diploid organism containing 36 chromosomes, the telomere bands smear detected here represent at least 144 telomeres of different sizes (1.5 to 3 kb). (B) Semiquantitative telomere specific PCR analysis. Equal amounts of genomic DNA(3 or 4 ng) from LdWT, Ku70(-), Ku70(-) cells grown in hygromycin containing medium (Ku70(-)hyg), Ku70(-)+Ku70, and Ku70/80(-) cells were used to set up the telomere specific PCR for a total of 15 cycles and separated in 3% agarose gel. Note, the typical telomere PCR bands (smears) were not detected in the tubes using H2O or plasmid DNA as the PCR template, similar telomere bands (smear) intensities were observed in those L. donovani WT and Ku-deficient cell lines. (C) A semiquantitative PCR to verify that equal amounts of genomic DNA were used in the telomere specific PCR in (B).
It is surprising that Ku’s function in telomere regulation could vary so greatly even in genetically closely related Trypansoma parasites. Interestingly, amino acid sequence alignments indicate that T. brucei Ku70 and Ku80 proteins are 160 to 220 amino acids smaller than L. donovani Ku proteins and they share only 26% amino acid identities. L. mexicana Ku proteins have similar sizes as L. donovani but they have 9% to 14% differences of amino acids. However, because there are many other genetic background variations between those Trypanosomatida parasites, it is not clear whether the differences present in the Ku proteins were directly responsible for the different involvement of Ku proteins in the telomere length maintenance observed among those parasites. Further studies are needed to determine whether L. donovani Ku has any direct or indirect interactions with the telomeres.
Leishmania Ku proteins are required for healthy proliferation of Leishmania promastigotes.
We next determined whether Ku deficiency could affect L. donovani promastigote proliferation in culture. As shown in Fig. 5, compared with WT cells, Ku70(-) promastigotes displayed a significant slower growth rate, adding back Ku70 gene was able to restore the growth rate close to the level of WT cells. Surprisingly, disruption of both Ku70 and Ku80 genes had less inhibition effect on cell growth than disrupting Ku70 gene alone. Adding back Ku70 gene in Ku70/80(-) mutant cells was not able to improve the growth rate but further inhibited the cell proliferation as seen in Ku70(-) cells. Because Ku70 and Ku80 normally form a stable heterodimer, Ku80 or Ku70 protein alone is not stable in the cell or may form Ku80 or Ku70 homodimers (7, 8, 57). Taken together, these data indicate that the Ku heterodimer is required to maintain normal growth of Leishmania promastigotes, Ku80 or Ku70 protein alone could be toxic to the cell (57).
FIG 5.

The growth curves of L. donovani WT and Ku-deficient promastigotes. Equal number of L. donovani promastigotes (1 million cells per mL) was inoculated in flask containing 4 mL culture medium. The parasite growth was monitored by microscope counting once a day for total 10 days. This is the representative data of three independent experiments.
Besides DSB ends, Ku could also bind other DNA structures and RNA stem loops (7). Ku may interact with other cellular proteins besides the NHEJ factors through the C terminal sequences of Ku70 or Ku80 proteins (7, 8). Aside from NHEJ and telomere maintenance, Ku has been implicated in other cellular processes, including DNA replication, transcription, and translation control (7, 8, 37–39). In this study, we have observed that Ku-deficient L. donovani promastigotes proliferated slower than the WT cells. A similar growth defect was also observed in Ku80 knockout L. mexicana mutants (45), suggesting Leishmania Ku could have a broader function in addition to its role in DSB repair. Thus, it will be interesting in future studies to investigate whether Leishmania Ku proteins interact with other cellular DNA, RNA structures, and proteins, and how those interactions impact their cellular function.
Reconstitution of Mycobacterium marinum nonhomologous end joining pathway in Leishmania.
Due to the presence of a competent NHEJ pathway, CRISPR gene targeting is very efficient in mammalian cells and in Toxoplasma gondii, making it possible to isolate the CRISPR mutated cell clones directly with no need to use selection marker donors, and to screen the genomes with CRISPR libraries (58–61). In comparison, due to absence of a functional NHEJ pathway, Leishmania depends on SSA and MMEJ to repair DSB, which often cause large deletions (see Fig. 3B) and fail (25, 26, 28). The mutation rate of CRISPR gene targeting is often less than 1% in Leishmania and antibiotic selection marker donors are typically required to facilitate isolating deletion mutants (25, 28–31).
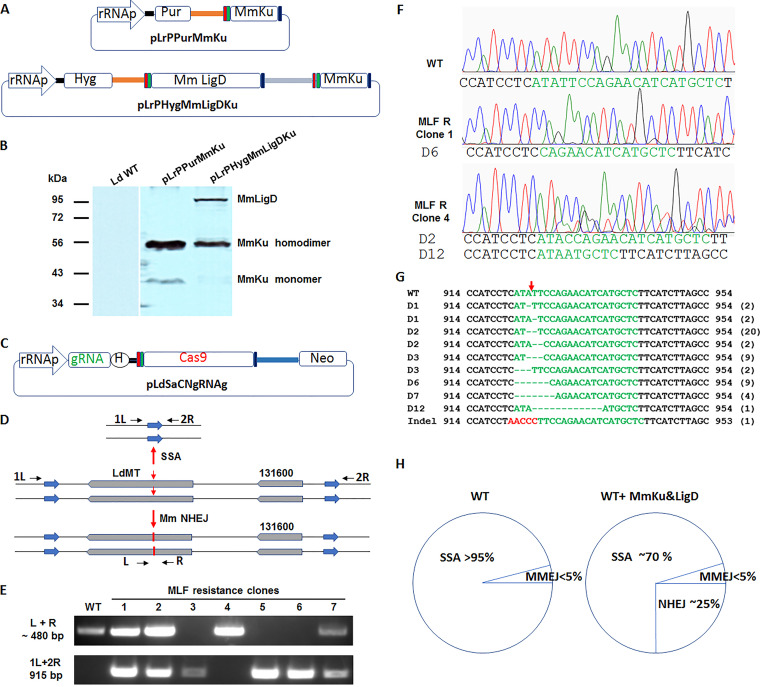
Unlike in eukaryotes, NHEJ pathway in mycobacteria is mainly composed of two proteins, Ku and Ligase D. The mycobacterial NHEJ pathway has been successfully reconstituted in yeast and in E. coli by simply introducing the Ku and Ligase D proteins (17–19). To improve DSB repair efficiency and specificity in Leishmania, we introduced the M. marinum NHEJ pathway into Leishmania. M. marinum Ku and Ligase D were found to be more efficient and accurate in joining DNA ends when expressed in E. coli than M. tuberculosis counterparts (19). M. marinum Ku (Mm Ku) and Ligase D genes were fused with SV40 and Nucleoplasmin nuclear localization signals at their 5′- and 3′-ends for proper nuclear localization and fused with Flag tag for detection in Leishmania. Mm Ligase D and Ku genes were ligated into the Leishmania expression vector pLrPHyg in tandem to generate plasmid pLrPHygMmLigDKu (Fig. 6A). We also constructed plasmid pLrPPurMmKu with a puromycin selection marker as an additional source to further increase Mm Ku expression in Leishmania (Fig. 6A). L. donovani promastigotes were individually or double transfected with these plasmids using hygromycin and puromycin selection. As shown in Fig. 6B, Mm Ligase D was expressed in Leishmania at expected 90 kDa size (19). Interestingly, Mm Ku was expressed in Leishmania as two bands, the light 38 kDa band is the expected size of a single Mm Ku protein. The dark large band around 56 kDa is most likely the Mm Ku homodimer, a ring shape structure which could migrate faster than linearized proteins (76 kDa), indicating the homodimer was very stable and resistant to separation under current SDS-PAGE running conditions.
FIG 6.
Reconstitution of Mycobacterium marinum NHEJ pathway in Leishmania. (A) The plasmids used to express Mm Ku and LigD in Leishmania. pLrPPurMmKu conferring puromycin resistance and expressing Mm Ku; pLrPHygMmLigDKu conferring hygromycin resistance and coexpressing Mm LigD and Ku. The rectangle in red, Flag tag; in green, SV40 NLS; in dark blue, Nucleoplasmin NLS. (B) Western blot analysis showing Mm LigD was expressed at the expected size and Mm Ku, however, mainly existed as homodimers in Leishmania. (C) The CRISPR vector pLdSaCNgRNAg used to generate DSBs in MT gene in the Mm Ku and LigD expressing Leishmania cells. (D) SSA and Mm NHEJ were likely the main repair mechanisms following CRISPR targeting of MT gene in the Mm Ku and LigD expressing Leishmania cells. Note, it is also possible that the DSB in one MT gene allele was repaired with Mm NHEJ but in another allele with SSA. The primers used to detect the Mm NHEJ events (L+R) and SSA events (1L + 2R) are also indicated. The expected PCR product is 480 bp for (L+R) primer pair and 9 kb for (1L + 2R) primer pair in the WT locus and 915 bp after SSA. (E) PCR analysis showing that Mm NHEJ repair (~480 bp bands), SSA (915 bp band) or both repair pathways could be detected in the Leishmania MLF resistance clones derived from the Mm Ku and LigD expressing cells. (F) Chromatograms showing the gRNAg targeting site (highlighted in green) in WT cells, and the typical small deletions resulted from Mm NHEJ repair in MLF resistance clone 1 and 4. Note, a 2-bp deletion in one MT gene allele and a 12-bp deletion in another MT gene allele have been detected in clone 4. (G) DNA sequence alignments showing all the Mm NHEJ mediated deletion and insertion events detected in Mm Ku and LigD expressing Leishmania cells following CRISPR targeting of MT gene with gRNAg. The number of times detected for a deletion or insertion event is indicated in parentheses after each sequence. Note, all the frequently detected NHEJ repairs D2(20), D3(9), D6(9), and D7(4) used 1 or 2 bp microhomology sequences to assist annealing. (H) The frequency of each repair pathway (SSA, MMEJ, and Mm NHEJ) in WT and Leishmania cells expressing Mm Ku and LigD. NHEJ could constitute around 25% of total repair events in Mm Ku and LigD expressing cells.
To determine whether Mm Ku and Ligase D could form a functional NHEJ pathway in Leishmania, WT and Leishmania cells expressing Mm Ku and Ligase D were transfected with the MT gene targeting CRISPR plasmid pLdSaCNgRNAg (Fig. 6C). Three- to 7-weeks posttransfection, the Leishmania cells were then placed in 96-well plates in medium containing MLF to detect the NHEJ mediated DSB repairs, and to determine whether Mm NHEJ improved the overall gene targeting efficiency in Leishmania. The genomic DNA extracted from the MLF resistance clones were subjected to PCR and DNA sequencing analysis. WT Leishmania cells mainly rely on SSA (>95% of times) and MMEJ (<5% of times) to repair DSBs in MT gene locus resulting in 9 kb deletions (26). It is therefore usually impossible to detect PCR bands near 480 bp in MLF resistance cells with primers LR unless the DSBs resulting from gRNAg targeting were repaired using Mm NHEJ pathway instead of SSA (Fig. 6D). As expected, we could not detect the 480 bp LR PCR band in WT Leishmania cells transfected with gRNAg CRISPR plasmid after MLF selection (data not shown). However, as shown in Fig. 6E as an example, the ~480 bp PCR products could be detected in more than half (50%) of the MLF resistance clones expressing Mm Ku and Ligase D. Sequencing analysis confirmed that the introduced Mm NHEJ pathway was functional in Leishmania and able to repair DSBs and cause the NHEJ distinctive small deletions and insertions (Fig. 6F and G). Interestingly, the most common deletions [D2 (20), D3 (9), D6 (9) and D7(4)] by Mm NHEJ all have used 1 bp (T or C) or 2 bp (TC) microhomology sequences between ends to assist annealing and repair (Fig. 6G). It is also interesting to note that while MT gene was mainly inactivated by frame shifts from the NHEJ deletions and insertions, deletion of the single amino acid isoleucine (I308) or phenylalanine (F309) in the MT protein caused by the D3 deletions was responsible for loss of MT activity.
Consistent with the above observation, there was an approximate 2-fold increase of MLF resistance rate in the Mm Ku and Ligase D expressing cells (2.3%) compared with in WT cells (1.2%). Both MT gene alleles are required to be mutated or deleted for the cell to become MLF resistance. We have observed that after CRISPR targeting, both MT gene alleles were mutated by Mm NHEJ (for example clone 4 in Fig. 6E and F) or deleted by SSA (for example, clones 3, 5, and 6 in Fig. 6E), or in combination, that is one allele was repaired and mutated by NHEJ while other allele was completely deleted by SSA mediated repair (for example, clones 1, 2, and 7 in Fig. 6E). As Mm NHEJ repairs were detected in approximately 50% of MLF resistance cells 7-weeks post-CRISPR plasmid transfection and considering that other MT gene allele could also be deleted by SSA mediated repair, we estimated that at least 25% of DSBs were repaired by NHEJ in the Mm Ku and Ligase D expressing Leishmania cells (Fig. 6H).
Because CRISPR cleavage by SpCas9 or SaCas9 generates blunt DNA ends, if Mm Ligase D can ligate those blunt DNA ends directly without any ends processing, those faithfully repaired DSBs will not be detected using MLF selection. To determine whether blunt DNA ends could be directly ligated by Mm Ligase D in Leishmania cells, a 70-bp double-strand DNA donor with blunt ends were transfected into Leishmania cells containing pLhygMmLigDKu and pLdSaCNgRNAg plasmids. Total genomic DNA were prepared 2 days later from those donor transfectants and subjected to PCR with primers (L+ Donor R; Donor F +R; L+ Donor F and Donor R+R) as indicated in Fig. 7A. The four PCR products (~130 bp and 440 bp) were then ligated into pSp72 vector. The individual clones were analyzed by DNA sequencing. Surprisingly, only three direct blunt ends ligations (5%) were detected among total 58 clones sequenced. Except for several small (1 to 2 bp) insertion mutations, most of the remaining clones display various length (1 to 27 bp) of deletions. Although NHEJ, as the name indicates, can join DNA ends without using any homologous sequences, many of those small NHEJ deletions occurred between 1 and 2 nt microhomology sequences (Fig. 6G; Fig. 7B).
FIG 7.
Direct ligation of blunt DNA ends without processing by Mm NHEJ was rarely detected in Mm Ku and LigD expressing Leishmania cells. (A) The strategy to create and detect the direct ligation events of blunt DNA ends by Mm NHEJ. (B) DNA sequencing analysis show direct ligation of blunt DNA ends by Mm NHEJ rarely occur in transgenic Leishmania. In over 58 clones sequenced, only three direct blunt ends ligation events (5%) were detected; see the top sequences in upper left and lower right panels. Again, except a few small (1 to 2 bp) insertion mutations (in red), most of the remaining clones display various length (1 to 27 bp) of small deletions, many of those deletions occurred between 1 and 2 bp microhomology sequences.
The Mm NHEJ DSB repair products with small deletions (1 to 27 bp) detected in Leishmania are similar to that observed in yeast expressing M. tuberculosis Ku and Ligase D (17), but different from M. marinum Ku and Ligase D expressing E. coli that were frequently much larger deletions (>1,000 bp) (19). Mm NHEJ in E. coli often used longer microhomology sequence (4 bp) for annealing (19), suggesting factors such as chromosome structure, nucleases, and polymerases from the receipting hosts could also be involved in Mm NHEJ repair.
Although Mm NHEJ mediated repairs can account for a quarter of the total DSB repair events in Mm Ku and Ligase D expressing Leishmania cells, further optimization of Mm NHEJ is needed to improve its DSB repair efficiency for CRISPR gene targeting without using a donor DNA in Leishmania. Depending on whether a heterologous protein is stable, harmless, or toxic, its expression level could vary greatly with time in Leishmania (62). It appears the proper expression level and ratio of Mm Ku and Ligase D are necessary for making efficient Mm NHEJ repairs since overexpression of Mm Ku alone may inhibit DSB repair (20, 34, 35, 63). Indeed, we have observed the total DSB repair rate could sometimes be lower in the Mm Ku and Ligase D expressing Leishmania cells (along with low NHEJ repair frequency) than in WT cells (data not shown). This could be due to several reasons. First, imbalanced overexpression of Mm Ku could lead to multiple Ku homodimers binding on DSB ends which may prevent proper ends processing and ligation by Ligase D (20, 63). Second, even after successful sealing of the break by Ligase D, Leishmania may have difficulty removing the extra trapped Ku homodimers on the DNA which may prevent DNA replication resulting in cell death because Leishmania may not have the specific and efficient mechanisms to remove Mm Ku promptly after end joining (36, 63, 64). Lastly, if an Mm NHEJ cannot be completed because of a low expression level of Ligase D, like Ku heterodimer in mammalian cells and Ld Ku shown in this study, Mm Ku binding could prevent 5′-end resections and inhibit DSB repairs by SSA and MMEJ (32–36). Moreover, we also observed that the DSB repair rates were 2 to 4 times lower in WT Leishmania cells than in LdKu70/80-deficient cells, when both cell lines were transfected with the Mm Ku and Ligase D expressing plasmids and targeted with MT targeting gRNAg or gRNAj. The Mm NHEJ-mediated repairs were rarely detected in those WT and LdKu70/80-deficient cells (data not shown). These may suggest that the balanced expression of Mm Ku and Ligase D was not achieved in those WT and LdKu70/80-deficient cells, and Mm Ku might assist Ld Ku to block 5′- to 3′-end resections. Further studies are therefore needed to find a way, for example by integrating Mm Ku and Ligase D genes into Leishmania chromosomes, which can express Mm Ku and Ligase D stably and in a proper ratio in Leishmania. Thus, before the current Mm NHEJ system in Leishmania can be optimized to improve efficiency, the most effective and specific CRISPR gene targeting method for Leishmania still requires the use of an oligo or selectable marker donors as demonstrated by disrupting the Ku70 and Ku80 genes in this study (25, 28–31).
Owing to the presence of a competent NHEJ pathway, CRISPR gene targeting is very efficient in Toxoplasma gondii with more than 90% gene mutation rate, making it possible to screen the genome with CRISPR libraries (61). In comparison, without using a donor, the mutation rate of CRISPR gene targeting in Leishmania is often less than 1% (25, 26, 28). Like Leishmania, NHEJ is not present in many eukaryotic parasites, including Entamoeba, Cryptosporidium, Theileria, Plasmodium, Giardia, Trichomonas, and Trypanosoma (65). Although further optimization is required, this study provides an example that the mycobacterial two components NHEJ pathway can be reconstituted in an eukaryotic parasite. It is possible that this M. marinum NHEJ may function more efficiently in a different parasite, which could be used to improve the CRISPR gene targeting (without using donors) specificity and efficiency in those NHEJ lacking parasites.
MATERIALS AND METHODS
Leishmania strains and culture medium.
L. donovani 1S/Cl2D promastigotes were cultured at 27°C in M199 medium (pH 7.4) supplemented with 10% heat-inactivated fetal bovine serum, 40 mM HEPES (pH 7.4), 0.1 mM adenine, 5 mg l−1hemin, 1 mg l−1 biotin, 1 mg l−1 biopterine, 50 U mL−1 penicillin, and 50 μg mL−1 streptomycin. Leishmania promastigotes were passaged to fresh medium at a 40-fold dilution once a week.
gRNA design and cloning.
The gRNAs were designed manually or with the aid of Eukaryotic Pathogen CRISPR guide RNA Design Tool (EuPaGDT) (http://grna.ctegd.uga.edu/) to identify off target sites in the genome and the microhomology sequences flanking the DSB. A single gRNA guide coding sequence was ordered as standard oligos with 5′-TTGT and 5′-AAAC overhangs. All oligos, primers, and oligo nucleotides donors used in this study were ordered from Alphaadn (http://alphaadn.com/). The optimal guide length is 19 or 20 nt for SpCas9 gRNA and 21 nt for SaCas9 gRNA. After phosphorylation and annealing, the gRNA guide coding sequences were ligated into the Bbs I site of pLdCN or pLdSaCN CRISPR vector as described (46).
The oligonucleotide sequences used for targeting Ld Ku70 gene (LdBPK_291140):
Ld291140 + 5′-TTGTCCCAGAGTGCTTCGCTGAAC
Ld291140- 5′-AAACGTTCAGCGAAGCACTCTGGG
Ld291140donor 5’TGGCCGCCTTTCCCGCATTCAAGTTCAGTGAGTGAATTCTAGCGGCAGCGAAGCACTCTGGGAGGCTCAG
Ld291140F 5′-TCGTGGCGATTGTGTTGTAC
Ld291140R 5′-TGGTTCGAAACGGCAAAGAG
Donorp 5′-CAGTGAGTGAATTCTAGCGG
The oligonucleotide sequences used for targeting Ld Ku80 gene (LdBPK_300340):
Ld300340 + 5′-TTGTCGGATTCACAGGGGAAGTTCA
Ld300340- 5′-AAACTGAACTTCCCCTGTGAATCCG
Ld300340donor:
5′-GCACCCACGGATTCACAGGGGAAGTCAGTGAGTGAATTCTAGCGGTCAACGAATTCACCGCAGGTGAAGC
Ld300340F 5′-ACTTCATCGACGCCCTTCA
Ld300340R 5′-GAGAGTGAACAGCTGCGTG
Donorp 5′-CAGTGAGTGAATTCTAGCGG
Expression vector construction.
NEBuilder HiFi DNA Assembly Master Mix from New England Biolabs was used to make all the expression constructs in this study. NEBuilder DNA Assembly Mix allows for directional and seamless assembly of DNA fragments with overlaps (15 to 30 bp), regardless of fragment length or end compatibility. The double-stranded fully sealed DNA vectors can be used for direct transformation of E coli. To facilitate assembly and ligation with NEBuilder DNA Assembly Mix, DNA fragments, including genes and intergenic sequences, were prepared using PCR primers containing additional 20- to 30-bp overlap sequences (see below underlined sequences in primers) with the vectors. The DNA assembly reactions were set up following the manufacture’s instruction. Briefly, the vector and insert sequences were purified from the agarose gel and mixed at 1:2 molar ratio (vector: insert). Five μL of the vector and insert sequences mix was then mixed with equal volume (5 μL) NEBuilder HiFi DNA Assembly Master Mix (total 10 μL). The assembly reaction was incubated at 50°C for 15 min; 5 μL of the assembly reaction was used for transformation of E coli. The correct clones were verified by size, restriction enzyme digestion, and DNA sequencing.
The Leishmania expression vector pLhyg was constructed as follows: (i) The 5′-intergenic sequence for LdCL351320 gene was amplified from L. donovani genomic DNA with primers LdCL3513205’F (5′-AGCACTCGTCCGGGATCATCATGATGCACGCAGCGCGTCG) and LdCL3513205’R (5’-GACTCACTATAGGGAGACCGGCAGATCTGATATCTTTGCTTGGTGGATGGAG). LdCL3513205’R primer also contains an EcoR V site (GATATC) to facilitate subsequent cloning. (ii) The PCR fragment (~700 bp) from step (i) was directly ligated with NEBuilder DNA Assembly Mix into Bgl II linearized pSPhyg vector downstream to the hygromycin resistance gene to generate a novel Leishmania expression vector pLhyg.
L. donovani Ku70 expression vector (pLhygLdKu70) was constructed as follows: (i) L. donovani Ku70 gene was PCR amplified from L. donovani genomic DNA with primer Ld291140F2 (5’-CTCCATCCACCAAGCAAAGATCAGCTCACCATGGATGAATTTG) and Ld291140R2 (5’-TAGGGAGACCGGCAGATCTGATATCAGGATGGCAGCGGAAAATGCTG) to get the 2950 bp gene fragment. (ii) The PCR fragment from step (i) was directly ligated into the EcoR V linearized pLhyg vector to generate pLhygLdKu70.
Mycobacterium marinum DNA Ligase D expression vector (pLhygMmLigD) was generated by overlap PCR and NEBuilder HiFi DNA Assembly as follows: (i) Three individual PCR fragments were first obtained. Primers MmLigDF1 (5’-CTCCATCCACCAAGCAAAGATGGACTATAAGGACCACGAC) and MmLigDR1 (5’-CAGGCCTGCGCCGAACCCATACCGACCTTCCGCTTCTT) with pLdCN vector as the template were used to get the ~150-bp sequence coding for 3X Flag and SV40 nuclear localization signal (NLS); Primers MmLigDF2 (5’-GAAGGTCGGTATGGGTTCGGCGCAGGCCTGG) and MmLigDR2 (5′-TCGTGGCCGCCGGCCTTTTTTCGCGCACCACCTCACTC) with pDUBmLig vector (19) as the template were used to get the 2,313 bp MmLigD gene fragment; Primers MmLigDF3 (5′-GAGTGAGGTGGTGCGCGAAAAAAGGCCGGCGGCCACGA) and MmLigDR3 (5′-TAGGGAGACCGGCAGATCTGATATCACTTTTTCTTTTTTGCCTGGC) with pLdCN as the template were used to get the ~90-bp sequence coding for nucleoplasmin NLS. (ii) The above three PCR products were mixed and used as the PCR templates with primers MmLigDF1 and MmLigDR3 to get the ~2,500-bp overlap fragment encoding MmLigD fused with Flag tag and two NLS sequences. (iii) The ~2,500-bp fragment from step (ii) was directly ligated into the EcoR V linearized pLhyg vector to generate pLhygMmLigD.
Mycobacterium marinum Ku expression vector (pLhygMmKu) was also generated by overlap PCR and NEBuilder HiFi DNA Assembly as follows: (i) Three individual PCR fragments were first obtained. Primers MmLigDF1 (5’-CTCCATCCACCAAGCAAAGATGGACTATAAGGACCACGAC) and MmKuR1 (5’-CCTTCCAGATGGAGCGCATACCGACCTTCCGCTTCTT) with pLdCN vector as the template were used to get the ~150-bp sequence coding for 3X Flag and SV40 nuclear localization signal (NLS); Primers MmKuF2 (5’-GAAGGTCGGTATGCGCTCCATCTGGAAGGGT) and MmKuR2 (5′-TCGTGGCCGCCGGCCTTTTGGACTTGGCAGCTTGTTTGG) with pDUBmKu vector (19) as the template were used to get the 876-bp MmKu gene fragment; Primers MmKuF3 (5′-CCAAACAAGCTGCCAAGTCCAAAAGGCCGGCGGCCACGA) and MmLigDR3 (5′-TAGGGAGACCGGCAGATCTGATATCACTTTTTCTTTTTTGCCTGGC) with pLdCN as the template were used to get the ~90-bp sequence coding for nucleoplasmin NLS. (ii) The above three PCR products were mixed and used as the PCR templates with primers MmLigDF1 and MmLigDR3 to get the ~1,000-bp overlap fragment encoding MmKu fused with Flag tag and two NLS sequences. (iii) The ~1,000-bp fragment from step (ii) was directly ligated into the EcoR V linearized pLhyg vector to generate pLhygMmKu.
Mycobacterium marinum DNA ligase D and Ku coexpression vector (pLhygMmLigDKu) was generated as follows: (i) The ~1,000-bp α tubulin intergenic sequence was amplified from L. donovani genomic DNA with primers TublinF2 (5′-CCAGGCAAAAAAGAAAAAGTGATGGTACACTCGTGCCGCGCGCTGA) and TublinR1 (5′-TAGGGAGACCGGCAGATCTGATATCCATGGCTGAAAAAGAAGAAAGAG). (ii) The 1,000-bp α tubulin intergenic sequence was directly ligated into the EcoR V linearized pLhygMmLigD vector downstream of LigD to generate the intermediate vector pLhygMmLigD-tubulin. (iii) Primers MmKuF1 (5′-CTCTTTCTTCTTTTTCAGCCATGGATTATAAGGACCACG) and MmLigDR3 with plasmid pLhygMmKu as the template were used to get the ~1,000-bp fragment encoding MmKu fused with Flag tag and two NLS sequences. (iv) The 1,000-bp Ku fragment from step (iii) was directly ligated into the EcoR V linearized pLhygMmLigD-tubulin vector to generate the coexpression vector pLhygMmLigDKu.
To improve expression of MmLigD and Ku in Leishmania, the L. donovani ribosome RNA promoter sequence was inserted upstream of the hygromycin resistance gene in pLhygMmLigDKu vector: (i) Primers LdrRNApF3 (5′-CTGCAGGTCGACTCTAGAGGATCCATTGCTTCGCGCCCCAGTACTGGAG) and LdrRNApR5 (5′-GAGAGAGCTCGGTACCCGGGGAGCGCGCTGCCACGTACAGTTG) with L. donovani genomic DNA as PCR template were used to get the ~340-bp L. donovani ribosome RNA promoter fragment. (ii) The ~340-bp ribosome RNA promoter fragment was directly ligated into the BamH I linearized pLhygMmLigDKu plasmid to generate pLrPhygMmLigDKu.
Mycobacterium marinum Ku expression vector (pLrPPurMmKu) with L. donovani ribosome RNA promoter and Puromycin resistance gene was generated as follows: (i) The ~450-bp DNA fragment containing L. donovani ribosome RNA promoter and a 96-bp pyrimidine track was derived from pLrPhygMmLigDKu with primers LdrRNApF3 (5′-CTGCAGGTCGACTCTAGAGGATCCATTGCTTC GCGCCCCAGTACTGGAG) and PurR1(5′-GTACTCGGTCATATCAATTCGAGCTCTGGAG). (ii) The 600-bp Puromycin resistance gene was derived from pGL58 plasmid with primers PurF1 (5′-GCTCGAATTGATATGACCGAGTACAAGCCCAC) and PurR2 (5′-GGAGCATATACGCCCGGAGCCGTCA GGCACCGGGCTTGCGGGTC). (iii) The fragments from step (i) and (ii) were joined by overlap PCR with primers LdrRNApF3 and PurR2. (iv) pLhygMmKu was digested with BamH I and Sac II to remove the hygromycin resistance gene. (v) The fragments from step (iii) and (iv) was joined by NEBuilder HiFi DNA Assembly mix to generate plasmid pLrPPurMmKu.
Parasite transfection.
Next, 2 × 107 Leishmania donovani promastigotes (middle log phase to early stationary phase) were harvested and washed once in 200 μL Tb-BSF buffer (90 mM Na2HPO4, 5 mM KCl, 0.15 mM CaCl2, 50 mM HEPES, pH 7.3), and resuspended in 100 μL Tb-BSF buffer in a 2-mm gap electroporation cuvette. Then, 2 to 5 μg CRISPR or other vectors in a volume < 20 μL were added and mixed. The transfection was performed with the LONZA Nucleofector 2b Device (program U33). The transfected Leishmania cells were selected with 50 μg/mL G418,100 μg/mL hygromycin, or 30 μg/mL puromycin the following day. Once the CRISPR vector transfected cell culture was established, those cells could be then transfected with the donors. To improve the targeting efficiency, 8 μL 100 μM oligonucleotide donor was used for each transfection every 2 days for total of 3 times before cloning the transfectants in 96-well plates. To reconstitute Mycobacterium marinum NHEJ pathway in Leishmania, the MmLigD and Ku coexpression plasmid pLrPhygMmLigDKu was first transfected into Leishmania cells with hygromycin selection, followed by transfections with plasmid pLrPPurMmKu using puromycin selection and a series of CRISPR vectors targeting the MT gene using G418 selection.
Determine MLF resistance rate and clone MLF resistant cells.
For Leishmania promastigotes transfected with various CRISPR vectors targeting MT gene, the MLF resistance rate was determined by limiting dilution culture in 96-well tissue culture plates containing 100 μL medium with 40 μM MLF. Depending on estimation of the MLF resistance rate, 2,000 to 8 million Leishmania promastigotes (100 μL per well) were inoculated into the first column of 96-well plates in quadruplicate for each transfected cell line, the cells in the first column were then serially 2-fold diluted until the last column in the plate. The plates were sealed and incubated in a 27°C incubator for 2 to 3 weeks. The MLF resistance rates (or the diallelic MT gene mutation frequency) were calculated by identifying the furthest MLF surviving wells. The surviving Leishmania cell clones in the farthest wells after MLF selection were directly subjected to genomic DNA extraction with M-Fast PCR Genotyping kit (ZmTech Scientifique, Montreal) or expanded in 24-well plates for subsequent genomic DNA extraction and PCR analysis.
Genomic DNA preparation and PCR analysis.
Parasite genomic DNA were extracted from WT Leishmania promastigotes and various MLF resistant cells with the minipreparation method (66) which includes phenol-chloroform extraction and ethanol precipitation. The purity and quantity of those genomic DNA were assessed by Nanodrop spectrophotometer. The parasite genomic DNA were also prepared with M-Fast PCR Genotyping kit (ZmTech Scientifique, Montreal). Briefly, 100 μL stationary phase Leishmania culture was harvested by centrifugation at 1,500 g for 5 min, and the cell pellet was resuspended in 15 μL reagent-A and incubated at 95°C for 30 min in a PCR apparatus. Once the tube cooled to room temperature, 3 μL reagent-B was added into the parasite tube and mixed well. Then, 1 μL lysate supernatant was added into 11 μL PCR mastermix for a total 12 μL reaction.
Primers were designed manually or using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/). Optimal primer length was 20 nucleotides with 60°C Tm. The primers used to detect the NHEJ and SSA specific bands in Fig. 6D and E are as follows: L, 5′-GCTGTTCCGAAACTTGAAGC; R, 5′-GTCGATCGGATTACCGAGAG. 1L, 5′-ACCTCCACCTAAACGGCTCT; 2R, 5′-TCGATCTCGTAATGCGTGAG. The various Taq DNA polymerases used in this study include all in one 2X Green PCR Mastermix (ZmTech Scientific, Montreal, Canada), 2X DreamTaq Green PCR Master MIX, and 2X Platinum SuperFi PCR Master Mix (Thermo Fisher Scientific). The PCR program was set up according to manufacturer’s instruction with variation in annealing temperature, extension time, and PCR cycles. The PCR products were separated in 1% to 1.5% agarose gel. The putative NHEJ, MMEJ, and SSA specific bands were extracted from the gel and sent to Genome Quebec Sequencing Center for sequencing confirmation.
Terminal restriction fragment Southern blot analysis.
The genomic DNA were extracted from WT, LdKu70(-), and Ku70 addback cell lines at 0, 3, 6, and 9 weeks of culture. The same amount of genomic DNA (10 μg) was digested to completion in 300 μL reaction with 50 units restriction enzyme Msp I at 37°C overnight. The Msp I digested genomic DNA were then ethanol precipitated and separated on 1% agarose gel. After denaturing in 1.5 M NaCl 0.5 N NaOH solution for 1 h and neutralization in 1.5 NaCl 1 M TRIS HCl for 30 min, the DNA were transferred to a nylon membrane with 20 × SSC (3M NaCl and 0.3 M Na3Citrate) and a stack of paper towels overnight. The membrane was UV-cross-linked for 5 min and baked at 80°C for 30 min. The membrane was prehybridized at 42°C for 1 h in 35 mL prehybridization solution consisting of 1% SDS, 1 M NaCl, and 10% dextran sulfate. The 24 nucleotides telomere oligo probe (5′ Biotin-TTAGGGTTAGGGTTAGGGTTAGGG-Biotin 3′) labeled with biotin at both 5′- and 3′-ends (http://alphaadn.com/) (50 nM) was then added into the prehybridization solution and incubated at 42°C for 2 h. The membrane was washed twice with 2 × SSC, 0.1% SDS for 10 min each at room temperature, and with 0.1 × SSC, 0.1% SDS once for 20 min at 65°C. The biotin-labeled DNA probe hybridized to the target DNA on the membrane was detected with alkaline phosphatase-conjugated streptavidin and the substrate, BCIP-T (5-bromo-4-chloro-3-indolylphosphate, p-toluidine salt) using the Biotin Chromogenic Detection Kit (Thermo Scientific).
Semiquantitative telomere specific PCR analysis.
The telomere specific PCR used for estimating telomere length was performed as described (50) with telomere specific primers: tel 1: 5′-GGTTTTTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGT; and tel 2: 5′-TCCCGACTATCCCTATCCCTATCCCTATCCCTATCCCTA. The PCR primers (Ld323480L, 5′-AGCTCCTGTGCACGCTTATT and Ld323480R, 5′-GCTGAATCGTCTCCTCGTTC) derived from a single copy gene LdBPK_323480 were used in a semiquantitative PCR to estimate that equal amounts of genomic DNA were used in those telomere specific PCR.
Western blot analysis.
Leishmania promastigotes were harvested by centrifuge at 1,300 g for 5 min and washed once with PBS. Parasites pellets were then resuspended in 1 × SDS sample loading buffer (22.5 mM Tris HCl pH 6.8, 5% Glycerol, 1% SDS, 2.5% β-mercaptoethanol, and 0.025% Bromphenol blue) to make the lysate concentration about 5 × 107 Leishmania cells per 20 μL, boiled in water bath for 3 min, and centrifuged at high speed for 10 min. The cell lysate supernatant (equals 5 × 107 promastigotes in 20 μL per lane) was separated in 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to 0.45 μM nitrocellulose membranes (GE) using Bio Rad semidry transfer cell. The membrane was blocked with 10 mL 10% skim dry milk in PBS-T buffer (0.1% Tween 20 in PBS) at room temperature for 1 h, then incubated with the primary anti-flag M2 antibody (Sigma-Aldrich) in 5% skim dry milk-PBS-T solution at 1:5,000 dilution for 2 h. After washing 3 times with PBS-T, the membrane was incubated with the secondary anti-mouse NA931V antibody (GE Healthcare), anti-mouse IgG HRP in 1:5000 dilution in 5% skim dry milk PBS-T solution for 1 h. Detection was performed with Spray-on ECL reagent (Cat# E208085 from ZmTech Scientifique, Montreal) according to manufacturer’s instructions.
Growth curves.
Leishmania promastigotes were seeded at the density of 1 × 106 promastigotes/mL in 4 mL M199 media in 25 cm2 flasks and counted by a microscope daily for total 10 days. A minimum of three biological replicates were evaluated.
ACKNOWLEDGMENTS
We thank Dr. Marc Ouellette and Dr. Barbara Papadopoulou for plasmids pSPneo and pSPhyg, and Dr J. C. Mottram for plasmid pGL58. This work was supported by grants from the Canadian Institute of Health Research to G.M. The funder had no role in study design and decision to submit the work for publication.
Contributor Information
Greg Matlashewski, Email: greg.matlashewski@mcgill.ca.
Ira J. Blader, University at Buffalo
REFERENCES
- 1.Ceccaldi R, Rondinelli B, D’Andrea AD. 2016. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol 26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang HHY, Pannunzio NR, Adachi N, Lieber MR. 2017. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Her J, Bunting SF. 2018. How cells ensure correct repair of DNA double-strand breaks. J Biol Chem 293:10502–10511. doi: 10.1074/jbc.TM118.000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karanam K, Kafri R, Loewer A, Lahav G. 2012. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell 47:320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao Z, Bozzella M, Seluanov A, Gorbunova V. 2008. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 7:1765–1771. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pannunzio NR, Watanabe G, Lieber MR. 2018. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem 293:10512–10523. doi: 10.1074/jbc.TM117.000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zahid S, Seif El Dahan M, Iehl F, Fernandez-Varela P, Le Du MH, Ropars V, Charbonnier JB. 2021. The multifaceted roles of Ku70/80. Int J Mol Sci 22:4134. doi: 10.3390/ijms22084134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbasi S, Parmar G, Kelly RD, Balasuriya N, Schild-Poulter C. 2021. The Ku complex: recent advances and emerging roles outside of non-homologous end-joining. Cell Mol Life Sci 78:4589–4613. doi: 10.1007/s00018-021-03801-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reginato G, Cejka P. 2020. The MRE11 complex: a versatile toolkit for the repair of broken DNA. DNA Repair (Amst) 91–92:102869. doi: 10.1016/j.dnarep.2020.102869. [DOI] [PubMed] [Google Scholar]
- 10.Sallmyr A, Tomkinson AE. 2018. Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J Biol Chem 293:10536–10546. doi: 10.1074/jbc.TM117.000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, Fazeli A, Feldman L, Nussenzweig A, Nussenzweig M, Alt FW. 2010. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci USA 107:3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. 2015. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, O'Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, Yusufzai T, D’Andrea AD. 2015. Homologous-recombination-deficient tumors are dependent on Polθ-mediated repair. Nature 518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinha S, Li F, Villarreal D, Shim JH, Yoon S, Myung K, Shim EY, Lee SE. 2017. Microhomology-mediated end joining induces hypermutagenesis at breakpoint junctions. PLoS Genet 13:e1006714. doi: 10.1371/journal.pgen.1006714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhargava R, Onyango DO, Stark JM. 2016. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet 32:566–575. doi: 10.1016/j.tig.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glickman MS. 2014. Double-strand DNA break repair in mycobacteria. Microbiol Spectr 2. doi: 10.1128/microbiolspec.MGM2-0024-2013. [DOI] [PubMed] [Google Scholar]
- 17.Della M, Palmbos PL, Tseng HM, Tonkin LM, Daley JM, Topper LM, Pitcher RS, Tomkinson AE, Wilson TE, Doherty AJ. 2004. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science 306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- 18.Malyarchuk S, Wright D, Castore R, Klepper E, Weiss B, Doherty AJ, Harrison L. 2007. Expression of mycobacterium tuberculosis Ku and Ligase D in Escherichia coli results in RecA and RecB-independent DNA end-joining at regions of microhomology. DNA Repair (Amst) 6:1413–1424. doi: 10.1016/j.dnarep.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright DG, Castore R, Shi R, Mallick A, Ennis DG, Harrison L. 2017. Mycobacterium tuberculosis and Mycobacterium marinum non-homologous end-joining proteins can function together to join DNA ends in Escherichia coli. Mutagenesis 232:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Öz R, Wang JL, Guerois R, Goyal G, Kk S, Ropars V, Sharma R, Koca F, Charbonnier JB, Modesti M, Strick TR, Westerlund F. 2021. Dynamics of Ku and bacterial non-homologous end-joining characterized using single DNA molecule analysis. Nucleic Acids Res 49:2629–2641. doi: 10.1093/nar/gkab083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burza S, Croft SL, Boelaert M. 2018. Leishmaniasis. Lancet 392:951–970. doi: 10.1016/S0140-6736(18)31204-2. [DOI] [PubMed] [Google Scholar]
- 22.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RM, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz-Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, et al. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, Peters N, Adlem E, Tivey A, Aslett M, Kerhornou A, Ivens A, Fraser A, Rajandream MA, Carver T, Norbertczak H, Chillingworth T, Hance Z, Jagels K, Moule S, Ormond D, Rutter S, Squares R, Whitehead S, Rabbinowitsch E, Arrowsmith C, White B, Thurston S, Bringaud F, Baldauf SL, Faulconbridge A, Jeffares D, Depledge DP, Oyola SO, Hilley JD, Brito LO, Tosi LR, Barrell B, Cruz AK, Mottram JC, Smith DF, Berriman M. 2007. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet 39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Passos-Silva DG, Rajão MA, Nascimento de Aguiar PH, Vieira-da-Rocha JP, Machado CR, Furtado C. 2010. Overview of DNA repair in Trypanosoma cruzi, Trypanosoma brucei, and Leishmania major. J Nucleic Acids 2010:840768. doi: 10.4061/2010/840768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang WW, Lypaczewski P, Matlashewski G. 2017. Optimized CRISPR-Cas9 genome editing for Leishmania and its use to target a multigene family, induce chromosomal translocation and study DNA break repair mechanisms. mSphere 2:e00340-16. doi: 10.1128/mSphere.00340-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang WW, Matlashewski G. 2019. Single-strand annealing plays a major role in double-strand DNA break repair following CRISPR-Cas9 cleavage in Leishmania. mSphere 4:e00408-19. doi: 10.1128/mSphere.00408-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldman-Pinkovich A, Kannan S, Nitzan-Koren R, Puri M, Pawar H, Bar-Avraham Y, McDonald J, Sur A, Zhang W-W, Matlashewski G, Madhubala R, Michaeli S, Myler PJ, Zilberstein D. 2020. Sensing host arginine is essential for Leishmania parasites' intracellular development. mBio 11:e02023-20. doi: 10.1128/mBio.02023-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang WW, Matlashewski G. 2015. CRISPR-Cas9-mediated genome editing in Leishmania donovani. mBio 6:e00861. doi: 10.1128/mBio.00861-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beneke T, Madden R, Makin L, Valli J, Sunter J, Gluenz E. 2017. A CRISPR Cas9 high-throughput genome editing toolkit for kinetoplastids. R Soc Open Sci 4 :170095. doi: 10.1098/rsos.170095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beneke T, Demay F, Hookway E, Ashman N, Jeffery H, Smith J, Valli J, Becvar T, Myskova J, Lestinova T, Shafiq S, Sadlova J, Volf P, Wheeler RJ, Gluenz E. 2019. Genetic dissection of a Leishmania flagellar proteome demonstrates requirement for directional motility in sand fly infections. PLoS Pathog 15:e1007828. doi: 10.1371/journal.ppat.1007828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adaui V, Kröber-Boncardo C, Brinker C, Zirpel H, Sellau J, Arévalo J, Dujardin JC, Clos J. 2020. Application of CRISPR/Cas9-based reverse genetics in Leishmania braziliensis: conserved Roles for HSP100 and HSP23. Genes 11:1159. doi: 10.3390/genes11101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wasko BM, Holland CL, Resnick MA, Lewis LK. 2009. Inhibition of DNA double-strand break repair by the Ku heterodimer in mrx mutants of Saccharomyces cerevisiae. DNA Repair (Amst)). 8:162–169. doi: 10.1016/j.dnarep.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang Z, Kumar V, Le Bouteiller M, Zurita J, Kenrick J, Lin SG, Lou J, Hu J, Ye AY, Boboila C, Alt FW, Frock RL. 2021. Ku70 suppresses alternative end joining in G1-arrested progenitor B cells. Proc Natl Acad Sci USA 118:e2103630118. doi: 10.1073/pnas.2103630118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castore R, Hughes C, Debeaux A, Sun J, Zeng C, Wang SY, Tatchell K, Shi R, Lee KJ, Chen DJ, Harrison L. 2011. Mycobacterium tuberculosis Ku can bind to nuclear DNA damage and sensitize mammalian cells to bleomycin sulfate. Mutagenesis 26:795–803. doi: 10.1093/mutage/ger049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao Z, Davis AJ, Fattah KR, So S, Sun J, Lee KJ, Harrison L, Yang J, Chen DJ. 2012. Persistently bound Ku at DNA ends attenuates DNA end resection and homologous recombination. DNA Repair (Amst)). 11:310–316. doi: 10.1016/j.dnarep.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee KJ, Saha J, Sun J, Fattah KR, Wang SC, Jakob B, Chi L, Wang SY, Taucher-Scholz G, Davis AJ, Chen DJ. 2016. Phosphorylation of Ku dictates DNA double-strand break (DSB) repair pathway choice in S phase. Nucleic Acids Res 44:1732–1745. doi: 10.1093/nar/gkv1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novac O, Matheos D, Araujo FD, Price GB, Zannis-Hadjopoulos M. 2001. In vivo association of Ku with mammalian origins of DNA replication. Mol Biol Cell 12:3386–3401. doi: 10.1091/mbc.12.11.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park S-J, Ciccone SLM, Freie B, Kurimasa A, Chen DJ, Li GC, Clapp DW, Lee S-H. 2004. A positive role for the Ku complex in DNA replication following strand break damage in mammals. J Biol Chem 279:6046–6055. doi: 10.1074/jbc.M311054200. [DOI] [PubMed] [Google Scholar]
- 39.Rampakakis E, Di Paola D, Zannis-Hadjopoulos M. 2008. Ku is involved in cell growth, DNA replication and G1-S transition. J Cell Sci 121:590–600. doi: 10.1242/jcs.021352. [DOI] [PubMed] [Google Scholar]
- 40.Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE. 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet 27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 41.Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ting NSY, Yu Y, Pohorelic B, Lees-Miller SP, Beattie TL. 2005. Human Ku70/80 interacts directly with hTR, the RNA component of human telomerase. Nucleic Acids Res 33:2090–2098. doi: 10.1093/nar/gki342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conway C, McCulloch R, Ginger ML, Robinson NP, Browitt A, Barry JD. 2002. Ku is important for telomere maintenance, but not for differential expression of telomeric VSG genes, in African trypanosomes. J Biol Chem 277:21269–21277. doi: 10.1074/jbc.M200550200. [DOI] [PubMed] [Google Scholar]
- 44.Anzen CJ, Lander F, Dreesen O, Cross GA. 2004. Telomere length regulation and transcriptional silencing in KU80-deficient Trypanosoma brucei. Nucleic Acids Res 32:6575–6584. doi: 10.1093/nar/gkh991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poláková E, Albanaz ATS, Zakharova A, Novozhilova TS, Gerasimov ES, Yurchenko V. 2021. Ku80 is involved in telomere maintenance but dispensable for genomic stability in Leishmania mexicana. PLoS Negl Trop Dis 15:e0010041. doi: 10.1371/journal.pntd.0010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang WW, Lypaczewski P, Matlashewski G. 2020. Application of CRISPR/Cas9-mediated genome editing in Leishmania. In Michels P., Ginger M., Zilberstein D. (ed), Trypanosomatids. Methods in Molecular Biology. pp 199–224. Humana, New York, NY. doi: 10.1007/978-1-0716-0294-2_14. [DOI] [PubMed] [Google Scholar]
- 47.Öbrich M, Jeggo P. 2017. A process of resection-dependent nonhomologous end joining involving the Goddess Artemis. Trends Biochem Sci 42:690–701. doi: 10.1016/j.tibs.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibata A, Jeggo P, Löbrich M. 2018. The pendulum of the Ku-Ku clock. DNA Repair (Amst) 71:164–171. doi: 10.1016/j.dnarep.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 49.de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 50.Porter SE, Greenwell PW, Ritchie KB, Petes TD. 1996. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res 24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boulton SJ, Jackson SP. 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res 24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, Kim SH, Li GC, Campisi J, Kohwi-Shigematsu T, Chen DJ. 2000. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev 14:2807–2812. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Ghosh G, Hendrickson EA. 2009. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci USA 106:12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melnikova L, Biessmann H, Georgiev P. 2005. The Ku protein complex is involved in length regulation of Drosophila telomeres. Genetics 170:221–235. doi: 10.1534/genetics.104.034538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cawthon RM. 2002. Telomere measurement by quantitative PCR. Nucleic Acids Res 30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poláková E, Záhonová K, Albanaz ATS, Butenko A, Lukeš J, Yurchenko V. 2021. Diverse telomeres in trypanosomatids. Parasitology 148:1254–1270. doi: 10.1017/S0031182021000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi YJ, Li H, Son MY, Wang X-H, Fornsaglio JL, Sobol RW, Lee M, Vijg J, Imholz S, Dollé MET, van Steeg H, Reiling E, Hasty P. 2014. Deletion of individual Ku subunits in mice causes an NHEJ-independent phenotype potentially by altering apurinic/apyrimidinic site repair. PLoS One 9:e86358. doi: 10.1371/journal.pone.0086358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, Lee H, Zhang F, Sharp PA. 2015. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sidik SM, Huet D, Ganesan SM, Huynh MH, Wang T, Nasamu AS, Thiru P, Saeij JPJ, Carruthers VB, Niles JC, Lourido S. 2016. A genome-wide CRISPR screen in toxoplasma identifies essential apicomplexan genes. Cell 166:1423–1435. doi: 10.1016/j.cell.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lypaczewski P, Zhang WW, Matlashewski G. 2021. Evidence that a naturally occurring single nucleotide polymorphism in the RagC gene of Leishmania donovani contributes to reduced virulence. PLoS Negl Trop Dis 15:e0009079. doi: 10.1371/journal.pntd.0009079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McGovern S, Baconnais S, Roblin P, Nicolas P, Drevet P, Simonson H, Piétrement O, Charbonnier JB, Le Cam E, Noirot P, Lecointe F. 2016. C-terminal region of bacterial Ku controls DNA bridging, DNA threading and recruitment of DNA ligase D for double strand breaks repair. Nucleic Acids Res 44:4785–4806. doi: 10.1093/nar/gkw149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Postow L. 2011. Destroying the ring: freeing DNA from Ku with ubiquitin. FEBS Lett 585:2876–2882. doi: 10.1016/j.febslet.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nenarokova A, Záhonová K, Krasilnikova M, Gahura O, McCulloch R, Zíková A, Yurchenko V, Lukeš J. 2019. Causes and effects of loss of classical nonhomologous end joining pathway in parasitic eukaryotes. mBio 10:e01541-19. doi: 10.1128/mBio.01541-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Medina-Acosta E, Cross GA. 1993. Rapid isolation of DNA from trypanosomatid protozoa using a simple ‘mini-prep’ procedure. Mol Biochem Parasitol 59:327–329. doi: 10.1016/0166-6851(93)90231-l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acids alignments of Ku70 and Ku80 proteins of human and Leishmania donovani and Ku protein of Mycobacterium marinum. (A) Amino acids alignment of human Ku70 and Leishmania donovani Ku70. (B) Amino acids alignment of human Ku80 and Leishmania donovani Ku80. (C) Amino acids alignment of Leishmania donovani Ku70 and Ku80. (D) Amino acids alignment of Leishmania donovani Ku70 and Mycobacterium marinum Ku. (E) Amino acids alignment of Leishmania donovani Ku80 and Mycobacterium marinum Ku. Download FIG S1, DOCX file, 0.1 MB (110.9KB, docx) .
Copyright © 2022 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Generation of L. donovani Ku70 and Ku80 double gene disruption mutant. (A) The CRISPR strategy used to disrupt both alleles of Ld Ku80 genes with stop codons containing oligo donors in Ku70 gene disruption mutant. pLdSaCH CRISPR vector expresses Staphylococcus aureus Cas9 and a gRNA targeting Ld Ku80 gene. The gRNA targeting site is highlighted as green, the SaCas9 PAM site highlighted as purple. (B) Chromatogram showing both Ld Ku80 gene alleles were disrupted as planned with the stop codons containing oligo donors. Download FIG S2, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2022 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.