ABSTRACT
Polyploidy is the state of having multiple copies of the genome within a nucleus or a cell, which has repeatedly evolved across the domains of life. Whereas most bacteria are monoploid, some bacterial species and endosymbiotic organelles that are derived from bacteria are stably polyploid. In the present study, using absolute quantitative PCR, we assessed the ploidy of Candidatus Carsonella ruddii (Gammaproteobacteria, Oceanospirillales), the obligate symbiont of the hackberry petiole gall psyllid, Pachypsylla venusta (Hemiptera, Psylloidea). The genome of this symbiont is one of the smallest known for cellular organisms, at 160 kb. The analysis revealed that Carsonella within a single bacteriocyte has ∼6 × 104 copies of the genome, indicating that some Carsonella cells can contain thousands or even tens of thousands of genomic copies per cell. The basis of polyploidy of Carsonella is unknown, but it potentially plays a role in the repair of DNA damage through homologous recombination.
IMPORTANCE Mitochondria and plastids are endosymbiotic organelles in eukaryotic cells and are derived from free-living bacteria. They have many highly reduced genomes from which numerous genes have been transferred to the host nucleus. Similar, but more recently established, symbiotic systems are observed in some insect lineages. Although the genomic sequence data of such bacterial symbionts are rapidly accumulating, little is known about their ploidy. The present study revealed that a bacterium with a drastically reduced genome is an extreme polyploid, which is reminiscent of the case of organelles.
KEYWORDS: ploidy, bacteriome-associated symbionts, small genome, insects
OBSERVATION
Polyploidy, the state where organisms have multiple copies of the genome within a nucleus or a cell, has repeatedly evolved across the domains of life (1). Whereas most bacteria are monoploid or mero-oligoploid only during fast growth, some species, including cyanobacteria and extremophiles, are stably polyploid (1–4). Mitochondria and plastids, endosymbiotic organelles that are derived from free-living bacteria, have highly reduced genomes and are also polyploid (5, 6). In this context, the ploidy of vertically transmitted organelle-like symbionts of insects attracts our interest. Various insect lineages harbor phylogenetically diversified bacterial symbionts within the specialized cells called bacteriocytes, which constitute the bacteriome organ (7). The bacteriome-associated symbionts have drastically reduced genomes like organelles (7), and their polyploidy has generally been suspected based on the strong signal intensity of fluorescent DNA staining. However, quantitative analyses of their ploidy have been reported for only two species thus far (8–10). Dot-blot hybridization, fluorimetry, and quantitative PCR showed that Buchnera aphidicola (Gammaproteobacteria, Enterobacterales; genome size: 640 kb) (11) of the pea aphid, Acyrthosiphon pisum (Hemiptera, Aphidoidea), had 10 to 600 genomic copies per cell depending on developmental stages and morphs of the host insect (8, 9). Digital PCR using four individual cells showed that Candidatus Sulcia muelleri (Bacteroidetes; genome size, 240 kb) of the green sharpshooter Draeculacephala minerva (Hemiptera, Membracoidea) had 200 to 900 genomic copies per cell (10).
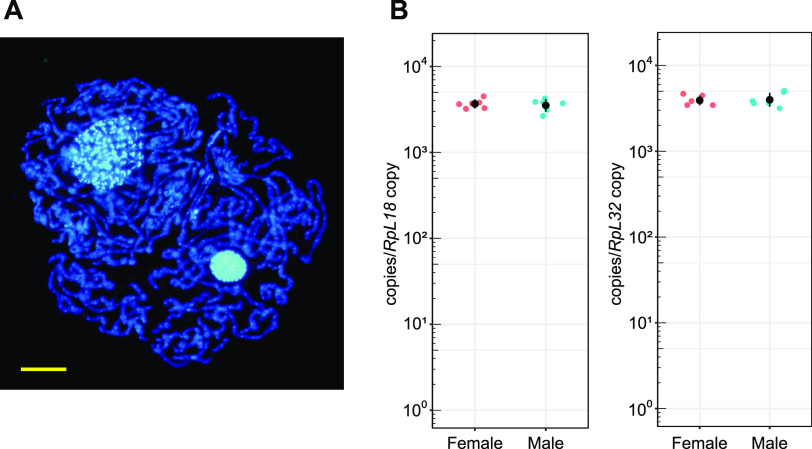
Candidatus Carsonella ruddii (Gammaproteobacteria, Oceanospirillales) (Fig. 1A), the obligate symbiont of the hackberry petiole gall psyllid, Pachypsylla venusta (Hemiptera, Psylloidea), has a genome of 160 kb, one of the smallest genomes known for cellular organisms (12). To measure the ploidy of bacteriome-associated symbionts, we performed absolute quantitative real-time PCR to estimate the ploidy of Carsonella harbored in uninucleate bacteriocytes (13–15). Whereas most psyllid species have another (secondary) bacterial symbiont in a syncytial region within the bacteriome (13, 16–18), P. venusta lacks secondary symbionts and the syncytium is rudimentary (12, 15, 19). Thus, we extracted DNA from the whole bacteriome isolated from male and female nymphs of P. venusta for use as templates in quantitative PCR. To assess copy numbers of the Carsonella genome, the 16S rRNA gene, a single-copy gene encoded in the Carsonella genome (12), was amplified with specific primers (Table 1). For calibration, the genes encoding ribosomal protein (Rp)L18 and RpL32, which are single-copy genes in the P. venusta genome (20), were also quantified. The results showed that the copy number of the 16S rRNA gene per copy of the RpL18 gene was 3690 ± 460 (mean ± standard deviation, n = 6) for females and 3571 ± 575 (n = 6) for male insects. When calibrated with the RpL32 gene, the values were 3942 ± 518 (n = 6) for females and 4040 ± 769 (n = 6) for male specimens (Fig. 1B). Because the bacteriocytes of P. venusta are 16-ploid (15), these data indicate that Carsonella within a single bacteriocyte has ∼6 × 104 copies (16 × ca. 4 × 103 copies) of the genome, assuming that all bacteriocytes are uninucleate. Because Carsonella is pleomorphic and usually tubular, the cell number within a single bacteriocyte varies (12, 14, 18). In some cases, Carsonella can be extremely long (greater than hundreds of micrometers), and only a few Carsonella cells are observed within a single bacteriocyte (Fig. 1A). Thus, a Carsonella cell can contain thousands or even tens of thousands of genomic copies. This is much more than observed in Buchnera or Sulcia (8–10) and analogous to the case of Epulopiscium sp. (Firmicutes), a giant bacterium observed in the fish gut, which contains tens of thousands of genomic copies per cell (21). There appears to be a tendency that bacteria with large cell sizes to be highly polyploid, as evidenced by a recent report that Candidatus Thiomargarita magnifica, a centimeter-long bacterium, has half a million copies of the genome (22).
FIG 1.
(A) Bacteriocytes stained with DAPI. Tubular Carsonella cells were squeezed out by applying gentle pressure on a coverslip. Extremely long Carsonella cells surrounding host nuclei show strong DAPI signals. Within Carsonella cells, numerous spots with higher signal intensities are observed. Bar, 10 μm. (B) Quantitative PCR analysis of Carsonella genomic copy number in the bacteriome of Pachypsylla venusta. Abundance values of the Carsonella 16S rRNA gene were normalized to the psyllid nuclear genes for RpL18 (left) and RpL32 (right). All data points (female, magenta; male, cyan) of six biological replicates of each sex are presented. Black dots and black bars represent means and standard deviations, respectively.
TABLE 1.
Gene-specific primers used in this study
| Target gene | Primer | Sequence | Product size |
|---|---|---|---|
| Carsonella 16S rRNA | CRPV_16S_10F | CATAGCTCAGATTGAACGCTGGTA | 89 |
| CRPV_16S_98R | CTCACCCGTTCGCTGCTAATAC | ||
| P. venusta RpL18 | PV_rpL18_420F | AGAACTGGACGAGAAGCCAACA | 81 |
| PV_rpL18_500R | TGATCTGACGTGTGCTTTTGTATG | ||
| P. venusta RpL32 | PV_rpL32_359F | GGTCACGCTGTGTCTTCAAA | 85 |
| PV_rpL32_443R | GGGCATGTCCATTGGTAAGT |
A potential cause of the polyploidy of Carsonella is mutational degradation of ancestral genes linking chromosome replication to cell division, resulting in extremely large cell sizes and polyploidy. However, a presumed benefit of polyploidy is to facilitate the mutualistic role of synthesizing essential amino acids that are required by the host psyllid (12) because polyploidy is assumed to increase metabolic output, diminishing the need to allocate resources and energy to cell division (1, 23). Additionally, the ploidy of Carsonella may have an evolutionary impact. Like other bacteriome-associated obligate symbionts of insects, Carsonella is confined in the bacteriocyte, and only a small part of the maternal population is transovarially transmitted to the next generation (18). This type of small, bottlenecked, and asexual population suffers from the accumulation of mildly deleterious mutations through the process known as Muller’s ratchet (24). Polyploidy could mask fresh mutations, and facilitate conventional repair processes within cells, such as the repair of double-strand breaks. However, for polyploidy to slow Muller’s ratchet, nonmutant sequences must be disproportionately favored as repair templates. A role of polyploidy in facilitating within-cell DNA repair is consistent with the conservation of recA, encoding the central repair enzyme, in sequenced Carsonella genomes (12, 16, 17, 25), though this role may not extend to all bacteriome-associated symbionts, many of which lack recA (7). Thus, further studies are required to assess ploidy and its evolutionary role in these symbionts.
MATERIALS AND METHODS
Absolute quantitative real-time PCR.
Absolute quantification was performed by real-time quantitative PCR targeting the Carsonella 16S rRNA gene and the P. venusta genes for RpL18 and RpL32. Galls containing 5th instar nymphs of the hackberry petiole gall psyllid, Pachypsylla venusta, were collected from hackberry trees, Celtis reticulata, in Tucson, AZ. Insects were removed from galls and dissected under a stereomicroscope. Individuals were sexed based on gonads therein, and bacteriomes were isolated and pooled from 3 to 5 individuals of either sex. DNA was extracted from the pooled bacteriomes using DNeasy blood and tissue kit (Qiagen). The quality of extracted DNA was assessed using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific) and the quantity was assessed using a Qubit 2.0 Fluorometer with a Qubit dsDNA HS assay kit (Thermo Fisher Scientific). Reference standards for quantification were constructed by PCR followed by TA-cloning as described previously (26). Briefly, PCR was performed using genomic DNA extracted from P. venusta and gene-specific primers (Table 1). The PCR products were cloned into the pGEM-T Easy vector (Promega) and amplified in Escherichia coli JM109. Inserts were sequenced following colony PCR using M13-F (5′-GTAAAACGACGGCCAG-3′) and M13-R (5′-CAGGAAACAGCTATGAC-3′) primers annealing to the vector. Plasmids with appropriate inserts were amplified in E. coli and purified using a Fast Plasmid Minikit (Eppendorf). PCRs were performed again using purified plasmids and M13-F and M13-R primers. Subsequently, PCR products were purified with a QIAquick PCR purification kit (Qiagen) and quantified with a Qubit 2.0 Fluorometer and Qubit dsDNA BR assay kit (Thermo Fisher Scientific). Copy numbers of the PCR products were calculated based on their concentration and molecular weights. For each target gene, 108, 107, 106, 105, 104, and 103 copies/μL of PCR product solutions were freshly prepared in LoBind tubes (Eppendorf) for use as reference standards. Real-time quantitative PCR was performed using the LightCycler instrument and FastStart DNA Master SYBR green I kit (Roche). Running parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 58°C for 3 s, and 72°C for 6 s. Signal intensity was measured at the end of each elongation phase. The absence of nonspecific products was confirmed by melting curve and electrophoretic analyses. Copy numbers of target genes in specimens were calculated based on standard curves generated with reference standards using the LightCycler software (ver. 3.0, Roche). The copy number of the Carsonella 16S rRNA gene was normalized to the copy numbers of the P. venusta genes for RpL18 and RpL32. Analyses were performed with six biological replicates for each sex and three technical replicates.
Microscopy.
Bacteriomes were dissected from 5th instar nymphs, fixed with fixation buffer (1% glutaraldehyde, 20 mM Tris-HCl [pH 7.65], 2.5 mM EDTA, 3.2 mM spermidine, 7 mM 2-mercaptoethanol, 0.4 mM phenylmethyl-sulfonyl fluoride), and stained with 4′,6-diamidino-2-phenylindole (DAPI). After repetitive pipetting, specimens were put on a glass slide and covered with a coverslip. The slides were examined by fluorescence microscopy (BX-53; Olympus).
ACKNOWLEDGMENTS
This study was supported by the Japan Society for the Promotion of Science (https://www.jsps.go.jp) KAKENHI grant numbers 21687020 and 20H02998 to A.N. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We declare no conflict of interest.
Contributor Information
Atsushi Nakabachi, Email: nakabachi.atsushi.ro@tut.jp.
Silvia Bulgheresi, University of Vienna.
REFERENCES
- 1.Van De Peer Y, Mizrachi E, Marchal K. 2017. The evolutionary significance of polyploidy. Nat Rev Genet 18:411–424. doi: 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- 2.Griese M, Lange C, Soppa J. 2011. Ploidy in cyanobacteria. FEMS Microbiol Lett 323:124–131. doi: 10.1111/j.1574-6968.2011.02368.x. [DOI] [PubMed] [Google Scholar]
- 3.Hansen MT. 1978. Multiplicity of genome equivalents in the radiation-resistant bacterium Micrococcus radiodurans. J Bacteriol 134:71–75. doi: 10.1128/jb.134.1.71-75.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohtani N, Tomita M, Itaya M. 2010. An extreme thermophile, Thermus thermophilus, is a polyploid bacterium. J Bacteriol 192:5499–5505. doi: 10.1128/JB.00662-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otten ABC, Smeets HJM. 2015. Evolutionary defined role of the mitochondrial DNA in fertility, disease and ageing. Hum Reprod Update 21:671–689. doi: 10.1093/humupd/dmv024. [DOI] [PubMed] [Google Scholar]
- 6.de Vries J, Archibald JM. 2018. Plastid genomes. Curr Biol 28:R336–R337. doi: 10.1016/j.cub.2018.01.027. [DOI] [PubMed] [Google Scholar]
- 7.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 8.Komaki K, Ishikawa H. 1999. Intracellular bacterial symbionts of aphids possess many genomic copies per bacterium. J Mol Evol 48:717–722. doi: 10.1007/pl00006516. [DOI] [PubMed] [Google Scholar]
- 9.Komaki K, Ishikawa H. 2000. Genomic copy number of intracellular bacterial symbionts of aphids varies in response to developmental stage and morph of their host. Insect Biochem Mol Biol 30:253–258. doi: 10.1016/s0965-1748(99)00125-3. [DOI] [PubMed] [Google Scholar]
- 10.Woyke T, Tighe D, Mavromatis K, Clum A, Copeland A, Schackwitz W, Lapidus A, Wu D, Mccutcheon JP, McDonald BR, Moran NA, Bristow J, Cheng J-FF. 2010. One bacterial cell, one complete genome. PLoS One 5:e10314. doi: 10.1371/journal.pone.0010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 12.Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 13.Profft J. 1937. Beiträge zur symbiose der aphiden und psylliden. Z Morph u Okol Tiere 32:289–326. doi: 10.1007/BF00403077. [DOI] [Google Scholar]
- 14.Thao ML, Moran NA, Abbot P, Brennan EB, Burckhardt DH, Baumann P. 2000. Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol 66:2898–2905. doi: 10.1128/AEM.66.7.2898-2905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakabachi A, Koshikawa S, Miura T, Miyagishima S. 2010. Genome size of Pachypsylla venusta (Hemiptera: Psyllidae) and the ploidy of its bacteriocyte, the symbiotic host cell that harbors intracellular mutualistic bacteria with the smallest cellular genome. Bull Entomol Res 100:27–33. doi: 10.1017/S0007485309006737. [DOI] [PubMed] [Google Scholar]
- 16.Sloan DB, Moran NA. 2012. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol 29:3781–3792. doi: 10.1093/molbev/mss180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakabachi A, Ueoka R, Oshima K, Teta R, Mangoni A, Gurgui M, Oldham NJ, Van Echten-Deckert G, Okamura K, Yamamoto K, Inoue H, Ohkuma M, Hongoh Y, Miyagishima S, Hattori M, Piel J, Fukatsu T. 2013. Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol 23:1478–1484. doi: 10.1016/j.cub.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 18.Dan H, Ikeda N, Fujikami M, Nakabachi A. 2017. Behavior of bacteriome symbionts during transovarial transmission and development of the Asian citrus psyllid. PLoS One 12:e0189779. doi: 10.1371/journal.pone.0189779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sloan DB, Nakabachi A, Richards S, Qu J, Murali SC, Gibbs RA, Moran NA. 2014. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol 31:857–871. doi: 10.1093/molbev/msu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Zhang B, Moran NA. 2020. The aphid X chromosome is a dangerous place for functionally important genes: diverse evolution of hemipteran genomes based on chromosome-level assemblies. Mol Biol Evol 37:2357–2368. doi: 10.1093/molbev/msaa095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendell JE, Clements KD, Choat JH, Angert ER. 2008. Extreme polyploidy in a large bacterium. Proc Natl Acad Sci USA 105:6730–6734. doi: 10.1073/pnas.0707522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volland J-M, Gonzalez-Rizzo S, Gros O, Tyml T, Ivanova N, Schulz F, Goudeau D, Elisabeth NH, Nath N, Udwary D, Malmstrom RR, Guidi-Rontani C, Bolte-Kluge S, Davies KM, Jean MR, Mansot J-L, Mouncey NJ, Angert E, Woyke T, Date SV. 2022. A centimeter-long bacterium with DNA compartmentalized in membrane-bound organelles. bioRxiv. doi: 10.1101/2022.02.16.480423. [DOI] [PubMed]
- 23.Comai L. 2005. The advantages and disadvantages of being polyploid. Nat Rev Genet 6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 24.Moran NA. 1996. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA 93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakabachi A, Piel J, Malenovský I, Hirose Y. 2020. Comparative genomics underlines multiple roles of Profftella, an obligate symbiont of psyllids: providing toxins, vitamins, and carotenoids. Genome Biol Evol 12:1975–1987. doi: 10.1093/gbe/evaa175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakabachi A, Fujikami M. 2019. Concentration and distribution of diaphorin, and expression of diaphorin synthesis genes during Asian citrus psyllid development. J Insect Physiol 118:103931. doi: 10.1016/j.jinsphys.2019.103931. [DOI] [PubMed] [Google Scholar]