Abstract
In a search for cosmopolitan phylogenetic clusters of freshwater bacteria, we recovered a total of 190 full and partial 16S ribosomal DNA (rDNA) sequences from three different lakes (Lake Gossenköllesee, Austria; Lake Fuchskuhle, Germany; and Lake Baikal, Russia). The phylogenetic comparison with the currently available rDNA data set showed that our sequences fall into 16 clusters, which otherwise include bacterial rDNA sequences of primarily freshwater and soil, but not marine, origin. Six of the clusters were affiliated with the α, four were affiliated with the β, and one was affiliated with the γ subclass of the Proteobacteria; four were affiliated with the Cytophaga-Flavobacterium-Bacteroides group; and one was affiliated with the class Actinobacteria (formerly known as the high-G+C gram-positive bacteria). The latter cluster (hgcI) is monophyletic and so far includes only sequences directly retrieved from aquatic environments. Fluorescence in situ hybridization (FISH) with probes specific for the hgcI cluster showed abundances of up to 1.7 × 105 cells ml−1 in Lake Gossenköllesee, with strong seasonal fluctuations, and high abundances in the two other lakes investigated. Cell size measurements revealed that Actinobacteria in Lake Gossenköllesee can account for up to 63% of the bacterioplankton biomass. A combination of phylogenetic analysis and FISH was used to reveal 16 globally distributed sequence clusters and to confirm the broad distribution, abundance, and high biomass of members of the class Actinobacteria in freshwater ecosystems.
During the last decade, rRNA-based methods have allowed studies of microbial diversity to move from a solely cultivation-based perspective to one that encompasses as yet uncultured microorganisms (42, 44). This development was promoted by the discovery of the potential of 16S rRNA for phylogenetic reconstructions and microbial diversity (42, 65). Global efforts have led to the 16S ribosomal DNA (rDNA) sequencing of nearly all validly described prokaryotes. This, plus the collection of environmental 16S rDNA clone libraries, has created one of the largest data sets for any individual gene (38).
Most 16S rRNA surveys so far have been performed in marine and soil habitats. Only a few are available for freshwater systems (33). Phylogenetic analysis of sequences from marine environments has revealed habitat-specific phylogenetic clusters. The most prominent are the SAR clusters, monophyletic lineages of solely marine 16S rDNA sequences (26, 41). Despite the smaller data set, some freshwater-specific clusters have been proposed, such as the freshwater clusters A, B, and C (66). All these marine and freshwater studies give good evidence for the postulated phyla or clusters based on careful phylogenetic analysis of the rDNA data sets, but little effort has been made to link these in silico results to the real environment by in situ methods. It is now necessary to bridge the gap between comparative sequence analysis and community composition with in situ detection methods like fluorescence in situ hybridization (FISH). FISH with rRNA-targeted oligonucleotide probes allows selective visualization of bacterial cells with defined phylogenetic affiliations (1, 5). In contrast to other quantitative methods, such as slot blot hybridization, FISH conserves the morphology and cell sizes of the targeted organisms (48, 50). In combination with image cytometry, this not only allows cell counts of defined phylogenetic groups but links them to biomass, an ecologically relevant measure (48). In this study, first the 16S rDNA data set was extended with 190 sequences retrieved from three different lakes (Lake Gossenköllesee, Lake Fuchskuhle, and Lake Baikal), and then a comparative sequence analysis was performed to identify widely distributed phylogenetic clusters of freshwater bacteria. Probes for one conspicuous new cluster within the class Actinobacteria were designed and successfully applied, linking phylogeny with community composition and succession.
MATERIALS AND METHODS
Description of sampling sites and sampling.
Lake Gossenköllesee is a small oligotrophic high-mountain lake in the Central Alps (Tyrol, Austria), situated at 2,417 m above mean sea level (AMSL) (20). Samples for FISH were collected monthly between 4 July 1996 and 25 June 1997 at the surface and at a depth of 4 m. Filtration, fixation, and storage were done as described earlier (27, 50). For DNA extraction, a sample was taken with sterile bottles from a 3-m depth in December 1995. In September 1997, a mixed sample from an 0- to 8-m depth was taken.
Lake Fuchskuhle is a meso- to acidotrophic forest lake in the Brandenburg-Mecklenburg lake district (Germany) at an altitude of 59 m AMSL. In 1990, it was artificially divided into four basins with different catchment areas (56). In May 1998, two independent samples for DNA extraction were taken with sterile bottles from the northeast and the southwest part at an 0.5-m depth.
Lake Baikal is the deepest (1,637 m) and, by volume, largest (23,000 km3) of all freshwater lakes. It is located in East Siberia between 51°29′N and 55°46′N latitude and 103°41′E and 109°57′E longitude at an altitude of 456 m AMSL (36). Samples for DNA extraction were collected from the South Basin at a 400- and a 1,200-m depth in December 1995 and at 25, 400, 1,200, and 1,400 m in August 1997. The Central Basin was sampled at a 25-, a 1,400-, and a 1,600-m depth in August 1997. Sampling was done with a metallic 10-liter sampler (batometer) prerinsed with lake water.
DNA extraction, amplification, and cloning.
For DNA extraction, water samples (100 ml to 1 liter) taken at Lake Gossenköllesee and Lake Fuchskuhle were prescreened (50-μm pore size) and subsequently filtered on hydrophilic filters (Durapore [pore size, 0.2 μm; diameter, 47 mm]; Millipore Corp., Bedford, Mass.) until the filters were completely clogged. During filtration, the samples were kept at ambient water temperature. The filters were cut into sections and stored at −20°C until further processing. DNA extraction was done according to the protocol of Fuhrman et al. (24). The December 1995 Lake Gossenköllesee sample was further processed for DNA extraction as described previously (50). Nearly full-length bacterial 16S rDNA fragments were amplified by PCR from the Lake Fuchskuhle and the September 1997 Lake Gossenköllesee samples with the general bacterial 16S rDNA primers 8F and 1492R (12). Amplification and cloning were performed as described elsewhere (29).
Treatment of the Lake Baikal samples.
Three liters of the water sample was immediately filtered through a 0.22-μm-pore-size polycarbonate filter (Millipore Corp.). Bacterioplankton were washed off the filters with 5 ml of a solution containing 10 mM Tris-HCl (pH 7.5) and 0.15 M NaCl and centrifuged. The pellet was frozen and transported to the laboratory. Chromosomal DNA was isolated using the QIAamp blood and tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Isolated DNA was amplified by PCR with the following primers: 500L (5′-CGTGCCAGCAGCCGCGGTAA-3′) and 1350R (5′-GACGGGCGGTGTGTACAAG-3′). PCR amplification and cloning were done as described in the work of Denisova et al. (15).
16S rDNA sequencing.
Most of the DNA sequencing was conducted on automated sequencers (ABI 377; Perkin-Elmer, Langen, Germany) by cycle sequencing with the chain termination technique using dye-labeled dideoxynucleotides (13), following the manufacturer's instructions. Manual sequencing of selected Lake Baikal clones was performed using a PCR Cyclist kit (Stratagene). Sequencing was done as recommended by the manufacturer. To screen out identical sequences, “single-letter” sequencing (ddGTP reactions only) was performed for all clones using primer 500L. For full sequencing, subfragments were amplified from the primary PCR products with the primer pairs 500L-1000R and 800L-1350R (15) and purified by agarose gel electrophoresis.
Data analysis.
The retrieved 16S rDNA sequences were added to the rDNA sequence database of the Technical University of Munich (release December 1998) using the program package ARB (Lehrstuhl für Mikrobiologie, Technische Universität München [http://www.mikro.biologie.tu-muenchen.de]). The tool ARB_ALIGN was used for automatic sequence alignment. The resulting alignments were checked and corrected manually, considering the secondary structure of the rRNA molecule. For a general phylogenetic comparison, the ARB database was supplemented by importing most of the available 16S rDNA sequences from freshwater habitats, including estuarine and coastal marine systems (February 2000). Phylogenetic trees were reconstructed based on distance matrix analyses of all rRNA primary structures in the database (more than 13,000). Tree topologies were evaluated by maximum parsimony, neighbor joining, and maximum likelihood analyses in combination with different filters excluding highly variable positions on a subset of 152 (FastDNAml) (43) and 269 (neighbor joining) (54) nearly full-length sequences. A consensus tree was constructed with topology corrected by consideration of the results of the various tree reconstruction algorithms. Partial sequences were inserted into the reconstructed tree by parsimony criteria, without allowing changes in the overall tree topology. The clone identifications, closest relatives, and similarity values are given in Table 1.
TABLE 1.
Clones of the different libraries, their affiliations, nearest relatives, percents similarity, and accession numbers
| Sample location (no. of clones) | Clone | Category | Nearest published relative (accession no.) | Similarity (%) | Accession no.a |
|---|---|---|---|---|---|
| Lake Gossenköllesee, December 1995 (8) | GKS59 | α-Proteobacteria | Rhodobacter sphaeroides (D16424) | 95.5 | AJ224988 |
| GKS69 | α-Proteobacteria | Clone T28 (Z93995); Sphingomonas sp. | 97.4 | AJ224989 | |
| GKS16, -70, -71 | β-Proteobacteria | Isolate 34-P (U14585); Rhodoferax sp. | 97.2 | AJ224987 | |
| GKS98, -35 | β-Proteobacteria | Alcaligenes denitrificans (D88005) | 96.5 | AJ224990 | |
| GKS61 | Actinobacteria | Clone ACK-M1 (U85190) | 92.5 | AJ290036 | |
| Lake Gossenköllesee, September 1997 (35) | GKS2-124 | α-Proteobacteria | Clone NO22 (AJ001344); Blastobacter sp. | 99.7 | AJ290027 |
| GKS2-215 | α-Proteobacteria | Beijerinckia indica (M59060) | 96.5 | AJ290032 | |
| GKS2-218 | α-Proteobacteria | Roseococcus thiosulfatophilus (X72908) | 92.7 | AJ290043 | |
| GKS2-122, -77, -187 | β-Proteobacteria | Clone T3 (Z93975); Rhodoferax sp. | 95.0 | AJ290026 | |
| GKS2-91 | γ-Proteobacteria | Isolate CDC658-79 (U88437); Rahnella sp. | 92.7 | AJ290038 | |
| GKS2-103, -38, -79, -176, -189 | Actinobacteria | Clone ACK-M1 (U85190) | 98.6 | AJ290024 | |
| GKS2-33, -7, -15, -19, -63, -88, -111, -112, -120, -121, -142, -155, -205, -207, -208, -232 | CFB | Spirosoma linguale (M27802) | 84.4 | AJ290035 | |
| GKS2-70 | CFB | Spirosoma linguale (M27802) | 87.7 | AJ290037 | |
| GKS2-106, -164 | CFB | Flavobacterium ferrugineum (M28237) | 92.2 | AJ290025 | |
| GKS2-216 | CFB | Flectobacillus major (M27800) | 93.5 | AJ290033 | |
| GKS2-217 | CFB | Haliscomenobacter hydrossis (M58790) | 94.6 | AJ290034 | |
| GKS2-30, -174 | Candidate division OP11 | Isolate Koll6 (AJ224539) | 78.4 | AJ290044 | |
| Lake Fuchskuhle, northeastern part (34) | FukuN22 | α-Proteobacteria | Clone T28 (Z93995); Sphingomonas sp. | 96.9 | AJ289994 |
| FukuN57 | α-Proteobacteria | Afipia genosp. 9 (U87777) | 94.0 | AJ290000 | |
| FukuN33, -2, -60, -66, -109 | β-Proteobacteria | Clone ACK-C4 (U85124); Polynucleobacter sp. | 99.7 | AJ289997 | |
| FukuN55, -107 | β-Proteobacteria | Clone T3 (Z93957); Rhodoferax sp. | 94.7 | AJ289999 | |
| FukuN65 | β-Proteobacteria | Alcaligenes denitrificans (D88005) | 96.4 | AJ290001 | |
| FukuN108 | β-Proteobacteria | Eikenella corrodens (M22467) | 89.1 | AJ289984 | |
| FukuN13 | γ-Proteobacteria | Isolate BB5.1 (AF016981); Methylobacter sp. | 96.6 | AJ290055 | |
| FukuN9, -97, -110 | δ-Proteobacteria | Bdellovibrio stolpii (M34125) | 96.1 | AJ290009 | |
| FukuN30, -8, -15, -44, -105 | Actinobacteria | Clone ACK-M1 (U85190) | 98.3 | AJ289996 | |
| FukuN101 | Actinobacteria | Rathayibacter tritici (X77438) | 95.5 | AJ289982 | |
| FukuN21 | CFB | Flavobacterium ferrugineum (M28237) | 90.8 | AJ289993 | |
| FukuN24, -3, -47, -50, -54, -104 | CFB | Flavobacterium ferrugineum (M28237) | 91.1 | AJ289995 | |
| FukuN36 | CFB | Flavobacterium aquatile (M28236) | 95.2 | AJ289998 | |
| FukuN63 | CFB | Clone WCHB1-53 (AF050539); Cytophaga sp. | 87.9 | AJ290056 | |
| FukuN18, -43, -106 | Verrucomicrobia | Clone LD29 (AF009975); Verrucomicrobium sp. | 92.2 | AJ289992 | |
| FukuN111 | Verrucomicrobia | Isolate 2 (X99390); Verrucomicrobium sp. | 94.6 | AJ289985 | |
| Lake Fuchskuhle, southwestern part (29) | FukuS56, -132, -182 | α-Proteobacteria | Sphingomonas sp. (AJ001051) | 97.5 | AJ290014 |
| FukuS110, -187 | α-Proteobacteria | Beijerinckia indica (M59060) | 96.5 | AJ289986 | |
| FukuS35, -12, -21, -36, -89, -99, -129, -183, -191 | β-Proteobacteria | Clone ACK-C4 (U85124); Polynucleobacter sp. | 99.4 | AJ290013 | |
| FukuS93, -68, -75, -135 | β-Proteobacteria | Isolate R6 (AJ002809); Alcaligenes sp. | 96.9 | AJ290018 | |
| FukuS94 | β-Proteobacteria | Rhodoferax fermentans (D16211) | 92.7 | AJ290057 | |
| FukuS5, -24, -98 | Actinobacteria | Isolate TM177 (X92701); Actinomycetales | 96.7 | AJ290022 | |
| FukuS81 | Actinobacteria | Clone ACK-M1 (U85190) | 92.5 | AJ290054 | |
| FukuS23 | CFB | Flexibacter sancti (M28057) | 89.2 | AJ290011 | |
| FukuS59, -20, -140, -188 | CFB | Flavobacterium ferrugineum (M28237) | 91.7 | AJ290042 | |
| FukuS27 | Verrucomicrobia | Clone T62 (Z94005); Prosthecobacter sp. | 98.0 | AJ290012 | |
| Lake Baikal, South Basin, 25 m (3) | B25-4-32 | β-Proteobacteria | Variovorax sp. strain WFF52 (AB003627) | 97.0 | AJ007642 |
| B25-4-10 | Actinobacteria | “Microthrix parvicella” (X89774) | 89.3 | AJ007641 | |
| B25-4-33 | Actinobacteria | Clone ACK-M1 (U85190) | 95.9 | AJ289950 | |
| Lake Baikal, Central Basin, 25 m (4) | B25-5-20 | β-Proteobacteria | Clone T3 (Z93957); Rhodoferax sp. | 98.6 | AJ289951 |
| B25-5-66, -64, -69 | Actinobacteria | Clone ACK-M1 (U85190) | 95.7 | AJ007644 | |
| Lake Baikal, South Basin, 400 m, 1995 (5) | B400-145 | α-Proteobacteria | Rhodobacter sphaeroides (D16425) | 96.4 | AJ001425 |
| B400-148 | α-Proteobacteria | Azospirillum lipoferum (M59061) | 87.0 | AJ001426 | |
| B400-149 | α-Proteobacteria | Acetobacter liquefaciens (X75617) | 94.4 | AJ001427 | |
| B400-1411 | Actinobacteria | Nocardioides sp. strain 4 (X94145) | 94.8 | AJ289954 | |
| B400-138 | Holophaga-Acidobacterium division | Clone II3-36 (Z95726) | 95.0 | Z95731 | |
| Lake Baikal, South Basin, 400 m, 1997 (9) | B404-5 | α-Proteobacteria | Acetobacter liquefaciens (X75617) | 92.3 | AJ289960 |
| B404-2 | Actinobacteria | Aeromicrobium erythreum (M37200) | 97.4 | AJ007645 | |
| B404-11, -23, -29 | Actinobacteria | “Microthrix parvicella” (X89560) | 85.3 | AJ289955 | |
| B404-57, -19, -451, -50 | Actinobacteria | Clone ACK-M1 (AJ007646) | 91.6 | AJ007646 | |
| Lake Baikal, South Basin, 1,200 m, 1995 (7) | B1200-9 | α-Proteobacteria | Sphingomonas macrogoltabidus (D13723) | 97.3 | X99989 |
| B1200-25 | α-Proteobacteria | Caulobacter subvibrioides (M83797) | 99.5 | X99988 | |
| B1200-63 | α-Proteobacteria | Clone NO22 (AJ001344); Blastobacter sp. | 99.5 | X99983 | |
| B1200-64 | α-Proteobacteria | Methylobacterium sp. (D32231) | 99.1 | X99984 | |
| B1200-34 | Planctomycetales | Planctomyces strain 130 (X81952) | 89.6 | X99985 | |
| B1200-12 | GPLGCb | Paenibacillus curdlanolyticus (D78466) | 98.3 | X99987 | |
| B1200-56 | Holophaga-Acidobacterium division | Clone MB1228 (Z95733) | 96.6 | X99986 | |
| Lake Baikal, South Basin, 1,200 m, 1997 (7) | B1204-15L | α-Proteobacteria | Clone OM75 (U70683); Rhodospirillum sp. | 88.8 | AJ289926 |
| B1204-53L | α-Proteobacteria | Beijerinckia indica (M59060) | 95.3 | AJ289931 | |
| B1204-33L | γ-Proteobacteria | Methylomonas methanica (L20840) | 92.1 | AJ289928 | |
| B1204-51L, -61L | δ-Proteobacteria | Desulfomonile tiedjei (M26635) | 88.2 | AJ289930 | |
| B1204-20L | Actinobacteria | Isolate TM213 (X92705) | 92.2 | AJ289927 | |
| B1204-39L | Actinobacteria | “Microthrix parvicella” (X89560) | 89.7 | AJ289929 | |
| Lake Baikal, South Basin, 1,400 m, 1997 (7) | B1404-8 | α-Proteobacteria | Isolate 23 (Y12598); Rhodopseudomonas sp. | 94.5 | AJ007647 |
| B1404-16 | α-Proteobacteria | “Nitrospira moscoviensis” (X82558) | 94.5 | AJ007648 | |
| B1404-44 | α-Proteobacteria | Acetobacter liquefaciens (X75617) | 94.5 | AJ289935 | |
| B1404-59 | α-Proteobacteria | Unidentified bacterium 22 (Y12596); Rhodospirillum sp. | 89.1 | AJ007657 | |
| B1404-25L | Actinobacteria | Clone ACK-M1 (U85190) | 88.4 | AJ289933 | |
| B1404-40 | Planctomycetales | Gemmata obscuriglobus (S39802) | 85.8 | AJ007656 | |
| B1404-27 | Holophaga-Acidobacterium division | Acidobacterium capsulatum (D26171) | 81.8 | AJ289934 | |
| Lake Baikal, Central Basin, 1,400 m, 1997 (15) | B1405-19 | α-Proteobacteria | Nitrospira sp. (AF35813) | 95.1 | AJ007652 |
| B1405-56, -60 | α-Proteobacteria | Isolate BF14 (Z23157); Blastobacter sp. | 98.6 | AJ289940 | |
| B1405-9, -79 | β-Proteobacteria | “Chromobacterium indigoferum” (U45995) | 91.0 | AJ007650 | |
| B1405-1, -2 | γ-Proteobacteria | Methylomonas methanica (L20840) | 93.3 | AJ007649 | |
| B1405-20 | γ-Proteobacteria | Isolate HTA580 (AB002659); Acinetobacter sp. | 99.3 | AJ007653 | |
| B1405-48 | γ-Proteobacteria | Methylobacter luteus (M95657) | 94.1 | AJ289939 | |
| B1405-10 | δ-Proteobacteria | Desulfomonile tiedjei (M26635) | 86.2 | AJ007651 | |
| B1405-72, -41 | Actinobacteria | Clone ACK-M1 (U85190) | 96.5 | AJ007655 | |
| B1405-49, -22 | Holophaga-Acidobacterium division | Clone C112 (AF013534) | 96.8 | AJ007654 | |
| B1405-62 | Holophaga-Acidobacterium division | Clone RB27 (Z95719) | 98.1 | AJ289942 | |
| Lake Baikal, Central Basin, 1,600 m, 1997 (7) | B1605-3 | α-Proteobacteria | Azospirillum irakense (Z29583) | 88.6 | AJ289947 |
| B1605-17 | β-Proteobacteria | Thiobacillus aquaesulis (U58019) | 92.8 | AJ289945 | |
| B1605-22 | β-Proteobacteria | Chromobacterium violaceum (M22510) | 89.1 | AJ289946 | |
| B1605-43 | γ-Proteobacteria | Methylobacter luteus (M95657) | 93.9 | AJ289949 | |
| B1605-59 | γ-Proteobacteria | Methylobacter sp. strain BB5.1 (AF016981) | 93.8 | AJ007658 | |
| B1605-35 | Actinobacteria | “Microthrix parvicella” (X89560) | 85.8 | AJ289948 | |
| B1605-10 | Actinobacteria | Clone ACK-M1 (U85190) | 94.3 | AJ289944 |
The accession number is that of the sequence of the first clone listed in each row. This corresponds generally with the full-length or long sequences determined.
GPLGC, gram-positive, low-G+C group.
We also used sequences from the following sources: Columbia River and estuary, Coastal Ocean, Oregon (14); Adirondack mountain lakes, New York (32); groundwater bacteria (A), Sweden (45); Toolik Lake, Alaska (6); gas vacuolate bacteria, Antarctica (31); activated sludge (T), Germany (58); negative controls (MT), California (61); Lake Loosdrecht (LD), The Netherlands (66); activated sludge (SMK), Germany (55); drinking water (B), Germany (35); activated sludge (Ben), Australia (10); subterranean groundwater bacteria (G), Africa (46); sequencing batch reactors (SBR), Australia (7); activated sludge (LDI), Germany (63); Stripa groundwater, Sweden (18); peat bog (TM), Germany (52); Carolina Bay, South Carolina (64); grassland soil, The Netherlands (21); blanket bog peat (MHP), United Kingdom (39); Mariana Trench mud (HTA), (60); freshwater enrichment (NO), Germany (28); borehole isolate (S), Sweden (47); subtropical soil (MC), Australia (59); contaminated aquifer (WCHB), Michigan (16); Wisconsin soil, Wisconsin (8); and Eastern Amazonian soil, Amazonia (9). All sequences are available in aligned ARB and GenBank format via our anonymous FTP server: ftp.mpi-bremen.de/pub/molecol_p/fog-aem/.
Probe design and FISH.
Using the PROBE_DESIGN tool of ARB (see above), two oligonucleotide probes and helper oligonucleotides (helper) (22) were constructed for the hgcI cluster (Fig. 1C). For the probes HGC236 and HGC664 (19), additional helpers were designed to enhance the signal intensities. Probes labeled with the indocarbocyanine dye Cy3 and unlabeled helpers were purchased from Interactiva (Ulm, Germany). All sequences and target positions of the probes and helpers are given in Table 2.
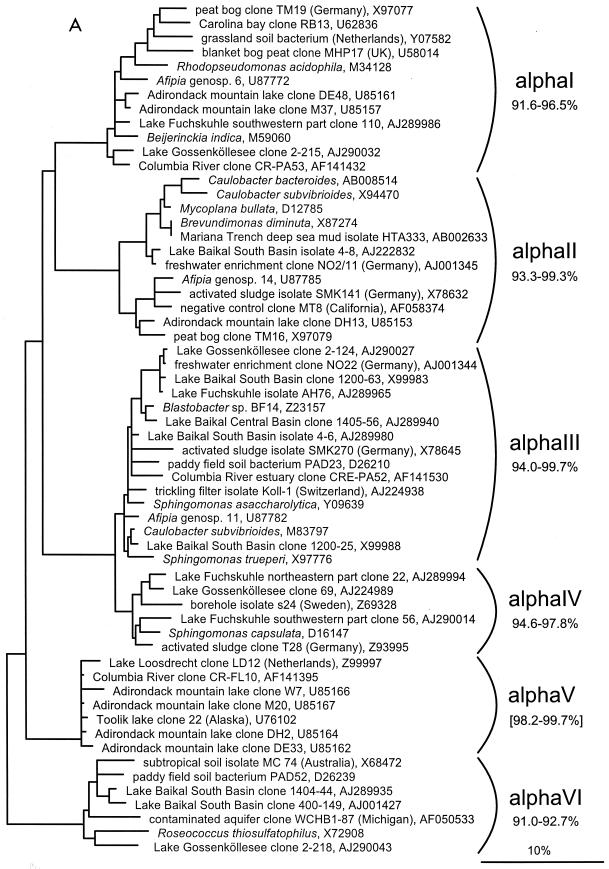
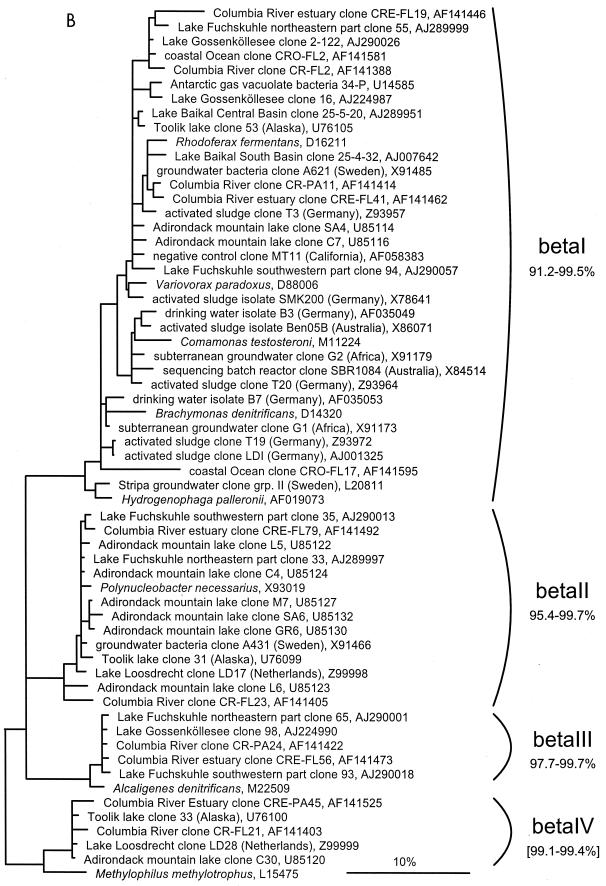
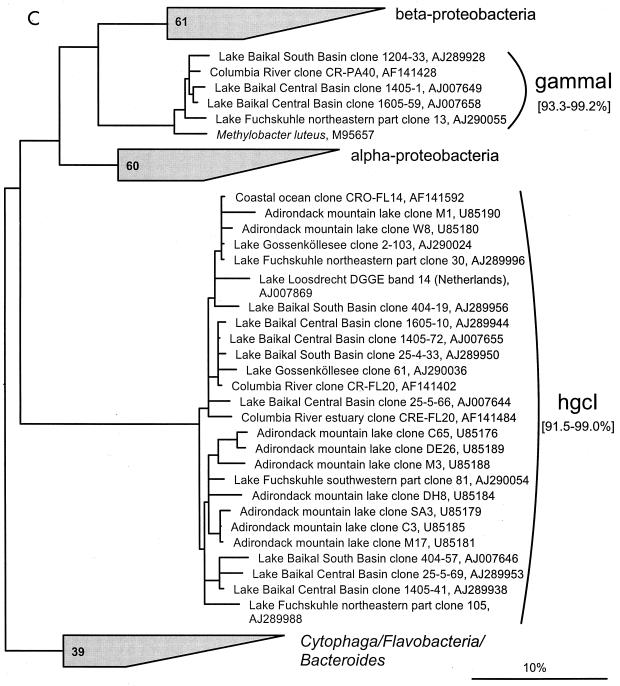
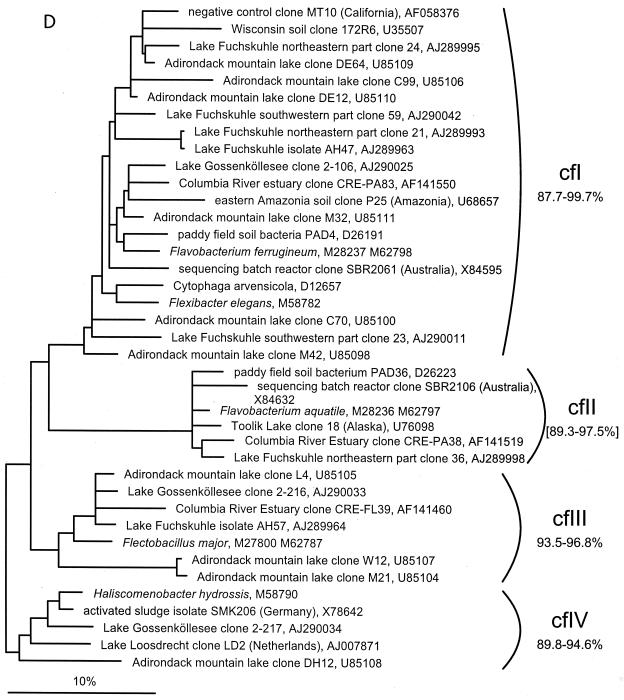
FIG. 1.
(A to D) Unrooted phylogenetic consensus tree based on maximum likelihood (FastDNAml) analysis showing the clones in the proposed clusters. (A) α-Proteobacteria; (B) β-Proteobacteria; (C) γ-Proteobacteria and Actinobacteria; (D) CFB phylum. Bifurcations indicate branchings which appeared stable and well separated from neighboring branchings in all cases. Multifurcations indicate tree topologies which could not be unambiguously resolved based on the available data set. For clarity, only the reported clusters are shown; all sequences between the clusters have been omitted. These are included in a more detailed version of the tree which is available at ftp.mpi-bremen.de/pub/molecol_p/fog-aem. The scale bar indicates 10% estimated sequence divergence.
TABLE 2.
Oligonucleotide probes used in this study
| Probe | Specificity | Sequence (5′ to 3′) of probe | Target site (rRNA positions)a | % FA in situb | Reference |
|---|---|---|---|---|---|
| EUB338 | Bacteria | GCTGCCTCCCGTAGGAGT | 16S, 338–355 | 0–35 | 4 |
| NON338 | ACTCCTACGGGAGGCAGC | 16S, 338–355 | 0–35 | 62 | |
| HGC69a | Actinobacteria | TATAGTTACCACCGCCGT | 23S, 1901–1981 | 20 | 53 |
| HGC236 | Actinobacteria | AACAAGCTGATAGGCCGC | 16S, 235–253 | 10 | 19 |
| HGC664 | Actinobacteria | AGGAATTCCAGTCTCCCC | 16S, 663–681 | 30 | 19 |
| HG1-840 | HG1 cluster of the class Actinobacteria | TCGCA(C/G)AAACCGTGGAAG | 16S, 840–846 | 10 | This study |
| HG1-654 | HG1 cluster of the class Actinobacteria (without FukuN105, 1605-10, 1405-72, and 25-4-33); “cross-reaction” with some CFBc” | AGTCTCCCCTACCAAACTC | 16S, 654–672 | 35 | This study |
| HGC270H | Helper for HGC236 | ACCCGTCG(T/A/C)(C/A)GCCTTGGT | 16S, 270–287 | 10 | This study |
| HGC697H | Helper for HGC664 | GTTCCTCCTGATATCTGCGCA | 16S, 697–717 | 30 | This study |
| HG1-810H | Helper for HGC840 | CTAG(C/T)GCCCA(C/T)CGTTTACGG | 16S, 810–829 | 10 | This study |
| HG1-820H | Helper for HGC840 | GTTC(G/C)CACAACTAG(C/T)GCCCA | 16S, 820–839 | 10 | This study |
| HG1-859H | Helper for HGC840 | GGGGC(G/A)CTTAATGCGTTAGCTG | 16S, 859–880 | 10 | This study |
E. coli numbering (11).
Percent formamide (FA) in in situ hybridization buffer.
FISH of filter sections with oligonucleotide probes, counterstaining with DAPI (4′,6′-diamidino-2-phenylindole), and mounting for microscopic evaluation were performed as described previously (27, 30). Epifluorescence microscopy was done on Zeiss Axioplan microscopes, equipped with HBO 100-W mercury lamps and 100× Plan Apochromat objectives. Between 500 and 1,000 DAPI-stained objects were evaluated per sample. Absolute densities of hybridized bacteria were calculated as the product of their relative abundances on filter sections (percentage of DAPI-stained objects) and the DAPI-stained direct cell counts. Since parallel counting on independent filters was not practicable, a general counting error of ±10% was calculated (49), which applies to all counts shown. Hybridization conditions for the probes HG1-654 and HG1-840 were adjusted by formamide series applied to different filters from Lake Gossenköllesee and evaluated by eye. Probes were also checked against the next nontarget organisms (Actinomadura glomerata, Terracoccus luteus, and Micromonospora aurantiaca) in the database which had one or two mismatches in the probe target region. Optimal formamide concentrations for all probes are given in Table 2.
Bacterial cell size determination.
On Lake Gossenköllesee samples from August 1996, November 1996, April 1997, and June 1997, size measurements of all DAPI-stained bacteria and of cells hybridizing with the probe HG1-840 were carried out as described earlier (34, 48). Double images of the Cy3 and DAPI fluorescence of individual cells in microscopic preparations were captured using a PC-based image analysis system. Between 500 and 1,000 hybridized cells were analyzed per sample (corresponding to 5 to 30 image pairs). Biomass was calculated as the product of mean cell size and abundance.
Nucleotide sequence accession numbers.
Accession numbers of the sequences determined in this study are AJ224988-90, AJ289926-61/63-65/80/82-99, AJ290000-80, AJ007641-42/44-58, and X99983-89.
RESULTS
Clone libraries.
A total of 190 clones from 13 independent 16S rDNA clone libraries were partially sequenced (∼400 to 500 nucleotides) and phylogenetically analyzed. Sequences with similarities greater than 97% were grouped together. Fifteen clones belonging to the Cyanobacteria phylum were rejected from further analysis because our focus was on heterotrophic bacteria. Furthermore, five chimeric sequences have been omitted. The value for sequence similarity to the next closest published sequence and phylogenetic affiliation of the remaining 170 clones are summarized in Table 1. From the Lake Gossenköllesee and Lake Fuchskuhle libraries, 43 clones representative of the different groups were selected for nearly full-length sequencing of 16S rDNA (∼1,400 nucleotides), whereas for 28 of the Lake Baikal clones 800- to 1,200-nucleotide-long sequences were determined. Although most of the clones fall into well-established groups, only 19 of the 71 long 16S rDNA sequences are more than 97% similar to published sequences. Clones affiliated with the class Actinobacteria (n = 37), the Cytophaga-Flavobacterium-Bacteroides (CFB) group (n = 35), the β subclass of the class Proteobacteria (n = 37), and the α subclass of Proteobacteria (n = 30) were about equally abundant and together accounted for almost three quarters of the 190 retrieved sequences. The sequences of 31 clones were affiliated with other groups: nine with γ-Proteobacteria, six each with the δ-Proteobacteria and the Holophaga-Acidobacterium group, five with the Verrucomicrobia, two each with the Planctomycetales and the candidate division OP11, and one with the low-G+C gram-positive bacteria.
Phylogenetic tree.
A phylogenetic comparison of the new sequences with published 16S rDNA sequences revealed 16 clusters. Five included only sequences from freshwater samples, seven included sequences from freshwater and freshwater-influenced coastal marine systems, and four clusters also contained sequences originating from soil samples. The criterion for defining a cluster was that at least three sequences from independent sources cluster together in all tree reconstructions. The clusters are based on phylogenetic affiliations and not on similarity thresholds because, in most cases, due to the sequencing of different 16S rDNA regions by various authors, no reasonable similarity values could be determined. With the exception of cluster αII, all described clusters are clearly separated from sequences found in the open ocean or marine sediments (Fig. 1).
α-proteobacterial 16S rDNA sequences.
Six clusters, αI to αVI, can be described in the α-Proteobacteria (Fig. 1A). The αV cluster has already been described as freshwater cluster A (66). The sequence similarity values within clusters range from 91.0 to 92.7% for αVI to 94.0 to 99.7% in αIII. These values were calculated only on sequences longer than 1,200 nucleotides, whereas the similarity value given in brackets for αV had to be calculated with truncated sequences (282 nucleotides). Based on the current data set, three of the clusters appear to be true freshwater sequence clusters (αII, αIV, and αV), although cluster αII includes a sequence derived from the Mariana Trench deep-sea mud (60) and a negative control clone sequence described as a common PCR contaminant (61). Cluster αIII also contains one sequence each from the Columbia River estuary and from paddy field soil, and clusters αI and αVI are intermingled with sequences originating from marine coastal regions and soil habitats. All clusters except αV include cultivated strains.
β-proteobacterial 16S rDNA sequences.
Among the β-proteobacterial sequences, four stable clusters could be identified, with in-cluster similarities ranging from 91.2 to 99.5% (βI) to 97.7 to 99.7% (βIII), calculated on nearly full-length sequences only (Fig. 1B). For βIV, the similarity of 99.1 to 99.4% is based on the sequences longer than 900 nucleotides. Sequences from coastal regions or estuaries are in all clusters, but no sequences from soil ecosystems can be found. With 35 sequences, βI is the largest cluster defined in this study. As sequences from additional environmental studies become available, subgroups already visible in the current topology might stabilize. Cluster βI contains numerous cultured bacteria, most formerly classified as pseudomonads, but now assembled in the genera Acidovorax, Comamonas, Hydrogenophaga, and Variovorax. βII, βIII, and βIV are comprised only of cloned sequences from environmental samples. The closest cultivated relatives of these clusters are Ralstonia eutropha, Alcaligenes denitrificans, and Methylophilus methylotrophus, with similarities of 91.9 to 93.4, 96.2 to 96.6, and 91.8 to 95.5%, respectively. βII and βIV have already been described by Zwart and coworkers as freshwater clusters B and C (66).
γ-proteobacterial 16S rDNA sequences.
Only one cluster was identified within the γ-Proteobacteria (γI), consisting currently only of cloned sequences (Fig. 1C). The in-cluster similarity is 93.3 to 99.2%, based on all sequences. The similarity to the next cultivated relative, Methylobacter luteus, ranges between 91.7 and 96.0%.
Actinobacteria-related 16S rDNA sequences.
The cluster hgcI is exceptional (Fig. 1C). It forms a stable monophyletic new branch within the class Actinobacteria, with a similarity of only 87.2 to 89.0% to the next cultivated relative, Rathayibacter rathayi. The similarity within the cluster ranges from 91.5 to 99.0%, calculated for the sequences which overlap at least in a 400-nucleotide-long, central region of the 16S rRNA (Escherichia coli position 502 to 961 [11]). Twenty-four of the 37 actinobacterial sequences retrieved in this study can be assigned to the hgcI cluster. This cluster consists mostly of cloned sequences from different freshwater habitats but also two sequences from estuary and coastal ocean systems. The uneven topology and the low similarity values within the cluster suggest that subgroups may be identifiable when more sequences become available. Remarkably, 16 out of 17 actinobacterial clone sequences from Adirondack mountain lakes, formerly described as the ACK-4 group by Hiorns et al. (32), belong to this cluster.
CFB sequences.
The four clusters proposed in the CFB phylum have a low in-cluster similarity of only 87.7 to 99.7% to 93.5 to 96.8% (Fig. 1D). The similarity values given for cluster cfII are calculated from an alignment truncated to the shortest sequence. Clusters cfI and cfII include sequences from different soil habitats and from the Columbia River estuary. In cluster cfI, the negative control clone MT10 described previously (61) is also present. Cluster cfIII consists mostly of freshwater sequences, with only one sequence originating from the Columbia River estuary, and cfIV consists only of freshwater sequences with low similarities ranging from 89.8 to 94.6% (calculated only on the sequences with more than 900 nucleotides). The low similarities within the whole CFB phylum imply a high unknown diversity and therefore affect the stability of the designated clusters. All clusters contain at least one pure culture.
FISH of hgcI.
To determine the abundance, biomass, and seasonal distribution of the organisms affiliated with the hgcI cluster, specific probes were designed and applied first to Lake Gossenköllesee samples (Fig. 2). Hybridizations performed on the filter samples from June 1997 yielded weak yet detectable signals from small coccoid rods (mean, 0.5 by 0.4 μm) with the probe HG1-654, but no signals with probe HG1-840. For the latter, oligonucleotide hybridization data on E. coli 16S rRNA indicate limited accessibility of this probe binding site (class IV-V according to reference 23). To enhance the FISH signal intensity, three helper oligonucleotides (HG1-810H, HG1-820H, and HG1-859H) were developed. The helpers were designed to be complementary to sites adjacent to the probe target site and to target at least the same organisms as the probe HG1-840. In addition, they were designed with theoretical thermal stabilities above that of the probe (22). A reevaluation of the June 1997 sample revealed detectable signals for the probe HG1-840 only when combined with all three helpers.
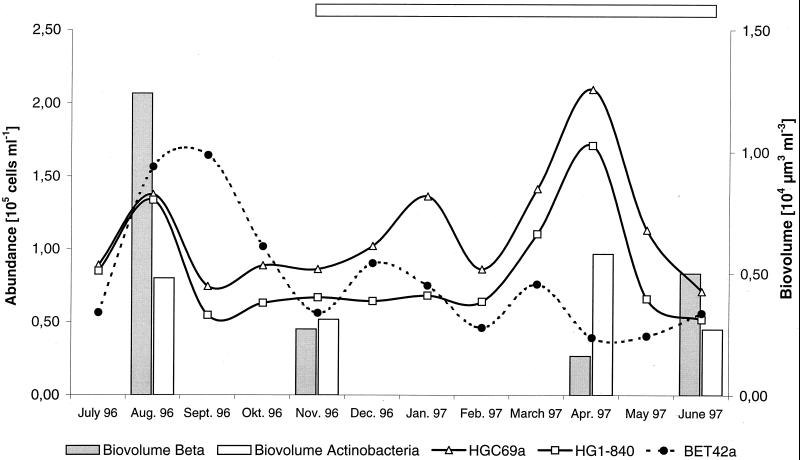
FIG. 2.
Seasonal dynamics of the abundance and biomass of the β-Proteobacteria and the Actinobacteria counted with probes HGC69a, HG1-840, and BET42a. The horizontal bar indicates the period of ice cover.
The probe HG1-840–helper mix was checked against the cultivated nontarget organism, Actinomadura glomerata, which had two mismatches. No signal could be detected even under nonstringent conditions. HG1-840 probe counts were supported with counts from the more general actinobacterial probes, HGC69a, HGC236, and HGC664 (Table 3). Although all probes showed detectable signals, helpers HGC270H and HGC697H were developed for the probes HGC236 and HGC664, respectively. A significant signal enhancement on the filters was observed for the combination of HGC236 and HGC270H, but no visible effect was seen for the pair HGC664-HGC697H.
TABLE 3.
DAPI and FISH counts
| Sampling site and date | Depth (m) | Total cell count (105 ml−1) | Fraction (%) of total cells detected with probea
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EUB338 | ALF1bb | BET42ab | CF319ab | HGC69a | HGC236 | HGC664 | HG1-840 | |||
| Lake Gossenköllesee | ||||||||||
| 4 July 1996 | 4 | 2.07 | 85 | 10 | 27 | NDc | 43 | 44 | 42 | 41 |
| 5 August 1996 | 4 | 3.93 | 94 | 18 | 40 | 2 | 35 | 33 | 31 | 34 |
| 3 September 1996 | 4 | 4.96 | 47 | 18 | 33 | 3 | 15 | 16 | 14 | 11 |
| 15 October 1996 | 4 | 2.86 | 89 | 18 | 36 | 5 | 31 | 25 | 21 | 22 |
| 13 November 1996 | 0 | 2.78 | 80 | 11 | 20 | 10 | 31 | 29 | 24 | 24 |
| 13 December 1996 | 0 | 3.78 | 76 | 6 | 24 | 2 | 27 | 26 | 18 | 17 |
| 15 January 1997 | 4 | 3.24 | 76 | 10 | 23 | 4 | 42 | 35 | 26 | 21 |
| 13 February 1997 | 4 | 2.78 | 63 | 11 | 17 | 6 | 31 | 33 | 32 | 23 |
| 5 March 1997 | 4 | 3.45 | 77 | 10 | 22 | 8 | 41 | 38 | 30 | 32 |
| 2 April 1997 | 4 | 3.49 | 82 | 8 | 11 | 3 | 60 | 45 | 56 | 49 |
| 29 April 1997 | 4 | 3.82 | 90 | 22 | 27 | 3 | 44 | 40 | 40 | 32 |
| 29 May 1997 | 4 | 2.36 | 79 | 16 | 17 | 3 | 48 | 40 | 40 | 28 |
| 25 June 1997 | 4 | 2.15 | 96 | 5 | 26 | 2 | 33 | 36 | 34 | 24 |
| Lake Fuchskuhle | ||||||||||
| Northeastern part, 9 September 1997 | 0.5 | 4.00 | 69 | ND | 6 | 1 | 25 | ND | 21 | 19 |
| Southwestern part, 23 July 1997 | 3.5 | 2.31 | 68 | ND | 31 | 3 | 19 | 18 | 14 | 15 |
| Lake Baikal | ||||||||||
| Central Basin, 13 July 1997 | 0 | 3.27 | 74 | ND | 17 | 21 | 4 | 3 | 0 | 0 |
| South Basin, 18 July 1997 | 0 | 8.55 | 71 | ND | 13 | 7 | 15 | 10 | 18 | 10 |
Percent detection of DAPI counts. Numbers have been corrected by subtracting NON338 counts.
Data for Lake Gossenköllesee were taken from the work of Pernthaler et al. (50).
ND, not done.
The seasonal distribution of members of the hgcI cluster was determined for Lake Gossenköllesee between July 1996 and June 1997 (Table 3). The abundances determined with the probe HG1-840–helper mix were high, up to 1.7 × 105 cells ml−1, or 49% of all DAPI-stained cells (mean, 28%; n = 13) (Fig. 2). A biphasic population development was observed, with the first peak in August 1996 and the second peak in April 1997 (Fig. 2). The general and the specific probes applied showed identical fluctuations, within the range of the estimated counting error, which indicates that hgcI is the main actinobacterial group in the bacterioplankton of this lake.
Comparison of the abundance and biomass of the two dominant groups in Lake Gossenköllesee, the β subclass of Proteobacteria and the class Actinobacteria, revealed important differences over the year. While β-Proteobacteria made up 61% of the bacterioplankton biomass (biovolume, 1.2 × 104 μm3 ml−3) in August 1996, 63% of the April 1997 biomass (biovolume, 0.6 × 104 μm3 ml−3) was accounted for by the Actinobacteria (Fig. 2). This is consistent with the development of the total abundances of the two populations where maxima for the β-Proteobacteria (1.6 × 105 cells ml−1) and Actinobacteria (2.1 × 105 cells ml−1) were reached in September 1996 and April 1997, respectively. When applying the actinobacterial probes to bacterioplankton samples, the strongest signals were obtained on cells that had been fixed in buffered paraformaldehyde solution and stored for more than 1 year at −20°C.
Several samples from Lake Baikal and Lake Fuchskuhle, hybridized with the set of actinobacterial probes, gave positive hybridization signals with cell counts ranging from 1.3 × 104 to 1 × 105 cells ml−1 for probe HGC69a and 0 to 7.6 × 104 cells ml−1 for probe HG1-840 (Table 3). The cell counts in general were moderately lower than those in Lake Gossenköllesee, especially in the Central Basin of Lake Baikal, where cell counts for the general actinobacterial probes ranged between 0 and 4% but no hgcI could be found. In this particular sample, the cells detected were large rods (2 by 1 μm) rather than the typical small rods or cocci (0.5 by 0.4 μm). Morphological diversity was also apparent in the Lake Fuchskuhle samples, where a fraction of bent rods could be detected with the general and specific actinobacterial probes.
DISCUSSION
The comparative analysis of 170 16S rDNA clones obtained from the bacterioplankton of three lakes showed that the vast majority affiliated in clusters that consisted only of sequences originating from freshwater, freshwater and soil, and, in some cases, from freshwater-influenced marine samples (Fig. 1). This clearly is suggestive evidence for the existence of phylogenetically coherent freshwater-continental clusters. A general bias for the selection of related sequences can be excluded, because the samples originated from spatially and ecologically diverse habitats and the strategies for retrieving the sequences were different in many ways. All freshwater-continental clusters are stable with the phylogenetic reconstruction methods applied and clearly separated from marine clusters like the SAR clusters (26, 41). Only cluster αII contains an isolate from a true marine habitat, the Mariana Trench sediment (60). It is noteworthy that the 16S rDNA sequence of this isolate is 100% identical with the sequence of Brevundimonas diminuta, which, together with several Caulobacter spp., was recently described as a member of a phylogenetically well-supported freshwater cluster (57).
Coherent clusters might also originate from a widespread use of contaminated PCR reagents. Some of the clusters proposed in this study, e.g., βI, αII, and cfI, indeed contain sequences retrieved from negative controls by Tanner and coworkers (61). Nevertheless, considering that most of the PCR reagents are low in ion concentration (e.g., the PCR water or the nucleotide mixtures), this does not necessarily falsify the clusters proposed. However, all the clusters, especially those consisting only of cloned sequences, have to be checked for occurrence in freshwater habitats by PCR-independent methods, e.g., FISH or slot blot hybridization.
Another intriguing observation was the presence of one sequence of different Afipia spp. in each of the clusters αI, αII, and αIII. This organism is described as a potential animal pathogen associated with cat scratch disease. More recent data have indicated that Bartonella henselae is the main causative agent, and therefore the real origin of Afipia sp. is unclear (25).
The 16S rDNA library data suggested that, in addition to members of the β subclass of Proteobacteria, members of the class Actinobacteria are abundant in limnic ecosystems. These results were quite unexpected in the light of the general perception that gram-positive bacteria are typical for soils and less common in aquatic habitats (37). FISH was an option for checking the 16S rDNA library data. For that purpose, a set of nested 16S and 23S rRNA-targeted probes specific for the class Actinobacteria, together with two probes specific for the hgcI cluster, was used. The cross-reaction of probe HGC654 to members of the Cytophaga-Flavobacterium group (Table 2) illustrates the importance of using multiple probes (2, 3). The morphological information obtained by FISH, i.e., the detection of filaments as well as the typical small, dim cells, was the key for detecting this cross hybridization.
In Lake Gossenköllesee, the two most abundant bacterial groups, the β-Proteobacteria and the Actinobacteria, may have distinct temporal niches. In a previous study, the β-Proteobacteria reached their greatest biomass after icebreak, between June and July, suggesting that they are the first of the studied groups to respond to thermal mixing and the allochthonous input of nutrients from the ice cover and surrounding rockland (50). The same study also described a second biomass maximum in late spring, in the deeper water layers, which at the time could not be assigned to a particular microbial group. In this study, we have identified this peak as actinobacteria, which show high abundance and biomass concentration in April, as well as in August (Fig. 2). The April maximum follows the spring phytoplankton bloom, which occurs in Lake Gossenköllesee between January and March (50), and unlike the August Actinobacteria maximum, it is not associated with a β-proteobacterial bloom. In contrast to the spring maximum, the August peak is primarily triggered by allochthonous inputs. In addition, the range of biomass fluctuation is much lower for Actinobacteria (2.1-fold) than for the β-Proteobacteria (7.8-fold). This leads us to the hypothesis that in this lake the β-Proteobacteria and Actinobacteria members inhabit separate functional niches: the β-Proteobacteria are able to rapidly utilize the main annual input, whereas the hgcI group more efficiently consumes lower levels of organic carbon at low temperature.
First results from Lake Baikal and Lake Fuchskuhle confirm a widespread occurrence of the hgcI cluster. Recently, a marine cluster of Actinobacteria has been described which is clearly separated from cluster hgcI (51). This supports our view that hgcI might be a typical freshwater cluster, comparable to the β-proteobacterial clusters (30, 40). Unfortunately, despite considerable efforts it was not possible to obtain pure cultures of hgcI cluster representatives, and therefore their physiology is still unknown. A screen of more than 200 Lake Fuchskuhle and Lake Baikal isolates by partial rDNA sequencing and FISH failed to identify any hgcI strains (data not shown).
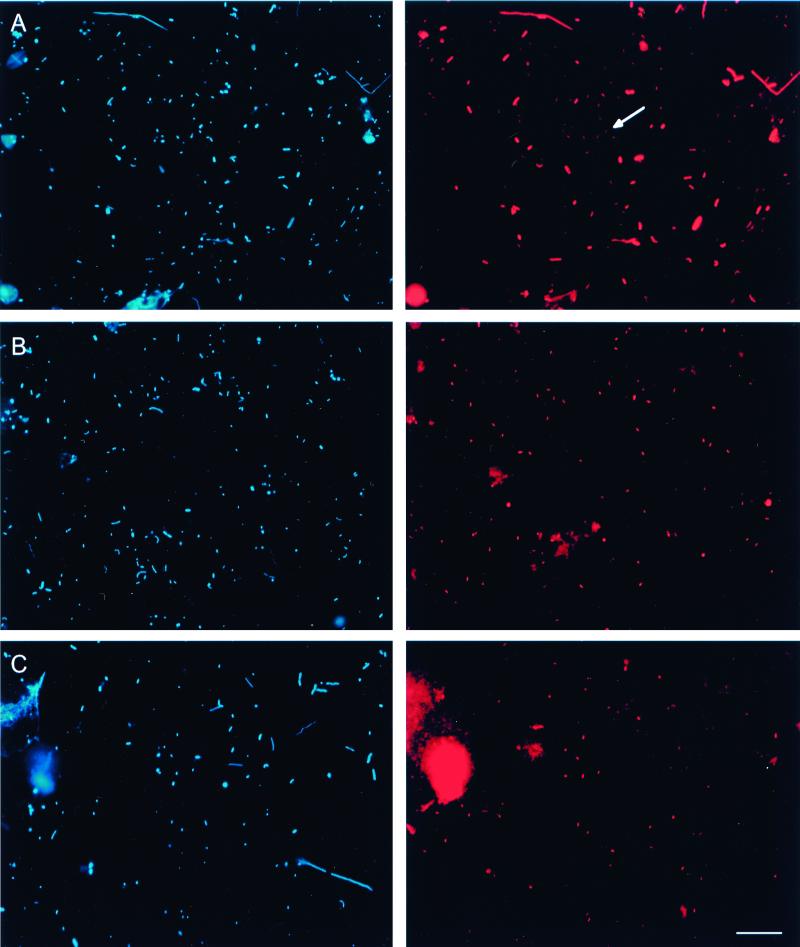
Despite the progress made with FISH, it is still a challenge to visualize small and dimly staining organisms like members of the hgcI cluster (Fig. 3A). To calculate this effect, we reevaluated the EUB338 counts reported by Pernthaler and coworkers (50). Detection yields relative to DAPI ranging from 47 to 96% (mean, 79%; n = 13) were obtained by including the small and dim cells that had been dismissed in the previous study. This is on average one-quarter more than determined earlier (50). By combining the reanalyzed cell counts for the group-specific probes ALF1b, BET42a, and CF319a with counts for the general actinobacterial probe HGC96a from this study, nearly all of the cells detected with probe EUB338 (mean, 99%; n = 12) could be accounted for (Table 3). In these particular samples, it seems that the currently available group-specific probes are adequate to further classify all bacteria detected by EUB338. Furthermore, the difference between the number of particles stained by DAPI and the sum of counts with the domain-specific probes EUB338 and ARCH915 (targeting Archaea) was reduced from 43% (range, 18 to 55%) (50) to 20% (range, 2 to 50%).
FIG. 3.
FISH of samples from Lake Gossenköllesee. DAPI (left) and epifluorescence (right) micrographs are shown for identical microscopic fields. (A) In situ hybridization with probe EUB338 labeled with Cy3. The arrow indicates a representative of a population of small, dim cells. (B) In situ hybridization with probe HGC664 labeled with Cy3. (C) In situ hybridization with probe HG1-840 labeled with Cy3. Bar, 10 μm (all panels).
A recent development to enhance signal intensities is the application of unlabeled helper oligonucleotides. Helpers that bind adjacent to the probe target site have been shown to enhance the fluorescence signals at nearly inaccessible sites in E. coli up to 25-fold (22). The general applicability of this new approach to environmental samples was demonstrated in this study. For probe HG1-840, all three helpers were needed to yield detectable signals. It remains unclear why the combination of probe HGC664 and helper HGC697H did not result in stronger signals, while the combination of probe HGC236 with helper HGC270H led to clearly visible signal enhancement. Perhaps the RNA-protein cross-linking sites at positions 695 and 703 (E. coli numbering) for the ribosomal proteins S11 and S21 prevent the hybridization of the helper (17). Due to the variable character of the adjacent regions it was not possible to design more than one helper with a reasonable number (less than 4) of wobble base pairs.
In this study, we show that a distinct cluster of small Actinobacteria members, hgcI, can become a dominant fraction of the bacterioplankton in freshwater lakes. The phylogenetic divergence indicated by the comparative 16S rDNA sequence analysis and the biotechnological potential of this class are good reasons for further efforts, both traditional and molecular, to learn more about these bacteria.
ACKNOWLEDGMENTS
This work has been supported by grants from the Deutsche Forschungsgemeinschaft (Am73/2-4), the Max Planck Society, and the Russian Foundation for Basic Research (N 96-04-50922 and N99-04-48571).
We thank Michail Grachev and Tamara Zemskaya from the Limnological Institute in Irkutsk for enabling the fieldwork at Lake Baikal, Stefanie Unterholzner for sampling in Lake Gossenköllesee, Jörg Wulf for expert technical assistance, Robert Erhart for making available probes HGC236 and HGC664 before publication, Wolfgang Ludwig for ARB and help in the reconstruction of phylogenetic trees, and Barbara MacGregor for helpful discussions.
REFERENCES
- 1.Amann R, Glöckner F O, Neef A. Modern methods in subsurface microbiology—in situ identification of microorganisms with nucleic acid probes. FEMS Microbiol Rev. 1997;20:191–200. [Google Scholar]
- 2.Amann R, Ludwig W. Typing in situ with probes. In: Priest F G, Ramos-Cormenzana A, Tindall B J, editors. Bacterial diversity and systematics. New York, N.Y: Plenum Press; 1994. pp. 115–135. [Google Scholar]
- 3.Amann R I. Fluorescently labelled, rRNA-targeted oligonucleotide probes in the study of microbial ecology. Mol Ecol. 1995;4:543–554. [Google Scholar]
- 4.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahr M, Hobbie J E, Sogin M L. Bacterial diversity in an arctic lake—a freshwater SAR11 cluster. Aquat Microb Ecol. 1996;11:271–277. [Google Scholar]
- 7.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borneman J, Skroch P W, O'Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia—evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford D, Hugenholtz P, Seviour E M, Cunningham M A, Stratton H, Seviour R J, Blackall L L. 16S rRNA analysis of isolates obtained from gram-negative, filamentous bacteria micromanipulated from activated sludge. Syst Appl Microbiol. 1996;19:334–343. [Google Scholar]
- 11.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 12.Buchholz-Cleven B, Rattunde B, Straub K. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst Appl Microbiol. 1997;20:301–309. [Google Scholar]
- 13.Chen E Y, Seeburg P H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985;4:165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- 14.Crump B, Armbrust E, Baross J. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol. 1999;65:3192–3204. doi: 10.1128/aem.65.7.3192-3204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denisova L Y, Bel'kova N L, Tulokhonov I I, Zaichikov E F. Bacterial diversity at various depths in the southern part of Lake Baikal as revealed by 16S rDNA sequencing. Microbiology. 1999;68:475–483. [PubMed] [Google Scholar]
- 16.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehresmann B, Ehresmann C, Romby P, Mougel M, Baudin F, Westhof E, Ebel J-P. Detailed structures of rRNAs: new approaches. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C.: American Society for Microbiology; 1990. pp. 148–159. [Google Scholar]
- 18.Ekendahl S, Arlinger J, Stahl F, Pedersen K. Characterization of attached bacterial populations in deep granitic groundwater from the Stripa research mine by 16S rRNA gene sequencing and scanning electron microscopy. Microbiology. 1994;140:1575–1583. doi: 10.1099/13500872-140-7-1575. [DOI] [PubMed] [Google Scholar]
- 19.Erhart R. Thesis. Munich, Germany: Technische Universität München; 1997. [Google Scholar]
- 20.Felip M, Sattler B, Psenner R, Catalan J. Highly active microbial communities in the ice and snow cover of high mountain lakes. Appl Environ Microbiol. 1995;61:2394–2401. doi: 10.1128/aem.61.6.2394-2401.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans A D L. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology. 1997;143:2983–2989. doi: 10.1099/00221287-143-9-2983. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs B M, Glöckner F O, Wulf J, Amann R. Unlabeled helper oligonucleotides increase the in situ accessibility of 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 2000;66:3603–3607. doi: 10.1128/aem.66.8.3603-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchs B M, Wallner G, Beisker W, Schwippl I, Ludwig W, Amann R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 1998;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuhrman J A, Comeau D E, Hagström A, Chan A M. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl Environ Microbiol. 1988;54:1426–1429. doi: 10.1128/aem.54.6.1426-1429.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giladi M, Avidor B, Kletter Y, Abulafia S, Slater L N, Welch D F, Brenner D J, Steigerwalt A G, Whitney A M, Ephros M. Cat scratch disease: the rare role of Afipia felis. J Clin Microbiol. 1998;36:2499–2502. doi: 10.1128/jcm.36.9.2499-2502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 27.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 28.Glöckner F O, Babenzien H-D, Amann R. Phylogeny and identification in situ of Nevskia ramosa. Appl Environ Microbiol. 1998;64:1895–1901. doi: 10.1128/aem.64.5.1895-1901.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glöckner F O, Babenzien H-D, Wulf J, Amann R. Phylogeny and diversity of Achromatium oxaliferum. Syst Appl Microbiol. 1999;22:28–38. doi: 10.1016/S0723-2020(99)80025-3. [DOI] [PubMed] [Google Scholar]
- 30.Glöckner F O, Fuchs B M, Amann R. Bacterioplankton compositions in lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gosink J J, Staley J T. Biodiversity of gas vacuolate bacteria from Antarctic Sea ice and water. Appl Environ Microbiol. 1995;61:3486–3489. doi: 10.1128/aem.61.9.3486-3489.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiorns W D, Methe B A, Nierzwicki-Bauer S A, Zehr J P. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl Environ Microbiol. 1997;63:2957–2960. doi: 10.1128/aem.63.7.2957-2960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hugenholtz P, Goebel B, Pace N. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jürgens K, Pernthaler J, Schalla S, Amann R. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl Environ Microbiol. 1999;65:1241–1250. doi: 10.1128/aem.65.3.1241-1250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalmbach S, Manz W, Szewzyk U. Isolation of new bacterial species from drinking water biofilms and proof of their in situ dominance with highly specific 16S rRNA probes. Appl Environ Microbiol. 1997;63:4164–4170. doi: 10.1128/aem.63.11.4164-4170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livingstone D M. Ice break-up on southern Lake Baikal and its relationship to local and regional air temperatures in Siberia and to the North Atlantic Oscillation. Limnol Oceanogr. 1999;44:1486–1497. [Google Scholar]
- 37.Madigan M T, Martinko J M, Parker J. Brock biology of microorganisms. 8th ed. Upper Saddle River, N.J: Prentice-Hall; 1997. [Google Scholar]
- 38.Maidak B L, Cole J R, Lilburn T G, Parker C T, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald I R, Hall G H, Pickup R W, Murrell J C. Methane oxidation potential and preliminary analysis of methanotrophs in blanket bog peat using molecular ecology techniques. FEMS Microbiol Ecol. 1996;21:197–211. [Google Scholar]
- 40.Methé B A, Hiorns W D, Zehr J P. Contrasts between marine and freshwater bacterial community composition—analyses of communities in Lake George and six other Adirondack lakes. Limnol Oceanogr. 1998;43:368–374. [Google Scholar]
- 41.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 42.Olsen G J, Lane D J, Giovannoni S J, Pace N R, Stahl D A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 43.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. FastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 44.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 45.Pedersen K, Arlinger J, Ekendahl S, Hallbeck L. 16S rRNA gene diversity of attached and unattached bacteria in boreholes along the access tunnel to the Äspö hard rock laboratory, Sweden. FEMS Microbiol Ecol. 1996;19:249–262. [Google Scholar]
- 46.Pedersen K, Arlinger J, Hallbeck L, Pettersson C. Diversity and distribution of subterranean bacteria in groundwater at Oklo in Gabon, Africa, as determined by 16S rRNA gene sequencing. Mol Ecol. 1996;5:427–436. doi: 10.1111/j.1365-294x.1996.tb00332.x. [DOI] [PubMed] [Google Scholar]
- 47.Pedersen K, Hallbeck L, Arlinger J, Erlandson A C, Jahromi N. Investigation of the potential for microbial contamination of deep granitic aquifers during drilling using 16S rRNA gene sequencing and culturing methods. J Microbiol Methods. 1997;30:179–192. [Google Scholar]
- 48.Pernthaler J, Alfreider A, Posch T, Andreatta S, Psenner R. In situ classification and image cytometry of pelagic bacteria from a high mountain lake (Gossenköllesee, Austria) Appl Environ Microbiol. 1997;63:4778–4783. doi: 10.1128/aem.63.12.4778-4783.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. Fluorescence in situ hybridization. Methods Microbiol., in press.
- 50.Pernthaler J, Glöckner F O, Unterholzner S, Alfreider A, Psenner R, Amann R. Seasonal community and population dynamics of pelagic Bacteria and Archaea in a high mountain lake. Appl Environ Microbiol. 1998;64:4299–4306. doi: 10.1128/aem.64.11.4299-4306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rappe M, Gordon D, Vergin K, Giovannoni S. Phylogeny of actinobacteria small subunit (SSU) rRNA gene clones recovered from marine bacterioplankton. Syst Appl Microbiol. 1999;22:106–112. [Google Scholar]
- 52.Rheims H, Rainey F A, Stackebrandt E. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol. 1996;17:159–169. [Google Scholar]
- 53.Roller C, Wagner M, Amann R, Ludwig W, Schleifer K-H. In situ probing of Gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 54.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 55.Schuppler M, Mertens F, Schon G, Gobel U B. Molecular characterization of nocardioform Actinomycetes in activated sludge by 16S rRNA analysis. Microbiology. 1995;141:513–521. doi: 10.1099/13500872-141-2-513. [DOI] [PubMed] [Google Scholar]
- 56.Simek K, Babenzien D, Bittl T, Koschel R, Macek M, Nedoma J, Vrba J. Microbial food webs in an artificially divided acidic bog lake. Int Rev Hydrobiol. 1998;83:3–18. [Google Scholar]
- 57.Sly L I, Cox T L, Beckenham T B. The phylogenetic relationships of Caulobacter, Asticcacaulis and Brevundimonas species and their taxonomic implications. Int J Syst Bacteriol. 1999;49:483–488. doi: 10.1099/00207713-49-2-483. [DOI] [PubMed] [Google Scholar]
- 58.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 60.Takami H, Inoue A, Fuji F, Horikoshi K. Microbial flora in the deepest sea mud of the Mariana trench. FEMS Microbiol Lett. 1997;152:279–285. doi: 10.1111/j.1574-6968.1997.tb10440.x. [DOI] [PubMed] [Google Scholar]
- 61.Tanner M A, Goebel B M, Dojka M A, Pace N R. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ-hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 63.Wallner G, Fuchs B, Spring S, Beisker W, Amann R. Flow sorting of microorganisms for molecular analysis. Appl Environ Microbiol. 1997;63:4223–4231. doi: 10.1128/aem.63.11.4223-4231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wise M G, McArthur J V, Shimkets L J. Bacterial diversity of a Carolina bay as determined by 16S rRNA gene analysis—confirmation of novel taxa. Appl Environ Microbiol. 1997;63:1505–1514. doi: 10.1128/aem.63.4.1505-1514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zwart G, Hiorns W D, Methe B A, Van Agterveld M P, Huismans R, Nold S C, Zehr J P, Laanbroek H J. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst Appl Microbiol. 1998;21:546–556. doi: 10.1016/S0723-2020(98)80067-2. [DOI] [PubMed] [Google Scholar]