ABSTRACT
Malassezia is a fungal genus found on the skin of humans and warm-blooded animals, with 18 species reported to date. In this study, we sequenced and annotated the genome of Malassezia arunalokei, which is the most recently identified Malassezia species, and compared it with Malassezia restricta, the predominant isolate from human skin. Additionally, we reanalyzed previously reported mycobiome data sets with a species-level resolution to investigate M. arunalokei distribution within the mycobiota of human facial skin. We discovered that the M. arunalokei genome is 7.24 Mbp in size and encodes 4,117 protein-coding genes, all of which were clustered with M. restricta. We also found that the average nucleotide identity value of the M. arunalokei genome was 93.5, compared with the genomes of three M. restricta strains, including M. restricta KCTC 27527. Our findings demonstrate that they indeed belong to different species and that M. arunalokei may have experienced specific gene loss events during speciation. Furthermore, our study showed that M. arunalokei was diverged from M. restricta approximately 7.1 million years ago and indicated that M. arunalokei is the most recently diverged species in the Malassezia lineage to date. Finally, our analysis of the facial mycobiome of previously recruited cohorts revealed that M. arunalokei abundance is not associated with seborrheic dermatitis/dandruff or acne, but was revealed to be more abundant on the forehead and cheek than on the scalp.
IMPORTANCE Malassezia is the fungus predominantly residing on the human skin and causes various skin diseases, including seborrheic dermatitis and dandruff. To date, 18 species have been reported, and among them, M. restricta is the most predominant on human skin, especially on the scalp. In this study, we sequenced and analyzed the genome of M. arunalokei, which is the most recently identified Malassezia species, and compared it with M. restricta. Moreover, we analyzed the fungal microbiome to investigate the M. arunalokei distribution on human facial skin. We found that M. arunalokei may have experienced specific gene loss events during speciation. Our study also showed that M. arunalokei was diverged from M. restricta approximately 7.1 million years ago and indicated that M. arunalokei is the most recently diverged species in the Malassezia lineage. Finally, our analysis of the facial mycobiome revealed that M. arunalokei has higher relative abundance on the forehead and cheek than the scalp.
KEYWORDS: genome, Malassezia arunalokei, Malassezia restricta, mycobiome, skin fungal community, skin mycobiome
INTRODUCTION
Malassezia is a fungal genus predominantly residing on the skin of humans and warm-blooded animals. A major physiological characteristic of Malassezia is its lipid dependency, which reflects the sebum-rich environment it inhabits. The lipophilic nature of Malassezia is due to the absence of a gene encoding fatty acid synthase in the genome of most Malassezia species (1). To date, 18 species have been reported, of which 10 (M. restricta, M. globosa, M. arunalokei, M. sympodialis, M. dermatis, M. slooffiae, M. furfur, M. obtusa, M. japonica, and M. yamatoensis) are found on human skin. Among the Malassezia species, Malassezia restricta is the most predominant on human skin, especially on the scalp. It is associated with skin diseases such as seborrheic dermatitis. In addition to M. restricta, Malassezia arunalokei is of particular interest because of its high internal transcribed spacer (ITS) sequence similarity to M. restricta (2).
M. arunalokei was first identified and reported in 2016 by Honnavar et al., who originally isolated it from the scalp and nasolabial fold of patients with seborrheic dermatitis/dandruff (SD/D) and healthy individuals in northwestern India (2). Honnavar et al. showed that the appearance of M. arunalokei colonies was slightly different from those of M. restricta and that the average single-cell size of M. arunalokei was also larger than that of M. restricta, with a mean area of 5.5 versus 3.2 μm2, respectively. Additionally, they showed that M. arunalokei is catalase negative. This phenotype was previously found in M. restricta, whose genome lacks a catalase gene, but was not observed in other Malassezia species (2). A comparison of ITS sequences between M. arunalokei and M. restricta showed 6.4% variation, and a fluorescent amplified fragment length polymorphism (FAFLP) analysis showed only 25% similarity to M. restricta, underpinning that M. arunalokei is a novel species and differentiated from M. restricta (2).
In this study, we sequenced and annotated the genome of the M. arunalokei strain NCCPF 127130 (CBS 13387) and compared it with M. restricta KCTC 27527 (3, 4). Moreover, we reanalyzed previously reported amplicon sequencing data sets at a species-level resolution to investigate the distribution of M. arunalokei within facial skin mycobiota of East Asian individuals (5, 6). We also estimated the time at which M. arunalokei diverged within the Malassezia genus.
RESULTS AND DISCUSSION
Analysis of the M. arunalokei genome.
The genomic DNA of M. arunalokei NCCPF 127130 (equivalent to CBS 13387 and MTCC 12054) was extracted and sequenced using an Illumina NextSeq. We obtained 19 contigs, resulting in a genome size of 7.24 Mbp. The results of BUSCO analysis are presented in Table 1; the genome assembly is high quality in terms of completeness and contamination compared to the completely assembled genome of M. restricta KCTC 27527 (3, 4). However, there are differences in the numbers and lengths of exon and introns, which may be attributed to the use of M. restricta's transcriptome sequencing (RNA-seq) data in gene prediction, which does not fully reflect all existing exon-intron structures. Genome annotation identified 4,117 protein-coding genes and 82 tRNA genes (see Table S3 in the supplemental material). To the best of our knowledge, this is the first study to present high-quality assembly and annotation of M. arunalokei, which represents an important contribution to the study of the Malassezia genus.
TABLE 1.
Summary statistics of the M. arunalokei genome assembly in comparison with the M. restricta genome
| Parameter | Result for: |
|
|---|---|---|
| M. arunalokei | M. restricta | |
| Status | Draft | Complete |
| Total genome size (bp) | 7,247,604 | 7,330,907 |
| No. of contigs | 19 | 9 (1)a |
| N50 (bp) | 771,082 | 1,222,814 |
| GC ratio (%) | 55.6 | 55.8 (31.4)a |
| Total no. of tRNAs | 82 | 74 (24)a |
| Total no. of CDSsb | 4,117 | 4,390 (16)a |
| CDS length (avg/median) | 1,525.4/1,287.0 | 1,473.8/1,236.0 |
| Exon length (avg/median) | 1,147.9/942.0 | 990.0/711.0 |
| Intron length (avg/median) | 68.1/40.0 | 37.2/30.0 |
| No. of introns | 1,354 | 2,150 |
| No. of exons | 5,471 | 6,556 |
| No. (%) of spliced genes | 858 (20.8) | 1,194 (27.2) |
| Gene density (genes/Mb) | 568.1 | 601.1 |
| No. (%) of BUSCOs: | ||
| Complete BUSCOs | 712 (93.9) | 715 (94.3) |
| Complete and single-copy BUSCOs | 711 (93.8) | 714 (94.2) |
| Complete and duplicated BUSCOs | 1 (0.1) | 1 (0.1) |
| Fragmented BUSCOs | 6 (0.8) | 8 (1.1) |
| Missing BUSCOs | 40 (5.3) | 35 (4.6) |
The value in parentheses shows the information for the mitochondrial genome.
CDSs, coding DNA sequences.
A previous study by Honnavar et al., which was based on ITS sequencing analysis, showed that M. arunalokei is phylogenetically clustered with M. restricta (2). In their study, M. restricta type strain CBS 7877 was used, and the percentage of ITS2 sequence differences between M. arunalokei NCCPF 127130 and M. restricta CBS 7877 was 8.8%, with eight deletions and 16 substitutions. Our study showed that the identity of the entire 757-bp ITS region, amplified using the primers ITS4 and ITS5 in the M. arunalokei genome, was 97%, with eight deletions and nine substitutions and eight deletions and 11 substitutions compared with both M. restricta KCTC27527 and M. restricta CBS 7877, respectively.
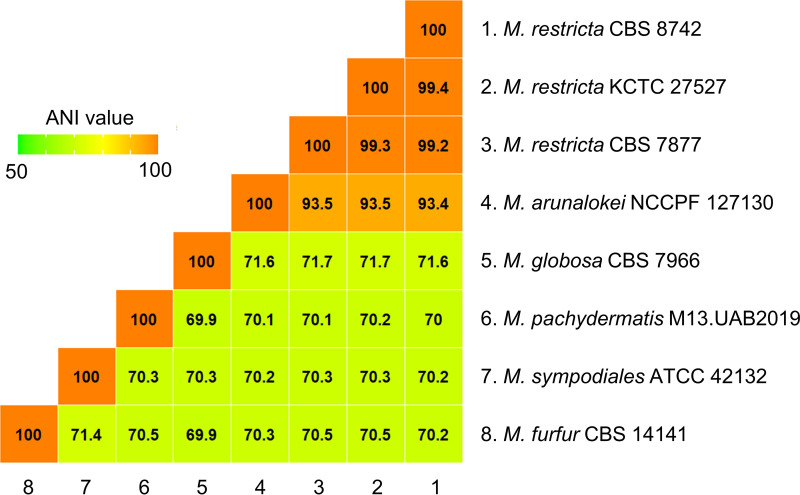
In our study, we extended the sequence comparisons between M. arunalokei and M. restricta to a genome-wide scale. The M. arunalokei NCCPF 127130 genome sequence obtained in the current study was compared with M. restricta KCTC 27527, which was previously analyzed by our group (3, 4). The results of the genome comparison showed that M. arunalokei and M. restricta possess highly conserved synteny sequence blocks (Fig. 1). Furthermore, we determined that the average nucleotide identity (ANI) value of the genome of M. arunalokei NCCPF 127130 compared with the genomes of the three M. restricta strains, including M. restricta KCTC 27527, was 93.5 indicating that they are different species (Fig. 2). The ANI value is widely used as a species boundary criterion in prokaryotes, whose genomes are generally comprised of about 10 to 20% intergenic regions (7). The intergenic and intronic sequences of eukaryotic genomes are likely to affect the calculation of the ANI value. Therefore, one may argue that evaluation of ANI values may not be applicable to the comparison of genomes of fungal organisms like the Malassezia species. However, we should note that Malassezia species have relatively small genomes compared to other fungi, and the proportions of intergenic regions in their genomes are similar to those observed in bacterial genomes. We note that intergenic regions in the Malassezia genome were included to calculate the ANI values in our study, and a similar methodology was successfully applied to analyze the evolutionary relatedness of Malassezia species in our previous study (4).
FIG 1.
MAUVE genome alignment between M. restricta KCTC 27527 and M. arunalokei NCCPF 127130. Red lines denote chromosomal or contig partition.
FIG 2.
Heat map visualization based on ANI values to find the distance among several representative species of Malassezia genus. ANI values were calculated by the ANIb method based on BLAST+ from JSpecies-WS.
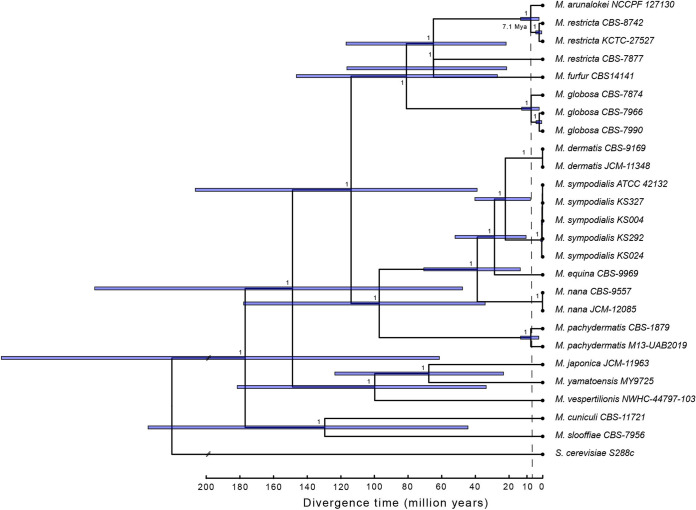
High levels of similarity between the M. arunalokei and M. restricta genomes led us to estimate divergence times within the Malassezia genus. We used seven highly conserved fungal barcoding genes to determine times of divergence, and the tree was produced using Bayesian inference (8). We found the crown age of all included Malassezia species for estimating the divergence times to be ~180 million years ago (mya), with the split between M. arunalokei NCCPF 127130 and the most closely related M. restricta strains (KCTC 27527 and CBS 8742) dating back to 7.1 mya (2.2 to ~13.1 mya) (Fig. 3). Furthermore, the results of our phylogenetic analysis indicated that M. arunalokei is the most recently diverged species in the Malassezia lineage currently known.
FIG 3.
Divergence time estimation. Shown are Bayesian divergence time estimates of Malassezia species using a node calibration for the split between M. furfur and M. restricta. Node numbers identify the posterior probability; blue bars indicate the 95% credible posterior age range. Saccharomyces cerevisiae was used as an outgroup (branch not to scale).
Previous phylogenetic analysis conducted by Honnavar et al. as well as other previous analyses using D1/D2 domains of large subunit (LSU) ribosomal DNA (rDNA) sequences showed that M. globosa, M. restricta, and M. arunalokei are closely clustered within the same clade in the phylograms (1, 2, 9), and our result was in accordance with these findings. However, we also observed two distinct subspecies groups in the M. restricta species (M. restricta CBS 8741 and KCTC 27527 versus M. restricta CBS 7877) in the phylogram we generated. The existence of subspecies groups (amplicon strain variants) of M. restricta on the human skin was also described elsewhere (5). To confirm the presence of subspecies groups in genetic levels in detail, more genomes of M. restricta strains should be fully sequenced.
Orthologous gene comparisons between M. arunalokei and M. restricta.
As indicated by the whole-genome alignment presented in Fig. 1, most M. arunalokei genes are expected to belong to M. restricta. To confirm this, we compared orthologous genes between M. arunalokei NCCPF 127130 and M. restricta KCTC 27527. Type strains of other Malassezia species, such as M. globosa strain CBS 7966, M. sympodiales ATCC 42132, and M. pachydermatis CBS 1879, the genomes of which were sequenced and annotated, were also included for more comprehensive analysis. As shown in Table 1, the genome of M. arunalokei NCCPF 127130 contains 4,117 protein-coding genes. Overall, most of orthologs in M. arunalokei were shared with M. restricta (Fig. 4A). Notably, 444 (11.4%) orthologs of M. arunalokei were shared with M. restricta only, a significantly higher proportion than the number of orthologs shared with other strains (Fig. 4B). In comparison with M. arunalokei and M. restricta alone, 214 M. restricta-specific orthologs were identified against 36 M. arunalokei-specific ones, implying that exclusive gene loss occurred in M. arunalokei during speciation (Fig. 4A; Table S4). (3, 4). We then closely examined the absent genes in the M. arunalokei genome compared with M. restricta. Gene Ontology (GO) analysis found no significant enrichment of any GO term in the categories of biological process and molecular function. However, regarding GO terms related to the cellular component category, we discovered that mitochondrial localization was the most enriched term (67 out of 214) among genes absent in the M. arunalokei genome compared with M. restricta. Although most of the deleted genes encode mitochondrial proteins, we found no defects in the growth of M. arunalokei in aerobic environments and under oxidative stress conditions (data not shown).
FIG 4.
Venn diagram of orthologous gene clusters by mcl algorithm. (A) Number of orthologs between M. restricta KCTC 27527, M. globosa CBS 7966, M. sympodiales ATCC 42132, M. pachydermatis CBS 1879, and M. arunalokei NCCPF 127130. (B) Venn diagram only showing the number of orthologs between M. restricta KCTC 27527 and M. arunalokei NCCPF 127130.
Among the genes that are absent in M. arunalokei, we paid particular attention to the ortholog of M. restricta MRET_0913, which encodes imidazole glycerol-phosphate dehydratase (IGPD) required for histidine biosynthesis. IGPD is encoded by HIS3 in the model yeast Saccharomyces cerevisiae, and deletion of the gene caused histidine auxotrophy of the fungus (10). On human skin, the proteogenic amino acid histidine plays an important role. Histidine is a natural moisturizing factor and is produced from the proteolysis of the histidine-rich protein filaggrin, which is mainly found in granular layer keratinocytes in the upper epidermis (11). Furthermore, studies have shown that histidine supplementation increases the barrier function of the skin and reduces atopic dermatitis symptoms (12, 13). Taken together, we hypothesize that the histidine-rich environment in the epidermal layer may have triggered the loss of the HIS3 homolog in the M. arunalokei lineage.
Such gene loss is consistent with regressive evolution frequently observed in parasitic organisms, which are themselves associated with hosts that show metabolic redundancy when parasitized (14). Gene loss upon environmental change or condition has also been observed, most commonly in unicellular organisms, including pathogenic bacteria and fungi. Furthermore, in some cases, gene loss influenced the pathogenicity of the organisms (14). Examples include loss of the functional BNA genes in a pathogenic yeast, Candida glabrata. The BNA genes are required for nicotinic acid (NA). Therefore, loss of the gene caused NA auxotrophy, which in turn increased expression of the EPA genes encoding a family of adhesin proteins and the pathogenicity of C. glabrata (15). However, we do not have additional evidence to determine whether gene loss in M. arunalokei, such as the HIS3 homolog, is associated with the pathogenicity of the fungus. This is mainly due to the lack of a tool for genetic manipulation.
M. arunalokei has high relative abundance on the forehead and cheek compared to the scalp.
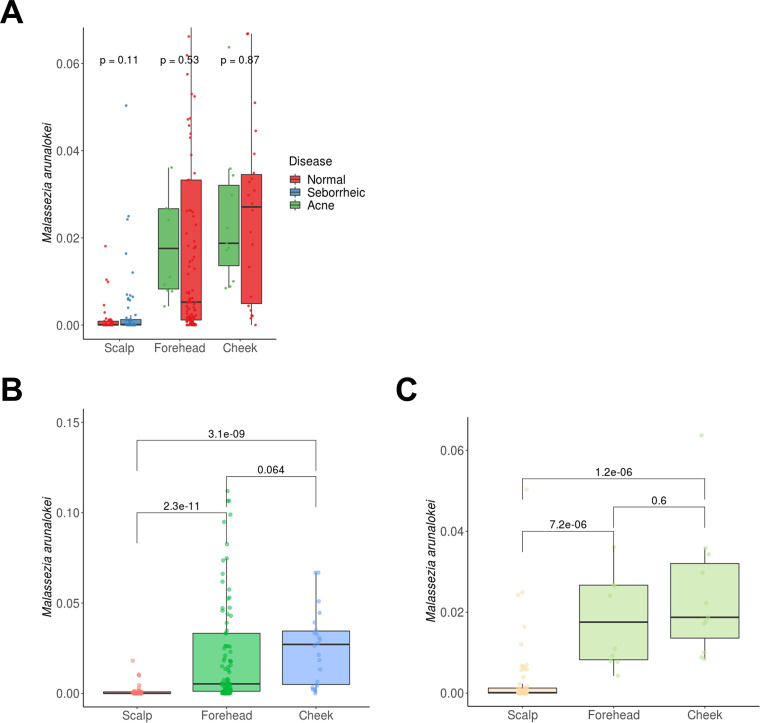
In our previous studies, we analyzed the mycobiomes on the scalp, forehead, and cheeks of healthy individuals and patients with SD/D or acne residing in Seoul, South Korea, using culture-independent amplicon sequencing (5, 6). These studies analyzed differences in the diversity of fungal communities between the patient groups and healthy groups, but the distribution of M. arunalokei among the fungal communities on the scalp, forehead, and cheeks could not be determined. Therefore, we reanalyzed the previously obtained mycobiome data sets at the species level in our current study in order to gain more precise information on the distribution of M. arunalokei on the scalp, forehead, and cheeks in our patient group and healthy group. Moreover, in addition to our own data sets, the amplicon sequencing data set generated by Tong et al. (16), who collected samples from multiple skin sites, including the forehead, on a healthy cohort residing Hong Kong, China, were included in the current study to expand the analysis (Table 2). A total of 356 species were detected in our mycobiome analysis. Among them, we assigned 346 species, while the remainder were unassigned (Table S5). The results of our mycobiome analysis showed that M. arunalokei was present in 80.7% of samples (209 out of 259), indicating that the fungus is universally present on the scalp, forehead, and cheeks, although its relative abundance is minimal compared with those of other Malassezia species (Fig. 5). We next performed a Wilcoxon rank sum test to determine the significance of the M. arunalokei abundance with respect to either disease status (SD/D or acne) or specific facial skin site (scalp, forehead, or cheeks). No significant differences in the abundance of M. arunalokei were observed between the patient group and the healthy group (Fig. 6A). However, we did find differences between M. arunalokei abundance and different skin sites (scalp versus forehead and cheeks). The abundance of M. arunalokei on the forehead and cheeks was significantly higher than that on the scalp in both the patient and healthy groups (Fig. 6B and C). These findings indicate that M. arunalokei abundance is not related to D/SD or acne but rather to the skin site. We also compared the number of nonchimeric reads with detection frequency of M. arunalokei and found no correlation between them (Fig. S1). This result further indicated that skin site has a greater influence on mycobiome composition than sequencing depth.
TABLE 2.
Occurrence of M. arunalokei in swab samples
| Parameter | Result for samples froma: |
||||||
|---|---|---|---|---|---|---|---|
| Seoul |
Hong Kong, forehead, normal | ||||||
| Scalp |
Forehead |
Cheek |
|||||
| Normal | SD/D | Normal | Acne | Normal | Acne | ||
| % of occurrences | 66.7 | 54.4 | 100 | 100 | 100 | 100 | 90.6 |
| No. of samples | 45 | 57 | 18 | 10 | 22 | 11 | 96 |
| No. of occurrences | 30 | 31 | 18 | 10 | 22 | 11 | 87 |
FIG 5.
Relative abundance of various fungal species (y axis) in samples from different skin sites of the face (labeled on the x axis). Each column represents either healthy or diseased individuals. Fungal species, skin site, and affected status are indicated in the legend on the right.
FIG 6.
Relative abundance of M. arunalokei at different facial skin sites in study cohorts. (A) Relative abundance of M. arunalokei between patient (seborrheic or acne) and healthy (normal) groups. (B and C) Relative abundance of M. arunalokei at different skin sites (scalp versus forehead and cheeks) in the (B) healthy and (C) patient groups. Values above the bar graphs are P values. The numbers of the samples used in the analysis are listed in Table 2.
Overall, our data showed that M. arunalokei is a fungal species possessing a genome highly homologous to that of M. restricta and indicated that it is the most recent species in the lineage of Malassezia that has adapted to the skin environment. Furthermore, our analysis of the facial mycobiome in humans showed that M. arunalokei had distinct niche preferences.
MATERIALS AND METHODS
Genomic DNA library construction and sequencing.
M. arunalokei NCCPF 127130 was obtained from the Westerdijk Fungal Biodiversity Institute (https://www.knaw.nl/en/institutes/westerdijkinstitute). The strain was cultured, and its genomic DNA was extracted as described previously (4). Genomic DNA was sheared using a g-TUBE device (Covaris, Woburn, MA, USA) to fragments 20 kb in size, and DNA library construction was then performed using the PacBio platform (Pacific Biosciences, Menlo Park, CA, USA) according to the manufacturer’s instructions. Sequencing was performed using a SMRT Cell and PacBio Sequel system (Pacific Biosciences). Additionally, an Illumina sequencing library was constructed using a NEBNext Ultra II FS DNA library prep kit for Illumina (E7805S; New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s instructions, followed by sequencing using a NextSeq 550 (Illumina, San Diego, CA, USA) to generate 2× 76-bp paired-end reads.
Genome assembly and analysis.
Generated raw sequences were assembled using the hybrid assembler Unicycler v.0.4.8 (17), and the general statistics are shown as Table S1 in the supplemental material. Gene prediction and annotation were performed using FunGAP pipeline v.1.1.0 using previous RNA sequencing data of M. restricta KCTC 27527 (3, 18). Protein sequences were clustered for an ortholog search using DIAMOND v.2.0.6, with the parameter “-id 50” (sequence identity) and mcl 14–137 with the parameter “-I 2.0” (inflation value, where higher values indicate more fine-grained clustering) (19). To evaluate the quality of genome assembly, we conducted an analysis using BUSCO v.5.3.0 (20) based on fungi_odb10 (13 December 2019). Genome sequences were aligned using MAUVE v.2.4.0, and their average nucleotide identity (ANI) values were calculated by ANIb method based on BLAST+ from JSpecies-WS (21–23).
Divergence time estimation.
We performed divergence time estimation based on Bayesian inference between species of the Malassezia genus. At first, we collected the genome sequences from the 25 strains belonging to 14 different species (Table S2). In addition, we used Saccharomyces cerevisiae S288c (BioProject accession no. PRJEB7245; assembly accession no. GCA_002057635.1) as an outgroup. Amino acid sequences of seven highly conserved fungal barcoding proteins were selected, namely, RPB2 (DNA-directed, RNA polymerase II subunit), phospholipase, lipase, actin, MCM7 (DNA replication licensing factor MCM7), TEF1 (elongation factor 1-alpha), and TUBB1 (beta-tubulin). We performed tBLASTn to target genomes using the barcoding protein sequences of M. arunalokei as queries and then extracted the resulting aligned sequences.
Amino acid sequences were aligned using MAFFT v.7.490 (24) with the “–maxiterate 1000” option used to refine the alignment. Aligned sequences were trimmed using CLIPKIT v.1.3.0 (25) using the smart-gap algorithm, which led to the removal of 110 sites, giving a final total alignment length of 4,753 sites.
BEAUti was used to define the parameter file for BEAST v.2.6.6 (26). We set the substitution model to Blosum62 with a gamma site model and estimated the substitution rate. The clock model was defined as strict. We used the Calibrated Yule model as the tree prior and estimated the birth rate. As an external calibration, we defined the split between M. furfur and M. restricta as ~50 million years ago (mya) (8) with a log-normal distribution. We centered the distribution at 4.1 (M) with a standard deviation of 0.4 (S) to reflect the uncertainty about the calibration. We set the birth rate and clock rate to a gamma distribution with the alpha and beta values set at 0.001 and 1,000, respectively. The Markov chain Monte Carlo (MCMC) was defined with a chain length of 10,000,000 and no pre-burn-in. We inspected BEAST output using Tracer v.1.7.4 (27) and checked whether the specified divergence prior matched the posterior distribution. We used TreeAnnotator v.2.6.6 (26) to identify the tree with the highest product of the posterior probability of all its nodes using the maximum clade credibility tree with mean heights as node heights. We visualized the final tree using FigTree v.1.4.4 (https://github.com/rambaut/figtree).
Skin mycobiome data analysis.
Facial skin mycobiome data based on ITS1 amplicon sequencing were retrieved from NCBI (Seoul, scalp, PRJNA335788; Seoul, forehead and cheek, PRJNA673754; Hong Kong, forehead, PRJNA421247) and processed in this study (5, 6, 16). Adapter and primer sequences (18S-F and 5.8S-1R) (28) in data sets were trimmed from raw paired-end reads using cutadapt (v.3.2) with default parameters (29). Length trimming was performed using forward and reverse reads to 250 bp and 180 bp, respectively.
To analyze the amplicon sequence variants (ASVs), the trimmed, paired-end reads were denoised and merged using the R package DADA2 (v.1.18.0) with a default parameter (30). A total of 5,955 ASVs with 13,595,931 reads remained after chimeras were removed using the “consensus” method. For taxonomic classification of the resulting ASVs, we applied the top BLASTN hits (NCBI-BLAST-2.10.1+) in the NCBI ITS database (NCBI Targeted Loci project, downloaded 1 December 2021) (31, 32) to discover M. arunalokei instead of the Bayesian classifier (33). The Bayesian classifier uses the bootstrap value from the Bayesian algorithm as the taxonomy confidence level. However, depending on the database, the bootstrap value may be biased to one side that has more data. The more M. restricta sequences in the NCBI database than M. arunalokei, the more the Bayesian classifier assigns a query sequence to M. restricta, although the query sequence has higher bitscore with M. arunalokei. For statistical analysis, the Wilcoxon rank sum test was performed using R (v.4.0.4).
Data availability.
The genome sequence of M. arunalokei NCCPF 127130 has been deposited in the National Center for Biotechnology Information (NCBI) under the accession no. PRJNA751706.
ACKNOWLEDGMENTS
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and Future Planning 2019R1A4A1024764, and 2021M3A9I4021431, and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI22C0894).
Footnotes
Supplemental material is available online only.
Contributor Information
Won Hee Jung, Email: whjung@cau.ac.kr.
Teresa R. O'Meara, University of Michigan
REFERENCES
- 1.Wu G, Zhao H, Li C, Rajapakse MP, Wong WC, Xu J, Saunders CW, Reeder NL, Reilman RA, Scheynius A, Sun S, Billmyre BR, Li W, Averette AF, Mieczkowski P, Heitman J, Theelen B, Schroder MS, De Sessions PF, Butler G, Maurer-Stroh S, Boekhout T, Nagarajan N, Dawson TL, Jr.. 2015. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet 11:e1005614. doi: 10.1371/journal.pgen.1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honnavar P, Prasad GS, Ghosh A, Dogra S, Handa S, Rudramurthy SM. 2016. Malassezia arunalokei sp. nov., a novel yeast species isolated from seborrheic dermatitis patients and healthy individuals from India. J Clin Microbiol 54:1826–1834. doi: 10.1128/JCM.00683-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho YJ, Park M, Jung WH. 2019. Resequencing the genome of Malassezia restricta strain KCTC 27527. Microbiol Resour Announc 8:e00213-19. doi: 10.1128/MRA.00213-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park M, Cho YJ, Lee YW, Jung WH. 2017. Whole genome sequencing analysis of the cutaneous pathogenic yeast Malassezia restricta and identification of the major lipase expressed on the scalp of patients with dandruff. Mycoses 60:188–197. doi: 10.1111/myc.12586. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Park T, Kim HJ, An S, Sul WJ. 2021. Inferences in microbial structural signatures of acne microbiome and mycobiome. J Microbiol 59:369–375. doi: 10.1007/s12275-021-0647-1. [DOI] [PubMed] [Google Scholar]
- 6.Park T, Kim HJ, Myeong NR, Lee HG, Kwack I, Lee J, Kim BJ, Sul WJ, An S. 2017. Collapse of human scalp microbiome network in dandruff and seborrhoeic dermatitis. Exp Dermatol 26:835–838. doi: 10.1111/exd.13293. [DOI] [PubMed] [Google Scholar]
- 7.Jain C, Rodriguez RL, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao R-L, Li G-J, Sánchez-Ramírez S, Stata M, Yang Z-L, Wu G, Dai Y-C, He S-H, Cui B-K, Zhou J-L, Wu F, He M-Q, Moncalvo J-M, Hyde KD. 2017. A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal Diversity 84:43–74. doi: 10.1007/s13225-017-0381-5. [DOI] [Google Scholar]
- 9.Ianiri G, Heitman J. 2020. Approaches for genetic discoveries in the skin commensal and pathogenic Malassezia yeasts. Front Cell Infect Microbiol 10:393. doi: 10.3389/fcimb.2020.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs NK. 2020. l-Histidine supplementation in adults and young children with atopic dermatitis (eczema). J Nutr 150:2576S–2579S. doi: 10.1093/jn/nxaa200. [DOI] [PubMed] [Google Scholar]
- 12.Tan SP, Brown SB, Griffiths CE, Weller RB, Gibbs NK. 2017. Feeding filaggrin: effects of l-histidine supplementation in atopic dermatitis. Clin Cosmet Invest Dermatol 10:403–411. doi: 10.2147/CCID.S146760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanifin JM, Thurston M, Omoto M, Cherill R, Tofte SJ, Graeber M, EASI Evaluator Group . 2001. The eczema area and severity index (EASI): assessment of reliability in atopic dermatitis. Exp Dermatol 10:11–18. doi: 10.1034/j.1600-0625.2001.100102.x. [DOI] [PubMed] [Google Scholar]
- 14.Albalat R, Canestro C. 2016. Evolution by gene loss. Nat Rev Genet 17:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 15.Domergue R, Castano I, De Las Penas A, Zupancic M, Lockatell V, Hebel JR, Johnson D, Cormack BP. 2005. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308:866–870. doi: 10.1126/science.1108640. [DOI] [PubMed] [Google Scholar]
- 16.Tong X, Leung MHY, Wilkins D, Cheung HHL, Lee PKH. 2019. Neutral processes drive seasonal assembly of the skin mycobiome. mSystems 4:e00004-19. doi: 10.1128/mSystems.00004-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min B, Grigoriev IV, Choi IG. 2017. FunGAP: Fungal Genome Annotation Pipeline using evidence-based gene model evaluation. Bioinformatics 33:2936–2937. doi: 10.1093/bioinformatics/btx353. [DOI] [PubMed] [Google Scholar]
- 19.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 20.Waterhouse RM, Seppey M, Simao FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol 35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richter M, Rossello-Mora R, Glockner FO, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steenwyk JL, Buida TJ, III, Li Y, Shen XX, Rokas A. 2020. ClipKIT: a multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biol 18:e3001007. doi: 10.1371/journal.pbio.3001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouckaert R, Vaughan TG, Barido-Sottani J, Duchene S, Fourment M, Gavryushkina A, Heled J, Jones G, Kuhnert D, De Maio N, Matschiner M, Mendes FK, Muller NF, Ogilvie HA, Du Plessis L, Popinga A, Rambaut A, Rasmussen D, Siveroni I, Suchard MA, Wu CH, Xie D, Zhang C, Stadler T, Drummond AJ. 2019. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol 15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Program N, Kong HH, Segre JA, NIH Intramural Sequencing Center Comparative Sequencing Program . 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 30.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruitt KD, Tatusova T, Maglott DR. 2005. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domingos P, Pazzani M. 1997. On the optimality of the simple Bayesian classifier under zero-one loss. Machine Learn 29:103–130. doi: 10.1023/A:1007413511361. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00506-22-s0001.pdf, PDF file, 6.8 MB (7MB, pdf)
Data Availability Statement
The genome sequence of M. arunalokei NCCPF 127130 has been deposited in the National Center for Biotechnology Information (NCBI) under the accession no. PRJNA751706.