ABSTRACT
A rapid and sensitive diagnosis is crucial for the management of tuberculosis (TB). A simple and label-free approach via homobifunctional imidoesters with a microfluidic platform (SLIM) assay showed a higher sensitivity than the Xpert MTB/RIF assay in the diagnosis of pulmonary TB (PTB). Here, we evaluated the efficacy of the SLIM assay for oral swab samples from cases of suspected PTB. Patients with clinically suspected PTB were prospectively enrolled and oral swab samples were processed using the SLIM assay and the attending physicians were blinded to the results of the SLIM assay. TB cases were defined as those treated with anti-TB chemotherapy for at least 6 months at the discretion of the specialists based on their clinical features and conventional laboratory results, including the Xpert assay. A total of 272 patients (with TB, n = 128 [47.1%]; without TB, n = 144 [52.9%]; mean age, 59.8 years) were enrolled. Overall, the sensitivity of the oral swab-based SLIM assay (65.6%) was higher than that of the sputum-based Xpert assay (43.4%; P = 0.001). Specifically, the SLIM oral swab assay showed a notably higher sensitivity in culture-negative TB cases compared with the Xpert assay (69.0% [95% CI: 49.2 to 84.7%] versus 7.4% [95% CI: 0.9 to 24.3%]; P = 0.001). The specificity of the SLIM and the Xpert assays was 86.1% (95% CI: 79.3 to 91.3%) and 100% (95% CI: 97.2 to 100%), respectively. When only culture-confirmed cases were analyzed, the SLIM oral swab was comparable to sputum Xpert in sensitivity (64.7% versus 54.3%, P = 0.26). The oral swab-based SLIM assay showed a superior sensitivity for TB diagnosis over the sputum-based Xpert assay, especially for culture-negative cases.
IMPORTANCE The development of a rapid, accessible, and highly sensitive diagnostic tool is a major challenge in the control and management of tuberculosis. Gene-based diagnostics is recommended for the rapid diagnosis of pulmonary tuberculosis (PTB), but its sensitivity, such as Xpert MTB/RIF assay (Xpert), drops in cases with a low bacterial load. It can only be applied to sputum samples, and it is quite difficult for some patients to produce an adequate amount of sputum. We evaluated the clinical validity of an oral swab-based microfluidic system, i.e., the SLIM assay. The SLIM assay showed a significantly higher sensitivity than the Xpert assay, especially in smear-negative TB cases. This non-sputum-based SLIM assay can be a useful diagnostic test by overcoming the limitations of conventional sputum-based tests in pulmonary TB.
KEYWORDS: tuberculosis, rapid diagnosis, oral swab, gene-based diagnosis
INTRODUCTION
Tuberculosis (TB) caused by infection with Mycobacterium tuberculosis (MTB) still ranks as a leading cause of death worldwide (1). Rapid and accurate diagnosis of pulmonary tuberculosis (PTB) in its early stage is vital for the successful control of the transmission of TB and for improving the treatment outcomes. In 2011, the World Health Organization (WHO) endorsed the use of the Xpert MTB/RIF assay (Xpert; Cepheid, Sunnyvale, CA, USA), a novel, rapid, automated, cartridge-based real-time PCR (PCR) method (2), for initial diagnosis of patients suspected of active PTB (3). In the landmark study, the Xpert assay showed a high sensitivity of 98.2% in acid-fast bacilli (AFB) smear-positive TB cases. However, the sensitivity was as low as 72.5% in smear-negative cases (4), and data from real-world settings reported a sensitivity of only around 60% to 74% (5, 6). Indeed, the sensitivity of Xpert for TB detection is inadequate when only a few bacilli are present in a clinical specimen.
The more recently developed Xpert-MTB/RIF Ultra assay showed a superior sensitivity to the Xpert assay (63% versus 46%) for diagnosing smear-negative PTB (7), but it was still not high enough. In addition, the Xpert assays can only be applied to sputum samples, which are occasionally hard to acquire from young children and asymptomatic patients with paucibacillary diseases. Furthermore, sputum collection is prone to producing potentially infectious aerosols that present a hazard for health care workers and fellow patients (8).
To overcome these limitations of the current TB diagnostics, we have developed a new assay for diagnosing PTB that involves simple and rapid pathogen enrichment by homobifunctional imidoesters (HIs) using a microfluidic system followed by conventional MTB PCR, i.e., the SLIM assay (9). The SLIM assay showed significantly better performance over the Xpert assay in terms of sensitivity (60%; 95% confidence interval [CI]: [47% to 72%] versus 37% [95% CI: 25% to 50%], P = 0.001) in the diagnosis of pulmonary TB (PTB) using sputum samples without a significant decrease in specificity (10).
To expand the clinical applicability of the SLIM assay, we investigated its performance for the diagnosis of PTB from oral swab samples. Specifically, this real-world, practice-based study was performed in patients with clinically suspected PTB in a country with an intermediate burden of TB and a low burden of the human immunodeficiency virus (HIV).
RESULTS
Participants.
A total of 272 patients suspected of PTB were enrolled in this study. The patients’ mean age was 58.8 ± 15.2 years and 174 (64.0%) were male. Malignant diseases (34.6%) and diabetes mellitus (19.9%) were the most common underlying diseases, followed by transplant status (4.8%) and liver cirrhosis (4.4%). Only one (0.4%) patient had an HIV infection (Table 1). A total of 52 patients (19.9%) had a history of previous pulmonary TB. Cough was the most common symptom (25%) Almost half of the participants (49.3%) had no specific respiratory symptoms and only had radiographic abnormalities.
TABLE 1.
Baseline characteristics of the participants
|
Characteristic |
Total | Treated as TB | Not TB | P value |
|---|---|---|---|---|
| (n = 272) | (n = 128) | (n = 144) | ||
| Age, yrs ± SDa | 58.8 ± 15.2 | 56.4 ± 16.0 | 60.9 ± 14.1 | 0.014 |
| Male sex, n (%) | 174 (64.0) | 85 (66.4) | 89 (61.8) | 0.43 |
| Symptoms, n (%) | ||||
| Cough | 68 (25.0) | 29 (22.7) | 39 (27.1) | 0.4 |
| Sputum | 47 (17.3) | 17 (13.3) | 30 (20.8) | 0.1 |
| Hemoptysis | 12 (4.4) | 4 (3.1) | 8 (5.6) | 0.33 |
| Fever | 28 (10.3) | 12 (9.4) | 16 (11.1) | 0.64 |
| Night sweat | 6 (2.2) | 2 (1.6) | 4 (2.8) | 0.69 |
| Dyspnea | 15 (5.5) | 7 (5.5) | 8 (5.6) | 0.98 |
| Chest pain | 8 (2.9) | 7 (5.5) | 9 (6.3) | 0.78 |
| General weakness | 16 (5.9) | 2 (1.6) | 6 (4.2) | 0.29 |
| Chest radiograph abnormality, only | 134 (49.3) | 56 (43.8) | 78 (54.2) | 0.09 |
| Previous TB history, n (%) | 52 (19.9) | 21 (16.4) | 31 (21.5) | 0.28 |
| AFB smear positive, n (%) | 46 (16.9) | 35 (27.3) | 11 (7.6) | <0.0001 |
| Mycobacterial culture, TB isolated, n (%) | 93 (34.2) | 93 (72.7) | 0 (0) | <0.0001 |
| IGRA, n (%) | 89 (49.2) | 45 (68.2) | 44 (38.3) | 0.0001 |
| Underlying disease, n (%) | ||||
| Malignant disease | 94 (34.6) | 31 (24.2) | 63 (43.8) | 0.001 |
| Diabetes mellitus | 54 (19.9) | 24 (18.8) | 30 (20.8) | 0.67 |
| Transplant recipient | 13 (4.8) | 8 (6.3) | 5 (3.5) | 0.28 |
| Liver cirrhosis | 12 (4.4) | 5 (3.9) | 7 (4.9) | 0.7 |
| Gastrectomy | 7 (2.6) | 1 (0.8) | 6 (4.2) | 0.13 |
| Rheumatoid disease | 3 (1.1) | 0 (0.0) | 3 (2.1) | 0.25 |
| HIV infection | 1 (0.4) | 1 (0.8) | 0 (0.0) | 0.47 |
SD, standard deviation; HIV, human immunodeficiency virus; TB, tuberculosis; IGRA, interferon-gamma release assay.
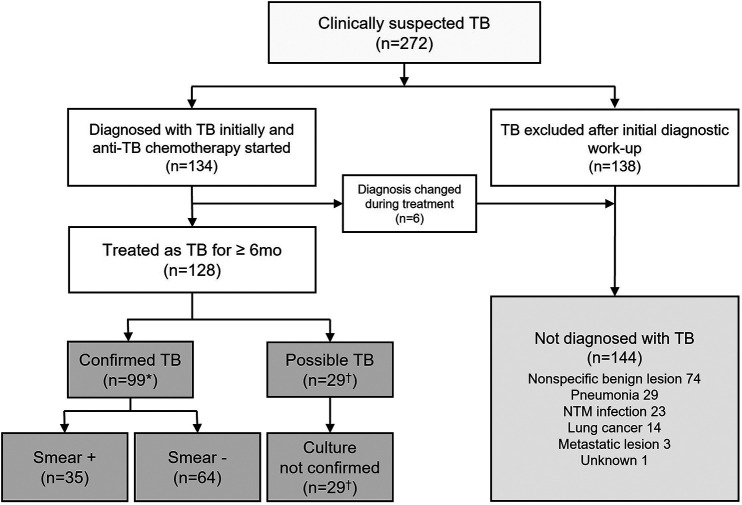
A total of 128 patients were finally treated as TB cases and planned to take full-course chemotherapy. They were categorized into smear-positive confirmed TB (n = 35), smear-negative confirmed TB (n = 64), and possible TB (n = 29). Among them, one participant (male, 57 years old) died of an underlying disease (neuroendocrine tumor with liver metastasis) after 3 months of TB treatment and could not complete the treatment. However, he was grouped as smear-negative culture-negative clinical TB because he had a good and persistent response to the TB treatment. The remaining 144 patients did not meet the criteria of TB diagnosis according to the study definition (Fig. 1).
FIG 1.
Diagnostic flow of the study patients. Among 272 patients with clinically suspected TB, 128 were finally treated for TB by respiratory or infection specialists who were blinded to the results of the SLIM oral swab assay. Confirmed TB was defined as culture-positive TB patients with at least one positive culture result for MTB from their sputum. Possible TB was defined as culture-negative TB patients with a high clinical likelihood of active TB and a negative mycobacterial culture finding in three or more sputum examinations, but with good clinical and radiographic responses to anti-TB treatment during follow-up without any evidence of an alternative diagnosis. *, six participants with TB not isolated in sputum but isolated from bronchial washing fluid were included. †, one participant who could not produce sputum was included. NTM, nontuberculous Mycobacterium.
Clinical validity of the assays.
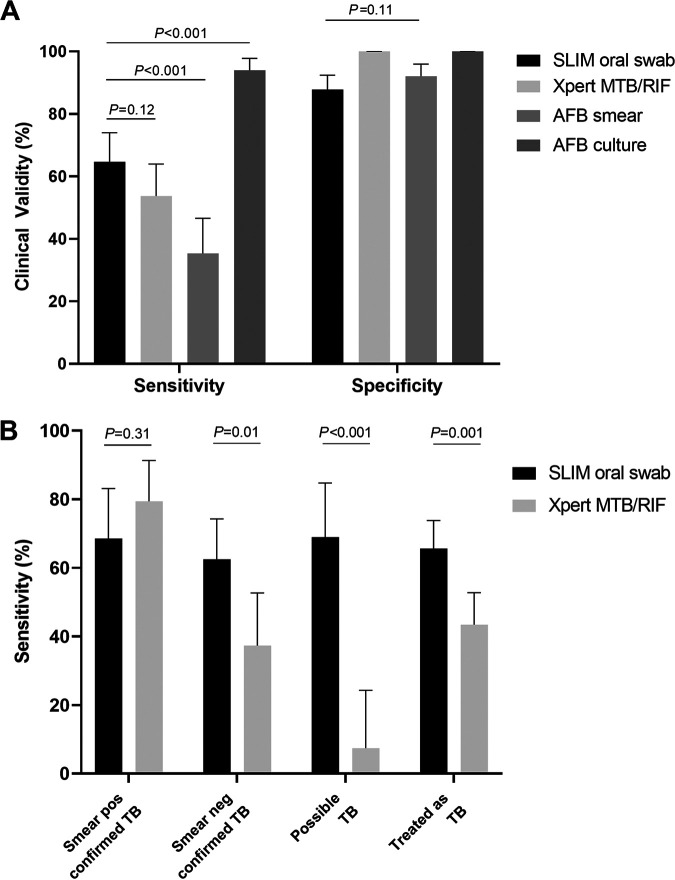
The results of the SLIM oral swab assay and the Xpert assay according to the clinical diagnosis stratified by the AFB smear and MTB culture results are in Table 2, and their diagnostic performances compared with the other tests are in Fig. 2 and Table 3. For confirmed TB, the sensitivity of the SLIM oral swab, Xpert, AFB smear and culture for TB were 64.7% (64/99; 95% CI: 54.4 to 73.8%), 53.7% (51/95; 95% CI: 43.2 to 64.0%), 35.4% (35/99; 95% CI: 26.0 to 46.6%) and 93.9% (93/99; 95% CI: 82.3 to 97.7%), respectively. The sensitivity of the SLIM oral swab assay was higher than that of the Xpert assay, but it did not reach statistical significance (P = 0.12). The specificity of the SLIM oral swab, Xpert, AFB smear, and MTB culture were 86.1% (124/144; 95% CI: 86.1 to 91.3%), 100% (130/130; 95% CI: 97.2 to 100.0%), 92.0% (127/138; 95% CI: 86.2 to 96.0%), and 100% (138/138; 95% CI: 97.4 to 100.0%), respectively.
TABLE 2.
Comparison of sputum exam results according to the categories of TB
| SLIM oral swab (n = 272) |
Xpert MTB/RIF (n = 252a) |
|||||
|---|---|---|---|---|---|---|
| Case definition | Positive | Negative | Total | Positive | Negative | Total |
| Confirmed TB | ||||||
| Smear positive | 24 | 11 | 35 | 27 | 7 | 34 |
| Smear negative | 40 | 24 | 64 | 24 | 37 | 61 |
| Possible TB | ||||||
| Culture negative | 20b | 9 | 29 | 2 | 25 | 27 |
| Not TB | 20c | 124 | 144 | 0 | 130 | 130 |
The results of Xpert MTB/RIF were not available for 20 patients.
One patient without a sputum exam was included. See Fig. 2 for more details.
See Table S1 for more details. Xpert MTB/RIF was not available for one participant.
FIG 2.
Clinical validity of the SLIM oral swab assay and the Xpert MTB/RIF assay for the diagnosis of TB. (A) Sensitivity and specificity of the five different types of assays for the diagnosis of confirmed TB. (B) Sensitivity of the SLIM oral swab assay and Xpert MTB/RIF according to the categories of TB.
TABLE 3.
Diagnostic performance of the TB assays according to the categories of TB
| Case definition | Sensitivity % (n/N, 95% CI) |
Specificity % (n/N, 95% CI) |
PPVa % (n/N, 95% CI) |
NPV % (n/N, 95% CI) |
Positive likelihood Ratio (95% CI) |
Negative likelihood Ratio (95% CI) |
|---|---|---|---|---|---|---|
| Confirmed TB (n = 99b)versus not TB (n = 144) | ||||||
| SLIM oral swab | 65 (64/99, 54–74) | 86 (124/144, 79–91) | 76 (64/84, 68–83) | 78 (124/159, 73–82) | 4.66 (3.02–7.17) | 0.41 (0.31–0.54) |
| Xpert MTB/RIF | 54 (51/95, 43–64) | 100 (130/130, 97–100) | 100 (51/51) | 75 (130/174, 70–79) | Not applicable | 0.46 (0.37–0.58) |
| AFB smear | 35 (35/99, 26–46) | 92 (127/138, 86–96) | 76 (35/46, 63–86) | 66 (127/191, 63–70) | 4.44 (2.37–8.30) | 0.70 (0.60–0.82) |
| MTB culture | 94 (93/99, 87–98) | 100 (138/138, 97–100) | 100 (93/93) | 96 (138/144, 91–98) | Not applicable | 0.06 (0.03–0.13) |
| Possible TB (n = 29) versus not TB (n = 144) | ||||||
| SLIM oral swab | 69 (20/29, 49–85) | 86 (124/144, 79–91) | 50 (20/40, 38–62) | 93 (124/133, 89–96) | 4.978 (3.09–7.98) | 0.36 (0.21–0.62) |
| Xpert MTB/RIF | 7 (2/27, 1–24) | 100 (130/130, 97–100) | 100 (2/2) | 84 (130/155, 82–85) | Not applicable | 0.93 (0.83–1.03) |
| AFB smear | 0 (0/28c, 0–12) | 92 (127/138, 86–96) | 0 (0/11) | 82 (127/155, 81–83) | 0.00 | 1.09 (1.04–1.14) |
| MTB culture | 0 (0/28c, 0–12) | 100 (138/138, 97–100) | Not applicable | 83 (138/166, 83–83) | Not applicable | 1.00 (1.00–1.00) |
PPV, positive predictive value; NPV, negative predictive value; CI, confidence interval; AFB, acid-fast bacilli; MTB, Mycobacterium tuberculosis.
Six participants with TB not isolated in sputum but isolated from bronchial washing fluid were included.
One participant without a sputum exam was included. See Fig. 2 for more details.
The sensitivity of the SLIM oral swab assay and the Xpert assay were further analyzed according to the four categories of TB: smear-positive confirmed, smear-negative confirmed, possible, and treated as TB (Fig. 2B). The sensitivity of the SLIM oral swab assay was not significantly different according to the TB categories and ranged from 62.5% to 69.0% (P = 0.55). In contrast, the sensitivity of the Xpert assay was the highest in smear-positive confirmed TB (79.4%) and was significantly lower in smear-negative confirmed TB (37.3%, P = 0.0002) and possible TB (7.4%, P < 0.0001). As such, whereas the two assays did not show significant differences in sensitivity for smear-positive PTB (P = 0.31), the SLIM oral swab assay had significantly higher sensitivity than the Xpert assay in both smear-negative confirmed TB (P = 0.01) and possible TB (P < 0.0001). The sensitivity of the SLIM oral swab relative to Xpert was significantly superior to that of Xpert relative to the SLIM oral swab (P = 0.039, Table S1).
A combination of the SLIM oral swab assay and Xpert assay was evaluated for its clinical usefulness. The SLIM oral swab found 31 additional patients with confirmed TB and 18 with possible TB. Accordingly, this combination showed a sensitivity of 86.3% (95% CI: 77.7% to 92.5%) and specificity of 85.4% (95% CI: 78.1% to 91.0%; Table S2).
Clinical characteristics of the patients with false-positive results on SLIM oral swab.
A total of 20 patients with positive SLIM oral swab results and the presence of MTB DNA confirmed by Sanger sequencing were not finally diagnosed with PTB according to the study definition. Among them, 10 (50.0%) patients had an inflammatory scar on the chest CT suspected of old inactive TB, and six of them had a history of TB treatment. Another five (25.0%) patients had lesions suspected of active PTB. However, they were regularly followed up without treatment because the physician considered that the clinical evidence for treatment was not sufficient. Three (15.0%) patients had pneumonic infiltration and were treated with broad-spectrum antibiotics, and the other two (10.0%) were diagnosed with NTM pulmonary disease. The detailed characteristics and representative chest images of these patients are provided in the online supplement (Table S3 and Fig. S2).
DISCUSSION
In this real-world practice setting study, we showed that the SLIM oral swab assay, a non-sputum-based diagnostic test, can detect PTB with high sensitivity, comparable to conventional sputum-based tests, such as mycobacterial culture and Xpert. The superiority of the SLIM assay was particularly pronounced for smear-negative PTB, especially culture-negative clinical PTB, which are cases with a low bacterial load.
The development of a rapid, accessible, and highly sensitive diagnostic tool is a major challenge in the control and management of TB. Among a total of 10 million new TB cases worldwide in 2019, as many as 2.9 million cases were estimated to have been not diagnosed or detected (1), which may be the main source of its transmission and morbidity. As an effort to overcome this diagnostic gap, the WHO recommended the use of the Xpert assay as the initial test for TB (11). However, the sensitivity of the Xpert assay is not high enough for paucibacillary TB cases (4, 12), and is thus limited for use in smear-negative TB cases that require more sensitive diagnostic methods. To meet this unmet clinical need, we applied the SLIM assay, a new generation pathogen enriching technique, and demonstrated its efficacy in PTB and other infectious diseases (9, 10, 13).
In conventional assays for DNA extraction, only a small volume (between 100 μL and 200 μL) of clinical samples is used for the detection of pathogens due to the capacity of the assays; in contrast, the SLIM assay can use both small volumes (between 100 μL and 1 mL) and large volume (more than 1 mL) samples by enabling simultaneous concentration and extraction of the pathogens in a single system. Due to this advantage, the sensitivity of the SLIM system for pathogen diagnosis is significantly higher than that of conventional assays (9, 10, 13).
In our previous study, SLIM assays with 2 mL sputum had a higher sensitivity than the Xpert assay for the diagnosis of culture-positive pulmonary TB (57% [95% CI: 39% to 73%] Xpert versus 91% [95% CI: 78% to 97%]) (SLIM 2 mL) (10). In addition, the SLIM system can minimize the time (<50 min for pathogen enrichment and DNA extraction), cost ($5 to $6), instrument requirements (centrifuges and vortexes), and additional reagents (antibodies) for sample processing considering the material required (9, 10).
Easier, safer, and more effective sampling methods are essential in TB diagnosis (14). Many patients struggle to produce an adequate amount of sputum for testing. Non-sputum-based samples, such as saliva, urine, blood, and exhaled breath concentrate, have been tested, but these samples are typically less useful than sputum (15–17). Recent studies have suggested the use of oral swab samples, which can easily be obtained through noninvasive, non-aerosol-producing methods. Previous studies have shown that MTB DNA can be detected in oral swabs from human and nonhuman primates (18–20). Wood et al. (21) reported 90% sensitivity of oral swab samples, although the number of participants was small, and more than half of them (60%) were smear-positive. Luabeya et al. (22) reported that oral swab samples had 92.8% sensitivity and 91.5% specificity. These two studies showed promising results, but both used two swabs instead of one and included participants with chronic respiratory symptoms, which might increase the sensitivity of those who could be distinguished as TB.
In our study, the SLIM assay was applied to detect PTB using a single oral swab sample per patient suspected of TB. This new method overcame the main limitation of the currently available diagnostics. It increased the sensitivity, which is relatively low in Xpert in cases of smear-negative PTB, and it showed the potential usefulness of non-sputum-based assays, whose efficacies are comparable or even superior to conventional sputum-based techniques. Non-sputum-based assays will be especially helpful in mass-screening large groups of people (e.g., school, prison, military base), in which obtaining sputum may be difficult, such as from children or those without symptoms (23, 24). It will also be useful in the setting of a TB outbreak investigation, in which many asymptomatic active cases may be present (25). The non-sputum-based sensitive examination will reduce the unnecessary spread of sputum production during mass screening by preselecting those few individuals who need a sputum exam.
There is also an unmet clinical need for diagnosing extrapulmonary TB and PTB in patients who cannot produce an adequate amount of sputum. The sensitivity of Xpert in the diagnosis of PTB is very low when using samples other than sputa, such as exhaled breath condensate (0%) and saliva (38.5%) (15). For other clinical samples, such as pleural fluid from TB pleurisy, the sensitivity was quite low at 30% (26). Other techniques showed the possibility of the detection of MTB DNA from plasma (27). Thus, further validation of the SLIM assay with various types of clinical samples and additional development of the system with automation can widen its clinical utility. We expect that our ongoing studies on the application of the SLIM assay to various specimens, such as cerebrospinal fluid (CSF), blood, and oral swabs will be able to provide useful data on this issue. These strengths, its high sensitivity, and application to various samples indicate the future direction and potential uses of the SLIM assay in TB diagnosis. The SLIM assay can increase the sensitivity and have a validated specificity by integrating with the Xpert platform. It can also be used as a rapid diagnostic for TB suspected patients without sputum. A more simplified method with automation will be a critical step to increase its utility independent of an extensive laboratory facility.
This study and the SLIM assay still have some limitations to overcome. First, the false-positive results should be further controlled. The reasons for this were not determined in this study. It may be related to the high sensitivity of the new method, the detection of remnant bacilli or DNA from previous infections, contamination of samples during collection or analysis, or combinations of these things. Further work is needed to discern these possibilities. As Xpert is a closed cartridge diagnostic system that uses automated PCR testing to detect TB with high specificity, the development of a closed SLIM assay may be a key factor in increasing the specificity of the SLIM assay for TB. The development of the automated SLIM system would reduce the problems that result from uncontrollable contaminants by minimizing the external exposure of the sample. Because the microfluidic chip of the SLIM assay is small and requires only an input system that can inject samples, buffers, and air, it can be easily combined with an automated detection system, such as Xpert, which does not have a pathogen concentration system, and it would be able to compensate for the shortcomings of low sensitivity.
Second, a comparison with the Xpert MTB/RIF Ultra assay, which was not available during the initiation step of the study period, should be carried out. Third, the sampling techniques for oral swabs were not compared because this study was targeted to examine the clinical utility of using a single sample in a real practice setting. The swabbing sites, times, and collection kit can affect the results (28, 29), and further study can elucidate if these factors affect the results, especially in this assay designed to increase the sensitivity.
Fourth, the sensitivity of Xpert in this study was relatively low compared with other studies (30, 31). The inclusion of a small number of smear-positive patients and many asymptomatic patients (49.3%) might be the reason. This study was performed in a metropolitan city where medical facilities are easy to access, and regular health checkup services are quite active. Therefore, this study setting makes it difficult to extrapolate our findings to countries with a high burden of TB, such as South Africa. The disease prevalence can affect the positive or negative predictive value (32). The utility of diagnostics should be considered in various clinical and geographical settings.
In conclusion, the sensitivity of the oral swab-based SLIM assay for the diagnosis of PTB was comparable to that of conventional sputum-based methods. The superiority of the SLIM assay in terms of sensitivity was more pronounced in cases of smear-negative PTB, for which tests with a higher sensitivity are critically needed. Further studies on the application of automation and the reduction of false-positive results will expand the role of the SLIM assay in various clinical settings by using both sputum and non-sputum-based samples.
MATERIALS AND METHODS
Participants.
Adult patients (>18 years of age) who were clinically suspected of active PTB were prospectively enrolled in two tertiary university-affiliated hospitals in Seoul, Republic of Korea (Asan Medical Center and Severance Hospital) from May 2019 to October 2020. The suspicion of PTB was based on the participants’ symptoms, history, and radiographic findings suggestive of TB (33, 34), and the enrollment was decided by three respiratory and infection specialists (SHK, YAK, and SWL) who each had experience in TB treatment for more than 15 years. Patients who could not understand the study design or the instructions for the sputum exam were excluded.
Study design and case definition.
After receiving informed consent from each patient and before starting the treatment, two trained researchers (YC and YAK) performed oral swabs using the OMNIgene.ORAL OMR-110 kit (DNA Genotek, Ottawa, Canada) according to the manufacturer’s instructions. In brief, the swabs were brushed in a back-and-forth motion along the participants’ palate, upper gum line, and tongue dorsum for about 10 s (5 to 6 times for each site, a total of 20 times), taking care not to reach back into the mouth. The swab was then inserted into a tube containing stabilizing liquid and the samples were immediately sent to the laboratory and kept at −80°C until analysis. All other steps were performed according to the routine practice of clinically suspected TB for these enrolled participants. Acid-fast bacilli (AFB) smears and mycobacterial cultures (both liquid and solid culture) were examined at least two times and the Xpert assay was performed according to the routine practice. AFB smear and cultures were examined by fluorochrome staining using auramine-rhodamine and culturing in a 3% Ogawa medium and mycobacteria growth-indicator tube medium (MGIT; Becton Dickson, NJ, USA) (34). If the patients could not produce a sufficient sputum sample, sputum induction with 3% normal saline nebulization was performed. The sputum samples were collected after taking the oral swab samples at the same visit from each patient.
TB cases were defined as those treated with anti-TB chemotherapy for at least 6 months according to the American Thoracic Society (ATS), Infectious Disease Society of America (IDSA), and Korean guidelines at the discretion of the respiratory and infectious specialists (SHK, YAK, and SWL) (34, 35) and the fulfillment of full-term treatment. ‘Confirmed TB’ was defined as culture-positive TB patients with at least one positive culture result for MTB from their sputum. Confirmed TB patients were considered smear-positive if they had at least one positive smear result (inclusive of scanty positive smears). ‘Possible TB’ was defined as culture-negative TB patients with a high clinical likelihood of active TB and good clinical and radiographic responses to anti-TB treatment during follow-up without any evidence of an alternative diagnosis, but the mycobacterial culture was not confirmed at the initial and follow-up sputum examinations. The specialists who decided on the TB treatments were blinded to the results of the SLIM assay. We intended to enroll more than 270 participants according to the estimation of the difference in sensitivity of 0.15 (36), a variance of 0.35, 95% CI, desired power of 0.8, and exclusion of 10% in the final analysis. The institutional review boards of Asan Medical Center (2018-0020) and Severance Hospital (4-2018-0029) approved this study, and the protocol of this study was registered at clinicaltrials.gov (NCT03423550).
Oral sample analysis.
Oral swabs were used for the SLIM assay (SLIM oral swab), and Fig. S1 depicts the overall workflow of the SLIM assay. The principle and the detailed structure of the SLIM assay have been described previously (9, 10, 13, 37). Briefly, the SLIM assay is based on a combination of a microfluidic platform with low-cost thin film and homobifunctional imidoesters (HIs) reagents for MTB enrichment and DNA extraction from the oral swab samples. HIs have two imido ester groups and act as cross-linkers. The imido ester groups of HI form an amidine bond with an amine group on the surface of the thin film. The remaining imido ester groups enable enrichment of MTB cells and extraction of MTB DNA based on electrostatic interactions with negatively charged MTB cells and DNA due to the positive charge of HIs and covalent bonding with the amine group of fragmented DNA due to the imido ester groups of HIs. The detailed processing techniques were described in the online supplement and all experiments were performed by a researcher (BK) blinded to the final diagnosis.
The extracted DNA was used to detect the IS6110 transposase and catalase-peroxidase (KatG) gene. The amplified DNA product from conventional PCR was purified, and TB was confirmed using Sanger sequencing. Diagnosis of TB was carried out according to the schematic flow. All conventional PCR (endpoint PCR) reactions were performed using the extracted DNA as a template from 272 oral swab samples, and all results were reported as “positive” or “negative” to determine TB (Fig. S3 and S4). Based on these results, the sensitivity and specificity were calculated as described in the next section.
Statistical analysis.
Baseline characteristics were compared by Student's t test for continuous variables or chi-square tests for categorical variables. To compare the clinical validity between tests, McNemar’s chi-square test was used. All data are expressed as mean ± standard deviation unless noted otherwise. All statistical analyses were performed using IBM SPSS Statistics for Windows, version 21.0 (IBM Corp., Armonk, NY, USA).
Data availability.
Individual participant data collected during the study, after deidentification, and the study protocols and statistical analysis code are available beginning 3 months and ending 2 years following article publication to researchers who provide a methodological sound proposal, with approval by an independent review committee. Data are available for analysis to achieve the aims stated in an approved proposal. Proposals should be directed to seiwon@amc.seoul.kr. To gain access, data requestors will need to sign a data access or material transfer agreement approved by Asan Medical Center and Severance Hospital.
ACKNOWLEDGMENTS
This work was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (KMDF_PR_20200901_0049), grants from the Asan Institute for Life Sciences (2019-7043) and the Ministry of Science, ICT, Future Planning (MSIP) through the National Research Foundation of Korea (NRF) (2020R1A2C2007148) and a grant of the Korea Health Promotion R&D Project, funded by the Ministry of Health & Welfare, Republic of Korea (HS21C0096). The funding organizations played no role in the design of the study, choice of enrolled patients, review, and interpretation of the data, preparation of the manuscript, or final approval of the manuscript.
We thank the patients and their families who were willing to participate in this study. We also appreciate the staff at Asan Medical Center and Severance Hospital who were involved in this study.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Yong Shin, Email: shinyongno1@yonsei.ac.kr.
Sei Won Lee, Email: seiwon@amc.seoul.kr.
Amit Singh, Indian Institute of Science Bangalore.
REFERENCES
- 1.World Health Organization. 2020. Global tuberculosis report 2020. https://www.who.int/tb/publications/global_report/en/. Accessed October 9, 2021.
- 2.World Health Organization. 2011. Policy statement: automated real-time nucleic acid amplification technology for rapid and simultaneous detection of tuberculosis and rifampicin resistance: Xpert MTB/RIF system. World Health Organization, Geneva, Switzerland. https://www.ncbi.nlm.nih.gov/books/NBK304235/. [PubMed] [Google Scholar]
- 3.World Health Organization. 2016. WHO treatment guidelines for drug-resistant tuberculosis, 2016 update. World Health Organization, Geneva, Switzerland. https://www.ncbi.nlm.nih.gov/books/NBK390455/. [Google Scholar]
- 4.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med 363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HS, Kee SJ, Shin JH, Kwon YS, Chun S, Lee JH, Won EJ, Choi HJ, Kim SH, Shin MG, Shin JH, Suh SP. 2019. Xpert MTB/RIF assay as a substitute for smear microscopy in an intermediate-burden setting. Am J Respir Crit Care Med 199:784–794. doi: 10.1164/rccm.201804-0654OC. [DOI] [PubMed] [Google Scholar]
- 6.Steingart KR, Schiller I, Horne DJ, Pai M, Boehme CC, Dendukuri N. 2014. Xpert MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev 1:CD009593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorman SE, Schumacher SG, Alland D, Nabeta P, Armstrong DT, King B, Hall SL, Chakravorty S, Cirillo DM, Tukvadze N, Bablishvili N, Stevens W, Scott L, Rodrigues C, Kazi MI, Joloba M, Nakiyingi L, Nicol MP, Ghebrekristos Y, Anyango I, Murithi W, Dietze R, Lyrio Peres R, Skrahina A, Auchynka V, Chopra KK, Hanif M, Liu X, Yuan X, Boehme CC, Ellner JJ, Denkinger CM, study team. 2018. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect Dis 18:76–84. doi: 10.1016/S1473-3099(17)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins CH, Grange JM. 1999. Tuberculosis acquired in laboratories and necropsy rooms. Commun Dis Public Health 2:161–167. [PubMed] [Google Scholar]
- 9.Jin CE, Koo B, Lee EY, Kim JY, Kim SH, Shin Y. 2018. Simple and label-free pathogen enrichment via homobifunctional imidoesters using a microfluidic (SLIM) system for ultrasensitive pathogen detection in various clinical specimens. Biosens Bioelectron 111:66–73. doi: 10.1016/j.bios.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SW, Kang YA, Jin CE, Kim HC, Noh GS, Lee HJ, Park JH, Koo YS, Shin Y, Kim SH. 2020. Gene-based diagnosis of tuberculosis with a new-generation pathogen enrichment technique. Eur Respir J 55:1901885. doi: 10.1183/13993003.01885-2019. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization. 2013. Automated real-time nucleic acid amplification technology for rapid and simultaneous detection of tuberculosis and rifampicin resistance: Xpert MTB/RIF assay for the diagnosis of pulmonary and extrapulmonary TB in adults and children: policy update. World Health Organization, Geneva, Switzerland. https://apps.who.int/iris/handle/10665/112472. [PubMed] [Google Scholar]
- 12.Boehme CC, Nicol MP, Nabeta P, Michael JS, Gotuzzo E, Tahirli R, Gler MT, Blakemore R, Worodria W, Gray C, Huang L, Caceres T, Mehdiyev R, Raymond L, Whitelaw A, Sagadevan K, Alexander H, Albert H, Cobelens F, Cox H, Alland D, Perkins MD. 2011. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet 377:1495–1505. doi: 10.1016/S0140-6736(11)60438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park JH, Jin CE, Koo B, Kwon JS, Cha HH, Kim JY, Noh GS, Koo YS, Jeon SB, Lee SA, Shin Y, Kim SH. 2019. A simple microfluidic assay for diagnosing tuberculous meningitis in HIV-uninfected patients. J Clin Microbiol 57:e01975-18. doi: 10.1128/JCM.01975-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fauci AS, Eisinger RW. 2018. Reimagining the research approach to tuberculosis. Am J Trop Med Hyg 98:650–652. doi: 10.4269/ajtmh.17-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shenai S, Amisano D, Ronacher K, Kriel M, Banada PP, Song T, Lee M, Joh JS, Winter J, Thayer R, Via LE, Kim S, Barry CE, 3rd, Walzl G, Alland D. 2013. Exploring alternative biomaterials for diagnosis of pulmonary tuberculosis in HIV-negative patients by use of the GeneXpert MTB/RIF assay. J Clin Microbiol 51:4161–4166. doi: 10.1128/JCM.01743-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicol MP, Spiers K, Workman L, Isaacs W, Munro J, Black F, Zemanay W, Zar HJ. 2013. Xpert MTB/RIF testing of stool samples for the diagnosis of pulmonary tuberculosis in children. Clin Infect Dis 57:e18-21–e21. doi: 10.1093/cid/cit230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah M, Hanrahan C, Wang ZY, Dendukuri N, Lawn SD, Denkinger CM, Steingart KR. 2016. Lateral flow urine lipoarabinomannan assay for detecting active tuberculosis in HIV-positive adults. Cochrane Database Syst Rev 5:CD011420. doi: 10.1002/14651858.CD011420.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engel GA, Wilbur AK, Westmark A, Horn D, Johnson J, Jones-Engel L. 2012. Naturally acquired Mycobacterium tuberculosis complex in laboratory pig-tailed macaques. Emerg Microbes Infect 1:e30. doi: 10.1038/emi.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilbur AK, Kubatko LS, Hurtado AM, Hill KR, Stone AC. 2007. Vitamin D receptor gene polymorphisms and susceptibility M. tuberculosis in native Paraguayans. Tuberculosis (Edinb) 87:329–337. doi: 10.1016/j.tube.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Wilbur AK, Engel GA, Rompis A, A Putra IGA, Lee B-H, Aggimarangsee N, Chalise M, Shaw E, Oh G, Schillaci MA, Jones-Engel L. 2012. From the mouths of monkeys: detection of Mycobacterium tuberculosis complex DNA from buccal swabs of synanthropic macaques. Am J Primatol 74:676–686. doi: 10.1002/ajp.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood RC, Luabeya AK, Weigel KM, Wilbur AK, Jones-Engel L, Hatherill M, Cangelosi GA. 2015. Detection of Mycobacterium tuberculosis DNA on the oral mucosa of tuberculosis patients. Sci Rep 5:8668. doi: 10.1038/srep08668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luabeya AK, Wood RC, Shenje J, Filander E, Ontong C, Mabwe S, Africa H, Nguyen FK, Olson A, Weigel KM, Jones-Engel L, Hatherill M, Cangelosi GA. 2019. Noninvasive detection of tuberculosis by oral swab analysis. J Clin Microbiol 57:e01847-18. doi: 10.1128/JCM.01847-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lima F, Santos AS, Oliveira RD, Silva CCR, Goncalves CCM, Andrews JR, Croda J. 2020. Oral swab testing by Xpert(R) MTB/RIF Ultra for mass tuberculosis screening in prisons. J Clin Tuberc Other Mycobact Dis 19:100148. doi: 10.1016/j.jctube.2020.100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicol MP, Wood RC, Workman L, Prins M, Whitman C, Ghebrekristos Y, Mbhele S, Olson A, Jones-Engel LE, Zar HJ, Cangelosi GA. 2019. Microbiological diagnosis of pulmonary tuberculosis in children by oral swab polymerase chain reaction. Sci Rep 9:10789. doi: 10.1038/s41598-019-47302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SW, Jang YS, Park CM, Kang HY, Koh WJ, Yim JJ, Jeon K. 2010. The role of chest CT scanning in TB outbreak investigation. Chest 137:1057–1064. doi: 10.1378/chest.09-1513. [DOI] [PubMed] [Google Scholar]
- 26.Huo ZY, Peng L. 2018. Is Xpert MTB/RIF appropriate for diagnosing tuberculous pleurisy with pleural fluid samples? A systematic review. BMC Infect Dis 18:284. doi: 10.1186/s12879-018-3196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Click ES, Murithi W, Ouma GS, McCarthy K, Willby M, Musau S, Alexander H, Pevzner E, Posey J, Cain KP. 2018. Detection of apparent cell-free M. tuberculosis DNA from plasma. Sci Rep 8:645. doi: 10.1038/s41598-017-17683-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deviaene M, Weigel KM, Wood RC, Luabeya AKK, Jones-Engel L, Hatherill M, Cangelosi GA. 2020. Sample adequacy controls for infectious disease diagnosis by oral swabbing. PLoS One 15:e0241542. doi: 10.1371/journal.pone.0241542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood RC, Andama A, Hermansky G, Burkot S, Asege L, Job M, Katumba D, Nakaye M, Mwebe SZ, Mulondo J, Bachman CM, Nichols KP, Le Ny AM, Ortega C, Olson RN, Weigel KM, Olson AM, Madan D, Bell D, Cattamanchi A, Worodria W, Semitala FC, Somoskovi A, Cangelosi GA, Minch KJ. 2021. Characterization of oral swab samples for diagnosis of pulmonary tuberculosis. PLoS One 16:e0251422. doi: 10.1371/journal.pone.0251422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G, Wang S, Jiang G, Yang X, Huang M, Huo F, Ma Y, Dai G, Li W, Chen X, Huang H. 2019. Xpert MTB/RIF Ultra improved the diagnosis of paucibacillary tuberculosis: a prospective cohort study. J Infect 78:311–316. doi: 10.1016/j.jinf.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Zifodya JS, Kreniske JS, Schiller I, Kohli M, Dendukuri N, Schumacher SG, Ochodo EA, Haraka F, Zwerling AA, Pai M, Steingart KR, Horne DJ. 2021. Xpert Ultra versus Xpert MTB/RIF for pulmonary tuberculosis and rifampicin resistance in adults with presumptive pulmonary tuberculosis. Cochrane Database Syst Rev 2:CD009593. doi: 10.1002/14651858.CD009593.pub5. [DOI] [PubMed] [Google Scholar]
- 32.Tenny S, Hoffman MR. 2017. Prevalence, StatPearls. www.ncbi.nlm.nih.gov/books/NBK430867/. Accessed March 26, 2022.
- 33.Fujita J, Higa F, Tateyama M. 2007. Radiological findings of mycobacterial diseases. J Infect Chemother 13:8–17. doi: 10.1007/s10156-006-0485-4. [DOI] [PubMed] [Google Scholar]
- 34.Joint Committee for the Revision of Korean Guidelines for Tuberculosis. 2020. Korean guidelines for tuberculosis, 4th ed. Korea Centers for Disease Control and Prevention, Cheongju. [Google Scholar]
- 35.Lewinsohn DM, Leonard MK, LoBue PA, Cohn DL, Daley CL, Desmond E, Keane J, Lewinsohn DA, Loeffler AM, Mazurek GH, O'Brien RJ, Pai M, Richeldi L, Salfinger M, Shinnick TM, Sterling TR, Warshauer DM, Woods GL. 2017. Official American Thoracic Society/Infectious Diseases Society of America/Centers for Disease Control and Prevention clinical practice guidelines: diagnosis of tuberculosis in adults and children. Clin Infect Dis 64:e1–e33. doi: 10.1093/cid/ciw694. [DOI] [PubMed] [Google Scholar]
- 36.Rosner B. 2015. Estimation of sample size and power for comparing two means, p 306–311. In Rosner B (ed), Fundamentals of Biostatistics, 8th ed Cengage Learning, Boston, MA, USA. [Google Scholar]
- 37.Jin CE, Koo B, Lee HJ, Park IJ, Kim SH, Shin Y. 2020. Bis(sulfosuccinimidyl)suberate-based helix-shaped microchannels as enhancers of biomolecule isolation from liquid biopsies. Anal Chem 92:11994–12001. doi: 10.1021/acs.analchem.0c02503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00207-22-s001.pdf, PDF file, 0.6 MB (639.9KB, pdf)
Data Availability Statement
Individual participant data collected during the study, after deidentification, and the study protocols and statistical analysis code are available beginning 3 months and ending 2 years following article publication to researchers who provide a methodological sound proposal, with approval by an independent review committee. Data are available for analysis to achieve the aims stated in an approved proposal. Proposals should be directed to seiwon@amc.seoul.kr. To gain access, data requestors will need to sign a data access or material transfer agreement approved by Asan Medical Center and Severance Hospital.