ABSTRACT
Higher-order spatial organization of the chromatin in the nucleus plays crucial roles in the maintenance of cell functions and the regulation of gene expression. Three-dimensional (3D) genome sequencing has been used to great effect in mammal and plants, but the availability of 3D genomes of filamentous fungi is severely limited. Here, we performed a chromosome-level genome assembly of Penicillium oxalicum through single-molecule real-time sequencing (Pacific Biosciences) and chromatin interaction mapping (Hi-C), with a scaffold N50 of 4.07 Mb and a contig N50 of 3.81 Mb, and further elucidated the 3D genome architecture of P. oxalicum. High-frequency interchromosomal contacts occurred within the centromeres and telomeres, as well as within individual chromosomes. There were 12,203 cis-interactions and 7,884 trans-interactions detected at a resolution of 1 kb. Moreover, a total of 1,099 topologically associated domains (or globules) were found, ranging in size from 2.0 to 76.0 kb. Interestingly, transcription factor-bound motifs were enriched in the globule boundaries. All the cellulase and xylanase genes were discretely distributed in the 3D model of the P. oxalicum genome as a result of few cis- and trans-interactions. Our results from this study provide a global view of chromatin interactions in the P. oxalicum genome and will act as a resource for studying spatial regulation of gene expression in filamentous fungi.
IMPORTANCE The spatial structure of chromatin plays important roles in normal cell functions and the regulation of gene expression. The three-dimensional (3D) architectures of the genomes of many mammals and plants have been elucidated, but corresponding studies on filamentous fungi, which play vital roles as decomposers of organic matter in the soil, are very limited. Penicillium oxalicum is one of the predominant cellulolytic aerobic fungi in subtropical and tropical forest soils and can secrete integrative cellulase and xylanase under integrated regulatory control, degrading plant biomass highly efficiently. In the present study, we employed Hi-C technology to construct the 3D genome model of P. oxalicum strain HP7-1 and to further investigate cellulase and xylanase as well as transcription factor genes in 3D genome. These results provide a resource to achieve a deeper understanding of cell function and the regulation of gene expression in filamentous fungi.
KEYWORDS: three-dimensional genome, Penicillium oxalicum, chromatin interaction, globule, cellulase
INTRODUCTION
Eukaryotic chromosomes are highly ordered and compactly packaged within the nucleus, an organization that is associated with important nuclear processes, including DNA replication and repair, gene transcription, and regulation of gene expression (1). Chromosome conformation capture (3C) combined with next-generation DNA sequencing (Hi-C) has been developed and widely used for establishment of three-dimensional (3D) genome sequences, revealing the true spatial structure and interactions, dynamic changes, and biological functions of chromatin (2). Studies of the 3D genome provide novel insights into cell biology, molecular biology, and genetics.
Eukaryotic genomic DNA is hierarchically folded into chromosome territories or subdomains, topologically associated domains (TADs), and chromatin loops at multiple scales. Among them, TADs are identified as functional genome-organizing units on a submegabase scale related to important structural and regulatory roles, which operate through an active and well controlled process rather than occurring randomly (3, 4). Both condensin and cohesin have been found to be implicated in TAD organization, although the mechanism has not been clarified yet (4). The role of TADs is well known in some eukaryotes, such as humans and Drosophila, but is less well understood in yeast and plants, which may be due to the lack of homologs of mammalian insulator proteins, such as the CCCTC-binding factor (1, 5). On the other hand, genome organization in the yeast Schizosaccharomyces pombe and Arabidopsis thaliana is associated with heterochromatic interaction (6, 7).
Although there is considerable literature on the 3D genome architecture in mammals and plants (1, 7, 8), less is known in filamentous fungi. In the sole report to date, the 3D organization of the Neurospora crassa genome is dominated by heterochromatic interactions, known as “heterochromatin bundling.” Blocks of heterochromatin divide the genome into numbers of TADs, also called “self-associated chromosomal domains” or “globules” (9). Fungal chromosomes and chromatin are highly diverse, even within a single species (10). Therefore, it will be of great value to understand the 3D genomic organization of other filamentous fungi.
Penicillium is widely distributed in various natural habitats, including terrestrial and marine environments (11–14), consisting of 483 accepted species (15). Among them, Penicillium oxalicum can secrete large amounts of carbohydrate-active enzymes (CAZymes) to digest plant biomass and produce secondary metabolites with a variety of biological activities, including antimicrobial and antitumor compounds (12), which have considerable potential for use in the biorefinery, bioremediation, and pharmaceutical industries.
The National Center for Biotechnology Information (NCBI) revealed that fewer than one-tenth of Penicillium strains have had their genome sequenced, with most of the sequenced genomes being at the draft level. Among the Penicillium strains that have been sequenced, six P. oxalicum strains (114-2, HP7-1, SYJ-1, JU-A10-T, TY02, and SGAir0226) are available (16–19), of which strain JU-A10-T was derived from wild-type 114-2 (20). However, the sequences obtained are insufficient to reach the requirements for analysis of the 3D chromatin architecture.
P. oxalicum strain HP7-1, isolated from a subtropical forest soil in China, generates high cellulase activity against pretreated sugarcane bagasse (17), a process that has industrial potential. In the present study, we reported a high-quality chromosome-level genome sequence of P. oxalicum HP7-1, using a combination of Pacific Biosciences (PacBio) sequencing and Hi-C chromosome conformation capture sequencing technology. Furthermore, the 3D structure of strain HP7-1 chromosomes was revealed, and the sequence interactions of cellulase and xylanase genes, as well as genes encoding putative transcription factors, were investigated.
RESULTS AND DISCUSSION
Chromosome-level genome assembly of P. oxalicum.
The genome sequence of P. oxalicum HP7-1 had previously been obtained using an Illumina HiSeq 2000 system (accession number JRVD00000000 in GenBank) with a scaffold N50 of 3.3 Mb and a contig N50 of 0.5 Mb, which is comparable to that of the genome sequence of other P. oxalicum strains, such as 114-2 (16) uploaded on NCBI. However, the quality of the previously reported P. oxalicum genome sequences was insufficient to be analyzed by Hi-C technology. In the current study, the genome of strain HP7-1 was further upgraded using the PacBio sequencing platform, with the assistance of the Hi-C technique for scaffolding. A total of 10.14 Gb clean subreads with an N50 size of 11.37 kb and average length of 9,888 bp was generated. The subread lengths ranged from 1.0 to 146.72 kb. After optimization of subread assembly by Falcon (version 0.3.0; https://github.com/PacificBiosciences/falcon) and Celera assembler (version 8.3; http://sourceforge.net/projects/wgs-assembler/files/wgs-assembler/wgs-8.3/), a 30.79-Mb genome of P. oxalicum, in accordance with previously reported genome sizes for this species (16–19), was generated, with 329-fold coverage. This initial genome was composed of 23 contigs with an N50 value of 3.81 Mb, with various lengths ranging from 4.18 to 5.87 Mb.
Hi-C technology was used to correct instances of misjoining and to reorder and reorientate the contig assembly. An average of 116 million valid reads was produced per replicate for use in further Hi-C analysis, accounting for 55.97% of the total clean reads mapped onto the genome of P. oxalicum. Analysis by Genome DISCO (20) revealed a correction coefficient of 0.976 between two replicates, thus suggesting that the Hi-C data were reproducible and hence suitable for further analyses. Finally, eight pseudochromosomes with lengths of 1.80 to 5.87 Mb were generated, assembled by using 13 contigs (Fig. 1A). The telomeric repeats (5′-TTAGGGG-3′)n were detected at the ends of all pseudochromosomes except for the 3′-end of chromosome VIII. The scaffold N50 reached 4.07 Mb, with a contig N50 of 3.81 Mb. The total length of all pseudochromosomes was 30.64 Mb, accounting for 99.51% of the estimated P. oxalicum genome. In addition, the average GC content of the P. oxalicum genome was 50.69% (Table 1). The Benchmarking Universal Single-Copy Orthologs (BUSCO) assay revealed a total of 286 (98.60%) complete gene models among 290 conserved genes in the fungi_odb9 data set (https://busco-archive.ezlab.org/v2/), suggesting a high level of contiguity and sequence quality of the final genome assembly.
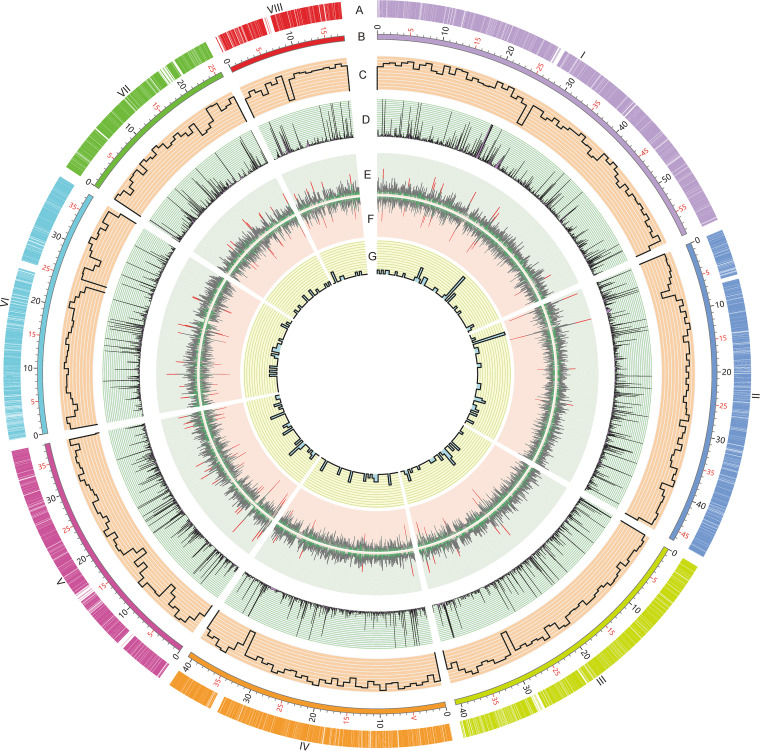
FIG 1.
Circular map indicating the genomic features of P. oxalicum genome. (A) Locations of predicted genes. The Roman numbers represent the chromosomes. (B) Schematic representation of chromosomal size (Mb scale). (C) Gene density represented by number of genes in 100-kb nonoverlapping windows. (D) Gene expression under Avicel induction. (E) Exon numbers in protein-coding genes. Red lines represent exon number (>10) in a gene, whereas green color indicates one exon contained. (F) Intron numbers in protein-coding genes. Red lines represent intron number (>10) in a gene, whereas green indicates no intron contained. (G) Number of tRNAs in 100-kb nonoverlapping windows.
TABLE 1.
Chromosome-level genome features of P. oxalicum strain HP7-1a
| Genome features | Value |
|---|---|
| Chromosome features | |
| No. of scaffolds | 8 |
| No. of contigs | 13 |
| Size of scaffolds (Mb) | 30.64 |
| GC content of scaffolds (%) | 50.69 |
| N50 scaffold length (bp) | 4,068,681 |
| N90 scaffold length (bp) | 2,593,408 |
| Maximum scaffold length (bp) | 5,870,233 |
| Minimum scaffold length (bp) | 1,795,220 |
| Avg no. of contigs per scaffold | 1.63 |
| Protein-coding genes of scaffolds | 9,728 |
| Avg gene length (bp) | 1,615.07 |
| Avg CDS length (bp) | 1,412.63 |
| GC content of CDS sequences (%) | 54.44 |
| Genome features | |
| Avg no. of exons per gene | 2.94 |
| Avg no. of introns per gene | 1.94 |
| Complete BUSCOs of genome (%) | 98.6 |
| rRNAs | 45 |
| tRNAs | 201 |
aBUSCO, Benchmarking Universal Single-Copy Orthologs; CDS, coding DNA sequence.
Genome annotation.
A combination of de novo and homology predictions was used for annotation of the P. oxalicum genome. A total of 9,728 protein-coding genes was predicted (Table S1). Of these genes, 5,359 (55.08%), 7,141 (73.40%), 3,148 (32.36%), 2,061 (21.19%), and 4,018 (41.30%) were annotated using the Gene Ontology (GO), InterPro, UniProt, euKaryotic Orthologous Groups of proteins, and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases, respectively. Relative to the proteins annotated by the Illumina HiSeq 2000 system (17), 85 novel hypothetical proteins were found. Notably, of the 9,728 proteins, 712 CAZymes, 496 putative transcription factors, and 47 clusters associated with secondary metabolism were included (Table S1), which are comparable with the previously reported (17, 21). Furthermore, 45 rRNAs (38 5S rRNAs, 2 5.8S rRNAs, 3 18S rRNAs, and 2 28S rRNAs) and 201 tRNAs were identified via rRNAmmer (version 1.2) and tRNAscan-SE (version 2.0.9), respectively (22, 23). The general features of P. oxalicum strain HP7-1 genome are presented in Table 1.
3D genome organization of P. oxalicum.
To understand the 3D genome architecture of P. oxalicum, Hi-C data were further analyzed. We generated genome-scale Hi-C interaction heat maps at resolutions of 20, 10, 2, and 1. The heat maps revealed much more frequent intrachromosomal interactions than interchromosomal interactions (Fig. 2A and B). Contact decay curves revealed fewer long-range interactions than short-range contacts within chromosomes (Fig. 2C). It was noted that high-frequency interchromosomal contacts occurred within the pericentromeric and subtelomeric regions, including centromeres and telomeres, of all eight chromosomes, as well as in intrachromosomal contacts (Fig. 2A and D), findings that are comparable with those from N. crassa (9). To further determine the chromosome-scale folding features, aggregate chromosome analysis indicated that the 3D genome architecture of P. oxalicum belonged to type I with the Rabl-like feature of centromere clustering, as described by Hoencamp et al. (24), clearly exhibiting enhanced contact frequency between centromeres and telomeres (Fig. 2D).
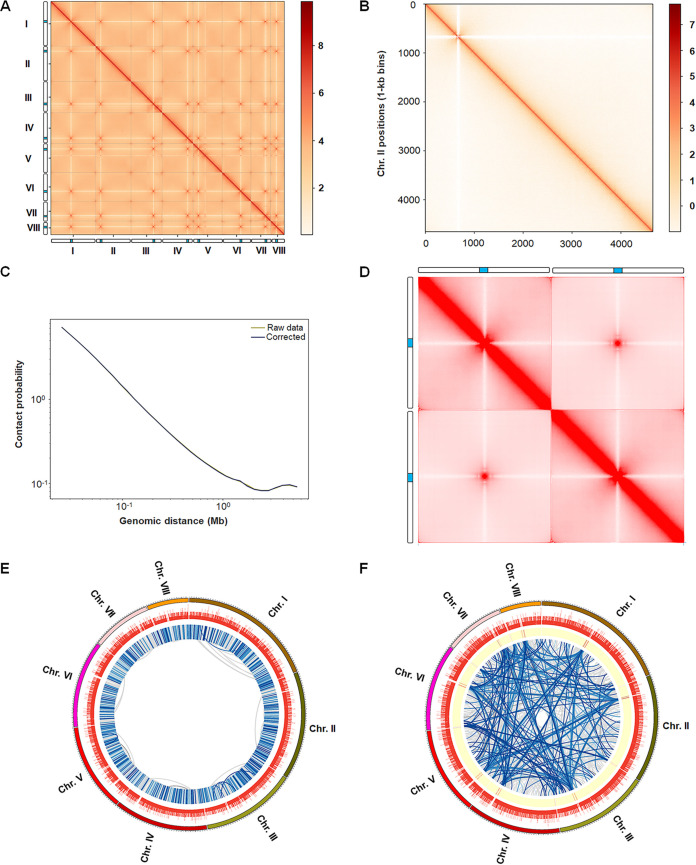
FIG 2.
Hi-C contact map and genome-wide interaction matrix of P. oxalicum. (A) Hi-C interactome (10-kb bins) within and among chromosomes (I to VIII). The color intensity represents the contact frequency. The columns beside the x and y axes indicate chromosomes, in which the blue color represents the predicted centromeres. (B) Heat map showing Hi-C interaction of chromosome II (1-kb min). (C) Genome-wide contact decay curve. (D) Aggregate chromosome analysis on Hi-C map of P. oxalicum, which helps highlight chromosome-scale features associated with a collection of chromosomes. This plot symmetrizes across all possible orders and orientations of individual chromosomes to generate representative isochromosome-by-isochromosome map showing the strong Rabl-like features. The columns beside the plot indicate virtual chromosomes, in which the blue color represents the centromeres. (E, F) Circos diagrams indicating the genome-wide significant cis- and trans-interactions, respectively. In panel E, the tracks (from outside to inside) indicate chromosome, gene number, and interaction links, the color of which becomes darker as the P value increases. In panel F, the tracks (from outside to inside) indicate chromosome, gene number, and enrichment degree of number of trans-interactions, where the red color indicates trans-interactions, and the interaction links, in which the color becomes darker as the number of reads increases.
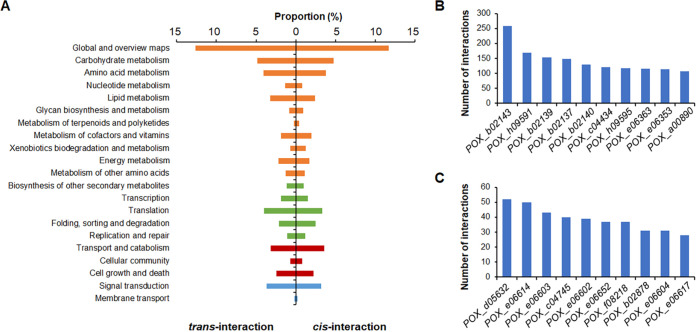
At a resolution of 1-kb, Fit-Hi-C analysis (25), a tool for assigning statistical confidence estimates to chromosomal contact heat maps, detected 12,203 cis-interactions (Fig. 2E; Table S2) and 7,884 trans-interactions (Fig. 2F; Table S3), with thresholds of q value ≤ 0.01 and number of interacted reads > 2. The frequency of cis-interaction was negatively correlated with DNA distance (Fig. S1). Furthermore, DNA fragments with cis-interactions and trans-interactions covered 5,738 and 1,324 genes, respectively. Of these genes, 967 genes exhibited both cis- and trans-interactions. KEGG annotation indicated that 30.7% of the 5,738 genes with cis-interactions were involved in metabolism, followed by genetic information processing (8.5%) and cellular processes (6.6%), whereas 33.3% of the 1,322 genes with trans-interactions were involved in metabolism. Comparative analysis revealed that the genes involved in nucleotide and energy metabolisms that exhibited cis-interactions were significantly fewer than those exhibiting trans-interactions, whereas the genes participating in xenobiotic biodegradation and metabolism that exhibited cis-interactions were more frequent than those with trans-interactions (Fig. 3A). Of these genes, the three most highly connected (Hub) genes with cis-interactions were POX_d05632 (52 interactions), POX_e06614 (50 interactions), and POX_e06603 (43 interactions), whereas the three most highly connected genes with trans-interactions were POX_b02143 (258 interactions), POX_h09591 (169 interactions), and POX_b02139 (154 interactions) (Fig. 3B and C). Of these genes, POX_d05632 and POX_e06614 encoded AMP-dependent synthetase/ligase and vacuolar fusion protein CCZ1, respectively, whereas POX_e06603 encoded a basic leucine zipper (bZIP) transcription factor. POX_b02143 and POX_h09591 encoded the ABC multidrug transporter AtrF and the AP-3 complex subunit involved in sorting of PQ-loop-family amino acid transporters to the vacuolar/lysosomal membrane, respectively (26), whereas POX_b02139 was annotated as a hypothetical protein (Table S1).
FIG 3.
Kyoto Encyclopedia of Genes and Genomes analysis of genes with cis- and trans-interactions (A) and the top 10 genes with the greatest number of cis- (B) and trans-interactions (C). In panel A, the orange bar charts indicate genes involved in metabolism; the green bar charts are genes participating in genetic information processing; the dark red bar charts are genes involved in environmental information processing; and the blue bar charts represent genes involved in cellular processes.
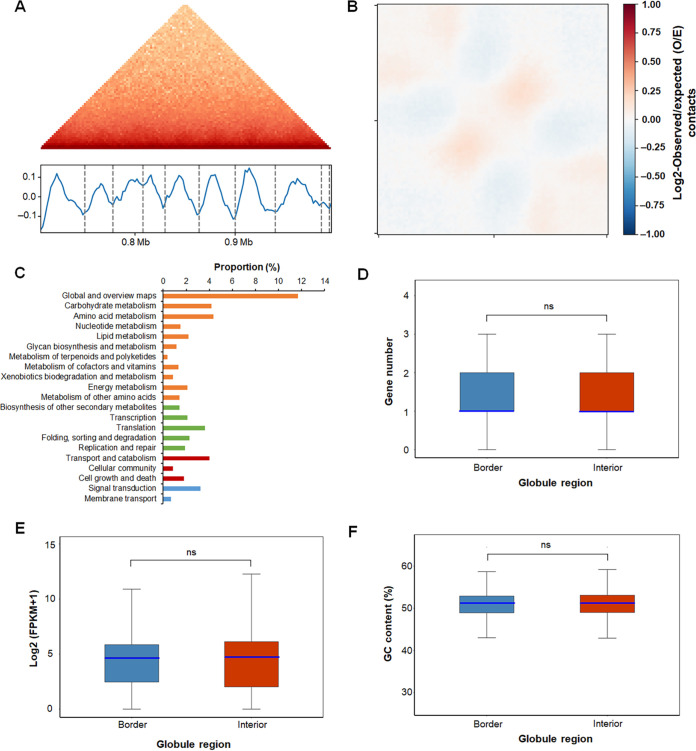
The TAD is considered to be the basic 3D genome-organizing unit in mammals and plants and is also called a “globule” in fungi, such as N. crassa and S. pombe. Commonly, TADs are defined as self-associating chromosomal domains at the submegabase scale, whereas globules are defined as locally self-interacting domains of 50 to 100 kb in size (9, 27, 28). Screening each chromosome at a resolution of 2 kb using insulation score (29) revealed 1,099 globules in the P. oxalicum 3D genome, ranging in size from 2.0 to 76.0 kb (Table S4; Fig. 4A). Polymeric analysis of globule structure (30) further showed that the self-interactions were separated by insulating boundaries (Fig. 4B). There were 1,282 and 9,476 genes localized in the boundaries and interiors of the globules, respectively, of which 1,051 genes were found across both the boundaries and the interiors. KEGG annotation indicated that 32.6% of these 1,051 genes were involved in metabolism, followed by genetic information processing (9.9%) and cellular processes (6.7%) (Fig. 4C). Neither the density (Fig. 4D) nor the transcription (Fig. 4E) of genes in each bin, including in globule boundary and interior regions or GC contents (Fig. 4F) was detected.
FIG 4.
Globule analysis in 3D architecture of P. oxalicum. (A) Hi-C interaction matrix of a region (Chr. III 704000 to 992000) indicating globules. (Top) Hi-C interaction matrix; (bottom) globule boundaries (vertical bars) and insulation scores. The vertical axis is the insulation score, whereas the gray dashed line represents the globule boundary. (B) Globule structure polymeric analysis. (C) Kyoto Encyclopedia of Genes and Genomes analysis of genes localized in globule boundaries. (D) Distribution of gene number in each bin included in globule interiors and borders. (E) Gene expression in globule interiors and borders through transcription analysis. (F) GC content of globule interiors and borders. In panels D to F, the light blue line represents the median. “ns” represents no significant difference between globule interior and border, analyzed by the Wilcoxon test.
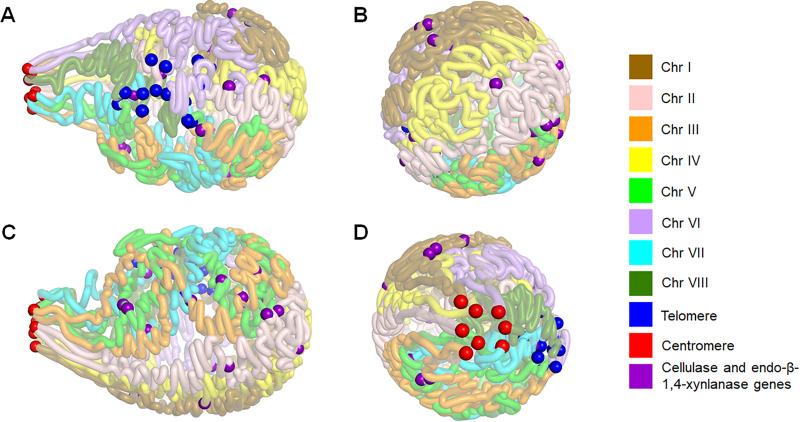
Based on the data above, we carried out in silico modeling of a 3D map of the P. oxalicum genome. The resulting map resembled a light bulb, comprising clustered centromeres as the “socket base” and 16 chromosome arms forming the “bulb.” All the telomeres were grouped together, localized on the side of the bulb. Each chromosome conformation supported a clothes-pin like structure, which was also observed in N. crassa chromosomes (9), with the centromere at one end of the folded chromosome and the telomeres almost at the opposite end (Fig. 5).
FIG 5.
3D model of P. oxalicum genome under Avicel induction. (A) Front view. (B) Right flank view. (C) Back view. (D) Left flank view. Red, blue, and purple balls represent the predicted centrosomes, telomeres, and cellulase and endo-β-1,4-xylanase genes, respectively.
3D organization of cellulolytic and xylanolytic enzyme genes in the P. oxalicum genome.
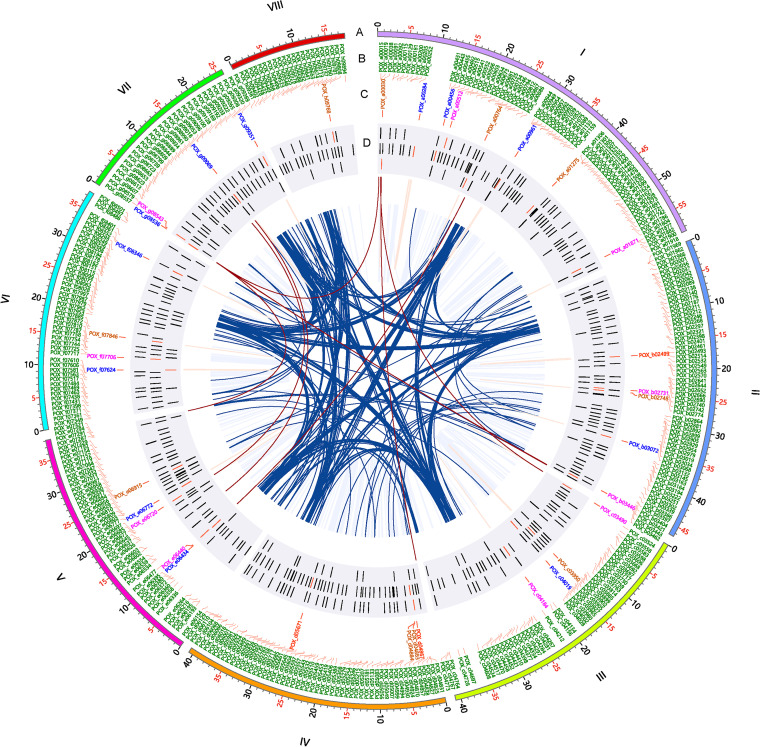
In the genome of P. oxalicum obtained, 25 cellulase (three cellobiohydrolases [CBHs], 10 endo-β-1,4-glucanases [EGs], and 12 β-glucosidases [BGLs]) and 10 endo-β-1,4-xylanase (XYNs) genes were annotated, a result that was comparable to that reported from the previously reported P. oxalicum genome (17). These genes were nonuniformly distributed across the eight chromosomes. Eight of the genes were localized on chromosome I, followed by five each on chromosomes II and V. The major cellobiohydrolase genes cbh1 and cbh2 were located on chromosome IV (Fig. 6). These cellulase and XYN genes were distributed into distinct globules, except for POX_g08536 and POX_g08543 in Globule 938 and POX_d4883 and POX_d4884 in Globule 516. Three genes, POX_a00284, POX_b03072, and POX_f07624, spanned globule boundaries and interiors (Table S1).
FIG 6.
Circos plot indicating interactions of cellulase and xylanase genes and transcription factor genes in P. oxalicum. (A) Schematic representation of chromosomal size (Mb scale). (B) Names of transcription factor genes. (C) Names of genes encoding cellulase and xylanase. (D) Location of the genes described above. The roman numbers represent the chromosomes. Red gene ID, cellobiohydrolase gene; brown gene ID, endo-β-1,4-glucanase; blue gene ID, β-glucosidase; pink gene ID, endo-β-1,4-xylanase. The brown and dark blue lines represent contacts among enzyme and transcription factor genes.
In addition, 15 genes, consisting of seven bgl, one cbh, five eg, and two xyns, exhibited cis- interactions. Six genes (eg gene Cel5C/POX_a00030; bgl genes POX_e06434, POX_g08536 and POX_g09069; and xyn genes xyn11A/POX_c03490 and POX_c03490) were found to exhibit both cis- and trans-interactions. cis-Interacted DNA fragments were localized in adjacent up- and/or down-flanking regions of each gene, including promoter and terminator regions. For example, the bgl gene POX_g09351 could make contact with DNA fragments associated with its predicted promoter and terminator regions, POX_g09352 and POX_g09357, respectively. With respect to the trans-interactions, the bgl gene POX_g08536 made contact with DNA fragments associated with POX_b03474, POX_b03469, and POX_e07327, and Chr I34,000 to 35,000. Of these, POX_b03469 and POX_e07327 were annotated as AMP-dependent synthetase/ligase and sulfite oxidase, respectively, whereas POX_b03474 was a hypothetical protein. In contrast, each of the other genes made contact with only one 1-kb DNA fragment. For example, the xyn gene POX_e06720/xyn11A interacted with a DNA fragment referred to as POX_g09033, encoding the translation initiation factor SUI1. In contrast, the key cellulase genes, such as POX_d04867/cbh1, POX_d05671/cbh2, and POX_d04883/eg1, did not form interactions. No interactions were found between cellulase genes and xylanase genes (Fig. 6; Tables S2 and S3). Those trans-interactions possibly resulted from overall genomic proximity of centromeres and telomeres, but it could not be excluded that they may be due to the induction in response to specific carbon source Avicel or coaffection by the proximity and induction. When cultivated in Avicel medium, major cellulase and xylanase genes in P. oxalicum were coexpressed (17, 31), but with a low-frequency DNA interaction, causing speculation that the biosynthesis of cellulases and xylanases was conditionally and specifically controlled, which might have happened during the transcription process resulted by a series of factors such as transcription factors (TFs).
3D organization of putative TF-encoding genes in P. oxalicum.
It is important to know how TF-encoding genes are spatially organized in P. oxalicum. TFs play essential roles in promoting the expression of cellulase and xylanase genes (32). We hypothesized that spatial interactions affected the expression of some TF-encoding genes. In the assembled genome, 496 TFs were predicted to be distributed into 430 globules. Among them, (partial) DNA sequences of 77 TF genes were localized at globule boundaries. There were 346 and 79 TF genes with cis- and trans-interactions, respectively, 64 of which shared both types of interaction (Tables S2 and S3).
The regulation of expression of the genes encoding the key TFs known to regulate the expression of cellulase and xylanase genes in P. oxalicum needs to be investigated for this biomass-degrading fungus, such as clrB/POX_b01969, cxrA/POX_a00763, cxrB/POX_f07563, atf1/POX_b03027, amyR/POX_f08097, xlnR/POX_d05133, cbh/POX_e06803, creA/POX_e07192, nsdD/POX_g08615, and mbf1/POX_g08739. Expression of these genes showed changes to various degrees at the transcriptional level in response to nonpreferred carbon sources, including Avicel, in comparison with that on glucose (31, 33–36). All the investigated genes, except atf1/POX_b03027 and amyR/POX_f08097, exhibited found cis-interactions with adjacent up- and/or down-flanking regions of each gene. In contrast, only two genes trans-interacted with DNA fragments, namely, cxrA/POX_a00763 with POX_c04329 and amyR/POX_f08097 with POX_b02143, POX_b02170, POX_e06384, POX_g09184, and POX_g09188, respectively (Fig. 6). Protein annotation revealed that POX_b02143, POX_g09184, and POX_g09188 are ABC multidrug transporter AtrF, glutathione transporter 1, and fatty acid synthase subunit alpha, respectively, whereas the others are hypothetical proteins (Table S1). Whether these interactions affect the expression of the TF-encoding genes described above needs to be further determined. In addition, it should be noted that the interactions involving TF-encoding genes were infrequent, comparable to the interactions involving cellulase and xylanase genes.
TF-binding motifs are enriched at the globule boundaries.
TAD/globule borders are generally rich in TF-binding specific motifs and heterochromatic marker-enriched regions, such as H3K9me3 and H3K27me2/3 (37). In mammalian genomes, the CCCTC-binding factor CTCF is responsible for TAD organization and is frequently colocalized in “convergent” CTCF sites with cohesin, one of the components of structural maintenance of chromosomes (SMC) complexes (38). However, the interaction of TFs, such as ACE2 and Ams2, with SMC complexes may predominantly contribute to higher-order genome architecture in yeasts because CTCF is absent (4, 39).
In the current study, the MEME Suite (http://meme-suite.org) was employed to screen motifs in the globule boundaries of P. oxalicum strain HP7-1, using the TF-binding profile of fungi in the JASPAR database (http://jaspar.genereg.net). A total of 163 candidate motifs were detected at globule boundaries, representing the potential binding sites of 163 TFs from S. cerevisiae and N. crassa. Unexpectedly, homologous alignment studies indicated the presence in P. oxalicum of 93 orthologs, with identities of 22% to 90% (Table S5).
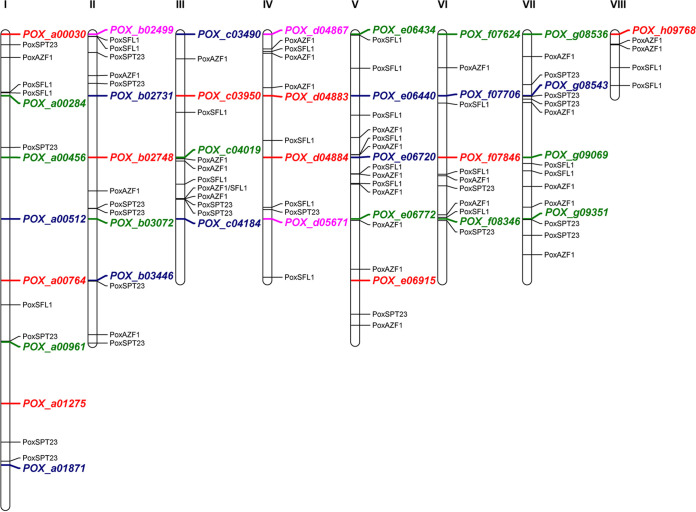
Of these proteins, POX_c04470, POX_c04124, and POX_b02187 ranked as the top three proteins in the “hierarchy” of the proportion of the TF motif-containing globule boundary number to the total globule boundary number, with values of 65.99, 53.25, and 50.69%, respectively. POX_c04470, POX_c04124, and POX_b02187 were identified as the homologous proteins of AZF1, SPT23, and SFL1 through NCBI BLASTP, with identities of 48.5, 28.9, and 45.4%, respectively, which were thereby redesignated PoxAZF1, PoxSPT23, and PoxSFL1. The function of AZF1, an asparagine-rich Zn2Cys6 protein, is dependent on carbon sources; AZF1 is required for the growth and division of S. cerevisiae in the presence of glucose, although it switches to the maintenance of cell wall integrity in the presence of glycerol-lactate (40). Recently, TrAZF1 has been shown to regulate the expression of major cellulase genes such as cbh1 and cel45a in the filamentous fungus Trichoderma reesei by directly binding to their promoters (41). MEME Suite screening detected the PoxAZF1-binding motif in the promoter regions (1.5 kb upstream) of several cellulase genes, including two cbh genes, POX_b02499 and cbh1/POX_d04867; four eg genes, Cel5C/POX_a00030, POX_e06915, POX_f07846, and Cel12A/POX_h09768; seven bgl genes, POX_b03072, POX_c04019, bgl1/POX_e06772, POX_f07624, POX_g08536, POX_g09069, and POX_g09351; and five xyn genes, POX_b02731, POX_c03490, POX_e06440, xyn11A/POX_e06720, and xyn11B/POX_g08543 (Fig. 7). These findings suggested that PoxAzf1 played an important role in the expression of cellulase and xylanase genes in P. oxalicum, as well as TrAZF1 in T. reesei (41).
FIG 7.
Motif enrichment analysis of three transcription factors, PoxAZF1, PoxSFL1, and PoxSPT23, in the promoter regions of cellulase and xylanase genes in P. oxalicum. These transcription factor-binding DNA motifs were found in the globule boundaries, and the numbers of motifs bound by them were ranked as the top three among all the identified transcription factors. Gene IDs in pink, red, green, and blue color represent cellobiohydrolase genes, endo-β-1,4-glucanase, β-glucosidase, and endo-β-1,4-xylanase genes, respectively. The roman numbers represent the chromosome number. The interval between each successive two genes indicates the 1.5-kb upstream sequence before the start codon (ATG) of the upper gene.
Interestingly, MEME Suite analysis showed that another two TFs, PoxSPT23 and PoxSFL1, were able to bind to the promoter regions of major cellulase and xylanase genes, such as cbh1 and eg1 (Fig. 7). SPT23 regulates lipid desaturation and ergosterol biosynthesis, playing an important role in the ability of yeast cells to adapt to environmental changes (42, 43). SFL1 is involved in acidic biofilm formation and hyphal development in the pathogenic yeast Candida albicans (44, 45) and in determining virulence and heat tolerance in the phytopathogenic filamentous fungus Magnaporthe oryzae (46). To date, there have been no reports of the effects of SPT23 and SFL1 on the expression of cellulase and xylanase genes in fungi, and further study is therefore needed.
Conclusions.
This study has provided the first opportunity to present a chromosomal-level genome of P. oxalicum and its 3D architecture. High-frequency interactions happened between the centromeres and the telomeres. There were 1,099 globules detected in the 3D genome sequence. Enriched at the globule boundaries were many motifs that bound TFs, such as PoxAZF1, PoxSPT23, and PoxSFL1. In addition, all the cellulase and xylanase genes were discretely distributed across the 3D genome model of P. oxalicum, with few cis- and trans-interactions, as well as the known TFs regulating the expression of cellulase and xylanase genes. This study has provided a global view of fungal chromatin interactions and a resource for studying spatial regulation of gene expression in filamentous fungi.
MATERIALS AND METHODS
Fungal materials and DNA extraction.
P. oxalicum strain HP7-1 was deposited in the China General Microbiological Culture Collection with the accession number 10781 and cultured on potato-dextrose-agar (PDA) at 4°C for temporary preservation. Asexual spores were collected using Tween 80 from a PDA plate inoculated with strain HP7-1 and cultured for 6 days at 28°C; spores were stored at –80°C in 50% (vol/vol) glycerol for long-term storage. To obtain the hyphae, fresh spores of strain HP7-1 were inoculated into glucose medium (1.0 g/liter glucose, 4.0 g/liter KH2PO4, 4.0 g/liter (NH4)2SO4, 0.6 g/liter MgSO4·7H2O, 0.6 g/liter CaCl2, 1.0 g/liter Tween 80, 100 μL trace elements, pH 5.5) in a flask at a final concentration of 1 × 106 spores/mL and cultivated in a shaker with 150 rpm at 28°C for 24 h. The collected hyphae were then inoculated into Avicel medium (1.0 g/liter Avicel, 4.0 g/liter KH2PO4, 4.0 g/liter (NH4)2SO4, 0.6 g/liter MgSO4·7H2O, 0.6 g/liter CaCl2, 1.0 g/liter Tween 80, 100 μL trace elements, pH 5.5) and cultured for 24 h (17). Fungal mycelium was harvested by filtration and used for total DNA extraction for Hi-C sequencing.
DNA extraction was carried out using the chemical method described by Zhao et al. (17). DNA concentration and purity were determined with a spectrophotometer, by measuring A260 and A260/A280, respectively, and DNA quality was determined by electrophoresis on 0.8% (wt/vol) agarose gels.
PacBio sequencing and assembly.
Fungal DNA was sequenced on a PacBio Sequel sequencer at Beijing Genomics Institute (BGI, Shenzhen, China). The raw reads were filtered to remove adapters and low-quality data to generate subreads that were used for further assembly with a serial of software Falcon version 0.3.0 (https://github.com/PacificBiosciences/falcon), proovread version 2.12 (https://github.com/BioInf-Wuerzburg/proovread), Celera assembler version 8.3 (http://sourceforge.net/projects/wgs-assembler/files/wgs-assembler/wgs-8.3/), GATK version 1.6-13 (http://www.broadinstitute.org/gatk/), and SSPACE_Basic version 2.0 (http://www.baseclear.com/genomics/bioinformatics/basetools/SSPACE). The contigs obtained were further reordered and integrated into the pseudochromosome assembly using the Juicebox assembly tool (47). BUSCO (48) was employed to evaluate the integrity of the genome assembly and the predicted genes.
Hi-C library construction and sequencing.
P. oxalicum mycelium was fixed in 1% (vol/vol) formaldehyde for 30 min at room temperature, and the cross-linking reaction was stopped by the addition of glycine solution. The cross-linked DNA of the fixed mycelium was extracted by the chemical method previously described (17) and subsequently cut with the restriction endonuclease MboI (New England BioLabs Inc., Ipswich, MA). The cohesive ends obtained were filled with the biotin marker to generate blunt ends, and then ligated by T4 DNA ligase (New England BioLabs Inc.). Proteinase K (Thermo Fisher, Waltham, MA) was employed to break DNA cross-linking. The purified DNA was sheared randomly into DNA fragments 300 to 500 bp long. Biotin-labeled DNA was purified by pulldown with streptavidin-coupled Dynabeads and then subjected to paired-end sequencing by BGI on the MGISEQ-2000 platform with PE150. Two biological replicates were used for each sample.
Reassembly of genome assisted by Hi-C technique.
Proximity-guided assembly (PGA) with Hi-C technique of P. oxalicum genome from PacBio sequencing by the 3D-DNA pipeline published previously (49). Briefly, the generated clean reads via Hi-C sequencing were mapped into the raw genome of P. oxalicum by PacBio sequencing and de novo assembly. Generally, the contigs/scaffolds localized in same chromosome present more Hi-C interactions than those in distinct chromosomes. Based on that, several groups containing numbers of contigs/scaffolds belonging to individual chromosome were clustered. In the individual chromosome, the contigs/scaffolds in close proximity exhibited improved interactions compared to those that were distant. In the obtained groups above, the contigs/scaffolds were reordered. Subsequently, the orientation of each contig/scaffold was determined according to the exaction positions of Hi-C interactions. Consequently, the assisted assembled genome was evaluated and corrected using software Juicer with Juicebox (50).
Genome annotation and functional analysis.
Gene prediction in the P. oxalicum genome and their functional annotation were carried out according to the method described by Zhao et al. (17). Numerous software packages were employed, including Augustus (http://bioinf.uni-greifswald.de/augustus/), GeneMark (http://exon.gatech.edu/), and GeneWise (https://www.ebi.ac.uk/Tools/psa/genewise/), as well as databases such as GeneOntology (GO; http://geneontology.org/), KEGG (https://www.genome.jp/kegg/), Cluster of Orthologous Groups of Proteins (COG; http://clovr.org/docs/clusters-of-orthologous-groups-cogs/), NR on NCBI (https://www.ncbi.nlm.nih.gov/), InterPro (http://www.ebi.ac.uk/interpro/), and dbCAN (https://bcb.unl.edu/dbCAN2/). Both rRNA and tRNA genes were predicted using RNAmmer (http://gtrnadb.ucsc.edu/). AntiSMASH (https://fungismash.secondarymetabolites.org/#!/start) was used to annotate secondary metabolism gene clusters. The rRNAs and tRNAs were screened by rRNAmmer (version 1.2) and tRNAscan-SE (version 2.0.9), respectively (22, 23).
Hi-C data analysis.
The sequenced raw reads were filtered and quality-evaluated using software Trimmomatic (51) and FastQC (52) with the default parameters to generate clean reads. The clean reads obtained were mapped onto the genome of P. oxalicum strain HP7-1 assembled as described above with the bowtie2 (53), and the valid pair reads were generated by the hiclib pipeline (54). The reliability of the Hi-C data from the two replicates was accessed with Genome DISCO (20). TADs/globules were identified by calculating the insulation score values using the software cworld-dekker (29). The intra- and interchromosomal interactions at the resolution of 10-kb bins were selected by Fit-Hi-C software (25), with thresholds of P value ≤ 0.01, q value ≤ 0.01, and contact count > 2. Motif scan was carried out in the JASPAR database (http://jaspar.genereg.net) using MEME Suite (http://meme-suite.org).
Aggregate chromosome analysis.
Aggregate chromosome analysis was performed by the method described by Hoencamp et al. (24). In brief, each chromosome arm was rescaled to a uniform length, and subsequently the signals from all inter- and intrachromosomal contacts were aggregated.
Prediction of TF-binding motifs in globule boundaries.
DNA motifs in the globule boundaries that were bound by TFs were predicted by Find Individual Motif Occurrences (FIMO) in MEME Suite (http://meme-suite.org) with default parameters. The TF-binding profiles of fungi from JASPAR CORE database (http://jaspar.genereg.net/) was used as the reference. Consequently, the homologous alignment was employed to search for the orthologs in P. oxalicum.
Establishment of 3D genomic model of P. oxalicum.
The 3D model of P. oxalicum genome from Hi-C data were predicted by the pastis software using a 20-kb resolution matrix with the multidimensional scaling (MDS) module (55). In the 3D model of P. oxalicum, the regions of centromeres were annotated based on Centurion developed by Varoquaux et al. (56) based on the obtained Hi-C data. In brief, we statistically analyzed the points of all trans-contacts. After removing those localized at the ends of chromosomes, the regions that most frequently interacted were selected as the centromeres.
Data availability.
The assembled genome sequence and Hi-C data of P. oxalicum strain HP7-1 were deposited at DDBJ/ENA/GenBank under the BioProject accession number PRJNA772803 and genome accession number JRVD00000000. The version described in this paper is version JRVD02000000. Transcriptome data of P. oxalicum on Avicel used in this study was available on SRA with an accession number GSE133258 (GSM3904303-05).
ACKNOWLEDGMENTS
We thank Xinqing Zhao from Shanghai Jiaotong University for constructive suggestions and the Beijing Genomics Institute (Shenzhen, China) for PacBio sequencing of the P. oxalicum strain genome.
This work was financially supported by grants U21A20178, 32060022, and 32060141 from the National Natural Science Foundation of China (to J.-X.F. and S.Z.), by grant AB21076010 from the Key Research and Development Program Project of Guangxi (to J.-X.F.), and by Training Program for 1000 Young and Middle-aged Key Teachers in Guangxi at 2019.
S.Z. and J.-X.F. conceived the study and were involved in data analysis and manuscript preparation. C.-X.L. prepared sequencing samples and analyzed genome data. L.L. provided support for Hi-C data analysis. T.Z. and X.-M.L. were involved in sample preparation. All authors read and approved the final version of the manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Shuai Zhao, Email: shuaizhao0227@gxu.edu.cn.
Kaustuv Sanyal, Jawaharlal Nehru Centre for Advanced Scientific Research.
REFERENCES
- 1.Ouyang W, Xiong D, Li G, Li X. 2020. Unraveling the 3D genome architecture in plants: present and future. Mol Plant 13:1676–1693. doi: 10.1016/j.molp.2020.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Li GL. 2020. Advances in technologies for 3D genomics research. Sci China Life Sci 63:811–824. doi: 10.1007/s11427-019-1704-2. [DOI] [PubMed] [Google Scholar]
- 3.Davidson IF, Peters JM. 2021. Genome folding through loop extrusion by SMC complexes. Nat Rev Mol Cell Biol 22:445–464. doi: 10.1038/s41580-021-00349-7. [DOI] [PubMed] [Google Scholar]
- 4.Kim KD, Tanizawa H, Iwasaki O, Noma KI. 2016. Transcription factors mediate condensin recruitment and global chromosomal organization in fission yeast. Nat Genet 48:1242–1252. doi: 10.1038/ng.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eres IE, Gilad YA. 2021. TAD skeptic: is 3D genome topology conserved? Trends Genet 37:216–223. doi: 10.1016/j.tig.2020.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kakui Y, Barrington C, Barry DJ, Gerguri T, Fu X, Bates PA, Khatri BS, Uhlmann F. 2020. Fission yeast condensin contributes to interphase chromatin organization and prevents transcription-coupled DNA damage. Genome Biol 21:272. doi: 10.1186/s13059-020-02183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pei L, Li G, Lindsey K, Zhang X, Wang M. 2021. Plant 3D genomics: the exploration and application of chromatin organization. New Phytol 230:1772–1786. doi: 10.1111/nph.17262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirny LA, Imakaev M, Abdennur N. 2019. Two major mechanisms of chromosome organization. Curr Opin Cell Biol 58:142–152. doi: 10.1016/j.ceb.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galazka JM, Klocko AD, Uesaka M, Honda S, Selker EU, Freitag M. 2016. Neurospora chromosomes are organized by blocks of importin alpha-dependent heterochromatin that are largely independent of H3K9me3. Genome Res 26:1069–1080. doi: 10.1101/gr.203182.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erlendson AA, Friedman S, Freitag M. 2017. A matter of scale and dimensions: chromatin of chromosome landmarks in the fungi. Microbiol Spectr 5:FUNK-0054–2017. doi: 10.1128/microbiolspec.FUNK-0054-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visagie CM, Houbraken J, Frisvad JC, Hong SB, Klaassen CH, Perrone G, Seifert KA, Varga J, Yaguchi T, Samson RA. 2014. Identification and nomenclature of the genus Penicillium. Stud Mycol 78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Assaf CEH, Zetina-Serrano C, Tahtah N, Khoury AE, Atoui A, Oswald IP, Puel O, Lorber S. 2020. Regulation of secondary metabolism in the Penicillium genus. Int J Mol Sci 21:9462. doi: 10.3390/ijms21249462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozlovsky AG, Kochkina GA, Zhelifonova VP, Antipova ТV, Ivanushkina NE, Ozerskaya SM. 2020. Secondary metabolites of the genus Penicillium from undisturbed and anthropogenically altered Antarctic habitats. Folia Microbiol 65:95–102. doi: 10.1007/s12223-019-00708-0. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Liu JL, Lan JY, Duan CJ, Ma QS, Feng JX. 2014. Predominance of Trichoderma and Penicillium in cellulolytic aerobic filamentous fungi from subtropical and tropical forests in China, and their use in finding highly efficient β-glucosidase. Biotechnol Biofuels 7:107. doi: 10.1186/1754-6834-7-107. [DOI] [Google Scholar]
- 15.Houbraken J, Kocsubé S, Visagie CM, Yilmaz N, Wang XC, Meijer M, Kraak B, Hubka V, Bensch K, Samson RA, Frisvad JC. 2020. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud Mycol 95:5–169. doi: 10.1016/j.simyco.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu G, Zhang L, Wei X, Zou G, Qin Y, Ma L, Li J, Zheng H, Wang S, Wang C, Xun L, Zhao GP, Zhou Z, Qu Y. 2013. Genomic and secretomic analyses reveal unique features of the lignocellulolytic enzyme system of Penicillium decumbens. PLoS One 8:e55185. doi: 10.1371/journal.pone.0055185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao S, Yan YS, He QP, Yang L, Yin X, Li CX, Mao LC, Liao LS, Huang JQ, Xie SB, Nong QD, Zhang Z, Jing L, Xiong YR, Duan CJ, Liu JL, Feng JX. 2016. Comparative genomic, transcriptomic and secretomic profiling of Penicillium oxalicum HP7-1 and its cellulase and xylanase hyper-producing mutant EU2106, and identification of two novel regulatory genes of cellulase and xylanase gene expression. Biotechnol Biofuels 9:203. doi: 10.1186/s13068-016-0616-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta AB, Gaultier NE, Aung NW, Purbojati RW, Oliveira EL, Wong A, Panicker D, Putra A, Uchida A, Drautz-Moses DI, Schuster SC. 2020. Complete genome sequence of Penicillium oxalicum strain SGAir0226 isolated from outdoor tropical air in Singapore. Mycopathologia 185:591–594. doi: 10.1007/s11046-019-00422-y. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Liu S, Sun H, Jiang Z, Zhou Z, Han X, Zhou Y, Sun H, Zhou W, Mao J. 2021. Enzyme production potential of Penicillium oxalicum M1816 and its application in ferulic acid production. Foods 10:2577. doi: 10.3390/foods10112577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ursu O, Boley N, Taranova M, Wang YXR, Yardimci GG, Noble WS, Kundaje A. 2018. GenomeDISCO: a concordance score for chromosome conformation capture experiments using random walks on contact map graphs. Bioinformatics 34:2701–2707. doi: 10.1093/bioinformatics/bty164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li CX, Zhao S, Zhang T, Xian L, Liao LS, Liu JL, Feng JX. 2017. Genome sequencing and analysis of Talaromyces pinophilus provide insights into biotechnological applications. Sci Rep 7:490. doi: 10.1038/s41598-017-00567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan PP, Lin BY, Mak AJ, Lowe TM. 2021. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res 49:9077–9096. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoencamp C, Dudchenko O, Elbatsh AMO, Brahmachari S, Raaijmakers JA, van Schaik T, Sedeño Cacciatore Á, Contessoto VG, van Heesbeen RGHP, van den Broek B, Mhaskar AN, Teunissen H, St Hilaire BG, Weisz D, Omer AD, Pham M, Colaric Z, Yang Z, Rao SSP, Mitra N, Lui C, Yao W, Khan R, Moroz LL, Kohn A, St Leger J, Mena A, Holcroft K, Gambetta MC, Lim F, Farley E, Stein N, Haddad A, Chauss D, Mutlu AS, Wang MC, Young ND, Hildebrandt E, Cheng HH, Knight CJ, Burnham TLU, Hovel KA, Beel AJ, Mattei PJ, Kornberg RD, Warren WC, Cary G, Gómez-Skarmeta JL, Hinman V, Lindblad-Toh K, et al. 2021. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science 372:984–989. doi: 10.1126/science.abe2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ay F, Bailey TL, Noble WS. 2014. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res 24:999–1011. doi: 10.1101/gr.160374.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llinares E, Barry A, André B. 2015. The AP-3 adaptor complex mediates sorting of yeast and mammalian PQ-loop-family basic amino acid transporters to the vacuolar/lysosomal membrane. Sci Rep 5:16665. doi: 10.1038/srep16665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Blüthgen N, Dekker J, Heard E. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuguchi T, Fudenberg G, Mehta S, Belton JM, Taneja N, Folco HD, FitzGerald P, Dekker J, Mirny L, Barrowman J, Grewal SI. 2014. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature 516:432–435. doi: 10.1038/nature13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. 2015. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kruse K, Hug CB, Vaquerizas JM. 2020. FAN-C: a feature-rich framework for the analysis and visualisation of chromosome conformation capture data. Genome Biol 21:303. doi: 10.1186/s13059-020-02215-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan YS, Zhao S, Liao LS, He QP, Xiong YR, Wang L, Li CX, Feng JX. 2017. Transcriptomic profiling and genetic analyses reveal novel key regulators of cellulase and xylanase gene expression in Penicillium oxalicum. Biotechnol Biofuels 10:279. doi: 10.1186/s13068-017-0966-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sukumaran RK, Christopher M, Kooloth-Valappil P, Sreeja-Raju A, Mathew RM, Sankar M, Puthiyamadam A, Adarsh VP, Aswathi A, Rebinro V, Abraham A, Pandey A. 2021. Addressing challenges in production of cellulases for biomass hydrolysis: targeted interventions into the genetics of cellulase producing fungi. Bioresour Technol 329:124746. doi: 10.1016/j.biortech.2021.124746. [DOI] [PubMed] [Google Scholar]
- 33.Li ZH, Yao GS, Wu RM, Gao LW, Kan QB, Liu M, Yang P, Liu GD, Qin YQ, Song X, Zhong YH, Fang X, Qu YB. 2015. Synergistic and dose-controlled regulation of cellulase gene expression in Penicillium oxalicum. PLoS Genet 11:e1005509. doi: 10.1371/journal.pgen.1005509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li CX, Zhao S, Luo XM, Feng JX. 2020. Weighted gene co-expression network analysis identifies critical genes for the production of cellulase and xylanase in Penicillium oxalicum. Front Microbiol 11:520. doi: 10.3389/fmicb.2020.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao S, Liao XZ, Wang JX, Ning YN, Li CX, Liao LS, Liu Q, Jiang Q, Gu LS, Fu LH, Yan YS, Xiong YR, He QP, Su LH, Duan CJ, Luo XM, Feng JX. 2019. Transcription factor Atf1 regulates expression of cellulase and xylanase genes during solid-state fermentation of ascomycetes. Appl Environ Microbiol 85:e01226-19. doi: 10.1128/AEM.01226-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li CX, Liao LS, Wan XD, Zhang FF, Zhang T, Luo XM, Zhao S, Feng JX. 2021. PoxCbh, a novel CENPB-type HTH domain protein, regulates cellulase and xylanase gene expression in Penicillium oxalicum. Mol Microbiol 116:140–153. doi: 10.1111/mmi.14696. [DOI] [PubMed] [Google Scholar]
- 37.Xiang JF, Corces VG. 2021. Regulation of 3D chromatin organization by CTCF. Curr Opin Genet Dev 67:33–40. doi: 10.1016/j.gde.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Q, Liu P, Wang L. 2020. Many facades of CTCF unified by its coding for three-dimensional genome architecture. J Genet Genomics 47:407–424. doi: 10.1016/j.jgg.2020.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Dai YL, Li G, Pei GS, Dong X, Ding GH, Zhao ZM, Li YX, Jia PL. 2018. Multiple transcription factors contribute to inter-chromosomal interaction in yeast. BMC Sys Biol 12:140. doi: 10.1186/s12918-018-0643-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slattery MG, Liko D, Heideman W. 2006. The function and properties of the Azf1 transcriptional regulator change with growth conditions in Saccharomyces cerevisiae. Eukaryot Cell 5:313–320. doi: 10.1128/EC.5.2.313-320.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antonieto ACC, Nogueira KMV, de Paula RG, Nora LC, Cassiano MHA, Guazzaroni M-E, Almeida F, da Silva TA, Ries LNA, de Assis LJ, Goldman GH, Silva RN, Silva-Rocha R. 2019. A novel Cys2His2 zinc finger homolog of AZF1 modulates holocellulase expression in Trichoderma reesei. mSystems 4:e00161-19. doi: 10.1128/mSystems.00161-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang M, Yu Q, Xiao C, Zhang K, Zhang D, Zhang B, Li M. 2018. Disruption of SPT23 results in increased heat sensitivity due to plasma membrane damage in Pichia pastoris. FEMS Yeast Res 18:foy015. [DOI] [PubMed] [Google Scholar]
- 43.Sinha A, Israeli R, Cirigliano A, Gihaz S, Trabelcy B, Braus GH, Gerchman Y, Fishman A, Negri R, Rinaldi T, Pick E. 2020. The COP9 signalosome mediates the Spt23 regulated fatty acid desaturation and ergosterol biosynthesis. FASEB J 34:4870–4889. doi: 10.1096/fj.201902487R. [DOI] [PubMed] [Google Scholar]
- 44.Unoje O, Yang M, Lu Y, Su C, Liu H. 2020. Linking Sfl1 regulation of hyphal development to stress response kinases in Candida albicans. mSphere 5:e00672-19. doi: 10.1128/mSphere.00672-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonçalves B, Bernardo R, Wang C, Schröder MS, Pedro NA, Butler G, Azeredo J, Henriques M, Pereira Mira N, Silva S. 2020. Effect of progesterone on Candida albicans biofilm formation under acidic conditions: a transcriptomic analysis. Int J Med Microbiol 310:151414. doi: 10.1016/j.ijmm.2020.151414. [DOI] [PubMed] [Google Scholar]
- 46.Li G, Zhou X, Kong L, Wang Y, Zhang H, Zhu H, Mitchell TK, Dean RA, Xu JR. 2011. MoSfl1 is important for virulence and heat tolerance in Magnaporthe oryzae. PLoS One 6:e19951. doi: 10.1371/journal.pone.0019951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL. 2016. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst 3:99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sim AFO, Waterhouse MR, Ioannidis P, Kriventseva VE, Zdobnov ME. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 49.Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, Aiden EL. 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, Aiden EL. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wingett SW, Andrews S. 2018. FastQ screen: a tool for multi-genome mapping and quality control. F1000Res 7:1338. doi: 10.12688/f1000research.15931.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. 2012. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods 9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Varoquaux N, Ay F, Noble WS, Vert JP. 2014. A statistical approach for inferring the 3D structure of the genome. Bioinformatics 30:i26–33. doi: 10.1093/bioinformatics/btu268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varoquaux N, Liachko I, Ay F, Burton JN, Shendure J, Dunham MJ, Vert JP, Noble WS. 2015. Accurate identification of centromere locations in yeast genomes using Hi-C. Nucleic Acids Res 43:5331–5339. doi: 10.1093/nar/gkv424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.02121-21-s001.xlsx, XLSX file, 2.7 MB (2.7MB, xlsx)
Supplemental material. Download spectrum.02121-21-s002.xlsx, XLSX file, 0.6 MB (589KB, xlsx)
Supplemental material. Download spectrum.02121-21-s003.xlsx, XLSX file, 0.3 MB (316.8KB, xlsx)
Supplemental material. Download spectrum.02121-21-s004.xlsx, XLSX file, 0.3 MB (347.2KB, xlsx)
Supplemental material. Download spectrum.02121-21-s005.xlsx, XLSX file, 0.02 MB (20.2KB, xlsx)
Supplemental material. Download spectrum.02121-21-s006.pdf, PDF file, 0.2 MB (208.6KB, pdf)
Data Availability Statement
The assembled genome sequence and Hi-C data of P. oxalicum strain HP7-1 were deposited at DDBJ/ENA/GenBank under the BioProject accession number PRJNA772803 and genome accession number JRVD00000000. The version described in this paper is version JRVD02000000. Transcriptome data of P. oxalicum on Avicel used in this study was available on SRA with an accession number GSE133258 (GSM3904303-05).