ABSTRACT
Populus euphratica Oliv. has a high tolerance for drought, salinity, and alkalinity. The main purpose of this study is to explore the effects of environments of different salinity intensities on endophytic community structure and the possible roles of endophytes in the tolerance of host plants. The characterization of endogenous bacteria in diversity has been investigated by using the Illumina high-throughput sequencing technique. The research showed that endophytic bacteria of P. euphratica in an extremely saline environment had low species diversity, especially in sap tissue. The dominant phyla in all groups were Proteobacteria, Actinobacteria, and Bacteroidetes. Notably, Firmicutes (relative abundance >5%) was a different dominant phylum in the samples from the high-saline environment compared with the relatively low-saline-environment group. The linear discriminant analysis effect size (LEfSe) analysis found that there were significant differences in different saline environments of Cytophagaceae (family), Rhodobacteraceae (family), and Rhodobacterales (order). These results indicated that the composition of the endogenous bacterial community was related to the growth environment of host plants. The predictive analysis of KEGG pathways and enzymes showed that the abundance of some enzymes and metabolic pathways of endophytes of P. euphratica increased with the increase of soil salinity, and most of the enzymes were related to energy metabolism and carbohydrate metabolism. These findings suggested that the endogenous bacteria of the host plant had different expression mechanisms under different degrees of stress, and this mechanism was very obvious in the distribution of endophytes, while the function of the endogenous bacteria needs to be further explored.
IMPORTANCE Euphrates poplar (Populus euphratica Oliv.), as the only tree species that grows in the desert, has tenacious vitality with the characteristics of cold tolerance, drought tolerance, salt-alkali tolerance, and wind-sand resistance. P. euphratica has a long growth cycle and a high growth rate, which can break wind, fix sand, green the environment, and protect farmland, making it an important afforestation tree species in arid and semiarid areas. The area of P. euphratica in Xinjiang accounts for 91.1% of its area in China. Studying the endophytic bacteria of P. euphratica can give people a systematic understanding of it and the adaptability of the endogenous flora to the host and special environments. In this study, by analyzing the endophytic bacteria of P. euphratica in different saline-alkali regions of Xinjiang, it was found that the bacteria in different tissues of P. euphratica changed with the change of soil salinity. Especially in the sap tissue of P. euphratica under extremely high salinity, the diversity of endogenous bacteria was significantly lower than that in other tissues. These differential bacteria under different salinities were mostly related to the stress resistance of themselves and the host. Not only that, we also selected a strain of Bacillus with high stress resistance from the tissues of P. euphratica, which can survive under the extreme conditions of 10% NaCl and pH 11. We obtained a genome completion map of this strain, named it Bacillus haynesii P19 (GenBank accession no. PRJNA648288), and tried to use it for fermentation but in a different work, so as to develop it into a promising industrial fermentation chassis bacterium. Therefore, this study was of great significance for the understanding of endophytic bacteria in P. euphratica and the acquisition of extremophilic microbial resources.
KEYWORDS: Populus euphratica, endophytic bacteria, saline environment
INTRODUCTION
Salt-affected soils (SAS) are a global issue. Data from the Food and Agriculture Organization of the United Nations (FAO) indicates that greater than 424 million hectares of topsoil (0 to 30 cm) and 833 million hectares of subsoil (30 to 100 cm) are salt affected: 85% of salt-damaged topsoils are saline, 10% are sodic, and 5% are saline-sodic, while 62% of salt-affected subsoils are saline, 24% are sodic, and 14% are saline-sodic (1). Among them, more than two-thirds of global SAS are found in arid and semiarid climatic zones, covering five continents except Antarctica: 37% of SAS are located in arid deserts and 27% of SAS are distributed in the arid steppe (half in the cold arid steppe and half in the hot arid steppe). These data also show that 20% to 50% of irrigated soils on all continents are too saline; this means that more than 1.5 billion people in the world are facing major challenges in food production due to soil degradation. Therefore, it is urgent for humankind to prevent soil salinization and improve biodiversity and the ecological environment.
For the prevention and control of SAS, one of the methods is to plant salt-tolerant plants (2). However, due to climate, topography, and landforms, the distribution area of SAS has little precipitation and considerable evaporation. The annual precipitation is not enough to wash away the accumulated salt on the surface of the soil. The accumulation of salt makes the vegetation that can grow very limited. Most plants are herbs and shrubs, and there are extremely few tall trees (3). For example, in Australia, the typical plants of Eucalyptus growing in the dry subtropical regions of the southeast and southwest mostly appear as bushes under the influence of geography and climate (4). The characteristic vegetation of the Nevada desert in the United States is the desert scrub Larrea tridentata-Ambrosia dumosa (5). Euphrates poplar (Populus euphratica Oliv.), as the only tree species that grows in the desert, has tenacious vitality with the characteristics of cold tolerance, drought tolerance, salt-alkali tolerance, and wind-sand resistance (6). P. euphratica has a long growth cycle and a high growth rate, which can break wind, fix sand, green the environment, and protect farmland, making it an important afforestation tree species in arid and semiarid areas (7).
In the past 30 years, the main research on plants in saline-alkali land has focused on the resistance of plants to salt stress. Due to the high adaptability of P. euphratica to the extreme desert environment, it has become an important model species for studying the effects of abiotic stresses on trees (8). Research on the molecular biology of P. euphratica has made progress in genetic diversity (9) and functional genes, genome, and transcriptomics (10). With the development of microbiology research, researchers found that the plant’s endophytic microorganisms have a close relationship with the host and have a positive impact on the adaptability to the host’s special environment. It has been demonstrated that in areas with abiotic stress factors, plants are more dependent on microorganisms that are able to enhance their ability to combat stress (11).
However, the research on the endogenous bacteria of P. euphratica mainly focuses on the isolation and identification of culturable endophytic microorganisms (12) and the performance of single bacteria isolated from Populus euphratica (13), while the understanding of the diversity and community structure of its endophytic bacteria is still lacking. In China, P. euphratica is primarily distributed in Xinjiang, Inner Mongolia, Qinghai, Gansu, and Ningxia. Among them, the area of P. euphratica growth in Xinjiang accounts for 91.1% of its growth in China. Studying the endophytic bacteria of P. euphratica can give people a systematic understanding of it and the adaptability of the endogenous flora to the host and special environments.
Therefore, this study analyzed the endophytic bacteria of P. euphratica on different saline-alkali soils in Xinjiang, observed whether the endophytic bacterium was stable or changed with the increase of the salt-alkali concentration, and clarified whether the endogenous bacteria of P. euphratica were related only to its own saline-alkali environment or would alter with variations in the external environment. If there is a change, is it related to the function of the endophytic bacteria? The findings could provide clues for the systematic recognition of endogenous bacterial groups in P. euphratica, especially the response of endophytic bacteria to the changes of salt environment, and the description of the interaction between the environment and the endogenous bacteria of the plant.
RESULTS AND DISCUSSION
Soil physicochemical properties of P. euphratica growth area.
We collected and detected the physicochemical properties of the soils where P. euphratica grows, and the results showed that SO42− and Cl− were the predominant anions in the soils from both areas. In addition, the main cations of the soils in Zephyr County were Na+ and K+, while the soils from the Darbancheng District had an individually high content of Ca2+ in addition to Na+ and K+. The pH values of soil samples ZP_8 and ZP_9 collected from Zephyr County were 9.11 and 8.81, respectively, and the electrical conductivity (ECe) values were 10.11 dS/m and 18.95 dS/m, respectively, while the pH values of soil samples DBC_8 and DBC_9 collected from the Darbancheng District were 7.13 and 7.09 and the ECe values were 3.69 dS/m and 6.58 dS/m, respectively (Table 1).
TABLE 1.
Soil physicochemical properties of soil samples
| Soil physicochemical property | Value for soil type and sample |
|||
|---|---|---|---|---|
| S_saline, ZP_9 | H_saline, ZP_8 | M_saline, DBC_9 | L_saline, DBC_8 | |
| Content (g/kg) | ||||
| CO32− | 0.061 | 0.014 | 0.000 | 0.000 |
| HCO3− | 0.366 | 0.202 | 0.328 | 0.236 |
| Cl− | 9.208 | 3.656 | 2.010 | 0.937 |
| Ca2+ | 0.600 | 1.350 | 4.340 | 2.670 |
| Mg2+ | 5.835 | 0.838 | 0.525 | 0.262 |
| SO42− | 7.685 | 1.567 | 2.683 | 2.016 |
| K+ | 0.523 | 8.049 | 0.158 | 0.129 |
| Na+ | 3.486 | 1.236 | 1.252 | 0.559 |
| pH | 8.810 | 9.110 | 7.090 | 7.130 |
| ECe (dS/m) | 18.950 | 10.110 | 6.580 | 3.690 |
| Salinity intensity (FAO) | Extreme | Very strong | Robust | Moderate |
The FAO considers soils with ECe values of >2 dS/m and pH >8.2 to be SAS. According to this standard, the soil samples we collected all belonged to SAS. Among them, an ECe value of >15 is extreme salinity intensity, and the ECe values between 8 and 15 are very strong salinity intensity. The ECe values between 4 and 8 are robust salinity intensity, and the ECe values between 2 and 4 are moderate salinity intensity. Therefore, we grouped ZP_9, ZP_8, DBC_9, and DBC_8 into S_saline, H_saline, M_saline, and L_saline. Since P. euphratica grows poorly in a hot, moist climate and heavy clay soil, it mostly lives in a dry climate (14). We did not observe the growth of P. euphratica in healthy soil under the natural environment of Xinjiang, so we could compare the differences in the endophytic microbial community of P. euphratica only under various salinities. There was no way to use P. euphratica grown in nonsaline soil as a control.
Alpha diversity of the microbial community.
In order to understand the endophytic microbial community of P. euphratica in different saline environments, we analyzed the alpha diversity index of each sample by MiSeq high-throughput sequencing, including the read numbers, coverage, Shannon index, and Chao index of each sample at a genetic distance of 3% (Table 2). The samples collected from the P. euphratica forest can be divided into three types: sap, xylem, and soil. Through alpha diversity analysis, we can get the relevant index reflecting the diversity and richness of the microbial community in the samples. Quality and chimera filtration of the raw data produced in total 680,897 high-quality sequencing Reads, ranging from 10,681 bp to 61,392 bp. The coverage values of all samples were greater than 0.99, which indicated that the sequencing depth was sufficient to cover most microorganisms.
TABLE 2.
Alpha diversity index of each sample of P. euphratica
| Group | Sample type | Sample identifier | No. of reads | Coverage | Alpha diversity |
|
|---|---|---|---|---|---|---|
| Shannon | Chao | |||||
| S_saline | ||||||
| S_saline_sap | Sap | ZP_1 | 10,681 | 0.99737 | 2.69762 | 140.0000 |
| ZP_2 | 17,325 | 0.9974 | 2.79639 | 230.6666 | ||
| S_saline_xylem | Xylem | ZP_4 | 31,909 | 0.99511 | 4.71727 | 927.9285 |
| ZP_5 | 32,844 | 0.99558 | 4.94794 | 815.3170 | ||
| S_saline_soil | Soil | ZP_9 | 47,914 | 0.99647 | 5.36877 | 1,295.3050 |
| H_saline | ||||||
| H_saline_sap | Sap | ZP_3 | 46,217 | 0.99645 | 4.44534 | 922.3589 |
| H_saline_xylem | Xylem | ZP_6 | 44,580 | 0.99748 | 3.84021 | 573.5384 |
| ZP_7 | 51,985 | 0.9978 | 4.27084 | 745.7205 | ||
| H_saline_soil | Soil | ZP_8 | 45,586 | 0.99618 | 4.55783 | 855.5816 |
| M_saline | ||||||
| M_saline_sap | Sap | DBC_1 | 35,509 | 0.99659 | 4.76852 | 691.4375 |
| M_saline_xylem | Xylem | DBC_5 | 26,235 | 0.99733 | 4.54128 | 491.5660 |
| M_saline_soil | Soil | DBC_9 | 32,787 | 0.99594 | 6.00210 | 1,387.0720 |
| L_saline | ||||||
| L_saline_sap | Sap | DBC_2 | 29,563 | 0.99644 | 4.31679 | 615.2222 |
| DBC_3 | 45,454 | 0.9967 | 4.91200 | 944.8829 | ||
| L_saline_xylem | Xylem | DBC_4 | 36,439 | 0.99596 | 4.91057 | 793.8658 |
| DBC_6 | 61,392 | 0.99747 | 5.14457 | 1,062.0670 | ||
| DBC_7 | 42,884 | 0.99706 | 4.92196 | 749.8088 | ||
| L_saline_soil | Soil | DBC_8 | 41,593 | 0.99689 | 4.89385 | 844.0153 |
The Chao index primarily reflects the number of operational taxonomic unit (OTU) species, while the Shannon index is relative to OTU diversity. The results showed that the Shannon index and Chao index of the sap tissue of P. euphratica under extremely high salinity were modest compared with other groups, indicating that its microbial number and diversity were both lower. Similar results were also found in saline samples from the Arava Valley, of which the microbial diversity was limited because of the extreme habitats (15). However, the Shannon and Chao indexes of xylem and soil samples of P. euphratica under different salinities did not differ greatly. Therefore, we can speculate that when the soil salinity is too high, it will affect the diversity of the endogenous bacteria of the sap tissue of P. euphratica but will not have much impact on the xylem.
Microbial community composition.
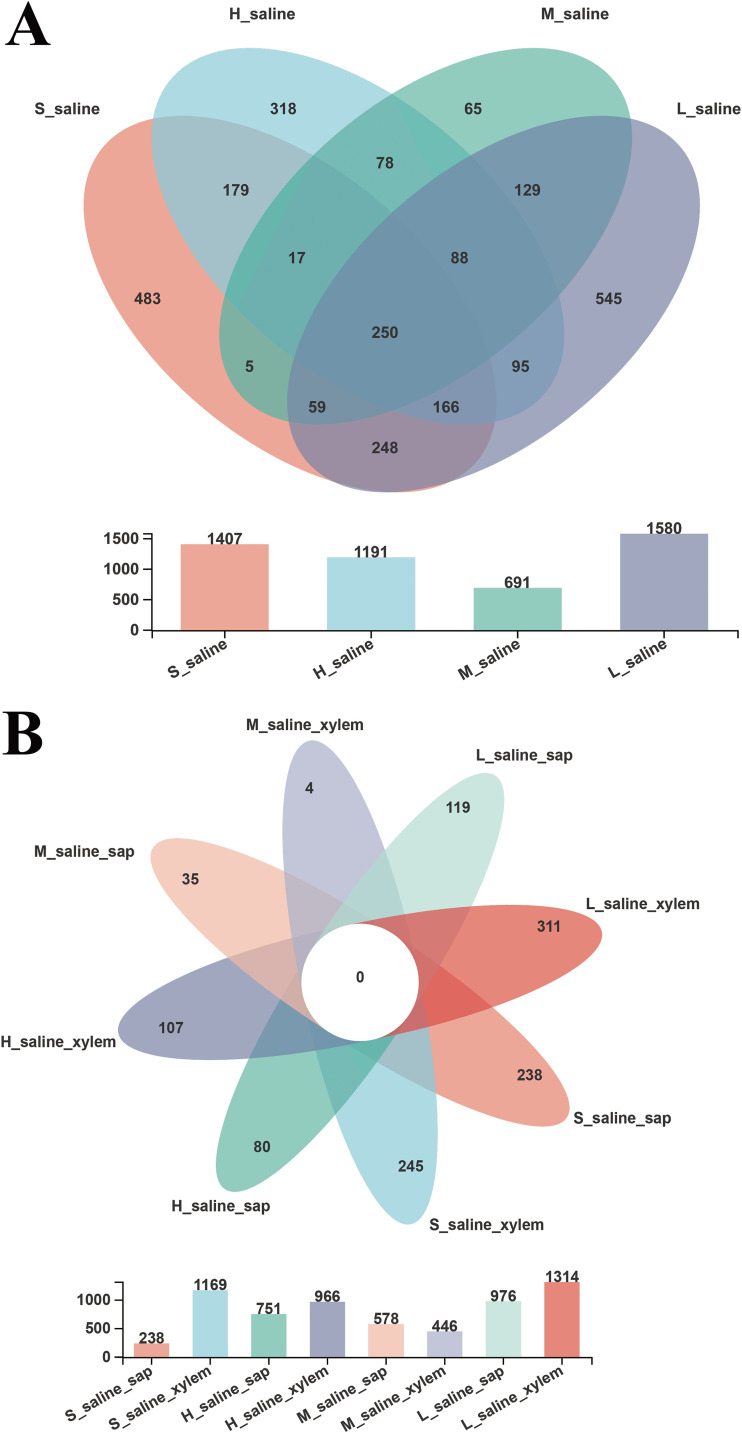
For further investigation of the composition of endophytic bacteria in each tissue sample of P. euphratica under different salinities, we constructed a Venn diagram to identify the number of OTUs presented in these groups (Fig. 1). Figure 1A showed that the numbers of group-specific OTUs of each group were 483, 318, 65, and 545 according to soil salinity from high to low. There were 250 shared OTUs in the four groups under different salinities, accounting for 5.13% of the total OTUs. These shared OTUs were composed of a number of bacterial groups, including Firmicutes, Proteobacteria, Chloroflexi, Actinobacteria, and Bacteroidetes. Previous studies also have shown that the majority of the isolated strains belonged to Firmicutes, Proteobacteria, and Actinobacteria (12). For the different tissues of P. euphratica under various salinities (Fig. 1B), the sap tissue of P. euphratica under extremely high salinity had the fewest OTUs (218), which was consistent with the results of alpha diversity analysis.
FIG 1.
Comparison of OTUs of P. euphratica under different salinities (A) and from different tissues (B) by Venn diagram (sequence similarity greater than 97% is classified as an OTU).
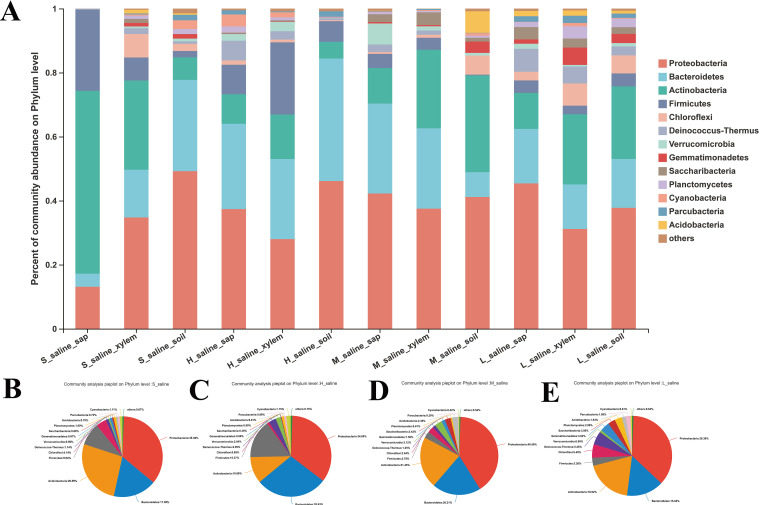
Then, we clustered these OTUs at the phylum level with a 70% threshold. The results were presented by bar and pie charts. All OTUs were identified as 37 prokaryotic phyla. Figure 2 showed the community composition at the phylum level in varied tissues (Fig. 2A) and different salinities (Fig. 2B to E) of P. euphratica (combining the species that account for less than 1% of the abundance in the sample as “others”).
FIG 2.
Microbiota composition of different tissues (A) and different salinities (B to E) of P. euphratica at phylum level (the phyla that account for less than 1% of the abundance in the sample are merged into “others”).
From the bar chart (Fig. 2A), we can see that the endophytes of P. euphratica were mainly concentrated in 13 phyla including Proteobacteria, Bacteroidetes, Actinobacteria, Firmicutes, Chloroflexi, Deinococcus-Thermus, Verrucomicrobia, Gemmatimonadetes, Saccharibacteria, Planctomycetes, Cyanobacteria, Parcubacteria, and Acidobacteria. Among them, Proteobacteria was the most abundant phylum across all tissues, and the relative abundance ranged from 13.12% to 49.22%. Most of the Proteobacteria in nature are Gram-negative bacteria. They are widely distributed and are important mediators of the nitrogen, sulfur, and carbon cycle in the ecosystem (16). The other two dominant phyla were Actinobacteria and Bacteroidetes. The relative abundance of the former ranged from 5.24% to 51%, and that of the latter ranged from 4.09% to 38.26%. Actinomycetes are known to colonize the internal tissues of plants and play a role in pathogen defense and growth regulation of their hosts (17). The Bacteroidetes are very diverse, and environmental Bacteroidetes can degrade complex organic matter (18).
It is worth noting that Firmicutes was another dominant phylum in the tissues of P. euphratica, and the relative abundance ranged from 0.25% to 25.36% (Fig. 2A). The relative abundances of the Firmicutes in the four groups under various salinities were quite different, 9.04% and 15.37% in the extreme saline group and the very robust saline group (Fig. 2B and C) and 2.79% and 3.26% in the robust saline group and the moderate saline group (Fig. 2D and E). Formation of endospores is a specific property of Firmicutes, which can resist excessive conditions by producing these spores (19). From the higher abundance of Firmicutes, it can be inferred that the high-saline environment will affect the composition of the endophytic microbiota of P. euphratica, and the Firmicutes with high stress resistance can survive to a greater degree in a high-pH and high-salt environment, so that P. euphratica can survive better under these conditions, too.
In previous research in the laboratory, 44 strains of cultivable endophytic bacteria were screened and identified from the tissue of P. euphratica by using LB medium with a salt content of 3%, and they belonged to 5 genera, namely, Bacillus, Planococcus, Nesterenkonia of Firmicutes, Oerskovia of Actinobacteria, and Halomonas of Proteobacteria. Among them, the number of cultivable bacteria belonging to Bacillus was the largest, accounting for 65.9% of the total. Not only that, a strain of Bacillus with strong saline and alkali tolerance was screened from the LB selective medium with high salinity and alkalinity which can survive under the extreme conditions of 10% NaCl and pH 11. We also obtained a genome completion map of this strain, named it Bacillus haynesii P19 (GenBank accession no. PRJNA648288), and tried to use it for fermentation but in a different work (see Fig. 6), so as to develop it into a promising industrial fermentation chassis bacterium (20). Therefore, we speculated that the Firmicutes containing Bacillus were of great significance for the endophytes of P. euphratica and the host’s resistance to a saline-alkali environment.
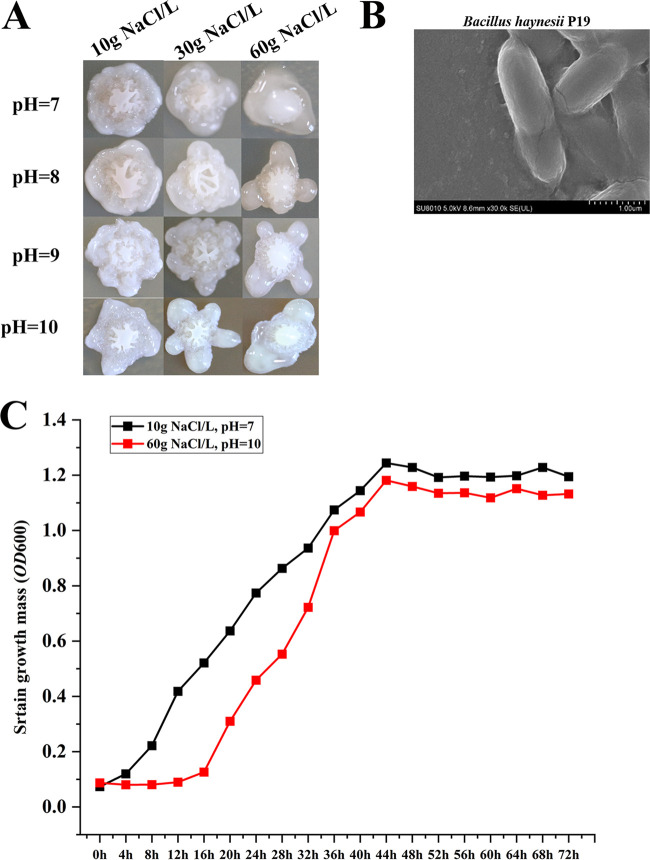
FIG 6.
Salt-alkali tolerance of Bacillus haynesii P19. (A) Colony growth of Bacillus haynesii P19 under different salinity and alkalinity levels. (B) Scanning electron microscope image of Bacillus haynesii P19 in a normal environment. (C) Growth curve of Bacillus haynesii P19 under different salinity and alkalinity conditions.
There was also a special phylum, Deinococcus-Thermus, in the tissues of P. euphratica. The comparative abundances of this phylum in the extreme saline group and very strong saline group were 1.14% and 2.99%, respectively (Fig. 2B and C), while those in the robust saline group and the moderate saline group were 1.25% and 5.45%, respectively (Fig. 2D and E). Moreover, its relative abundance was the highest in the sap and xylem of the moderate saline group of P. euphratica, reaching 7.15% and 5.32%, respectively (Fig. 2A). Deinococcus-Thermus is recognized as one of the most extremophilic phyla of bacteria and is divided into the orders Deinococcales and Thermales (21). Sayeh et al. obtained an enriched culture of Deinococcus-Thermus from Tunisian geothermal springs in the temperature range of 50 to 75°C (22). Deinococcus-Thermus are not only resistant to high temperature and drought but also resistant to radiation. Chanal et al. exposed a sand sample to 15 kGy in a 60Co source from the desert of Tataouine and obtained two radiation-tolerant isolates, which were affiliated with the Thermus-Deinococcus phylum (23). The high abundance of Deinococcus-Thermus in the tissues of P. euphratica in semiarid areas with moderate salinity indicated that this phylum was drought tolerant but not saline tolerant.
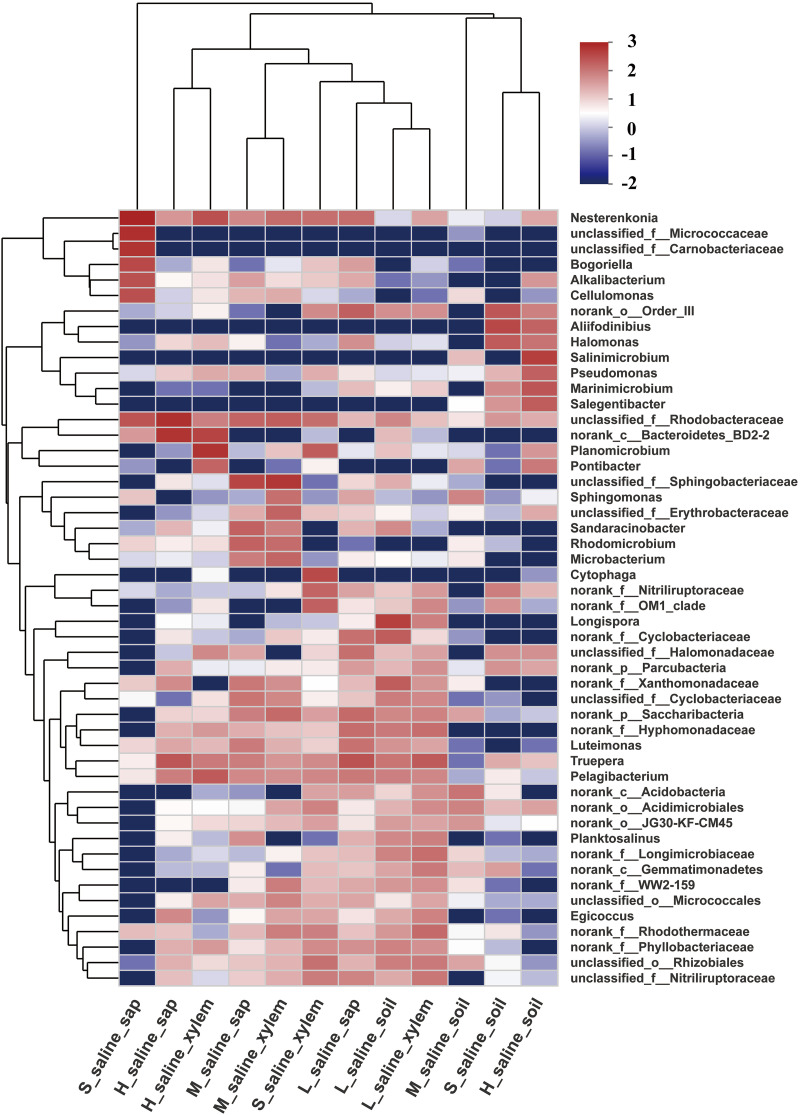
For more details, the hierarchical clustering heatmap showed the distribution of microbial communities at the genus level in different tissues and under different salinities (Fig. 3). The top 50 classified genera and 12 sample types were both hierarchically clustered based on the Bray-Curtis similarity index. Cluster analysis showed that endophytic flora of the tissues of P. euphratica under the same salinity had a correlation. The dominant genera of sap of P. euphratica under extreme salinity were very different from those of other groups, including Nesterenkonia, unclassified_f_Micrococcaceae, unclassified_f_Carnobacteriaceae, Bogoriella, Alkalibacterium, and Cellulomonas. Among them, Nesterenkonia is a genus of Actinobacteria, Micrococcineae, and Micrococcaceae. The strains of this genus are aerobic, mesophilic, moderately halophilic, or halotolerant. Some species are also alkaliphilic or alkali tolerant (24). Strains have been isolated not only from some extreme environments, such as soda lakes (25), saline and alkaline soils (26), a hypersaline lake (27), Antarctic soil (28), desert soil (29), and so on, but also from human beings (30). Among the 44 strains of cultivable endophytic bacteria screened above, there were also bacteria of Nesterenkonia (20). Groth et al. isolated a new alkaliphilic actinomycete of the genus Bogoriella from a soda lake in Africa (31). Cellulomonas in sap of P. euphratica under extreme salinity can produce xylanase to degrade cellulose (32).
FIG 3.
Heatmap of selected most differentially abundant features at the genus level. The top 50 classified genera and 12 sample types were both hierarchically clustered based on Bray-Curtis similarity index.
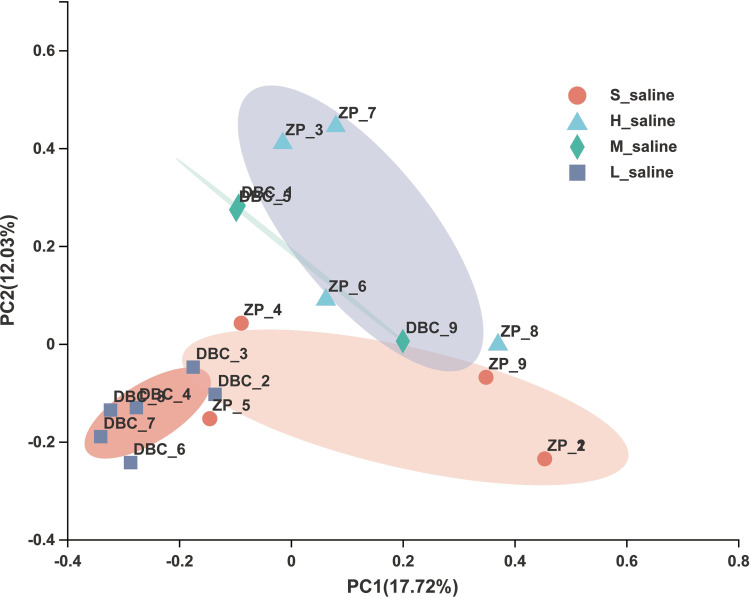
It can also be seen from the principal-coordinate analysis (PCoA) results that the microbial community of P. euphratica under different salinities had an obvious trend of separation, indicating that with the change of salinity of soil, the microbial community will change accordingly (Fig. 4). The separation trend of sap samples ZP_1 and ZP_2 of P. euphratica under extreme salinity compared with other samples was very obvious, indicating that the community composition structure was very different, and the same was true for the alpha diversity (Table 2) and community composition at the phylum level (Fig. 2A) and the genus level (Fig. 3). This may be due to the special growth environment of P. euphratica. In order to adapt to the high-saline-alkali environment, P. euphratica removes the excess salt absorbed in the soil into white crystals on the trunk or branches to maintain its own osmotic balance. Therefore, the salinity of the sap of P. euphratica is relatively high, resulting in a great difference between the microbial community structure of the sap and that of other tissues.
FIG 4.
Principal-coordinate analysis (PCoA) using Bray-Curtis metric distances of beta diversity in the samples of P. euphratica from different salinities at the OTU level.
However, although the diversity of the tissue of P. euphratica was reduced under extremely high salinity, most of the surviving bacteria were associated with salinity tolerance. As shown in Fig. 3, Nesterenkonia and Bogoriella, which were highly abundant under extremely high salinity, had been documented to be related to salt tolerance. Culturable bacteria such as Bacillus and Halomonas screened from the tissue of P. euphratica in the early stage of the laboratory work also had literature showing that they can improve the salt tolerance of plants as plant growth-promoting rhizobacteria (33). For example, Upadhyay and Singh found that the inoculation of Bacillus aquimaris could increase the N content of wheat leaves under salt stress (34). Mahgoub et al. applied salt-tolerant endophytic bacteria Bacillus subtilis and Bacillus thuringiensis isolated from halophytes to broad beans under salt stress and found that the bacteria could induce salinity tolerance, enhance nutrient uptake, and promote broad bean growth under salinity stress (35). Zhang et al. treated cotton seeds with rhizosphere salt-tolerant growth-promoting bacteria including Bacillus and Halomonas, which were isolated from Kalidium foliatum, and effectively improved the growth of cotton under saline-alkali stress (36). Therefore, based on the data of this study and combined with the reported data, we support the idea that endophytes with low diversity can still improve the salinity tolerance of P. euphratica under extremely high salinity.
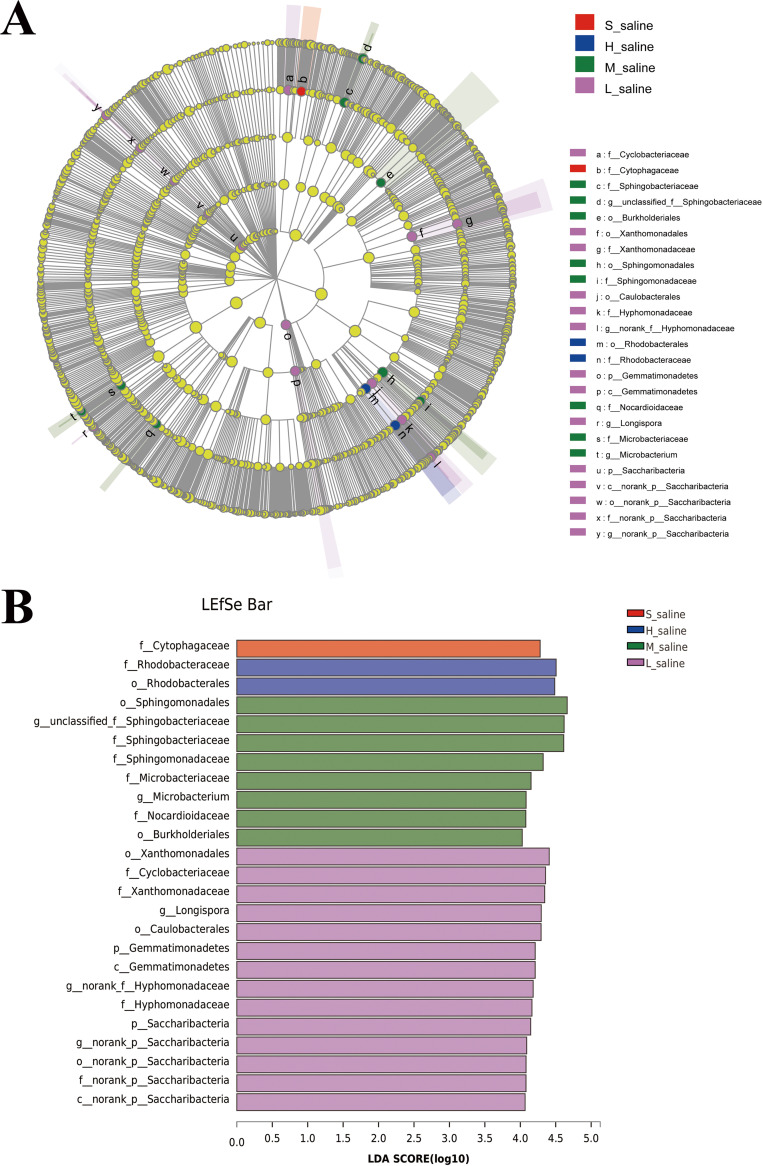
Significant differences in microbial communities.
To determine the classified bacterial taxa with significant divergences in abundance between different groups under different salinities, we used the linear discriminant analysis (LDA) effect size (LEfSe) method to analyze biomarkers. As shown in Fig. 5A, there were 25 bacterial clades that were statistically significantly different with an LDA threshold of 4 (Fig. 5B). The highest number of bacteria were significantly enriched in the groups of robust and moderate salinity, while only 1 clade showed the advantage of abundance in the group under extreme salinity, which was Cytophagaceae (family). Chaudhary et al. isolated two novel psychrotolerant members of the family Cytophagaceae from Arctic soil (37). Some researchers have pointed out that the Cytophagaceae are widely present in soil, fresh water, and oceans and are the main degraders of cellulosic polysaccharides (38). Therefore, we can use low-cost raw materials containing cellulosic polysaccharides to ferment the strains of this family so as to reduce the cost of industrial production. At the same time, this family is resistant to not only cold but also drought and extreme salinity and has high robustness to maintain its own characteristics without weakening, so that it can better grow in the fermentation system. There were 2 clades that showed the advantage of abundance in the group under very strong salinity, which were Rhodobacteraceae (family) and Rhodobacterales (order). In previous studies, most Rhodobacterales originate from marine habitats but some originate from (hyper-)saline lakes or soil (39).
FIG 5.
LEfSe analysis of microbial abundance of samples under different salinities. (A) Cladogram of microbial communities. (B) LDA score identified the size of differentiation between saline and mild saline with a threshold value of 4.
The above results suggested that different salinity will affect the composition of the endogenous microbial community of P. euphratica. Some stress-resistant bacteria in P. euphratica can better help it resist the external stress environment. For example, the highly abundant Firmicutes of P. euphratica under high salinity can help it resist extreme conditions by producing endospores. The high-abundance Deinococcus-Thermus in tissues of P. euphratica with moderate salinity are not salt tolerant but are tolerant of drought, high temperature, and radiation. Some salt-tolerant or alkali-tolerant bacteria such as Nesterenkonia, Bogoriella, Alkalibacterium, Cellulomonas, and Cytophagaceae that exist in tissues of P. euphratica under extreme salinity will have potential application value if they can be used well.
Functional prediction of the microbiota.
Since the endogenous bacterial community of P. euphratica would change with the change of environmental salinity, we used the PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) tool to predict the function of endophytic bacteria of P. euphratica and compared the microbial metabolic functional abundances in different salinity groups.
Based on the results, we can see that the abundance of some predicted enzymes and metabolic pathways of endophytes of P. euphratica changed with the change of soil salinity (Tables 3 and 4). These enzymes that were upregulated with increasing salinity were involved in microbial metabolism in diverse environments, such as 3-oxoadipate coenzyme A (CoA)-transferase, phenol 2-monooxygenase, and rhamnulose-1-phosphate aldolase. Mannitol-1-phosphate 5-dehydrogenase and rhamnulose-1-phosphate aldolase were involved in the metabolism of fructose and mannose. There were also enzymes involved in amino acid metabolism such as putrescine oxidase and choline oxidase. In particular, amino acid metabolism and sugar metabolism were very important for endophytic bacteria and the host to resist the external stress environment.
TABLE 3.
Abundances of enzymes in different salinity groups
| Enzyme | Description | Abundance |
|||
|---|---|---|---|---|---|
| S_saline | H_saline | M_saline | L_saline | ||
| 2.8.3.6 | 3-Oxoadipate CoA-transferase | 6,580.22 | 5,828.69 | 4,821.95 | 4,703.5 |
| 1.1.1.17 | Mannitol-1-phosphate 5-dehydrogenase | 4,179.15 | 2,651.65 | 1,661.91 | 1,453.26 |
| 1.1.1.306 | S-(Hydroxymethyl)mycothiol dehydrogenase | 3,463.98 | 3,187.88 | 3,056.98 | 1,950.35 |
| 1.8.1.15 | Mycothione reductase | 2,753.88 | 2,171.89 | 1,987.81 | 1,188.31 |
| 1.14.13.7 | Phenol 2-monooxygenase | 2,713.43 | 2,663.21 | 2,122.94 | 1,123.9 |
| 2.8.4.2 | Arsenate-mycothiol transferase | 2,658.63 | 2,006.08 | 1,015.02 | 662.22 |
| 1.4.3.10 | Putrescine oxidase | 2,636.6 | 2,128.5 | 2,037.33 | 886.65 |
| 4.1.2.19 | Rhamnulose-1-phosphate aldolase | 2,599.36 | 1,951.45 | 1,267.72 | 900.38 |
| 1.1.3.17 | Choline oxidase | 2,537 | 1,749.25 | 884.01 | 612.83 |
| 4.1.2.40 | Tagatose-bisphosphate aldolase | 2,398.51 | 1,228.64 | 1,141.33 | 531.6 |
TABLE 4.
Abundance of KEGG pathways in different salinity groups
| Pathway level 1 | Pathway level 2 | Pathway level 3 | Description | Abundance |
|||
|---|---|---|---|---|---|---|---|
| S_saline | H_saline | M_saline | L_saline | ||||
| Genetic information processing | Transcription | ko03022 | Basal transcription factors | 4,580.44 | 4,444.06 | 4,238.44 | 2,539.97 |
| Metabolism | Metabolism of terpenoids and polyketides | ko00253 | Tetracycline biosynthesis | 1,046.96 | 887.62 | 537.79 | 107.78 |
For example, 3-oxoadipate CoA-transferase can oxidize 3-oxoadipate to 3-oxoadipate-CoA and then through acetyl-CoA C-acyltransferase and 3-oxoadipyl-CoA thiolase to generate succinyl-CoA, which participates in the tricarboxylic acid cycle and provides significant energy for bacteria and plants to resist saline stress. Similarly, mannitol-1-phosphate 5-dehydrogenase is involved in the conversion process between fructose and mannose. The degradation of fructose and mannose is also an important part of glycolysis. Glycolysis can transfer the released free energy to ATP to provide the body with the energy needed for life activities. Phenol 2-monooxygenase can convert phenol to catechol and participate in benzoate degradation. The produced catechol can finally produce succinyl-CoA and acetyl-CoA through a series of degradations and enter the tricarboxylic acid cycle (40). Rhamnulose-1-phosphate aldolase can generate glycolaldehyde, and glycolaldehyde is widely present in the solvent system of organisms. It can react with acrolein to generate ribose, which is very important for the synthesis of RNA in organisms. Rhamnulose-1-phosphate aldolase also participates in fructose and mannose metabolism, which is an important part of glycolysis. Tagatose-bisphosphate aldolase participates in the pentose phosphate pathway and is also one of the main pathways of carbohydrate metabolism in the body. In addition, putrescine oxidase can catalyze the production of putrescine. Putrescine is found in Gram-negative bacteria or fungi and exists in different species in high concentrations. Putrescine is the precursor of spermidine and spermine metabolism. It can control cell pH by increasing or decreasing its content. Therefore, putrescine plays an important role in microbial metabolism. These enzymes highly expressed in a saline environment play a very important role in the resistance of endophytes and plants to salt stress.
Salt-alkali tolerance of Bacillus haynesii P19.
As mentioned above, we screened a salt-tolerant strain, Bacillus haynesii P19, from the tissue of P. euphratica. In order to test its salt and alkali tolerance, we carried out liquid shake flask fermentation and solid plate spotting, and the results are shown in Fig. 6. Figure 6A shows the colony growth of Bacillus haynesii P19 on the LB solid plate under different salt concentrations and different pH conditions. It can be seen that the colony color was milky white, the shape was irregular, and the surface was raised with grooves. The colony could still grow under the conditions of 60 g/L NaCl and pH 10. Figure 6B shows the scanning electron microscope (SEM) image of Bacillus haynesii P19, whose cells are rod shaped and 2 to 3 μm in size. Figure 6C is the growth curve test of Bacillus haynesii P19 in normal LB liquid medium (pH 7, 10 g/L NaCl) and high-salinity LB liquid medium (pH 10, 60 g/L NaCl). It can be seen that under high pH and in a high-salt environment, Bacillus haynesii P19 took more time to adapt to the saline-alkali environment during the adaptation period, and when it reached the plateau period, the growth density of the bacteria was only slightly lower than that in the normal environment. Although Bacillus haynesii P19 could survive under the extreme condition of 10% NaCl and pH 11, its growth was poor, so it was not shown in Fig. 6. Since these results showed that Bacillus haynesii P19 had a high salt-alkali tolerance, we can try to use it as a chassis strain for research and industrial production.
Conclusions.
In this study, we report the endogenous bacterial community structure of P. euphratica in different saline environments. Although the habitats were different, the compositions and structures of endophytic bacterial communities were similar, mainly including Proteobacteria, Actinobacteria, Bacteroidetes, and Firmicutes.
We compared the differences of microorganisms in different salinities; alpha diversity showed that the endogenous bacterial diversity of P. euphratica in different saline environments and different tissues was varied; in particular, the Shannon index and Chao index of the sap of P. euphratica under extreme salinity were lower than those in other groups. At the phylum level, in the endophytes of P. euphratica, the high-abundance Firmicutes under high salinity can form endospores to resist extreme conditions. The high-abundance Deinococcus-Thermus under moderate salinity were drought tolerant but not saline tolerant. At the genus level, the dominant genera of sap of P. euphratica under extreme salinity were very different from those of other groups, almost all of which were related to saline-alkali tolerance. The LEfSe analysis suggested that the endogenous bacteria in P. euphratica were significantly varied at the different taxonomic levels, indicating that the different growth environments of P. euphratica can affect the abundance of its endogenous bacteria. These clades that showed the advantage of abundance in the groups under extreme and very strong salinities not only were resistant to salt-alkali stress but also had certain advantages in other aspects: for example, Cytophagaceae are the main degraders of cellulosic polysaccharides.
By predicting the function of endophytic bacteria of P. euphratica, we found that the abundance of some enzymes and metabolic pathways of endophytes of P. euphratica increased with the increase of soil salinity. Most of the enzymes were related to energy metabolism and carbohydrate metabolism, such as the tricarboxylic acid cycle and glycolysis.
The saline-alkali-tolerant strain Bacillus haynesii P19 screened from the tissue of P. euphratica can still grow well under the conditions of 60 g/L NaCl and pH 10. Under these conditions, in addition to the adaptation period and the time to reach the plateau phase of the strain being longer than those in the normal environment, the bacterial growth density in the plateau phase was only slightly lower than that in the normal environment. This indicated that Bacillus haynesii P19 has great potential to be developed as an industrial fermentation chassis bacterium.
Taken together, this study has systematically described the endogenous bacterial flora of P. euphratica and compared the microbial divergences in different salinities. We found that different saline environments will affect the composition of the endogenous flora of P. euphratica. These findings may provide valuable information for understanding the relationship between the environment and endophytes of a plant. On the other hand, we found that the endophytes of P. euphratica may be involved in some of the metabolic pathways that tolerate salt stress. These provided evidence that the endogenous bacteria of the host plant had different expression mechanisms under different degrees of stress, and this mechanism was very obvious in the distribution of endophytes. However, due to the limitation of conditions, it was impossible to determine which specific metabolic pathways and physiological systems were regulated by the predicted microbial functions alone in a saline-alkali environment. Therefore, further exploration through metagenomics is needed in the future.
MATERIALS AND METHODS
Sample collection.
We collected four samples of P. euphratica xylem, three samples of P. euphratica sap, and two samples of rhizosphere soil from a P. euphratica forest (high-saline soil) in Zephyr, Xinjiang, China (E76°57.935′, N38°01.848′), and collected four samples of P. euphratica xylem, three samples of P. euphratica sap, and two rhizosphere soil samples as controls from a P. euphratica forest (mild saline soil) in Darbancheng, Xinjiang, China (E87°91.531′, N43°52.026′). To ensure the least damage to Populus euphratica, we used a scalpel to scrape 10 to 15 g of the lower xylem tissue and collected the sap from the incision of the tree trunk and soil samples at a depth of 5 cm in the corresponding area. The above samples were collected in October 2019. Since the location of the sample collection was in the P. euphratica forest reserve, in order to prevent the destruction of P. euphratica vegetation, we had no way to use traditional methods to collect samples, such as the five-point method, so we could select only typical samples to collect.
When collecting each sample, three parallel samples were taken and mixed into one sample, which was stored in a 20- by 20-cm sterile zip-lock bag and placed in an incubator with an ice bag for transportation to the laboratory. Prior to DNA extraction, samples were stored at −80°C.
Determination of soil physicochemical and biological parameters.
We dried, crushed, and sieved (2-mm-pore-size sieve) the soil samples; estimated soil parameters with a soil-to-water ratio of 1:5 (wt/vol); and used a conductivity meter (DDSJ-318; Lei-Ci, China) to measure the total amount of water-soluble salt. The pH of the soil was measured with a pH meter (FE20; Mettler-Toledo Instruments, China). Soil salt content (grams per kilogram) was calculated as the total mass fraction of cations (K+, Na+, Ca2+, and Mg2+) and anions (CO32−, HCO3−, Cl−, and SO42−), in which K+ and Na+ were determined by flame photometry, Ca2+ and Mg2+ were determined by EDTA complex metric titration, CO32− and HCO3− were determined by double indicator neutralization titration, Cl− was determined by silver nitrate titration, and SO42− was determined by EDTA indirect titration. The specific determination method of each indicator refers to Soil and Agricultural Chemistry Analysis (41).
DNA extraction and PCR amplification.
Total microbial DNA was extracted from all samples using the E.Z.N.A. soil DNA kit (Omega Bio-Tek, Norcross, GA, USA) according to manufacturer protocols. Subsequently, DNA concentration and purity were determined with a NanoDrop 2000 UV-visible (UV-vis) spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
The hypervariable region V3-V4 of the bacterial 16S rRNA gene was amplified with primer pairs 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) with an ABI GeneAmp 9700 PCR thermocycler (ABI, CA, USA) (42).
PCRs were performed in triplicate as follows: 4 μL of 5× FastPfu buffer, 2 μL of 2.5 mM deoxynucleoside triphosphates (dNTPs), 0.8 μL of each primer (5 μM), 0.4 μL FastPfu polymerase, 0.2 μL bovine serum albumin (BSA), 10 ng template DNA, and double-distilled water (ddH2O) were combined into a total volume of 20 μL. The PCR amplifications were performed as follows: 95°C for 3 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s, and a final extension of 72°C for 10 min. The PCR products (3 μL) were detected using 2% agarose gels containing ethidium bromide, purified with the AxyPrep DNA gel extraction kit (Axygen Biosciences, Union City, CA, USA), and quantified using QuantiFluor-ST (Promega, USA).
Illumina MiSeq sequencing.
Purified amplicons were pooled in equimolar amounts and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, CA, USA) according to standard protocols of Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
16S rRNA gene sequence analysis.
The raw 16S rRNA gene sequencing reads were demultiplexed, quality filtered by Trimmomatic, and merged by FLASH with the indicated criteria. (i) The 300-bp reads were truncated at any site receiving an average quality score of <20 over a 50-bp sliding window, the truncated reads shorter than 50 bp were discarded, and reads containing ambiguous characters were also discarded. (ii) Only overlapping sequences longer than 10 bp were assembled according to their overlapped sequence. The maximum mismatch ratio of the overlapping region is 0.2. Reads that could not be assembled were discarded. (iii) Samples were distinguished according to the barcode and primers, and the sequence direction was adjusted; the barcode matching had to be exact, but a 2-nucleotide mismatch was allowed in primer matching.
Statistical analysis.
We used Uparse (version 7.1) to analyze the data of the 16S rRNA gene. The obtained sequences were denoised, unified, and aligned into OTUs using definitions of ≥97% sequence similarity. The taxonomy of each OTU representative sequence was analyzed by RDP (Ribosomal Database Project) Classifier (v.2.11) against the 16S rRNA database (Silva SSU138) using the confidence threshold of 0.7.
At the same time, Mothur (v.1.30.2) was used to calculate the alpha diversity (including Shannon and Chao indexes) to reflect the species richness and diversity in the samples. Principal-coordinate analysis (PCoA) was done in R using the ape package from the Bray-Curtis distance matrix to analyze the beta diversity to compare the community compositions of the tested samples. R language (version 3.3.1) was used to analyze the endophytic bacterial composition of the samples.
In the LEfSe software (http://huttenhower.sph.harvard.edu/galaxy/root?tool_id=lefse_upload), the nonparametric Kruskal-Wallis (KW) rank sum test was first used to detect the differences in species abundance between the two groups and obtain significantly different species. Then, the Wilcoxon rank sum test was used to test the difference and consistency of the different species in the subgroups between different groups. Finally, LDA was used to estimate the impact of these different species on the difference between groups.
The PICRUSt2 tool (43) was used for functional gene annotation of the 16S rRNA amplicon sequences with reference to the KEGG (Kyoto Encyclopedia of Genes and Genomes) database. This approach was used to search for enzymes involved in functional KEGG pathways, which included biological reaction and regulation of gene expression associated with metabolism, genetic and environmental information processing, cellular processes, and human disease.
Salt and alkali tolerance test of Bacillus haynesii P19.
LB liquid medium was prepared with different salt concentrations and different pH values (10 g/L peptone; 5 g/L yeast extract powder; 10 g/L, 30 g/L, and 60 g/L sodium chloride; pH 7.0, 8.0, 9.0, and 10.0), and LB solid medium was prepared with 19 g/L agar powder added to liquid medium. Bacillus haynesii P19 was inoculated into LB liquid medium with an inoculum of 3% and cultured for 72 h. The absorbance of the strain at 600 nm was measured with a microplate reader every 4 h, and Origin was used to plot the values. At the same time, Bacillus haynesii P19 was spotted on LB solid plates with different concentrations of NaCl and pH values, incubated at 37°C for 24 h, and photographed.
Then, the bacteria after centrifugal cleaning were smeared on the glass slide, dried naturally, fixed on the scanning electron microscope (SEM) copper plate with liquid conductive glue on the back side up, sprayed with gold, and then placed on the SEM stage to collect images.
Data availability.
The sequence of Bacillus haynesii P19 has been deposited in GenBank under accession no. PRJNA648288.
ACKNOWLEDGMENTS
This work was supported by the Chinese National Natural Science Foundation (grant no. U2003305, 31860018, and 31500075), the Tianshan Innovation Team Project of Xinjiang Autonomous Region (grant no. 2020D14022), and the Outstanding Young Scientific and Technological Talents Training Program of Xinjiang Autonomous Region (grant no. 2020Q02), and thanks to Huakun Yue for helping to collect poplar samples.
Contributor Information
Haitao Yue, Email: yuehaitao@tsinghua.org.cn.
Jing Han, State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences.
REFERENCES
- 1.Food and Agriculture Organization of the United Nations (FAO). 2021. Global map of salt-affected soils (GSASmap). Food and Agriculture Organization of the United Nations (FAO), Rome, Italy. [Google Scholar]
- 2.Gao Z, Tong R, Fan C, Cui Y, Wang L. 2019. Research structure based on the characteristics of salinized plants in different soils in northeast China, p 234–237. In Proceedings of 2019 Asia-Pacific Conference on Emerging Technologies and Engineering (ACETE 2019). Francis Academic Press, Beijing, China. [Google Scholar]
- 3.De Jie Y, Jie Z, Rui J, Li D. 2018. Relationships between plant community and soil chemical factors in coastal saline area of Shandong, China. J Appl Ecol 29:3521–3529. [DOI] [PubMed] [Google Scholar]
- 4.Ellis MV, Taylor JE, Rayner L. 2017. Growth characteristics of Eucalyptus camaldulensis trees differ between adjacent regulated and unregulated rivers in semi-arid temperate woodlands. For Ecol Manage 398:1–9. doi: 10.1016/j.foreco.2017.05.004. [DOI] [Google Scholar]
- 5.Koyama A, Harlow B, Evans RD. 2019. Greater soil carbon and nitrogen in a Mojave Desert ecosystem after 10 years exposure to elevated CO2. Geoderma 355:e113915. doi: 10.1016/j.geoderma.2019.113915. [DOI] [Google Scholar]
- 6.Nekoa HT, Shirvany A, Assareh MH, Naghavi MR, Pessaraklie M, Pourmeidani A. 2018. Effects of NaCl on growth, yield and ion concentration of various Populus euphratica Oliv. ecotypes in Iran. Desert 23:189–198. [Google Scholar]
- 7.Hao JQ, Yue N, Zheng CX. 2017. Analysis of changes in anatomical characteristics and physiologic features of heteromorphic leaves in a desert tree, Populus euphratica. Acta Physiol Plant 39:160. doi: 10.1007/s11738-017-2467-9. [DOI] [Google Scholar]
- 8.Qiu Q, Ma T, Hu QJ, Liu BB, Wu YX, Zhou HH, Wang Q, Wang J, Liu JQ. 2011. Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol 31:452–461. doi: 10.1093/treephys/tpr015. [DOI] [PubMed] [Google Scholar]
- 9.Meng KB, Wu YX. 2017. Molecular evolution of glutathione peroxidase (Gpx) family genes in desert poplar (Populus euphratica Oliv.). Tree Genet Genomes 13:42. doi: 10.1007/s11295-017-1127-y. [DOI] [Google Scholar]
- 10.Tian QQ, Chen JH, Wang D, Wang HL, Liu C, Wang S, Xia XL, Yin WL. 2017. Overexpression of a Populus euphratica CBF4 gene in poplar confers tolerance to multiple stresses. Plant Cell Tissue Organ Cult 128:391–407. doi: 10.1007/s11240-016-1118-y. [DOI] [Google Scholar]
- 11.Timmusk S, Abd El-Daim IA, Copolovici L, Tanilas T, Kannaste A, Behers L, Nevo E, Seisenbaeva G, Stenström E, Niinemets Ü. 2014. Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments: enhanced biomass production and reduced emissions of stress volatiles. PLoS One 9:e96086. doi: 10.1371/journal.pone.0096086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ju XA, Ouyang LA, Zhang LB. 2014. Endophytes community of Populus euphratica Oliv and the strains improving the salt tolerance of plant. J Pure Appl Microbiol 8:43–52. [Google Scholar]
- 13.Anwar N, Abaydulla G, Zayadan B, Abdurahman M, Hamood B, Erkin R, Ismayil N, Rozahon M, Mamtimin H, Rahman E. 2016. Pseudomonas populi sp. nov., an endophytic bacterium isolated from Populus euphratica. Int J Syst Evol Microbiol 66:1419–1425. doi: 10.1099/ijsem.0.000896. [DOI] [PubMed] [Google Scholar]
- 14.Yang YK, Yan ZL. 2012. Analysis of the correlation between the geographical distribution of Populus euphratica Oliv. and the climatic environment. Agric Sci Technol 13:1450–1455. [Google Scholar]
- 15.Bachran M, Kluge S, Lopez-Fernandez M, Cherkouk A. 2019. Microbial diversity in an arid, naturally saline environment. Microb Ecol 78:494–505. doi: 10.1007/s00248-018-1301-2. [DOI] [PubMed] [Google Scholar]
- 16.İnceoğlu Ö, Salles JF, van Overbeek L, van Elsas JD. 2010. Effects of plant genotype and growth stage on the betaproteobacterial communities associated with different potato cultivars in two fields. Appl Environ Microbiol 76:3675–3684. doi: 10.1128/AEM.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coombs JT, Franco CMM. 2003. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl Environ Microbiol 69:5603–5608. doi: 10.1128/AEM.69.9.5603-5608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. 2011. Environmental and gut bacteroidetes: the food connection. Front Microbiol 2:93. doi: 10.3389/fmicb.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galperin MY. 2013. Genome diversity of spore-forming Firmicutes. Microbiol Spectr 1:(2). doi: 10.1128/microbiolspectrum.TBS-0015-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang D, Yue HT, Wu JY, Zhao LY, Xing XX, Guo F, Yang J. 2022. Diversity and biological function of endophytic bacteria in Populus euphratica leaves and phloem. Acta Microbiol Sin 62:1–14. [Google Scholar]
- 21.Theodorakopoulos N, Bachar D, Christen R, Alain K, Chapon V. 2013. Exploration of Deinococcus-Thermus molecular diversity by novel group-specific PCR primers. Microbiologyopen 2:862–872. doi: 10.1002/mbo3.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayeh R, Birrien JL, Alain K, Barbier G, Hamdi M, Prieur D. 2010. Microbial diversity in Tunisian geothermal springs as detected by molecular and culture-based approaches. Extremophiles 14:501–514. doi: 10.1007/s00792-010-0327-2. [DOI] [PubMed] [Google Scholar]
- 23.Chanal A, Chapon V, Benzerara K, Barakat M, Christen R, Achouak W, Barras F, Heulin T. 2006. The desert of Tataouine: an extreme environment that hosts a wide diversity of microorganisms and radiotolerant bacteria. Environ Microbiol 8:514–525. doi: 10.1111/j.1462-2920.2005.00921.x. [DOI] [PubMed] [Google Scholar]
- 24.Machin EV, Asem MD, Salam N, Iriarte A, Langleib M, Li WJ, Menes RJ. 2019. Nesterenkonia natronophila sp. nov., an alkaliphilic actinobacterium isolated from a soda lake, and emended description of the genus Nesterenkonia. Int J Syst Evol Microbiol 69:1960–1966. doi: 10.1099/ijsem.0.003409. [DOI] [PubMed] [Google Scholar]
- 25.Delgado O, Quillaguamán J, Bakhtiar S, Mattiasson B, Gessesse A, Hatti-Kaul R. 2006. Nesterenkonia aethiopica sp. nov., an alkaliphilic, moderate halophile isolated from an Ethiopian soda lake. Int J Syst Evol Microbiol 56:1229–1232. doi: 10.1099/ijs.0.63633-0. [DOI] [PubMed] [Google Scholar]
- 26.Li WJ, Zhang YQ, Schumann P, Liu HY, Yu LY, Zhang YQ, Stackebrandt E, Xu LH, Jiang CL. 2008. Nesterenkonia halophila sp. nov., a moderately halophilic, alkalitolerant actinobacterium isolated from a saline soil. Int J Syst Evol Microbiol 58:1359–1363. doi: 10.1099/ijs.0.64226-0. [DOI] [PubMed] [Google Scholar]
- 27.Collins MD, Lawson PA, Labrenz M, Tindall BJ, Weiss N, Hirsch P. 2002. Nesterenkonia lacusekhoensis sp. nov., isolated from hypersaline Ekho Lake, East Antarctica, and emended description of the genus Nesterenkonia. Int J Syst Evol Microbiol 52:1145–1150. doi: 10.1099/00207713-52-4-1145. [DOI] [PubMed] [Google Scholar]
- 28.Finore I, Orlando P, di Donato P, Leone L, Nicolaus B, Poli A. 2016. Nesterenkonia aurantiaca sp. nov., an alkaliphilic actinobacterium isolated from Antarctica. Int J Syst Evol Microbiol 66:1554–1560. doi: 10.1099/ijsem.0.000917. [DOI] [PubMed] [Google Scholar]
- 29.Li WJ, Chen HH, Kim CJ, Zhang YQ, Park DJ, Lee JC, Xu LH, Jiang CL. 2005. Nesterenkonia sandarakina sp. nov. and Nesterenkonia lutea sp. nov., novel actinobacteria, and emended description of the genus Nesterenkonia. Int J Syst Evol Microbiol 55:463–466. doi: 10.1099/ijs.0.63281-0. [DOI] [PubMed] [Google Scholar]
- 30.Chander AM, Nair RG, Kaur G, Kochhar R, Dhawan DK, Bhadada SK, Mayilraj S. 2017. Genome insight and comparative pathogenomic analysis of Nesterenkonia jeotgali strain CD08_7 isolated from duodenal mucosa of celiac disease patient. Front Microbiol 8:129. doi: 10.3389/fmicb.2017.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groth I, Schumann P, Rajney FA, Martin K, Schuetze B, Augsten K. 1997. Bogoriella caseilytica gen. nov., sp. nov., a new alkaliphilic actinomycete from a soda lake in Africa. Int J Syst Bacteriol 47:788–794. doi: 10.1099/00207713-47-3-788. [DOI] [PubMed] [Google Scholar]
- 32.Cayetano-Cruz M, Caro-Gómez LA, Plascencia-Espinosa M, Santiago-Hernández A, Benítez-Cardoza CG, Campos JE, Hidalgo-Lara ME, Zamorano-Carrillo A. 2021. Effect of the single mutation N9Y on the catalytical properties of xylanase Xyn11A from Cellulomonas uda: a biochemical and molecular dynamic simulation analysis. Biosci Biotechnol Biochem 85:1971–1985. doi: 10.1093/bbb/zbab124. [DOI] [PubMed] [Google Scholar]
- 33.Liu SF, Wang RY. 2019. Advance in research on plant salt tolerance improved by plant-growth-promoting rhizobacteria. J Desert Res 39:1–12. [Google Scholar]
- 34.Upadhyay SK, Singh DP. 2015. Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol (Stuttg) 17:288–293. doi: 10.1111/plb.12173. [DOI] [PubMed] [Google Scholar]
- 35.Mahgoub HA, Fouda A, Eid AM, Ewais EED, Hassan SED. 2021. Biotechnological application of plant growth-promoting endophytic bacteria isolated from halophytic plants to ameliorate salinity tolerance of Vicia faba L. Plant Biotechnol Rep 15:819–843. doi: 10.1007/s11816-021-00716-y. [DOI] [Google Scholar]
- 36.Zhang ZD, Gu MY, Tang QY, Chu M, Zhu J, Sun J, Yang R, Xu W. 2021. Screening of salt-tolerant and growth-promoting bacteria in the rhizosphere of Kalidium foliatum and the functional identification in pot experiments. J Agric Sci Technol 23:186–192. [Google Scholar]
- 37.Chaudhary DK, Dahal RH, Kim J. 2020. Dyadobacter psychrotolerans sp. nov. and Dyadobacter frigoris sp. nov., two novel psychrotolerant members of the family Cytophagaceae isolated from Arctic soil. Int J Syst Evol Microbiol 70:569–575. doi: 10.1099/ijsem.0.003796. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Ma LJ, Long ZH, Min W, Hou ZA. 2020. Effects of straw biochar on soil microbial metabolism and bacterial community composition in drip-irrigated cotton field. Environ Sci 41:410–490. [DOI] [PubMed] [Google Scholar]
- 39.Simon M, Scheuner C, Meier-Kolthoff JP, Brinkhoff T, Wagner-Döbler I, Ulbrich M, Klenk HP, Schomburg D, Petersen J, Göker M. 2017. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J 11:1483–1499. doi: 10.1038/ismej.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie NZ, Huang YY, Li JX, Guo L, Li Y, Wang QY, Chen D, Du QS, Huang RB. 2014. Research progress in benzoate degradation pathways and transformation for cis, cis-muconic acid. J Guangxi Acad Sci 30:74–81. [Google Scholar]
- 41.Bao SD. 2000. Soil and agricultural chemistry analysis. China Agriculture Press, Beijing, China. [Google Scholar]
- 42.Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J. 2014. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. 2020. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence of Bacillus haynesii P19 has been deposited in GenBank under accession no. PRJNA648288.