LETTER
The high-risk clone ST167 associated with blaNDM-5 resistance determinant is currently recognized to be a source of public health concern worldwide (1–4), since it has been identified even beyond hospital borders, in companion animals, wastewater, rivers, and wildlife (5–8).
In this work, we characterized an NDM-5-producing Escherichia coli ST167 collected in Italy from a liver sample of a 4-month-old cat who died from parvovirus hemorrhagic enteritis. The E. coli strain 167624 was tested for antibiotic susceptibility and sequenced using both Illumina and Nanopore technologies.
Bacterial identification and antibiotic susceptibility tests were performed with the semiautomated system MicroScan autoSCAN4 (Beckman Coulter); results were interpreted according to EUCAST guidelines (v10.0-2020, http://www.eucast.org). The E. coli 167624 strain showed a multidrug-resistant (MDR) profile, being resistant to all the antibiotics tested, with the exception of colistin, amikacin, and fosfomycin (Table 1).
TABLE 1.
Antimicrobial susceptibility profile of the ECO167624 strain
| Antibiotic | MICa (μg/mL) | Interpretation |
|---|---|---|
| AMK | ≤8 | S |
| AMP | >8 | R |
| AMC | >8|4 | R |
| AZT | >4 | R |
| FEP | >8 | R |
| CTX | >16 | R |
| CAZ | >8 | R |
| CIP | >1 | R |
| LEV | >1 | R |
| GNT | >4 | R |
| COL | ≤2 | S |
| FOS | ≤32 | S |
| ERT | >1 | R |
| MER | >8 | R |
| PTZ | >16 | R |
| SXT | >4|76 | R |
| PIP | >16 | R |
| TBR | >4 | R |
AMK, amikacin; AMP, ampicillin; AMC, amoxicillin/clavulanate; AZT, aztreonam; FEP, cefepime; CTX, cefotaxime; CAZ, ceftazidime; CIP, ciprofloxacin; LEV, levofloxacin; GNT, gentamicin; COL, colistin; FOS, fosfomycin; ERT, ertapenem; MER, meropenem; PTZ, piperacillin-tazobactam; SXT, trimethoprim-sulfamethoxazole; PIP, piperacillin; TBR, tobramycin; S, susceptible; R, resistant. Susceptibility results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2020) criteria.
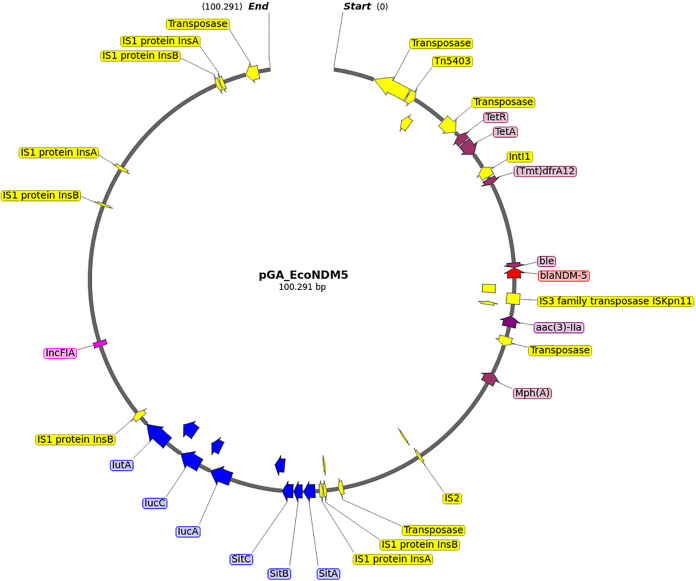
Genomic DNA was sequenced via both Oxford Nanopore MinION, with library FLO-MIN106 (rapid barcoding kit SQK-RBK004), and Illumina MiSeq platform (Nextera XT library preparation kit, with a 2 × 250 paired-end run), after extraction with DNeasy blood and tissue kit (Qiagen). A complete hybrid genome was obtained (genome size of 5,141,416 bp, chromosome sequence of 4,849,672 bp) using Unicycler v0.4.8-beta (9). A main plasmid, pGA_EcoNDM5 (size of 100,291 bp), harboring the blaNDM-5 gene was detected and annotated (Fig. 1, and see supplemental material).
FIG 1.
Graphical representation of the pGA_EcoNDM5 plasmid sequence. Colored arrows represent genes or coding regions: red, blaNDM-5 gene; purple, antimicrobial resistance genes; yellow, insertion sequences (IS) and transposons; blue, aerobactin operon and virulence genes; fuchsia, incompatibility group.
In silico multilocus sequence type (MLST) analysis showed that the strain ECO16724 belonged to the high-risk clone ST167 (MLST Achtman scheme), phylogroup A, and serotype O101:H9.
Investigation of the resistance genes content highlighted the copresence of multiple β-lactamase determinants, including the plasmid-borne blaNDM-5 and blable, as well as blaAmpH and blaAmpC1 on the chromosome. In addition, virulence factors associated with flagellar motility (Fli/Flg family), fimbriae (fimF, fimG), and siderophore (ybtT, iucA) were detected on the chromosome and on the pGA_NDM5 plasmid. Resistance determinants included blaNDM-5, blable, blaAmpH, blaAmpC1, gyrA (S83L, D87N), parC (S80 I), parE (S458A), mph(A), tet(A), tet(R), aac(3)-Ila, aadA2, sul1, and dfrA12. Virulence determinants included fliN, fliM, fliL, fliJ, fliA, flgH, flgG, flgD, flgC, flgB, fimF, fimG, ybtT, iucA, cea, capU, fyuA, gad, hra, irp2, and aerobactin operon. The pGA_EcoNDM5 belonged to the IncFIA with an identity score of 99.48%.
To place the ECO16724 isolate within the proper taxonomic context, a coreSNP phylogeny was inferred (see supplemental material). The phylogenetic analysis (Fig. 2) showed ECO167624 to be part of a clade including blaNDM-5-positive strains: four from human and dog sources in Switzerland (2017 to 2018) and one, LR880734.1, from a dog in Italy (2019).
FIG 2.
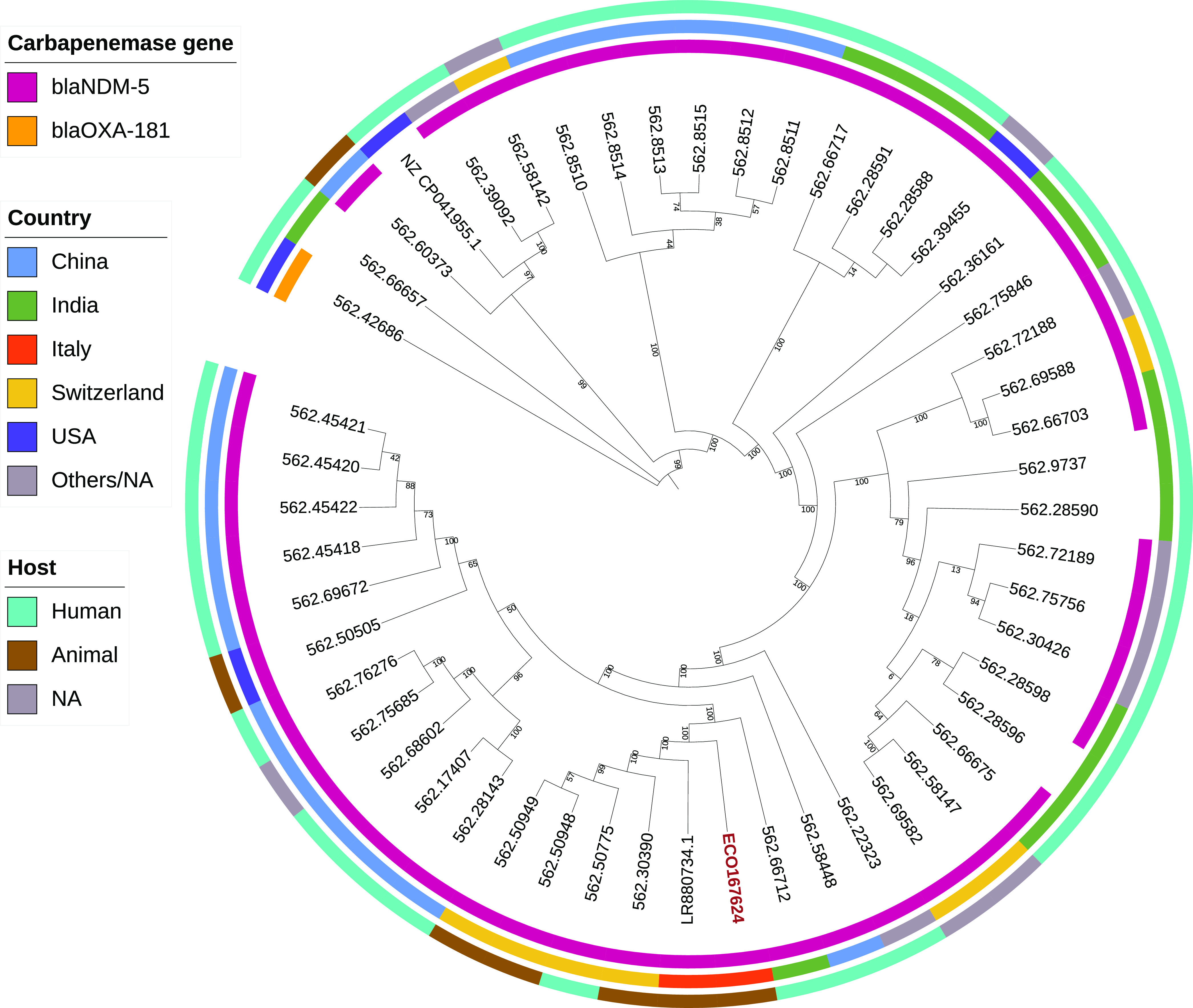
CoreSNP-based phylogeny of the 50 E. coli strains closest to ECO167624 retrieved from PATRIC database.
The comparison of the blaNDM-5 genetic environment among the plasmids of the strains within this clade highlighted a high similarity, showing the same NDM-carrying integron (Fig. S1).
Transmission between animals and humans of ST167 NDM-5-producing E. coli has been already demonstrated in a familiar context (7). Although we were not able to trace the origin of the here-presented ECO167624 strain, a human-animal transmission event could be hypothesized. In Italy, the blaNDM-5 gene is to date associated mainly with human clinical cases (1, 2), but our results raise the hypothesis that community could represent a hidden reservoir of NDM-5-producing ST167 high-risk clone.
The ability to trace rapidly the source of infection is of particular relevance in a globalized world, where the boundaries among the different settings (humans, environment, animals) are continuously crossed by bacteria. Hence, the standardization of tools and user-friendly platforms for the genomic surveillance, such as Pathogenwatch and BacWGSTdb 2.0 (10, 11), is acquiring an increasingly pivotal role.
The increased reports of MDR clones in the hospital, community, and environment surely sound like an alarm bell, suggesting the appropriateness of the “One-Health” approach.
Data availability.
The nucleotide sequence of the strain ECO167624 was submitted to NCBI with the following accession codes: BioProject ID PRJNA816063 and BioSample SAMN26656496.
ACKNOWLEDGMENT
We declare no conflicts of interest or funding sources.
Footnotes
Supplemental material is available online only.
Contributor Information
Aurora Piazza, Email: aurora.piazza@unipv.it.
Sadjia Bekal, Institut National de Santé Publique du Québec.
REFERENCES
- 1.Corbellini S, Scaltriti E, Piccinelli G, Gurrieri F, Mascherpa M, Boroni G, Amolini C, Caruso A, De Francesco MA. 2022. Genomic characterisation of Escherichia coli isolates co-producing NDM-5 and OXA-1 from hospitalised patients with invasive infections. J Glob Antimicrob Resist 28:136–139. doi: 10.1016/j.jgar.2021.12.018. [DOI] [PubMed] [Google Scholar]
- 2.Giufrè M, Errico G, Accogli M, Monaco M, Villa L, Distasi MA, Del Gaudio T, Pantosti A, Carattoli A, Cerquetti M. 2018. Emergence of NDM-5-producing Escherichia coli sequence type 167 clone in Italy. Int J Antimicrob Agents 52:76–81. doi: 10.1016/j.ijantimicag.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 3.Weingarten RA, Johnson RC, Conlan S, Ramsburg AM, Dekker JP, Lau AF, Khil P, Odom RT, Deming C, Park M, Thomas PJ, Henderson DK, Palmore TN, Segre JA, Frank KM, NISC Comparative Sequencing Program . 2018. Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio 9:e02011-17. doi: 10.1128/mBio.02011-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou H, Jia X, Liu H, Li S, Wu X, Huang S. 2020. Emergence of NDM-5-producing Escherichia coli in a teaching hospital in Chongqing, China: IncF-Type plasmids may contribute to the prevalence of blaNDM-5. Front Microbiol 11:334. doi: 10.3389/fmicb.2020.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleichenbacher S, Stevens MJA, Zurfluh K, Perreten V, Endimiani A, Stephan R, Nüesch-Inderbinen M. 2020. Environmental dissemination of carbapenemase-producing Enterobacteriaceae in rivers in Switzerland. Environ Pollut 265:115081. doi: 10.1016/j.envpol.2020.115081. [DOI] [PubMed] [Google Scholar]
- 6.Alba P, Taddei R, Cordaro G, Fontana MC, Toschi E, Gaibani P, Marani I, Giacomi A, Diaconu EL, Iurescia M, Carfora V, Franco A. 2021. Carbapenemase IncF-borne blaNDM-5 gene in the E. coli ST167 high-risk clone from canine clinical infection, Italy. Vet Microbiol 256:109045. doi: 10.1016/j.vetmic.2021.109045. [DOI] [PubMed] [Google Scholar]
- 7.Grönthal T, Österblad M, Eklund M, Jalava J, Nykäsenoja S, Pekkanen K, Rantala M. 2018. Sharing more than friendship - transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Euro Surveill 23:1700497. doi: 10.2807/1560-7917.ES.2018.23.27.1700497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterhans S, Stevens M, Nüesch-Inderbinen M, Schmitt S, Stephan R, Zurfluh K. 2018. First report of a blaNDM-5-harbouring Escherichia coli ST167 isolated from a wound infection in a dog in Switzerland. J Glob Antimicrob Resist 15:226–227. doi: 10.1016/j.jgar.2018.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argimón S, David S, Underwood A, Abrudan M, Wheeler NE, Kekre M, Abudahab K, Yeats CA, Goater R, Taylor B, Harste H, Muddyman D, Feil EJ, Brisse S, Holt K, Donado-Godoy P, Ravikumar KL, Okeke IN, Carlos C, Aanensen DM, NIHR Global Health Research Unit on Genomic Surveillance of Antimicrobial Resistance . 2021. Rapid genomic characterization and global surveillance of Klebsiella using Pathogenwatch. Clin Infect Dis 73:S325–S335. doi: 10.1093/cid/ciab784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Zou S, Chen H, Yu Y, Ruan Z. 2021. BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res 49:D644–D650. doi: 10.1093/nar/gkaa821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00832-22-s0001.pdf, PDF file, 0.5 MB (507.1KB, pdf)
Data Availability Statement
The nucleotide sequence of the strain ECO167624 was submitted to NCBI with the following accession codes: BioProject ID PRJNA816063 and BioSample SAMN26656496.