ABSTRACT
Immunocompromised hosts with prolonged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections have been implicated in the emergence of highly mutated SARS-CoV-2 variants. Spike mutations are of particular concern because the spike protein is a key target for vaccines and therapeutics for SARS-CoV-2. Here, we report the emergence of spike mutations in two immunocompromised patients with persistent SARS-CoV-2 reverse transcription (RT)-PCR positivity (>90 days). Whole-genome sequence analysis of samples obtained before and after coronavirus disease 2019 (COVID-19) treatment demonstrated the development of partial therapeutic escape mutations and increased intrahost SARS-CoV-2 genome diversity over time. This case series thus adds to the accumulating evidence that immunocompromised hosts with persistent infections are important sources of SARS-CoV-2 genome diversity and, in particular, clinically important spike protein diversity.
IMPORTANCE The emergence of clinically important mutations described in this report highlights the need for sustained vigilance and containment measures when managing immunocompromised patients with persistent COVID-19. Even as jurisdictions across the globe start lifting pandemic control measures, immunocompromised patients with persistent COVID-19 constitute a unique group that requires close genomic monitoring and enhanced infection control measures, to ensure early detection and containment of mutations and variants of therapeutic and public health importance.
KEYWORDS: SARS-CoV-2, immunocompromised hosts, prolonged COVID-19
OBSERVATION
Immunocompromised hosts with prolonged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection have been implicated in the emergence of highly mutated SARS-CoV-2 variants (1–12). Unlike immunocompetent hosts, patients with compromised immunity from underlying medical conditions or immunosuppressive therapies have been reported to have high SARS-CoV-2 burdens for protracted periods (1, 3, 7, 9–11, 13). Accumulating evidence also suggests that these immunocompromised patients are at significantly increased risk of severe coronavirus disease 2019 (COVID-19), necessitating aggressive prophylaxis and therapy (14–18). The prolonged infection and consequent viral replication in immunocompromised hosts, combined with selection pressure exerted by therapies, constitute a rich reservoir from which therapeutic and immune escape mutations can emerge and accumulate. The intrahost evolution of the SAR-CoV-2 virus within this subgroup of patients might have played an important role in the emergence of the currently predominant B.1.1.529 (Omicron) variant and other variants, as evidenced by the multitude of mutations found in a single patient with advanced human immunodeficiency virus (HIV) disease (8, 13). The highly mutated Omicron variant, which partially evades Pfizer BNT162b2 vaccine and multiple therapeutic SARS-CoV-2 antibodies (19–23), has caused massive waves of infection globally since it was first described in November 2021 (24).
Therefore, immunocompromised patients with persistent COVID-19 constitute a unique patient group that requires close genomic monitoring and enhanced infection control measures, even as jurisdictions across the globe start embracing SARS-CoV-2 endemicity (25). The utility of near-real-time genomic monitoring of this high-risk patient group is 2-fold. First, the genomic data directly benefit individual patients by allowing the attending clinical team to tailor therapeutics based on any potential therapeutic escape mutations. Second, genomic surveillance allows emergent variants to be monitored to safeguard public health. Therefore, our institution has in place an on-site genomic surveillance program to enable near-real-time sequencing of SARS-CoV-2 samples. Here, we describe the emergence of multiple partial therapeutic escape spike mutations in two immunocompromised patients with persistent SARS-CoV-2 reverse transcription (RT)-PCR positivity, who were identified through our on-site genomic surveillance program.
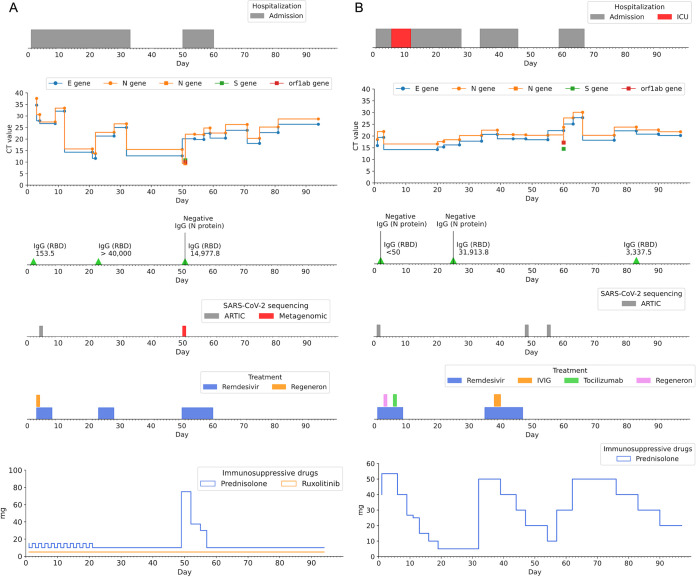
Patient A is a fully COVID-19-vaccinated (two doses of Pfizer-BioNTech vaccine) 53-year-old man with a history of acute myeloid leukemia (AML-M2). He received an allogeneic stem cell transplant in January 2019. This was complicated by pulmonary chronic graft-versus-host disease (cGVHD). He had multiple opportunistic infections in the past, including Cytomegalovirus (CMV) retinitis, pulmonary aspergillosis and pulmonary nocardiosis. He was taking ruxolitinib (5mg daily) and prednisolone (15mg/10mg daily on alternate days) for the cGVHD when he caught Covid 19. His other significant comorbidities include visual impairment and end-stage renal failure requiring dialysis, secondary to diabetic nephropathy. Patient A was initially admitted on 15 November 2021 (day 1) for inpatient hemodialysis support when his primary caregiver tested positive for COVID-19. He was nursing a cough at admission and tested negative for SARS-CoV-2 by RT-PCR, as well as daily rapid antigen testing (SD Biosensor Standard Q COVID-19 antigen test) until day 3 of admission. On day 3, his chest X-ray showed right lower zone opacities, and he tested positive for SARS-CoV-2 by RT-PCR (Xpert Xpress SARS-CoV-2; Cepheid, USA). In view of his International Severe Acute Respiratory and Emerging Infections Consortium (ISARIC) 4C mortality score of 10 (26), he was promptly started on one dose of casirivimab (REGN10933) and imdevimab (REGN10987) cocktail (REGN-COV2), and a 5-day course of remdesivir, for SARS-CoV-2 pneumonia. He remained minimally symptomatic for COVID-19 but continued to test positive on SARS-CoV-2 RT-PCR. On day 21 of admission, a sudden drop in the RT-PCR cycle threshold (CT) value was observed (from an E gene CT value of 32.1 on day 12 to an E gene CT value of 14.3 on day 21), suggesting an acute increase in SARS-CoV-2 viral burden. He was started on a second 5-day course of remdesivir, and his prednisolone dose was reduced to 10 mg daily. He remained in clinically stable and was discharged on day 34. On day 50, he was re-admitted due to new-onset fever and worsening cough and breathlessness. SARS-CoV-2 RT-PCR testing on admission showed low CT values for the E gene (CT value of 14.3) and N gene (CT value of 15.7) targets, and the patient was started on a third course of remdesivir (10-day course) and monitored as an inpatient. His chest X-ray showed no new acute changes and resolution of the previously seen right lower zone opacities. His respiratory symptoms improved significantly on day 54, and he was discharged on day 61. He continued to test positive on RT-PCR to the time of writing, on day 94. Nasopharyngeal swab samples from day 4 (PA_D4) and day 50 (PA_D50) underwent whole-genome sequencing to investigate for possible immune and/or therapeutic escape mutations. Figure 1A summarizes the timeline, relevant investigations and treatments, and key clinical events.
FIG 1.
Summary and timeline of SARS-CoV-2 RT-PCR positivity, serology results, treatments, and key clinical events for patient A (A) and patient B (B). SARS-CoV-2 RT-PCR E gene and N gene CT values were obtained during routine diagnostic testing of nasopharyngeal swab samples using the Cepheid Xpert Xpress SARS-CoV-2 assay. The TaqPath Combo kit (Thermo Fisher Scientific) additionally detected S gene and ORF1ab targets in both patients’ samples (circles, Xpert Xpress SARS-CoV-2 assay; squares, TaqPath Combo kit). Serology tests for IgG against the SARS-CoV-2 RBD and N protein were performed during routine diagnostic testing using the chemiluminescent microparticle immunoassay (CMIA) on the Architect i2000SR system (Abbott Laboratories). Hydrocortisone (patient A, days 50 to 55) and dexamethasone (patient B, days 2 to 11) dosages were converted to prednisolone-equivalent doses for ease of visualization. Data used to generate this figure are available in Supplemental tables 1A and 1B.
Patient B is a fully COVID-19-vaccinated (two doses of Pfizer-BioNTech vaccine) 67-year-old woman who was recently diagnosed with splenic marginal zone lymphoma, status post splenectomy in March 2021. She has a longstanding history of Evans syndrome, which has been complicated by recurrent admissions for symptomatic autoimmune hemolytic anemia, for which she received multiple courses of steroids and intravenous rituximab therapy in 2018. Prior to her admission for COVID-19, she had been managed on prednisolone (40 mg daily) and was receiving acyclovir and trimethoprim-sulfamethoxazole prophylaxis. Patient B presented to the emergency department on 13 November 2021 (day 1) for worsening cough and breathlessness. Chest X-rays obtained at admission showed bilateral perihilar and left lower zone consolidation with air space opacity. The patient tested positive for SARS-CoV-2 by RT-PCR (Xpert Xpress SARS-CoV-2) and was started on remdesivir and dexamethasone on the same day. One dose of REGN-COV2 was given on day 3. On day 5, the patient clinically deteriorated with episodes of hypotension and hypoxemia. She was transferred to the high-dependency unit and was started on high-flow oxygen supplementation. She progressed to type 1 respiratory failure, and she was intubated and started on mechanical ventilation on the following day (day 6). Endotracheal aspirate cultures grew Aspergillus fumigatus complex, and the patient was initially treated with voriconazole for COVID-19-associated pulmonary aspergillosis. While she was in the intensive care unit, her hypotension responded to fluid resuscitation and noradrenaline, and her oxygenation improved progressively. She was given a dose of tocilizumab while she was in the intensive care unit. On day 11, she was extubated and weaned down to 2 L/min of oxygen via nasal cannula. During her 6-day stay in the intensive care unit, she was noted to have arrhythmia and deranged liver function test results; as a result, remdesivir treatment was stopped. The patient completed a 7-day course of remdesivir. She was transferred out of the intensive care unit to the general ward on day 12. Computed tomography scans of the chest performed on day 14 showed diffuse ground-glass changes in bilateral lungs and bilateral segmental and subsegmental pulmonary embolisms, for which the patient was started on a therapeutic regimen of enoxaparin. She persistently tested positive for SARS-CoV-2 throughout her admission, with low CT values for E gene (CT value of 16.2) and N gene (CT value of 18.4) targets on day 27. Her prednisolone dose was tapered progressively to 5 mg daily in an attempt to reduce the degree of immunosuppression, to facilitate clearance of SARS-CoV-2. She was discharged from inpatient service to telemedicine monitoring on day 28. She was subsequently readmitted on day 34 for symptomatic anemia secondary to a flare of autoimmune hemolytic anemia, likely contributed by the relatively low dose of prednisolone (5 mg) on which she had been maintained since discharge. Upon admission, she was started on high-dose prednisolone (50 mg). Her SARS-CoV-2 RT-PCR results continued to be positive (E gene CT value of 17.8 and N gene CT value of 20.2), and she was started on a second (10-day) course of remdesivir. Her prednisolone dose was again tapered to 10 mg to facilitate viral clearance, but she was admitted again for symptomatic anemia on day 59. She was discharged on day 67 with high-dose prednisolone (50 mg), which was subsequently tapered gradually to 20 mg at the time of writing. On day 97, her SARS-CoV-2 RT-PCR results remained positive. Nasopharyngeal swab samples from day 1 (PB_D1), day 48 (PB_D48), and day 55 (PB_D55) underwent whole-genome sequencing to investigate for possible immune and/or therapeutic escape mutations. Figure 1B summarizes the timeline, relevant investigations and treatments, and key clinical events.
Whole-genome sequencing of samples from patient A (PA_D4 and PA_D50) and patient B (PB_D1, PB_D48, and PB_D55) was first performed on a MinION MK1b system (Oxford Nanopore Technologies [ONT], Oxford, UK) in accordance with the ARTIC Network protocol v3 (27). The RAMPART protocol (https://github.com/artic-network/rampart) was used to monitor the depth of coverage for each sample and to construct a draft genome. Sample PA_D50 was also sequenced using a metagenomic approach to improve coverage of the SARS-CoV-2 genome. All samples sequenced with the ARTIC Network protocol (PA_D4, PB_D1, PB_D48, and PB_D55) achieved coverage of >97% and an average depth of >462×. For all samples, total nucleic acid extraction of the chemically inactivated remnant samples was performed on the Promega Maxwell RSC 48 instrument, using the Maxwell RSC viral total nucleic acid purification kit (Promega, USA). cDNA synthesis was performed using the LunaScript RT SuperMix (New England BioLabs, USA) according to the ARTIC Network protocol (27). For metagenomic sequencing of PA_D50, library preparation was performed using the SQK-LSK 109 ligation sequencing kit (ONT) according to the manufacturer’s instructions. PA_D50 was monoplex sequenced on an FLO-MIN106D flow cell (R9.4.1) for 72 h and achieved 100% coverage and an average depth of 173×.
Guppy v5.0.7 (in super accuracy mode) was used for the basecalling of all samples. Consensus sequences were generated using the ARTIC Network pipeline for SARS-CoV-2 v1.2.1 (https://github.com/artic-network/artic-ncov2019). Within the ARTIC Network pipeline, Medaka v1.5.0 and Longshot v0.4.1 were chosen for variant calling. For PA_D50, reads that were aligned to the host genome (GRCh38) using minimap2 v2.17 were filtered out, and the remaining reads were aligned to the SARS-CoV-2 reference genome (GenBank accession number NC_045512.2). Allele frequency information was extracted from the VCF files before the final filtering step, to preserve variants with alternate allele frequencies of less than 0.5. Lineages of all consensus sequences were determined based on PANGO v3.1.17. Lists of mutations were retrieved from CalmBelt, our in-house tool for analyzing SARS-CoV-2 consensus sequences (28). This study was approved by the SingHealth Centralized Institutional Review Board (protocol 2013/397/F).
Consensus sequences of initial samples from patients A and B (PA_D4 and PB_D1, respectively) indicated that both patients were infected with the B.1.617.2 (Delta) variant. No signature Omicron single-nucleotide variations (SNVs) were found to have emerged over the course of investigation of these two cases. Since both patients had received two doses of Pfizer-BioNTech vaccines more than 4 weeks prior to being diagnosed with COVID-19 and had detectable anti-receptor-binding domain (RBD) IgG titers, we looked for and found the putative vaccine escape mutation L452R (29, 30) in the initial samples from both patients (PA_D4 and PB_D1).
For both patients, nasopharyngeal swab samples were sequenced before (PA_D4 and PB_D1) and after (PA_D50, PB_D48, and PB_D55) REGN-COV2 and remdesivir treatments. Compared to the initial sample for patient A (PA_D4), the day 50 sample (PA_D50) had five additional nonsynonymous mutations. The G5736T (A1006V) mutation occurred in the region encoding nonstructural protein 3 (NSP3), while the G27509T (T39I) mutation was found in the accessory protein open reading frame 7a (ORF7a) gene. The G15451A (G671S) mutation occurred in the NSP12 gene, which encodes a subunit of the RNA-dependent RNA polymerase (RdRp). Of note, two mutations (G22895T [V445F] and G22989A [G476D]) were found in the spike protein S1 subunit RBD. The mutations and associated allele frequencies of the samples from patient A are illustrated in Fig. 2A.
FIG 2.
(A) Heatmap showing the position and frequency of viral genome variations in the day 4 (PA_D4) and day 50 (PA_D50) samples for patient A, compared to the reference genome (GenBank accession number NC_045512.2). The intensity of the heatmap illustrates the variant frequencies. Data used to generate this figure are available in Data Set S2 in the supplemental material. (B) Day 1 (PB_D1), day 48 (PB_D48), and day 55 (PB_D55) samples for patient B, compared to the reference genome. Data used to generate this figure are available in Data Set S2. (C) Schematic displaying locations of deletions and mutations of SARS-CoV-2 genomes based on consensus sequences. The samples from patient A (PA) and patient B (PB), as well as other published reports of persistent infections in immunocompromised hosts (2, 5, 9, 11, 13), are aligned to illustrate the accumulation of mutations in the RBD region. Red lines, deletions; black lines, substitutions; blue lines, emergent mutations not found in the patient’s initial samples; *, mutation not found in GISAID sequences uploaded from Singapore between 1 November 2021 and 28 February 2022. NTD, N-terminal domain; FP, fusion peptide; TM, transmembrane domain; CT, cytoplasmic tail.
The day 48 (PB_D48) and day 55 (PB_D55) samples for patient B similarly showed accumulating mutations over time, compared to the initial sample (PB_D1). Similar to the findings for patient A, nonsynonymous mutations emerged in NSP3 (G4352A [E545K]) and the spike protein. Three spike protein mutations occurred in the S1 subunit RBD (G22899T [G466V], G22992T [S477I], and C23039G [Q493E]), and one was found in the S2 subunit heptapeptide repeat sequence 1 (HR1) region (C24378T [S939F]). One additional nonsynonymous mutation was observed in NSP2 (C1404T [P200L]). The mutations and associated allele frequencies of the samples from patient B are illustrated in Fig. 2B.
Whole-genome sequence comparison of samples from the patients in this case series showed the emergence and accumulation of mutations within a host over time. Despite differences in their clinical courses, both patients’ later samples demonstrated increased intrahost SARS-CoV-2 genome diversity. Of note, for both patients, more than one new major allele was found in the region encoding the S1 subunit RBD (Fig. 2A and B). This case series thus adds to the fast-accumulating evidence that immunocompromised hosts with persistent infections are important sources of SARS-CoV-2 genome diversity and, in particular, S gene diversity. Although patients in this case series were infected with the delta variant, our findings are in keeping with other recently reported cases of immunocompromised hosts with persistent SARS-CoV-2 infections caused by other variants (Fig. 2C).
The accumulation of mutations in the spike RBD is of particular concern because S protein is a key target for vaccines and therapeutics for SARS-CoV-2. Collectively, the patients in this study had new mutations at positions 445, 446, 476, 477, and 493 of the spike protein. Among these, substitutions at residue 477 were shown to be associated with broad resistance to monoclonal antibody panels (31). Substitutions at position 446 were reported to affect neutralization by both monoclonal antibodies and antibodies present in polyclonal sera (31–33), whereas substitutions at positions 445, 446, and 493 were reported to confer various degrees of monoclonal antibody neutralization resistance (34, 35). The day 50 sample for patient A had V445F and G476D substitutions, which were found to be partial escape mutations for REGN10987 (imdevimab) and REGN10933 (casirivimab), respectively, by deep mutational scanning (35). The day 55 sample for patient B similarly developed partial escape mutations G446V and Q493E for REGN10987 (imdevimab) and REGN10933 (casirivimab), respectively (35). These REGN-COV2 partial escape mutations in major alleles identified after REGN-COV2 therapy may be a consequence of SARS-CoV-2 intrahost evolution under selection pressure.
In this case series, both patients received multiple courses of remdesivir. Therefore, we examined the genome sequences for known remdesivir resistance mutations. No known or putative mutation conferring remdesivir resistance was found (36).
To understand whether patients A and B might have acquired a new SARS-CoV-2 infection in the period between the initial and subsequent samples, we examined the frequency of their new mutations in the GISAID database (https://www.gisaid.org). Among 4,255 GISAID sequences uploaded from Singapore between 1 November 2021 and 28 February 2022, no other sequence harbors the C5736T, G22895T, or G22989A mutations found in the day 50 sample for patient A. Similarly, the G4352A and C23039G mutations in the samples from patient B were not found in the same data set. Among >3 million GISAID sequences uploaded globally between 1 November 2021 and 28 February 2022, the occurrence of these mutations ranged from 0.001% to 0.017%. The data suggest that patients A and B were unlikely to have acquired a new infection with a separate SARS-CoV-2 strain with these mutations.
There are several limitations to this study. Although prolonged SARS-CoV-2 RT-PCR positivity was established, viral culture of the samples was not performed to confirm virus viability or infectivity. Furthermore, the application of whole-genome analysis provided a static glimpse of the dynamic viral subpopulations in the hosts. ONT sequencing was used in our setting within a hospital diagnostic environment, to reduce costs and turnaround time. Although it has been established that SARS-CoV-2 ONT sequencing has high accuracy, compared to short-read sequencing, ONT sequencing generally achieves lower sequencing depth, compared to short-read sequencing (37).
Nonetheless, this case series adds to the literature two cases of prolonged SARS-CoV-2 infection, with the emergence of clinically important mutations, in immunocompromised hosts. Relatively low-cost and simple on-site SARS-CoV-2 whole-genome sequence analysis identified putative vaccine escape mutations in the initial samples, as well as the emergence of partial therapeutic escape mutations in the subsequent samples. This emergence and accumulation of escape mutations, as well as the prolonged course of infection, put individual vulnerable hosts at risk of therapeutic failures and severe infections. Furthermore, the emergence and spread of such variants harboring vaccine and therapeutic escape mutations have important public health and infection control implications. Although some jurisdictions have progressively lifted pandemic restrictions, tertiary centers caring for immunocompromised patients need to exercise continued vigilance and containment measures against such emergent variants. Routine and near-real-time SARS-CoV-2 genomic surveillance in this subgroup is necessary for early detection and containment of mutations and variants of therapeutic and public health importance.
Data availability.
Sequence data were submitted to GenBank with the following accession numbers: OM881791 (PB_D55), OM881787 (PB_D48), OM881786 (PA_D50), OM877509 (PB_D1), and OM877508 (PA_D4).
ACKNOWLEDGMENTS
We thank B. H. Tan for his foresight and T. H. Koh for his valuable comments.
The sequencing of SARS-CoV-2 samples was funded by Singapore General Hospital. K.K.K.Ko. is supported by the Singapore National Medical Research Council Research Training Fellowship. This work was also supported by funding from A*STAR and Chiang Mai University.
Footnotes
Supplemental material is available online only.
Contributor Information
Karrie K. K. Ko, Email: karrie.ko.k.k@singhealth.com.sg.
Heba H. Mostafa, Johns Hopkins Hospital
REFERENCES
- 1.Avanzato VA, Matson MJ, Seifert SN, Pryce R, Williamson BN, Anzick SL, Barbian K, Judson SD, Fischer ER, Martens C, Bowden TA, de Wit E, Riedo FX, Munster VJ. 2020. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 183:1901–1912.e9. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, Solomon IH, Kuo H-H, Boucau J, Bowman K, Adhikari UD, Winkler ML, Mueller AA, Hsu TY-T, Desjardins M, Baden LR, Chan BT, Walker BD, Lichterfeld M, Brigl M, Kwon DS, Kanjilal S, Richardson ET, Jonsson AH, Alter G, Barczak AK, Hanage WP, Yu XG, Gaiha GD, Seaman MS, Cernadas M, Li JZ. 2020. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kemp SA, Collier DA, Datir RP, Ferreira IATM, Gayed S, Jahun A, Hosmillo M, Rees-Spear C, Mlcochova P, Lumb IU, Roberts DJ, Chandra A, Temperton N, CITIID-NIHR BioResource COVID-19 Collaboration, COVID-19 Genomics UK (COG-UK) Consortium, Sharrocks K, Blane E, Modis Y, Leigh KE, Briggs JAG, van Gils MJ, Smith KGC, Bradley JR, Smith C, Doffinger R, Ceron-Gutierrez L, Barcenas-Morales G, Pollock DD, Goldstein RA, Smielewska A, Skittrall JP, Gouliouris T, Goodfellow IG, Gkrania-Klotsas E, Illingworth CJR, McCoy LE, Gupta RK. 2021. SARS-CoV-2 evolution during treatment of chronic infection. Nature 592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corey L, Beyrer C, Cohen MS, Michael NL, Bedford T, Rolland M. 2021. SARS-CoV-2 variants in patients with immunosuppression. N Engl J Med 385:562–566. doi: 10.1056/NEJMsb2104756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weigang S, Fuchs J, Zimmer G, Schnepf D, Kern L, Beer J, Luxenburger H, Ankerhold J, Falcone V, Kemming J, Hofmann M, Thimme R, Neumann-Haefelin C, Ulferts S, Grosse R, Hornuss D, Tanriver Y, Rieg S, Wagner D, Huzly D, Schwemmle M, Panning M, Kochs G. 2021. Within-host evolution of SARS-CoV-2 in an immunosuppressed COVID-19 patient as a source of immune escape variants. Nat Commun 12:6405. doi: 10.1038/s41467-021-26602-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark SA, Clark LE, Pan J, Coscia A, McKay LGA, Shankar S, Johnson RI, Brusic V, Choudhary MC, Regan J, Li JZ, Griffiths A, Abraham J. 2021. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell 184:2605–2617.e18. doi: 10.1016/j.cell.2021.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Zody MC, Di Germanio C, Martinelli R, Mediavilla JR, Cunningham MH, Composto K, Chow KF, Kordalewska M, Corvelo A, Oschwald DM, Fennessey S, Zetkulic M, Dar S, Kramer Y, Mathema B, Germer S, Stone M, Simmons G, Busch MP, Maniatis T, Perlin DS, Kreiswirth BN. 2021. Emergence of multiple SARS-CoV-2 antibody escape variants in an immunocompromised host undergoing convalescent plasma treatment. mSphere 6:e00480-21. doi: 10.1128/mSphere.00480-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cele S, Karim F, Lustig G, San JE, Hermanus T, Tegally H, Snyman J, Moyo-Gwete T, Wilkinson E, Bernstein M, Khan K, Hwa S-H, Tilles SW, Singh L, Giandhari J, Mthabela N, Mazibuko M, Ganga Y, Gosnell BI, Karim SSA, Hanekom W, Van Voorhis WC, Ndung’u T, COMMIT-KZN Team, Lessells RJ, Moore PL, Moosa M-YS, de Oliveira T, Sigal A. 2022. SARS-CoV-2 prolonged infection during advanced HIV disease evolves extensive immune escape. Cell Host Microbe 30:154–162.e5. doi: 10.1016/j.chom.2022.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffman SA, Costales C, Sahoo MK, Palanisamy S, Yamamoto F, Huang C, Verghese M, Solis DA, Sibai M, Subramanian A, Tompkins LS, Grant P, Shafer RW, Pinsky BA. 2021. SARS-CoV-2 neutralization resistance mutations in patient with HIV/AIDS, California, USA. Emerg Infect Dis 27:2720–2723. doi: 10.3201/eid2710.211461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truong TT, Ryutov A, Pandey U, Yee R, Goldberg L, Bhojwani D, Aguayo-Hiraldo P, Pinsky BA, Pekosz A, Shen L, Boyd SD, Wirz OF, Röltgen K, Bootwalla M, Maglinte DT, Ostrow D, Ruble D, Han JH, Biegel JA, Li M, Huang C, Sahoo MK, Pannaraj PS, O'Gorman M, Judkins AR, Gai X, Dien Bard J. 2021. Increased viral variants in children and young adults with impaired humoral immunity and persistent SARS-CoV-2 infection: a consecutive case series. EBioMedicine 67:103355. doi: 10.1016/j.ebiom.2021.103355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bazykin GA, Stanevich O, Danilenko D, Fadeev A, Komissarova K, Ivanova A, Sergeeva M, Safina K, Nabieva E, Klink G, Garushyants S, Zabutova J, Kholodnaia A, Skorokhod I, Ryabchikova VV, Komissarov A, Lioznov D. 2021. Emergence of Y453F and Δ69-70HV mutations in a lymphoma patient with long-term COVID-19. Virologicalorg. https://virological.org/t/emergence-of-y453f-and-69-70hv-mutations-in-a-lymphoma-patient-with-long-term-covid-19/580. [Google Scholar]
- 12.Leung WF, Chorlton S, Tyson J, Al-Rawahi GN, Jassem AN, Prystajecky N, Masud S, Deans GD, Chapman MG, Mirzanejad Y, Murray MCM, Wong PHP. 2022. COVID-19 in an immunocompromised host: persistent shedding of viable SARS-CoV-2 and emergence of multiple mutations: a case report. Int J Infect Dis 114:178–182. doi: 10.1016/j.ijid.2021.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karim F, Moosa MYS, Gosnell BI, Cele S, Giandhari J, Pillay S, Tegally H, Wilkinson E, San JE, Msomi N, Mlisana K, Khan K, Bernstein M, Manickchund N, Singh L, Ramphal U, COMMIT-KZN Team, Hanekom W, Lessells RJ, Sigal A, de Oliveira T. 2021. Persistent SARS-CoV-2 infection and intra-host evolution in association with advanced HIV infection. medRxiv. doi: 10.1101/2021.06.03.21258228. [DOI] [Google Scholar]
- 14.Fung M, Babik JM. 2021. COVID-19 in immunocompromised hosts: what we know so far. Clin Infect Dis 72:340–350. doi: 10.1093/cid/ciaa863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Messika J, Eloy P, Roux A, Hirschi S, Nieves A, Le Pavec J, Sénéchal A, Saint Raymond C, Carlier N, Demant X, Le Borgne A, Tissot A, Debray M-P, Beaumont L, Renaud-Picard B, Reynaud-Gaubert M, Mornex J-F, Falque L, Boussaud V, Jougon J, Mussot S, Mal H, French Group of Lung Transplantation. 2021. COVID-19 in lung transplant recipients. Transplantation 105:177–186. doi: 10.1097/TP.0000000000003508. [DOI] [PubMed] [Google Scholar]
- 16.Graff K, Smith C, Silveira L, Jung S, Curran-Hays S, Jarjour J, Carpenter L, Pickard K, Mattiucci M, Fresia J, McFarland EJ, Dominguez SR, Abuogi L. 2021. Risk factors for severe COVID-19 in children. Pediatr Infect Dis J 40:e137–e145. doi: 10.1097/INF.0000000000003043. [DOI] [PubMed] [Google Scholar]
- 17.Goldman JD, Robinson PC, Uldrick TS, Ljungman P. 2021. COVID-19 in immunocompromised populations: implications for prognosis and repurposing of immunotherapies. J Immunother Cancer 9:e002630. doi: 10.1136/jitc-2021-002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair V, Jandovitz N, Hirsch JS, Nair G, Abate M, Bhaskaran M, Grodstein E, Berlinrut I, Hirschwerk D, Cohen SL, Davidson KW, Dominello AJ, Osorio GA, Richardson S, Teperman LW, Molmenti EP. 2020. COVID-19 in kidney transplant recipients. Am J Transplant 20:1819–1825. doi: 10.1111/ajt.15967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, Nehlmeier I, Graichen L, Moldenhauer A-S, Winkler MS, Lier M, Dopfer-Jablonka A, Jäck H-M, Behrens GMN, Pöhlmann S. 2022. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell 185:447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, Bolland W-H, Porrot F, Staropoli I, Lemoine F, Péré H, Veyer D, Puech J, Rodary J, Baele G, Dellicour S, Raymenants J, Gorissen S, Geenen C, Vanmechelen B, Wawina-Bokalanga T, Martí-Carreras J, Cuypers L, Sève A, Hocqueloux L, Prazuck T, Rey F, Simon-Loriere E, Bruel T, Mouquet H, André E, Schwartz O. 2022. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 21.Liu L, Iketani S, Guo Y, Chan JF-W, Wang M, Liu L, Luo Y, Chu H, Huang Y, Nair MS, Yu J, Chik KK-H, Yuen TT-T, Yoon C, To KK-W, Chen H, Yin MT, Sobieszczyk ME, Huang Y, Wang HH, Sheng Z, Yuen K-Y, Ho DD. 2022. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 22.Dejnirattisai W, Huo J, Zhou D, Zahradník J, Supasa P, Liu C, Duyvesteyn HME, Ginn HM, Mentzer AJ, Tuekprakhon A, Nutalai R, Wang B, Dijokaite A, Khan S, Avinoam O, Bahar M, Skelly D, Adele S, Johnson SA, Amini A, Ritter TG, Mason C, Dold C, Pan D, Assadi S, Bellass A, Omo-Dare N, Koeckerling D, Flaxman A, Jenkin D, Aley PK, Voysey M, Costa Clemens SA, Naveca FG, Nascimento V, Nascimento F, Fernandes da Costa C, Resende PC, Pauvolid-Correa A, Siqueira MM, Baillie V, Serafin N, Kwatra G, Da Silva K, Madhi SA, Nunes MC, Malik T, Openshaw PJM, Baillie JK, Semple MG, Townsend AR, Huang K-YA, Tan TK, Carroll MW, Klenerman P, et al. 2022. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cele S, Jackson L, Khoury DS, Khan K, Moyo-Gwete T, Tegally H, San JE, Cromer D, Scheepers C, Amoako DG, Karim F, Bernstein M, Lustig G, Archary D, Smith M, Ganga Y, Jule Z, Reedoy K, Hwa S-H, Giandhari J, Blackburn JM, Gosnell BI, Abdool Karim SS, Hanekom W, NGS-SA, COMMIT-KZN Team, von Gottberg A, Bhiman JN, Lessells RJ, Moosa MS, Davenport MP, de Oliveira T, Moore PL, Sigal A. 2022. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.WHO Team Emergency Response. 2022. Weekly epidemiological update on COVID-19: 8 February 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---8-february-2022.
- 25.De Foo C, Grépin KA, Cook AR, Hsu LY, Bartos M, Singh S, Asgari N, Teo YY, Heymann DL, Legido-Quigley H. 2021. Navigating from SARS-CoV-2 elimination to endemicity in Australia, Hong Kong, New Zealand, and Singapore. Lancet 398:1547–1551. doi: 10.1016/S0140-6736(21)02186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight SR, Ho A, Pius R, Buchan I, Carson G, Drake TM, Dunning J, Fairfield CJ, Gamble C, Green CA, Gupta R, Halpin S, Hardwick HE, Holden KA, Horby PW, Jackson C, Mclean KA, Merson L, Nguyen-Van-Tam JS, Norman L, Noursadeghi M, Olliaro PL, Pritchard MG, Russell CD, Shaw CA, Sheikh A, Solomon T, Sudlow C, Swann OV, Turtle LC, Openshaw PJ, Baillie JK, Semple MG, Docherty AB, Harrison EM, ISARIC4C Investigators. 2020. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C mortality score. BMJ 370:m3339. doi: 10.1136/bmj.m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Artic Network. 2020. SARS-CoV-2. https://artic.network/ncov-2019.
- 28.Yingtaweesittikul H, Ko K, Abdul Rahman N, Tan SYL, Nagarajan N, Suphavilai C. 2021. CalmBelt: rapid SARS-CoV-2 genome characterization for outbreak tracking. Front Med (Lausanne) 8:790662. doi: 10.3389/fmed.2021.790662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, COVID-19 Genomics UK (COG-UK) Consortium, Peacock SJ, Robertson DL. 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R, Chen J, Gao K, Wei G-W. 2021. Vaccine-escape and fast-growing mutations in the United Kingdom, the United States, Singapore, Spain, India, and other COVID-19-devastated countries. Genomics 113:2158–2170. doi: 10.1016/j.ygeno.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, VanBlargan LA, Bloyet L-M, Rothlauf PW, Chen RE, Stumpf S, Zhao H, Errico JM, Theel ES, Liebeskind MJ, Alford B, Buchser WJ, Ellebedy AH, Fremont DH, Diamond MS, Whelan SPJ. 2021. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 29:477–488.e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, Bloom JD. 2021. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, Zhao C, Zhang Q, Liu H, Nie L, Qin H, Wang M, Lu Q, Li X, Sun Q, Liu J, Zhang L, Li X, Huang W, Wang Y. 2020. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann H-H, Michailidis E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Luchsinger L, Hillyer CD, Caskey M, Robbiani DF, Rice CM, Nussenzweig MC, Hatziioannou T, Bieniasz PD. 2020. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, Li JZ, Bloom JD. 2021. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Focosi D, Maggi F, McConnell S, Casadevall A. 2022. Very low levels of remdesivir resistance in SARS-COV-2 genomes after 18 months of massive usage during the COVID19 pandemic: a GISAID exploratory analysis. Antiviral Res 198:105247. doi: 10.1016/j.antiviral.2022.105247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bull RA, Adikari TN, Ferguson JM, Hammond JM, Stevanovski I, Beukers AG, Naing Z, Yeang M, Verich A, Gamaarachchi H, Kim KW, Luciani F, Stelzer-Braid S, Eden J-S, Rawlinson WD, van Hal SJ, Deveson IW. 2020. Analytical validity of Nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat Commun 11:6272. doi: 10.1038/s41467-020-20075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00791-22-s001.xlsx, XLSX file, 0.1 MB (88KB, xlsx)
Supplemental material. Download spectrum.00791-22-s002.xlsx, XLSX file, 0.01 MB (12.7KB, xlsx)
Supplemental material. Download spectrum.00791-22-s003.xlsx, XLSX file, 0.01 MB (15.1KB, xlsx)
Data Availability Statement
Sequence data were submitted to GenBank with the following accession numbers: OM881791 (PB_D55), OM881787 (PB_D48), OM881786 (PA_D50), OM877509 (PB_D1), and OM877508 (PA_D4).