ABSTRACT
Chikungunya virus (CHIKV) is a reemerging alphavirus causing chikungunya disease (CHIKD) and is transmitted to humans by Aedes mosquitoes. The virus establishes an intricate balance of cellular interactions that ultimately helps in its replication and dodges cellular immune response. In an attempt to identify cellular host factors required during CHIKV replication in Aag2 cells, we performed global transcriptomics of CHIKV-infected Aag2 cells, and further, we compared this library with the Drosophila RNAi Screening Center (DRSC) database and identified transcripts that were regulated in Aedes aegypti during CHIKV infection. These analyses revealed specific pathways, such as ubiquitin-related pathways, proteolysis pathways, protein catabolic processes, protein modification, and cellular protein metabolic processes, involved during replication of the virus. Loss-of-function assays of selected candidates revealed their proviral or antiviral characteristics upon CHIKV infection in A. aegypti-derived Aag2 cells. Further validations identified that the ubiquitin proteasomal pathway is required for CHIKV infection in A. aegypti and that an important member of this family of proteins, namely, AeCullin-3 (Aedes ortholog of human cullin-3), is a proviral host factor of CHIKV replication in Aag2 cells.
IMPORTANCE Arboviruses cause several diseases in humans and livestock. Vector control is the main strategy for controlling diseases transmitted by mosquitoes. In this context, it becomes paramount to understand how the viruses replicate in the vector for designing better transmission blocking strategies. We obtained the global transcriptome signature of A. aegypti cells during CHIKV infection, and in order to obtain the maximum information from these data sets, we further utilized the well-characterized Drosophila system and arrived upon a set of transcripts and their pathways that affect A. aegypti cells during CHIKV infection. These analyses and further validations reveal that important pathways related to protein degradation are actively involved during CHIKV infection in A. aegypti and are mainly proviral. Targeting these molecules may provide novel approaches for blocking CHIKV replication in A. aegypti.
KEYWORDS: Chikungunya virus, Aedes aegypti, vector–virus interactions, ubiquitin proteasomal pathway
INTRODUCTION
Chikungunya virus (CHIKV) is one of the reemerging zoonotic viruses endemic mainly to tropical and sub-Saharan regions. The virus is transmitted by mosquitoes belonging to the genus Aedes, with Aedes aegypti serving as the favored vector in the tropics and Aedes albopictus in temperate regions (1). Increased travel and trade have led to reemergence and large-scale epidemics in India, Africa, and Europe (1–3). Infection with CHIKV has been associated with extensive morbidity causing severe and self-limiting arthralgia in affected individuals that is sometimes chronic (4). Several approaches are under way for the development of an effective treatment against the infection. Despite the efforts, no effective therapeutics are developed that could limit pathogen spread and infectivity. Disease control strategies pertain mostly to demolishing vector breeding grounds and usage of insecticides (5, 6). Newer approaches involve developing genetically modified mosquitoes that could block viral transmission (7–9). Therefore, increased understanding of vector-virus interactome will help design newer and more effective strategies to restrict viral spread.
Host proteins play a significant role during viral replication that can be categorized as either proviral or antiviral depending on whether they facilitate or counter the establishment of virus in the host/vector. A number of elaborated studies in the last decade that were focused on CHIKV and host/vector interactions recognized a plethora of cellular processes and pathways that were of significance during infection (10–14). These included components of stress granules (15), mitochondria (13), and endoplasmic reticulum (16). Thus, by virtue of modulating these essential cellular proteins, the virus becomes capable of hijacking cellular responses enabling its efficient replication.
High-throughput screening and computational analyses are some of the tactics that are routinely employed to identify cellular factors. These studies provide a global perspective for the role of the host genes in viral infection. The present study is one such study where we identified A. aegypti host factors involved in CHIKV replication. We first performed global transcriptomic analysis of an A. aegypti cell line post CHIKV infection and identified transcripts that were regulated during CHIKV replication. In addition, we selected known genes from the Drosophila RNAi Screening Center (DRSC) database and performed a hybrid analysis to identify transcripts that may be involved in arboviral replication in the vector. The identified transcripts were evaluated for their role in CHIKV replication through a double-stranded RNA (dsRNA)-mediated RNAi screen. Finally, A. aegypti ortholog of human cullin-3 protein, referred to here as AeCullin-3, was found to be proviral during CHIKV replication.
RESULTS
Identification of Aedes aegypti host factors involved during infection with CHIKV.
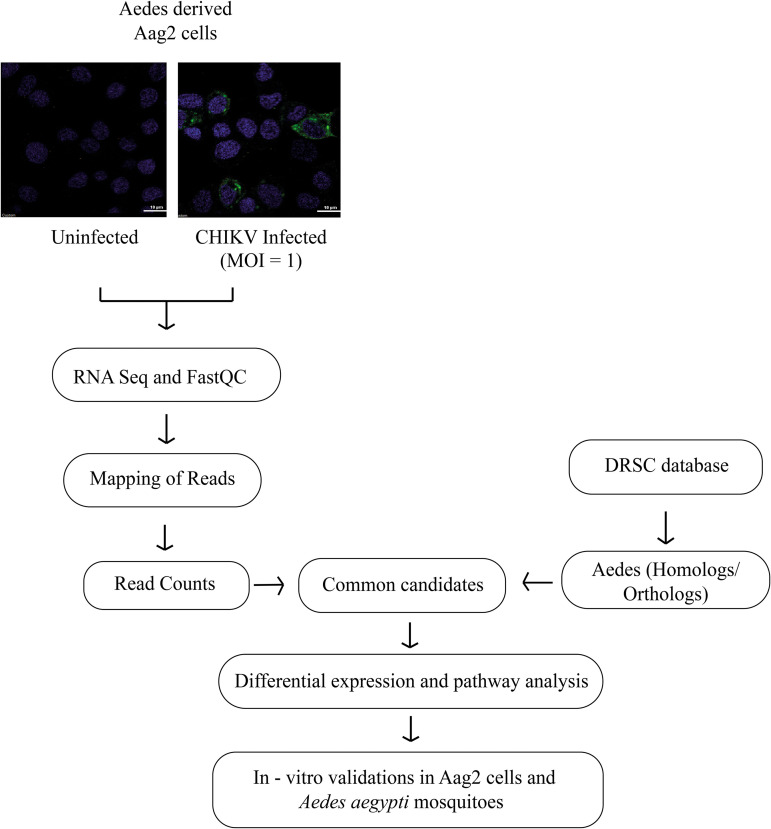
The pipeline used for the identification of A. aegypti host factors is depicted in Fig. 1.
FIG 1.
Pipeline used for the identification of Aedes aegypti host factors during infection with CHIKV.
RNA-Seq data acquired by processing of the uninfected and infected libraries (done in triplicates) were processed for quality trimming and removal of adapter sequences before proceeding with mapping to A. aegypti genome. The mapping percentage for all samples was found to be above 80%, and the total number of transcripts identified in each individual library varied between 12,000 to 15,000 (Table 1, Fig. 2a).
TABLE 1.
RNA-Seq data processing: reads obtained after trimming and mapping percentage and number of identified transcripts
| Sample name | No. of reads after trimming (bp) | Mapping percentage (%) | No. of identified transcripts |
|---|---|---|---|
| Aag2_infected_1 | 32,897,025 | 84.39% | 14,179 |
| Aag2_uninfected_1 | 67,948,278 | 84.15% | 14,893 |
| Aag2_infected_2 | 20,229,369 | 85.33% | 12,716 |
| Aag2_uninfected_2 | 24,229,683 | 84.10% | 12,987 |
| Aag2_uninfected_3 | 22,723,606 | 87.73% | 12,687 |
| Aag2_infected_3 | 20,948,346 | 85.31% | 12,704 |
FIG 2.
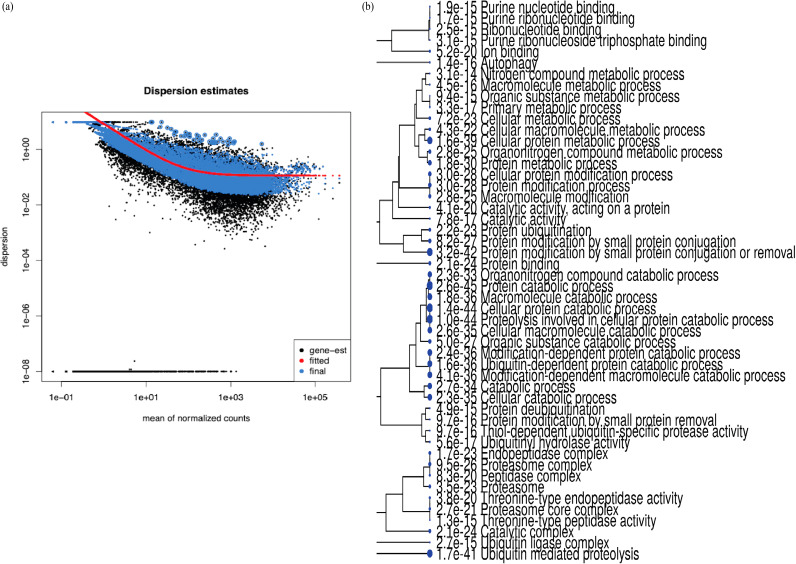
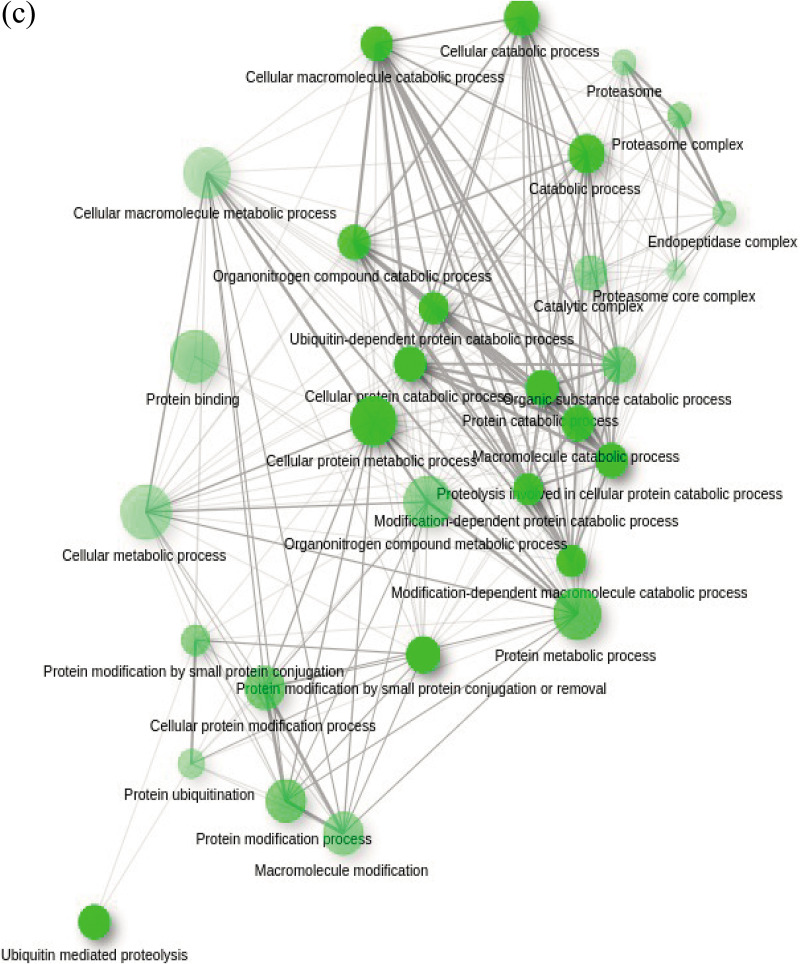
Identification of putative A. aegypti host factors upon CHIKV infection. (a) Representation of differentially expressed genes between two libraries. Black dots represent overall differentially expressed genes while blue dots represent significantly expressed genes (Fold change ± 1.5 and P value < 0.05). (b) Pathway analysis of all 8,608 transcripts found common between lab-generated transcriptomics libraries and DRSC database. Transcripts from pathways such as ubiquitin-related pathways, proteolysis pathways, protein catabolic processes, protein modification, and cellular protein metabolic processes were significantly affected upon CHIKV infection of Aag2 cell line. The sizes of the blue dots represent significance, with bigger dots having a lower P value. (c) Functional analysis of identified orthologous 346 genes found from significant selected pathways. Darker nodes are more significantly enriched gene sets. Bigger nodes represent larger gene sets. Thicker edges represent more overlapped genes.
To further explore the vector-virus interactome within the host cell, we utilized the extensive information available on the model insect, namely, Drosophila, to better annotate the Aedes genes. From the early 1900s to the present, Drosophila melanogaster has been central to groundbreaking research in diverse organisms ranging from humans to insects (17–19). Previous studies have performed hybrid high-throughput screens of Drosophila genes, including RNAi screens, to understand host-pathogen interactions in their organisms of interest and have established different protocols to perform the same (17, 20). In the present study, using a similar approach, we compared our transcriptome libraries with Drosophila RNAi Screening Center (DRSC) database. Comparison of the Drosophila data sets with our transcriptome data presented a total of 8,608 transcripts that were found to be common and were taken for further analysis. Pathway analysis revealed that ubiquitin-related pathways, proteolysis pathways, protein catabolic processes, protein modification, and cellular protein metabolic processes were significantly affected (Fig. 2b and c). Differential expression analysis revealed 346 genes to be differentially expressed, of which 34 genes were found to be significant (P value of <0.05 and/or fold change difference of ±1.5) (Data set S1). This set of genes was further reviewed for genes’ pathways, and 7 genes were taken for downstream validation.
dsRNA-based screening to elucidate the proviral or antiviral role of identified cellular factors.
In addition to the above 7 genes, we performed a literature search for putative host factors in insects that played a role in infection and cellular physiology and selected 23 additional genes for a dsRNA-based screening to evaluate their role in CHIKV infection in A. aegypti cells (Table 2). The final set of 30 transcripts for dsRNA screening were involved in cellular processes, including metabolism, biosynthesis, and oxidative pathways (AAEL001895 [21], AAEL013361 [22, 23], AAEL013603 [24], AAEL003977, AAEL014840 [25], AAEL001292 [26], AAEL012868 [27]), protein modification and proteolysis (AAEL011287, AAEL007187, AAEL019450, AAEL006797 [28], AAEL017567 [29], AAEL003104 [30], AAEL010641, AAEL001112, AAEL012337), transcriptional/translational regulatory and nucleic acid binding proteins (AAEL019736 [31], AAEL019431, AAEL008073, AAEL007945, AAEL001612), and ion channels, receptors, cell adhesion proteins, and transmembrane proteins (AAEL002922, AAEL009229, AAEL012421, AAEL005681 [32], AAEL005133, AAEL008641, AAEL006685). One transcript was found to be involved in endocytosis (AAEL007041 [32]). Apart from these, one uncharacterized transcript was also included in the study (AAEL024345).
TABLE 2.
Detailed information on the putative host factors selected for dsRNA-based screening; the description shown is acquired from Uniprot (83) and Vectorbase (72)
| Gene ID | Gene name | Gene ontology |
|---|---|---|
| AAEL011287 | Ubiquitin specific protease 1 | Protein deubiquitination |
| AAEL014840 | Short change dehydrogenase | Oxidoreductase activity |
| AAEL001895 | Alpha-1,4-galactosyltransferase | Glycan biosynthesis and metabolism |
| AAEL001112 | Ubiquitin carboxyl-terminal hydrolase | Protein deubiquitination |
| AAEL019736 | HTH CENPB-type domain-containing protein | DNA binding |
| AAEL007187 | Cullin ubiquitin ligase | E3 ubiquitin ligase |
| AAEL019431 | Ecdysone receptor | DNA-binding transcription factor activity |
| AAEL002922 | Ionotropic receptor 8a | Ion channel |
| AAEL019450 | Peptidase_S8 domain-containing protein | Protease |
| AAEL008641 | GTP-binding protein (o) alpha subunit, gnao | G protein coupled receptor |
| AAEL008073 | Putative mRNA binding protein | RNA binding |
| AAEL013603 | Short-chain dehydrogenase | Oxidoreductase activity |
| AAEL013361 | Lipase | Hydrolase activity, lipid catabolic process |
| AAEL009229 | Mitochondrial citrate transport protein, putative | Transmembrane transport |
| AAEL003977 | Very-long-chain 3-oxoacyl-CoA synthase | Fatty acid biosynthesis |
| AAEL012421 | Cadherin | Cell-cell adhesions |
| AAEL010641 | SUMO-activating enzyme subunit | Protein sumoylation |
| AAEL006797 | F-box and leucine-rich repeat protein 7 | Ubiquitination |
| AAEL005681 | GPRHIS | G protein coupled receptor |
| AAEL007945 | Eukaryotic translation initiation factor 3 subunit H | Translational initiation factor activity |
| AAEL001612 | Dicer-1 | Posttranscriptional gene silencing by RNA |
| AAEL017567 | Metalloprotease | Hydrolase/metallopeptidase activity, proteolysis |
| AAEL024345 | Uncharacterized protein | Uncharacterized protein |
| AAEL012337 | Goliath E3 ubiquitin ligase | E3 ubiquitin ligase |
| AAEL012868 | Cmp-n-acetylneuraminic acid synthase | Metabolism |
| AAEL001292 | Cytochrome P450 | Oxidoreductase activity |
| AAEL003104 | Tripartite motif protein trim 2,3 | Metal ion binding, ubiquitination |
| AAEL005133 | Tetratricopeptide repeat protein, tpr | Nonmotile cilium assembly |
| AAEL007041 | Low-density lipoprotein receptor (ldl) | Calcium ion binding, endocytosis |
| AAEL006685 | Guanine nucleotide-binding protein subunit gamma | G protein coupled receptor |
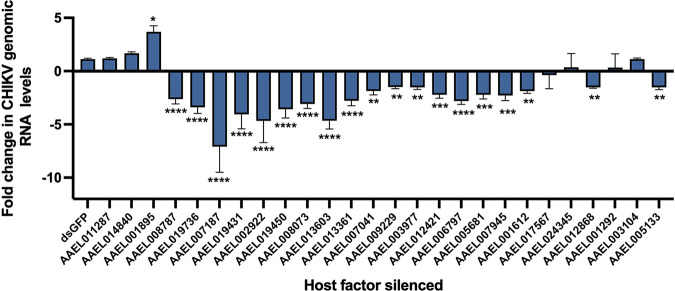
The selected transcripts were silenced using dsRNA after estimating their silencing efficiencies (14). dsRNAs showing a silencing efficiency of >50% were taken further for the loss-of-function studies (data not shown). A known host factor, V-type proton ATPase catalytic subunit A, that was previously reported to be involved in arboviral infections was also taken and served as the positive control (AAEL008787) (33), and dsRNA against GFP served as the negative control. Five genes (AAEL012337, AAEL006685, AAEL008641, AAEL010641, AAEL001112) could not be optimized for silencing and were therefore not included further. Screening of the selected transcripts during CHIKV infection using dsRNA-mediated silencing in A. aegypti-derived Aag2 cells revealed that CHIKV viral RNA levels differed significantly among cells with silenced transcripts compared to those in the negative control (GFP dsRNA-transfected cells) upon CHIKV infection. The relative CHIKV genomic RNA level for AAEL011287, AAEL014840, and AAEL001895 were found to be higher than that of the negative control. Among these factors, AAEL001895 (4-alpha-galactosyltransferase) was the most significant one exhibiting antiviral activity. Viral genomic RNA levels showed no significant change for six genes relative to negative control, namely, AAEL011287, AAEL014840, AAEL017567, AAEL024345, AAEL001292, and AAEL003104. Genes AAEL019736, AAEL007187, AAEL019431, AAEL002922, AAEL019450, AAEL008073, AAEL013603, AAEL013361, AAEL007041, AAEL009229, AAEL003977, AAEL012421, AAEL006797, AAEL005681, AAEL007945, AAEL001612, AAEL012868, and AAEL005133 all showed reduction in relative CHIKV genomic RNA level, suggesting potential proviral activity. Among these factors, AAEL007187 (AeCullin-3) was the most significant factor showing the proviral activity for CHIKV (Fig. 3).
FIG 3.
RNAi screening for A. aegypti host factors in CHIKV replication. Quantitative real-time PCR (qRT-PCR) analysis of CHIKV genomic RNA levels upon dsRNA-mediated silencing followed by CHIKV infection. Rps17 was used as an endogenous control and data are analyzed using 2−ΔΔCT. Data are expressed as mean ± SD; ****, P < 0.0001 versus control group; ***, P < 0.001 versus control group; **, P < 0.01 versus control; *, P < 0.1 versus control.
Proteasomal pathway is required for CHIKV replication in Aag2 cells post entry.
Previous studies have established a functional role for ubiquitin proteasomal machinery during viral replication in host cells (as reviewed in references 34 and 35). Studies done on alphaviruses have employed proteasome inhibitors to decipher the function of ubiquitin proteosome pathway (UPP) machinery (36–38). Global proteomic analysis of CHIKV-infected host cells has also revealed several components of the proteasome to be differentially regulated (39). In our initial analysis, we found the protein modification pathway to be evidently involved during the process of CHIKV replication. Transcripts for proteins such as proteases, ubiquitin ligases, and deubiquitinases were found to be significantly regulated in Aag2 cells upon infection. E3 ubiquitin ligase belonging to the TRIM family (AAEL003104) showed no significant impact on CHIKV replication upon being silenced. The F-box and leucine-rich repeat protein 7 (which functions as an adaptor for SCF E3 ligase; AAEL006797) along with cullin-3 (now referred to as AeCullin-3) were found to be proviral for CHIKV replication.
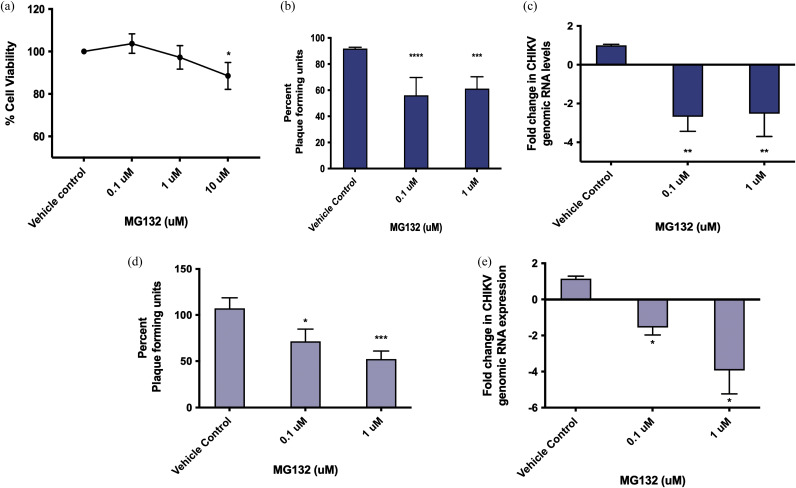
To elucidate if there was any role of proteasomal pathway in CHIKV replication, we used a well-known proteasome inhibitor MG132 in pre and posttreatment assays. MG132 has also previously been employed as a proteasome inhibitor in Aag2 cells (40, 41). Prior to the assay, cell viability upon MG132 treatment was tested in Aag2 cells, and it was found that the cell viability remained above 80% at the three drug concentrations tested, namely, 0.1 μM, 1 μM, and 10 μM (Fig. 4a). Based on the results, 0.1 μM and 1 μM were used for the inhibitor assays. Pretreatment assay determined whether proteasomal pathway had a significant involvement during early stages of viral entry and internalization. A significant reduction of approximately 40% (P < 0.0001) was observed in PFU at both 0.1 μM and 1 μM concentrations of MG132 (Fig. 4b). These results were also reproduced using quantitative real-time-PCR (qRT-PCR), and a 2-fold reduction (P < 0.01) was observed at both these concentrations (Fig. 4c). A posttreatment assay, on the other hand, determined the possible involvement at a postentry stage of viral life cycle within the cells. It was observed that during posttreatment of the drug, viral inhibition was dose dependent. While at 0.1 μM concentration a 30% reduction was obtained in plaque forming units (PFU/μL; P < 0.05), we observed a 50% reduction in viral genomic RNA levels at 1 μM concentration of MG132 (P < 0.001) (Fig. 4d). Similar results were also obtained in qRT-PCR, showing a 2-fold reduction at 0.1 μM (P < 0.05) and 4-fold reduction at 1 μM concentration of MG132 (P value < 0.05) (Fig. 4e).
FIG 4.
AeCullin-3 is proviral during CHIKV replication in Aag2 cells. (a) MTT assay to determine percentage of viable cells in the presence of MG132 at 0.1 μM, 1 μM, and 10 μM. One percent DMSO was used as vehicle control. (b) Plaque assay for viral titer determination in MG132 - CHIKV pretreatment assay. (c) qRT-PCR for CHIKV genomic RNA levels in MG132-CHIKV pretreatment assay. (d) Plaque assay for viral titer determination in MG132-CHIKV post-treatment assay. (e) qRT-PCR for CHIKV genomic RNA levels in MG132-CHIKV posttreatment assay. Rps17 was used as an endogenous control in qRT-PCR, and the results were analyzed using 2−ΔΔCT. The experiments were performed a minimum of three times, with each experiment set in triplicates. Data are expressed as mean ± SD. ****, P value < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.1 (versus control).
Role of AeCullin-3 during CHIKV infection in A. aegypti.
In order to further validate the relevance of AeCullin-3 in CHIKV infection, we performed a dsRNA-based silencing in A. aegypti mosquitoes. First, expression levels of AeCullin-3 were determined using qRT-PCR in normal blood-fed and CHIKV-spiked blood-fed female A. aegypti individuals. AeCullin-3 expression levels increased to up to 10-fold at 48 h and starting at 72 h postinfection gradually returned to normalcy (Fig. 5a).
FIG 5.
Role of AeCullin-3 during CHIKV infection in A. aegypti. (a) A. aegypti mosquitoes were fed with either uninfected blood (control group) or CHIKV-spiked blood (106 PFU/mL), a pool of n = 5 mosquitoes was collected at indicated time points in TRIzol for RNA extractions. AeCullin-3 gene expression was quantified in the two groups by qRT-PCR. The experiments were performed a minimum of three times, with each experiment set in triplicates. Fold change in expression in CHIKV-spiked blood-fed mosquitoes is compared to that in uninfected blood-fed mosquitoes. (b) Experimental plan for dsRNA-mediated knockdown of AeCullin-3 in A. aegypti mosquitoes. (c) Mosquitoes were intrathoracically injected with either dsRNA against GFP (dsGFP, control group) or dsRNA against AeCullin-3 (dsCUL3 group), and the mosquitoes (n = 5) from both groups were then collected and AeCullin-3 gene expression was estimated in each individual mosquito using qRT-PCR. (d) dsGFP- and dsCUL3-injected mosquitoes were subjected to CHIKV-spiked blood feeding 24 h post nanoinjection. Mosquitoes (n = 5) were then collected and CHIKV genomic RNA levels were analyzed in each individual mosquito. (a to d) Rps17 served as endogenous control, and qRT-PCR analysis for experiments indicated in this figure was done using 2−ΔCT method. Fold change in expression in dsCUL3-injected mosquitoes is compared to that in dsGFP-injected mosquitoes. Data are expressed as mean ± SD. ***, P < 0.001; **, P < 0.01; *, P < 0.1 (versus control).
We then went on to further characterize the role of AeCullin-3 by performing a loss-of-function assay on the mosquitoes (Fig. 5b). Four- to six-day-old female A. aegypti individuals were injected with dsRNA against green fluorescent protein (GFP; control) or AeCullin-3. As seen in Fig. 5c, a 2-fold reduction in AeCullin-3 expression levels was obtained in all the time points tested (from 24 h to 120 h post nanoinjection). Based on this, 24-h post nanoinjection of both GFP and AeCullin-3, the mosquitoes were subjected to CHIKV infectious blood meal (106 PFU/mL). In dsAeCullin-3-injected mosquitoes, a reduction of more than 3-fold in CHIKV genomic RNA levels (P < 0.0001) was obtained compared to those in the GFP control group (Fig. 5d), thereby confirming that AeCullin-3 acts as a proviral CHIKV host factor in A. aegypti mosquitoes.
DISCUSSION
While much research has been executed to identify cellular factors of the mammalian host that play important roles in CHIKV replication (42–45), limited literature is available with respect to host factors regulated upon CHIKV infection in Aedes (13, 46–48). A deficit Aedes-specific database for genes/proteins is one of the major reasons for the dearth of in-depth research in this area. Despite the lack of a proper repertoire, RNA-Seq-based studies on A. aegypti (49–51) as well as A. albopictus (52) have often been employed to study vector-virus infection. These studies have helped in deciphering transcript patterns in response to monoinfections and coinfections (51) in midgut upon infection (49) and also in salivary glands (50).
Researchers have also resorted to computational analyses and better studied insect systems such as Drosophila to identify orthologs and Aedes factors that could affect alphavirus infection (19). Combining the knowledge available on the Drosophila system with lab-generated A. aegypti transcriptome data, and with further validations, our study unravels the role of the protein modification pathways such as ubiquitin-proteasome pathway (UPP) during CHIKV infection in Aag2 cells. Other than UPP, protein catabolic processes as well as cellular protein metabolism were found to be significantly affected during CHIKV infection, as has been emphasized by previous reports (11, 36, 39). In our analysis, we found that a major proportion of genes were part of the UPP, the pathway that regulates cellular protein turnover by an intricate interplay of three diverse enzymes, E1 ubiquitin activating enzyme, E2 ubiquitin conjugating enzyme, and E3 ubiquitin ligase. Briefly, the E1 enzyme activates ubiquitin by ATP to a high-energy intermediate followed by transfer of ubiquitin from E1 to substrate which is mediated by the E2 and E3 enzymes. Cellular proteins with covalently attached ubiquitin are targeted for degradation in the 26S proteasome (53).
Ubiquitin proteasomal pathway is considered to be one of the most important regulatory machineries in the cellular system. The machinery governs biological processes such as cell cycle regulation, apoptosis, transcription, translation, and cell signaling and therefore serves as an attractive target for viral manipulation (54). Viruses are often found to subvert ubiquitin proteasomal machinery by redirecting the E3 ubiquitin ligases to degrade host proteins in order to ensure survival and promote viral replication and dissemination (as reviewed in references 55 and 56). Using both proteasomal inhibitors and performing loss-of-function studies, our present study corroborates with these earlier studies and identified AeCullin-3 to be an important proviral host factor during CHIKV infection in both Aag2 cells and A. aegypti mosquitoes.
AeCullin-3 is known to serve as a scaffolding protein for the cullin-RING E3 ubiquitin ligase (CRUL). CRULs are the largest family of E3 ubiquitin ligases and are distinguished by the presence of a structural motif called RING domain that forms the interface for recruiting E2-conjugating enzymes (57). There are eight mammalian homologues for cullin, each pertaining to different substrate specificities. Viral proteins are often found to interact with CRULs (cullin-RING ubiquitin ligases) to subvert the machinery to newer cellular targets or to protect existing targets thus modulating protein turnover (55). Elaborate studies in the past two decades have established a number of downstream effects of CRULs subversion that include (i) degradation of proteins of host defense mechanism, (ii) degradation of cellular proteins to promote oncogenic transformation, and (iii) degradation of cell cycle regulatory proteins.
Viruses are known to encode proteins that interact with CRULs, as seen in Ectromelia virus and Vaccinia virus, where they encode proteins that contain domains structurally similar to the one present in cullin-3 adapters, and by virtue of the domain (BTB Kelch), viral proteins are able to interact with this sophisticated machinery and bring about changes in cellular physiology (58, 59). Similarly, viral proteins are known to participate in the subversions as reported in paramyxoviruses (60–63). All these studies, irrespective of the cullin homolog involved in the process, seem to target the STAT proteins. STAT dimerization is an important messenger for downstream activation of interferon (IFN) response, which might explain the subversion of different cullin homologues during virus replication. In the case of arboviruses, a study suggests that West Nile virus, when present in Culex mosquitoes, also adopts this subversion mechanism of CRULs for STAT degradation (64). However, unlike in the case of flaviviruses, immunity during an alphavirus infection in A. aegypti is imparted mainly by RNAi defenses, and other pathways such as JAK STAT and Toll/Imd have limited roles to play (65). Based on these findings, it would be interesting to identify whether subversion of A. aegypti cullin (AeCullin 3)-based E3 ubiquitin ligases is responsible for the nominal involvement of JAK STAT pathway during alphavirus infections. Apart from STAT proteins, viruses also divert CRULs to target other proteins of host defense, as well, such as in the case of HBV, where HBx protein mediates the degradation of antiviral protein through cullin-4-based CRULs (66). During retroviral infection, as well, HIV vif protein hijacks cullin 1 machinery to degrade APOBEC3G and promote viral replication (67). Oncogenic viruses also utilize cullin homologues to induce transformations; a well-known example includes SV40 LT antigen, which inhibits cullin-7 and increases downstream signaling pathway, ultimately leading to tumorigenic transformation (68). Thus, based on their requirements, viruses of different families often interact with CRULs in order to facilitate the degradation of host defense proteins or, in the case of retroviruses, promote oncogenic transformation. In the present study, we observed that within the A. aegypti-derived Aag2 cells, presence of AeCullin-3 promoted viral replication. We do hypothesize that CHIKV might be subverting CRUL-3 for degradation of host defense proteins; however, further studies are required to decipher the exact mechanism of action.
Conclusion.
In summary, based on integrative analysis using global transcriptomics of A. aegypti-derived Aag2 cells upon CHIKV infection and Drosophila database screening, AeCullin-3 was identified and validated as one of the major and important host factors differentially regulated upon CHIKV infection in A. aegypti. Our study also revealed that the novel host factor has a proviral role during the postentry stage of viral replication.
MATERIALS AND METHODS
Cell culture and virus maintenance.
A. aegypti cell line (Aag2) was a kind gift from Alain Kohl (University of Glasgow, Scotland, UK). The cells were maintained at 28°C in Leibovitz medium (L-15; cat no. AL011S, HiMedia) supplemented with glutamine, tryptose phosphate broth, penicillin/streptomycin, and 10% fetal bovine serum (FBS). Vero cells (ATCC-CCL-81) and C6/36 cells were used for chikungunya virus propagation and were maintained in Dulbecco’s modified Eagle medium (DMEM; cat no. AL007A, HiMedia) supplemented with 10% FBS, penicillin/streptomycin, and glutamine at 37°C and 5% CO2.
Chikungunya virus (accession no. JF950631.1) was isolated from patient sample collected during the 2010 outbreak in India (69) and was propagated in alternate cycles in C6/36 cells and Vero cells. Virus propagated in Vero cells was collected after 48 h postinfection for determination of titer and subsequently used for infection in Aag2 cells.
Sample preparation and RNA sequencing.
Briefly, Aag2 cells were seeded in two 6-well plates. Cells from one plate were incubated with CHIKV at a multiplicity of infection (MOI) of 1, and cells from the other were used as uninfected control. Twenty-four hours postinfection, cells were collected in TRIzol reagent (Invitrogen). RNA isolation was subsequently performed using TRIzol method (70). For RNA-Seq, three samples from uninfected control cells and three samples from CHIKV-infected cells were used. RNA-Seq was performed on total RNA using Illumina HiSeq 4000 at NGB Diagnostics Pvt. Ltd. (Noida, UP, India).
Transcriptome assembly and read mapping.
The sequences from the reads of all the libraries were trimmed using Cutadapt (71) and FASTX toolkit (version 0.013) (http://hannonlab.cshl.edu/fastx_toolkit). Further quality check was performed on the trimmed sequences using FastQC tool (https://www.bioinformatics.babraham.ac.uk/index.html). The high-quality reads with a quality score of ≥20 were retained for further analysis. High-quality reads were mapped on A. aegypti genome (AaegL5.3) downloaded from VectorBase (https://vectorbase.org/vectorbase/app) (72) using STAR tool (73) to align the RNA-Seq reads. featureCounts (74) and the DESEQ2 package (75) were used for further analyses like count identification and differential expression analysis studies. The annotations of identified transcripts were also fetched from the VectorBase database using BioMart tool.
Identification of putative host factors from analysis of public databases.
The RNA-Seq data were supplemented with publicly available data from the Drosophila DRSC (76) to identify well-annotated host factors of A. aegypti upon CHIKV infection. (Fig. 1b). The gene/transcript IDs of Drosophila were converted into A. aegypti ID using g:Profiler toolkit https://biit.cs.ut.ee/gprofiler/gost (77). Host factors from data mining and differential expression of Aag2 cells from RNA-Seq analyses were compiled and common significant transcripts were selected for gene ontology analysis, performed using g:profiler, ShinyGO (v0.741) (78), and webgestalt 2019 (http://www.webgestalt.org) (79). The databases and the toolkits were accessed during January 2021 to November 2021. Further, based on the functional annotation and characterization, putative host factors for CHIKV replication were selected for dsRNA screening.
Double-stranded RNA preparation and transfection.
For in vitro transcription (IVT), T7 and SP6 polymerase were used for dsRNA preparation. pGEMT-easy vector was used for cloning genes for generating dsRNA. After IVT, the dsRNAs were purified using the TRIzol method. Purified dsRNAs were then used for transfections in Aag2 cell lines using transfection reagent Attractene (Qiagen). For transfections, 2 × 104 to 8 × 104 cells were seeded into 24-well plates in replicates. In each well, 400 ng of each dsRNA was transfected when 70% confluence was reached, along with negative control. After 4 h of incubation, the transfection medium was changed and fresh medium was added to the wells. Post 24 h, cells from one set of plates were collected to check silencing of the factors. The other plates were infected with CHIKV at an MOI of 1 and further processed for primary and secondary screening. dsRNAs were designed using snapdragon (80) primer details in Data set S2.
RNAi screening of putative host factors using qRT-PCR.
Silencing efficiencies of dsRNA were estimated before setting up the experiments by transfecting cells with dsRNA and analyzing gene expression of the target transcript using qRT-PCR. Cells were transfected with 400 ng dsRNA 24 h prior to CHIKV infection and collected at 24 h postinfection. Further, RNA was isolated using TRIzol reagent (Invitrogen) per manufacturer’s instructions and was used for qRT-PCR analysis using QuantiTect SYBR green PCR kit (Qiagen, Germany). To estimate CHIKV genomic RNA, E1 gene was used and Rps17 served as endogenous control. Experiments were conducted a minimum of three times, with each experiment set up in triplicates. Expression levels were calculated using the 2−ΔΔCT method (81).
Cell viability using MTT assay.
Aag2 cells were seeded in a 96-well format at a density of 20,000 cells per well and incubated at 28°C overnight. MG132 (Sigma; dissolved in dimethyl sulfoxide [DMSO]) was used at 0.1, 1, and 10 μM (final concentration of DMSO, 1%) in serum-free L-15 medium and incubated at 28°C for 24 h. Cells were washed with phosphate-buffered saline (PBS), and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) reagent (0.5 mg/mL) was added to each well, after which the cells were incubated for 4 h at 37°C. Cells were again washed, 100 μL DMSO was added to each well to dissolve MTT crystals, and cells were incubated at 37°C for 30 min. Absorbance was recorded at 570 nm and was used to calculate percent cell viability.
Pretreatment and posttreatment assays with proteasomal inhibitor.
MG132 (Sigma) proteasome inhibitor was used for suppression of proteasomal machinery. For pretreatment assay, cells were initially treated with 0.1 μM and 1 μM drug concentrations with 1% DMSO as a vehicle control and incubated at 28°C for 24 h in 2% FBS-containing L-15 medium, which was followed by washing and CHIKV infection (MOI = 1) for 2 h. Cells and supernatants were collected at 24 h for qRT-PCR (viral genome quantification using primers for CHIKV E1 gene) and plaque assay (determination of viral titer), respectively.
For posttreatment assay, cells were infected with CHIKV (MOI = 1) for 2 h, washed, and then treated with MG132 at 0.1 μM and 1 μM concentrations again using 1% DMSO as vehicle. After a 24-h incubation, cells and supernatant were collected for qRT-PCR and plaque assays, respectively.
Plaque assay.
Vero cells were plated in a 96-well format in DMEM containing 10% FBS and were incubated overnight such that the cells formed a monolayer the following day. The virus was used starting at a dilution of 1:200 and was subsequently double diluted. Cells were incubated with the viral particles for 2 h at 37°C for virus absorption in serum-free medium. The medium was then removed and a final volume of 200 μL containing equal parts 2% carboxymethyl cellulose (CMC) and FBS-containing medium was used to overlay. Cells were further incubated for 72 h for the plaques to develop. Five percent formaldehyde was used as a fixative for 1 h and 0.25% crystal violet (in 30% methanol) was used for staining cells. The plates were washed three times with 1× PBS, and finally, the plaques obtained were calculated as follows to determine viral titer: PFU/mL = no. of plagues/dilution factor × volume of diluted virus per well.
Mosquito maintenance, blood feeding, and nanoinjections.
A. aegypti mosquitoes were maintained as described previously (82). Blood feeding was performed by initially starving female mosquitoes for 4 h. Uninfected rabbit blood or blood spiked with CHIKV (106 PFU/mL) was then fed to mosquitoes for a period of 1 h using membrane feeders. Fully fed mosquitoes were distinguished by presence of engorged midgut and separated.
dsRNA injections were performed by injecting 800 ng of total dsRNA into female mosquitoes. For experiments with subsequent blood feeding, injected mosquitoes were fed with CHIKV-infected blood 24 h after nanoinjection. Fed mosquitoes were separated from unfed mosquitoes. A total of 5 individual mosquitoes were then collected in TRIzol and analyzed for cullin-3 and CHIKV genomic RNA levels in qRT-PCR using primers against E1 gene.
qRT-PCR.
A total of 2 × 106 cells were collected in TRIzol reagent (Invitrogen) for total RNA isolation. Gene expression levels were determined using specific primers with qRT-PCR using QuantiTect SYBR green PCR kit (Qiagen) following manufacturer’s instructions.
For mosquito experiments, 4- to 5-day-old female A. aegypti mosquitoes were fed with either CHIKV-spiked blood (106 PFU/mL) or uninfected blood. Fully engorged mosquitoes were separated. A pool of 5 mosquitoes were then collected in TRIzol every 24 h for 5 days. RNA was extracted from the pool of mosquitoes subjected to qRT-PCR using QuantiTect SYBR green PCR kit (Qiagen) with AeCullin-3-specific primers. Rps17 was used as endogenous control. The experiment was performed three times with three technical replicates. Data were analyzed by 2−ΔCT method and 2−ΔΔCT and expressed as mean ± standard deviation (SD).
Primer details for qRT-PCR have been listed in Data set S2.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism 6. All experiments were performed a minimum of three times with three technical replicates. Data are expressed as mean ± SD. The statistical significance of primary screening, MG132 treatment assays, and expression profile for cullin in infected Aedes aegypti mosquitoes was done using one-way analysis of variance (ANOVA) with Dunnett’s multiple-comparison test. Statistical significance of expression profile for viral genomic RNA levels in AeCullin-3-silenced mosquitoes was performed using unpaired Student’s t tests. P values of <0.05 were considered significant and are represented with an asterisk in the figures.
Data availability.
Transcriptomics data sets from the current study have been submitted to National Center for Biotechnology Information accession number GSE195852.
ACKNOWLEDGMENTS
We thank Jatin Shrinet, postdoctoral associate, Florida State University, Tallahassee, Florida for the critical readings and insights he provided during the study. We also thank Dinesh Singh for his assistance with all the mosquito experiments.
The study has been funded by Department of Science and Technology, Government of India grant SB/SO/BB-23/2011 granted to S.S. D.M. and A.H. are supported by PhD fellowships from Council for Scientific and Industrial Research (CSIR) and Department of Biotechnology (DBT).
The authors declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Sujatha Sunil, Email: sujathasunil13@gmail.com.
Bo Zhang, Wuhan Institute of Virology.
REFERENCES
- 1.Burt FJ, Rolph MS, Rulli NE, Mahalingam S, Heise MT. 2012. Chikungunya: a re-emerging virus. Lancet 379:662–671. doi: 10.1016/S0140-6736(11)60281-X. [DOI] [PubMed] [Google Scholar]
- 2.Chretien J-P, Linthicum KJ. 2007. Chikungunya in Europe: what’s next? Lancet 370:1805–1806. doi: 10.1016/S0140-6736(07)61752-8. [DOI] [PubMed] [Google Scholar]
- 3.Staples JE, Breiman RF, Powers AM. 2009. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin Infect Dis 49:942–948. doi: 10.1086/605496. [DOI] [PubMed] [Google Scholar]
- 4.Borgherini G, Poubeau P, Jossaume A, Gouix A, Cotte L, Michault A, Arvin-Berod C, Paganin F. 2008. Persistent arthralgia associated with chikungunya virus: a study of 88 adult patients on reunion island. Clin Infect Dis 47:469–475. doi: 10.1086/590003. [DOI] [PubMed] [Google Scholar]
- 5.Powers AM. 2018. Vaccine and therapeutic options to control Chikungunya virus. Clin Microbiol Rev 31. doi: 10.1128/CMR.00104-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benelli G, Jeffries CL, Walker T. 2016. Biological control of mosquito vectors: past, present, and future. Insects 7:52. doi: 10.3390/insects7040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores HA, O’Neill SL. 2018. Controlling vector-borne diseases by releasing modified mosquitoes. Nat Rev Microbiol 16:508–518. doi: 10.1038/s41579-018-0025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kistler KE, Vosshall LB, Matthews BJ. 2015. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep 11:51–60. doi: 10.1016/j.celrep.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Yang T, Bui M, Gamez S, Wise T, Kandul NP, Liu J, Alcantara L, Lee H, Edula JR, Raban R, Zhan Y, Wang Y, DeBeaubien N, Chen J, Sanchez CH, Bennett JB, Antoshechkin I, Montell C, Marshall JM, Akbari OS. 2021. Suppressing mosquito populations with precision guided sterile males. Nat Commun 12:5374. doi: 10.1038/s41467-021-25421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meshram CD, Agback P, Shiliaev N, Urakova N, Mobley JA, Agback T, Frolova EI, Frolov I. 2018. Multiple host factors interact with the hypervariable domain of chikungunya virus nsP3 and determine viral replication in cell-specific mode. J Virol 92. doi: 10.1128/JVI.00838-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourai M, Lucas-Hourani M, Gad HH, Drosten C, Jacob Y, Tafforeau L, Cassonnet P, Jones LM, Judith D, Couderc T, Lecuit M, Andre P, Kummerer BM, Lotteau V, Despres P, Tangy F, Vidalain PO. 2012. Mapping of Chikungunya virus interactions with host proteins identified nsP2 as a highly connected viral component. J Virol 86:3121–3134. doi: 10.1128/JVI.06390-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tchankouo-Nguetcheu S, Bourguet E, Lenormand P, Rousselle JC, Namane A, Choumet V. 2012. Infection by chikungunya virus modulates the expression of several proteins in Aedes aegypti salivary glands. Parasit Vectors 5:264. doi: 10.1186/1756-3305-5-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubey SK, Shrinet J, Sunil S. 2019. Aedes aegypti microRNA, miR-2944b-5p interacts with 3'UTR of chikungunya virus and cellular target vps-13 to regulate viral replication. PLoS Negl Trop Dis 13:e0007429. doi: 10.1371/journal.pntd.0007429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubey SK, Shrinet J, Jain J, Ali S, Sunil S. 2017. Aedes aegypti microRNA miR-2b regulates ubiquitin-related modifier to control chikungunya virus replication. Sci Rep 7:17666. doi: 10.1038/s41598-017-18043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholte FE, Tas A, Albulescu IC, Zusinaite E, Merits A, Snijder EJ, van Hemert MJ. 2015. Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication. J Virol 89:4457–4469. doi: 10.1128/JVI.03612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joubert PE, Werneke S, de la Calle C, Guivel-Benhassine F, Giodini A, Peduto L, Levine B, Schwartz O, Lenschow D, Albert ML. 2012. Chikungunya-induced cell death is limited by ER and oxidative stress-induced autophagy. Autophagy 8:1261–1263. doi: 10.4161/auto.20751. [DOI] [PubMed] [Google Scholar]
- 17.Schneider D. 2000. Using Drosophila as a model insect. Nat Rev Genet 1:218–226. doi: 10.1038/35042080. [DOI] [PubMed] [Google Scholar]
- 18.Markow TA. 2015. The secret lives of Drosophila flies. Elife 4. doi: 10.7554/eLife.06793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yasunaga A, Hanna SL, Li J, Cho H, Rose PP, Spiridigliozzi A, Gold B, Diamond MS, Cherry S. 2014. Genome-wide RNAi screen identifies broadly-acting host factors that inhibit arbovirus infection. PLoS Pathog 10:e1003914. doi: 10.1371/journal.ppat.1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohr S, Bakal C, Perrimon N. 2010. Genomic screening with RNAi: results and challenges. Annu Rev Biochem 79:37–64. doi: 10.1146/annurev-biochem-060408-092949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramasamy R. 2021. Mosquito vector proteins homologous to alpha1–3 galactosyl transferases of tick vectors in the context of protective immunity against malaria and hypersensitivity to vector bites. Parasit Vectors 14:303. doi: 10.1186/s13071-021-04801-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arrese EL, Patel RT, Soulages JL. 2006. The main triglyceride-lipase from the insect fat body is an active phospholipase A1: identification and characterization. J Lipid Res 47:2656–2667. doi: 10.1194/jlr.M600161-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Christeller JT, Poulton J, Markwick NM, Simpson RM. 2010. The effect of diet on the expression of lipase genes in the midgut of the lightbrown apple moth (Epiphyas postvittana Walker; Tortricidae). Insect Mol Biol 19:9–25. doi: 10.1111/j.1365-2583.2009.00924.x. [DOI] [PubMed] [Google Scholar]
- 24.Mayoral JG, Leonard KT, Nouzova M, Noriega FG, Defelipe LA, Turjanski AG. 2013. Functional analysis of a mosquito short-chain dehydrogenase cluster. Arch Insect Biochem Physiol 82:96–115. doi: 10.1002/arch.21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonizzoni M, Dunn WA, Campbell CL, Olson KE, Marinotti O, James AA. 2012. Strain variation in the transcriptome of the dengue fever vector, Aedes aegypti. G3-Genes Genom Genet 2:103–114. doi: 10.1534/g3.111.001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishak IH, Kamgang B, Ibrahim SS, Riveron JM, Irving H, Wondji CS. 2017. Pyrethroid resistance in Malaysian populations of dengue vector Aedes aegypti is mediated by CYP9 family of cytochrome P450 genes. PLoS Negl Trop Dis 11:e0005302. doi: 10.1371/journal.pntd.0005302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cime-Castillo J, Delannoy P, Mendoza-Hernández G, Monroy-Martínez V, Harduin-Lepers A, Lanz-Mendoza H, Hernández-Hernández F. d l C, Zenteno E, Cabello-Gutiérrez C, Ruiz-Ordaz BH. 2015. Sialic acid expression in the mosquito Aedes aegypti and its possible role in dengue virus-vector interactions. Biomed Res Int 2015:504187. doi: 10.1155/2015/504187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. 2009. Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science 324:258–261. doi: 10.1126/science.1171400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kantor AM, Dong S, Held NL, Ishimwe E, Passarelli AL, Clem RJ, Franz AW. 2017. Identification and initial characterization of matrix metalloproteinases in the yellow fever mosquito, Aedes aegypti. Insect Mol Biol 26:113–126. doi: 10.1111/imb.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martins M, Ramos LFC, Murillo JR, Torres A, de Carvalho SS, Domont GB, de Oliveira DMP, Mesquita RD, Nogueira FCS, Maciel-de-Freitas R, Junqueira M. 2021. Comprehensive quantitative proteome analysis of Aedes aegypti identifies proteins and pathways involved in Wolbachia pipientis and Zika virus interference phenomenon. Front Physiol 12:642237. doi: 10.3389/fphys.2021.642237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mateo L, Gonzalez J. 2014. Pogo-like transposases have been repeatedly domesticated into CENP-B-related proteins. Genome Biol Evol 6:2008–2016. doi: 10.1093/gbe/evu153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tree MO, Londono-Renteria B, Troupin A, Clark KM, Colpitts TM, Conway MJ. 2019. Dengue virus reduces expression of low-density lipoprotein receptor-related protein 1 to facilitate replication in Aedes aegypti. Sci Rep 9:6352. doi: 10.1038/s41598-019-42803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunt SR, Hernandez R, Brown DT. 2011. Role of the vacuolar-ATPase in Sindbis virus infection. J Virol 85:1257–1266. doi: 10.1128/JVI.01864-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo H. 2016. Interplay between the virus and the ubiquitin-proteasome system: molecular mechanism of viral pathogenesis. Curr Opin Virol 17:1–10. doi: 10.1016/j.coviro.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi AG, Wong J, Marchant D, Luo H. 2013. The ubiquitin-proteasome system in positive-strand RNA virus infection. Rev Med Virol 23:85–96. doi: 10.1002/rmv.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur P, Lello LS, Utt A, Dutta SK, Merits A, Chu JJH. 2020. Bortezomib inhibits chikungunya virus replication by interfering with viral protein synthesis. PLoS Negl Trop Dis 14:e0008336. doi: 10.1371/journal.pntd.0008336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karpe YA, Pingale KD, Kanade GD. 2016. Activities of proteasome and m-calpain are essential for Chikungunya virus replication. Virus Genes 52:716–721. doi: 10.1007/s11262-016-1355-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llamas-Gonzalez YY, Campos D, Pascale JM, Arbiza J, Gonzalez-Santamaria J. 2019. A functional ubiquitin-proteasome system is required for efficient replication of New World Mayaro and Una alphaviruses. Viruses 11:370. doi: 10.3390/v11040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thio CL, Yusof R, Abdul-Rahman PS, Karsani SA. 2013. Differential proteome analysis of chikungunya virus infection on host cells. PLoS One 8:e61444. doi: 10.1371/journal.pone.0061444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Troupin A, Londono-Renteria B, Conway MJ, Cloherty E, Jameson S, Higgs S, Vanlandingham DL, Fikrig E, Colpitts TM. 2016. A novel mosquito ubiquitin targets viral envelope protein for degradation and reduces virion production during dengue virus infection. Biochim Biophys Acta 1860:1898–1909. doi: 10.1016/j.bbagen.2016.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schauer S, Burster T, Spindler-Barth M. 2012. N- and C-terminal degradation of ecdysteroid receptor isoforms, when transiently expressed in mammalian CHO cells, is regulated by the proteasome and cysteine and threonine proteases. Insect Mol Biol 21:383–394. doi: 10.1111/j.1365-2583.2012.01144.x. [DOI] [PubMed] [Google Scholar]
- 42.Sukkaew A, Suksatu A, Roytrakul S, Smith DR, Ubol S. 2020. Proteomic analysis of CHIKV-infected human fibroblast-like synoviocytes: identification of host factors potentially associated with CHIKV replication and cellular pathogenesis. Microbiol Immunol 64:445–457. doi: 10.1111/1348-0421.12793. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka A, Tumkosit U, Nakamura S, Motooka D, Kishishita N, Priengprom T, Sa-Ngasang A, Kinoshita T, Takeda N, Maeda Y. 2017. Genome-wide screening uncovers the significance of N-sulfation of heparan sulfate as a host cell factor for Chikungunya virus infection. J Virol 91. doi: 10.1128/JVI.00432-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pott F, Postmus D, Brown RJP, Wyler E, Neumann E, Landthaler M, Goffinet C. 2021. Single-cell analysis of arthritogenic alphavirus-infected human synovial fibroblasts links low abundance of viral RNA to induction of innate immunity and arthralgia-associated gene expression. Emerg Microbes Infect 10:2151–2168. doi: 10.1080/22221751.2021.2000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meertens L, Hafirassou ML, Couderc T, Bonnet-Madin L, Kril V, Kummerer BM, Labeau A, Brugier A, Simon-Loriere E, Burlaud-Gaillard J, Doyen C, Pezzi L, Goupil T, Rafasse S, Vidalain PO, Bertrand-Legout A, Gueneau L, Juntas-Morales R, Ben Yaou R, Bonne G, de Lamballerie X, Benkirane M, Roingeard P, Delaugerre C, Lecuit M, Amara A. 2019. FHL1 is a major host factor for chikungunya virus infection. Nature 574:259–263. doi: 10.1038/s41586-019-1578-4. [DOI] [PubMed] [Google Scholar]
- 46.Lee RC, Chu JJ. 2015. Proteomics profiling of chikungunya-infected Aedes albopictus C6/36 cells reveal important mosquito cell factors in virus replication. PLoS Negl Trop Dis 9:e0003544. doi: 10.1371/journal.pntd.0003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monteiro VVS, Navegantes-Lima KC, de Lemos AB, da Silva GL, de Souza Gomes R, Reis JF, Rodrigues Junior LC, da Silva OS, Romao PRT, Monteiro MC. 2019. Aedes-Chikungunya virus interaction: key role of vector midguts microbiota and its saliva in the host infection. Front Microbiol 10:492. doi: 10.3389/fmicb.2019.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McFarlane M, Almire F, Kean J, Donald CL, McDonald A, Wee B, Laureti M, Varjak M, Terry S, Vazeille M, Gestuveo RJ, Dietrich I, Loney C, Failloux AB, Schnettler E, Pondeville E, Kohl A. 2020. The Aedes aegypti Domino Ortholog p400 regulates antiviral exogenous small interfering RNA pathway activity and ago-2 expression. mSphere 5. doi: 10.1128/mSphere.00081-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong S, Behura SK, Franz AWE. 2017. The midgut transcriptome of Aedes aegypti fed with saline or protein meals containing chikungunya virus reveals genes potentially involved in viral midgut escape. BMC Genomics 18:382. doi: 10.1186/s12864-017-3775-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao L, Alto BW, Jiang Y, Yu F, Zhang Y. 2019. Transcriptomic analysis of Aedes aegypti innate immune system in response to ingestion of Chikungunya virus. Int J Mol Sci 20:3133. doi: 10.3390/ijms20133133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shrinet J, Srivastava P, Sunil S. 2017. Transcriptome analysis of Aedes aegypti in response to mono-infections and co-infections of dengue virus-2 and chikungunya virus. Biochem Biophys Res Commun 492:617–623. doi: 10.1016/j.bbrc.2017.01.162. [DOI] [PubMed] [Google Scholar]
- 52.Vedururu RK, Neave MJ, Tachedjian M, Klein MJ, Gorry PR, Duchemin JB, Paradkar PN. 2019. RNASeq analysis of Aedes albopictus mosquito midguts after Chikungunya virus infection. Viruses 11:513. doi: 10.3390/v11060513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ciechanover A. 1994. The ubiquitin-proteasome proteolytic pathway. Cell 79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 54.Ciechanover A, Schwartz AL. 1998. The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc Natl Acad Sci USA 95:2727–2730. doi: 10.1073/pnas.95.6.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahon C, Krogan NJ, Craik CS, Pick E. 2014. Cullin E3 ligases and their rewiring by viral factors. Biomolecules 4:897–930. doi: 10.3390/biom4040897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barry M, Fruh K. 2006. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci STKE 2006:pe21. doi: 10.1126/stke.3352006pe21. [DOI] [PubMed] [Google Scholar]
- 57.Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 58.Gao C, Pallett MA, Croll TI, Smith GL, Graham SC. 2019. Molecular basis of cullin-3 (Cul3) ubiquitin ligase subversion by vaccinia virus protein A55. J Biol Chem 294:6416–6429. doi: 10.1074/jbc.RA118.006561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilton BA, Campbell S, Van Buuren N, Garneau R, Furukawa M, Xiong Y, Barry M. 2008. Ectromelia virus BTB/kelch proteins, EVM150 and EVM167, interact with cullin-3-based ubiquitin ligases. Virology 374:82–99. doi: 10.1016/j.virol.2007.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Precious B, Young DF, Andrejeva L, Goodbourn S, Randall RE. 2005. In vitro and in vivo specificity of ubiquitination and degradation of STAT1 and STAT2 by the V proteins of the paramyxoviruses simian virus 5 and human parainfluenza virus type 2. J Gen Virol 86:151–158. doi: 10.1099/vir.0.80263-0. [DOI] [PubMed] [Google Scholar]
- 61.Nishio M, Tsurudome M, Ito M, Garcin D, Kolakofsky D, Ito Y. 2005. Identification of paramyxovirus V protein residues essential for STAT protein degradation and promotion of virus replication. J Virol 79:8591–8601. doi: 10.1128/JVI.79.13.8591-8601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elliott J, Lynch OT, Suessmuth Y, Qian P, Boyd CR, Burrows JF, Buick R, Stevenson NJ, Touzelet O, Gadina M, Power UF, Johnston JA. 2007. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J Virol 81:3428–3436. doi: 10.1128/JVI.02303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ulane CM, Horvath CM. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160–166. doi: 10.1006/viro.2002.1773. [DOI] [PubMed] [Google Scholar]
- 64.Paradkar PN, Duchemin JB, Rodriguez-Andres J, Trinidad L, Walker PJ. 2015. Cullin4 Is pro-viral during West Nile virus infection of culex mosquitoes. PLoS Pathog 11:e1005143. doi: 10.1371/journal.ppat.1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McFarlane M, Arias-Goeta C, Martin E, O'Hara Z, Lulla A, Mousson L, Rainey SM, Misbah S, Schnettler E, Donald CL, Merits A, Kohl A, Failloux AB. 2014. Characterization of Aedes aegypti innate-immune pathways that limit Chikungunya virus replication. PLoS Negl Trop Dis 8:e2994. doi: 10.1371/journal.pntd.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minor MM, Hollinger FB, McNees AL, Jung SY, Jain A, Hyser JM, Bissig KD, Slagle BL. 2020. Hepatitis B virus HBx protein mediates the degradation of host restriction factors through the cullin 4 DDB1 E3 ubiquitin ligase complex. Cells 9:834. doi: 10.3390/cells9040834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 68.Hartmann T, Xu X, Kronast M, Muehlich S, Meyer K, Zimmermann W, Hurwitz J, Pan ZQ, Engelhardt S, Sarikas A. 2014. Inhibition of Cullin-RING E3 ubiquitin ligase 7 by simian virus 40 large T antigen. Proc Natl Acad Sci USA 111:3371–3376. doi: 10.1073/pnas.1401556111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shrinet J, Jain S, Sharma A, Singh SS, Mathur K, Rana V, Bhatnagar RK, Gupta B, Gaind R, Deb M, Sunil S. 2012. Genetic characterization of Chikungunya virus from New Delhi reveal emergence of a new molecular signature in Indian isolates. Virol J 9:100. doi: 10.1186/1743-422X-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rio DC, Ares M, Jr, Hannon GJ, Nilsen TW. 2010. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc 2010:pdb prot5439. doi: 10.1101/pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- 71.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17. [Google Scholar]
- 72.Giraldo-Calderon GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, Ho N, Gesing S, VectorBase C, Madey G, Collins FH, Lawson D, VectorBase Consortium . 2015. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res 43:D707–D713. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 75.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davis MN, Horne-Badovinac S, Naba A. 2019. In-silico definition of the Drosophila melanogaster matrisome. Matrix Biol Plus 4:100015. doi: 10.1016/j.mbplus.2019.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. 2019. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ge SX, Jung D, Yao R. 2020. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. 2019. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu Y, Comjean A, Roesel C, Vinayagam A, Flockhart I, Zirin J, Perkins L, Perrimon N, Mohr SE. 2017. FlyRNAi.org-the database of the Drosophila RNAi screening center and transgenic RNAi project: 2017 update. Nucleic Acids Res 45:D672–D678. doi: 10.1093/nar/gkw977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 82.Sirisena P, Kumar A, Sunil S. 2018. Evaluation of Aedes aegypti (Diptera: Culicidae) life table attributes upon Chikungunya virus replication reveals impact on egg-laying pathways. J Med Entomol 55:1580–1587. doi: 10.1093/jme/tjy097. [DOI] [PubMed] [Google Scholar]
- 83.The UniProt Consortium. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00595-22-s001.xlsx, XLSX file, 0.04 MB (37.6KB, xlsx)
Supplemental material. Download spectrum.00595-22-s002.xlsx, XLSX file, 0.01 MB (11KB, xlsx)
Data Availability Statement
Transcriptomics data sets from the current study have been submitted to National Center for Biotechnology Information accession number GSE195852.